Abstract
The heat shock response is an evolutionarily conserved, stress-responsive signaling pathway that adapts cellular proteostasis in response to pathologic insult. In metazoans, the heat shock response primarily functions through the posttranslational activation of heat shock factor 1 (HSF1), a stress-responsive transcription factor that induces the expression of cytosolic proteostasis factors including chaperones, cochaperones, and folding enzymes. HSF1 is a potentially attractive therapeutic target to ameliorate pathologic imbalances in cellular proteostasis associated with human disease, although the underlying impact of stress-independent HSF1 activation on cellular proteome composition remains to be defined. Here, we employ a highly controllable, ligand-regulated HSF1 that activates HSF1 to levels compatible with those that could be achieved using selective small molecule HSF1 activators. Using a combination of RNAseq and quantitative proteomics, we define the impact of stress-independent HSF1 activation on the composition of the cellular proteome. We show that stress-independent HSF1 activation selectively remodels cytosolic proteostasis pathways without globally influencing the composition of the cellular proteome. Furthermore, we show that stress-independent HSF1 activation decreases intracellular aggregation of a model polyglutamine-containing protein and reduces the cellular toxicity of environmental toxins like arsenite that disrupt cytosolic proteostasis. Collectively, our results reveal a proteome-level view of stress-independent HSF1 activation, providing a framework to establish therapeutic approaches to correct pathologic imbalances in cellular proteostasis through the selective targeting of HSF1.
Intracellular protein homeostasis (or proteostasis) is maintained by stress-responsive signaling pathways such as the heat shock response (HSR).1,2 The HSR is an evolutionarily conserved, stress-responsive signaling pathway that adapts the composition of cytosolic proteostasis pathways in response to environmental, genetic, or aging-related stress.3,4 The HSR is induced by the accumulation of misfolded proteins within the cytosol, resulting in the activation of stress-responsive transcription factors that induce transcriptional programs including proteostasis network components such as chaperones, cochaperones, folding enzymes, and quality control factors. In mammals, the HSR primarily functions through activation of the winged-helix transcription factor heat shock factor 1 (HSF1).3,4 During heat shock, HSF1 activity is regulated through a complex, posttranslational mechanism involving nuclear localization, trimerization, and numerous modifications including phosphorylation and SUMOylation.3−5 Once activated, trimeric HSF1 induces a transcriptional program that remodels the cytosolic proteostasis network in an effort to resolve proteotoxic stress.6−8
HSF1 directly affects cytosolic proteostasis at the level of gene regulation, making HSF1 an attractive therapeutic target to intervene in human diseases etiologically linked to imbalances in cellular proteostasis.1,2,9−12 HSF1 overexpression attenuates the pathologic intracellular aggregation of several misfolding-prone proteins, including mutant huntingtin (a protein whose aggregation is pathologically associated with Huntington’s disease).13−16 Furthermore, overexpression of HSF1 in mice attenuates symptoms associated with Alzheimer’s disease (AD),17 a neurodegenerative disorder that also involves imbalances in intracellular proteostasis.18−20 The ability of HSF1 overexpression to attenuate pathologic imbalances in intracellular proteostasis associated with these diseases has led to significant effort to identify small-molecule activators of HSF1.12,21−23 Despite advances in high throughput screening efforts to discover HSF1 activators, further efforts are necessary to hone the response and selectivity of HSF1 activation by small molecules.
In the absence of selective and stress-independent small-molecule HSF1 activators, current approaches have either employed HSF1 overexpression or cell-based assays using heat shock promoter reporter constructs to explore the therapeutic potential for HSF1 activation in ameliorating pathologic imbalances in cytosolic proteostasis.24,25 HSF1 overexpression often leads to sustained extremely high levels of HSF1 activity, whereas cell-based assays for the heat shock response may identify compounds that cause proteotoxic damage and mimic cell stress. Consequently, critical questions remain regarding the therapeutic potential for HSF1 activation to treat human disease. Exactly how would sustained stress-independent HSF1 activation similar to what can be achieved using small-molecule HSF1 activators remodel cellular proteostasis pathways? What effects (if any) does stress-independent HSF1-dependent remodeling of cellular proteostasis pathways have on the composition and function of the endogenous cellular proteome not transcriptionally targeted by HSF1? Is there a therapeutic window for HSF1 activation to correct imbalances in cellular proteostasis without resulting in negative physiologic consequences of HSF1 hyperactivation associated with increased tumorigenesis?6,9,11,26
Here, we evaluate the impact of stress-independent HSF1 activation on cellular proteostasis capacity and global proteome composition. We recently described a ligand-regulated, constitutively active HSF1 fused to a destabilized mutant of FKBP (FKBP·cHSF1) that allows dose-dependent, ligand-mediated induction of HSF1 target genes in the absence of stress.27 We now show that ligand-dependent FKBP·cHSF1 activation induces the expression of HSF1 target genes to levels nearly identical to those observed during heat shock and significantly less than the levels obtained using more traditional tetracycline (tet)-inducible cHSF1 overexpression. This shows that stress-independent FKBP·cHSF1 activation allows for the increased expression of HSF1 target genes to levels consistent with those that could be obtained using selective HSF1 activators. We employ FKBP·cHSF1 to characterize the remodeling of cellular proteostasis networks induced by stress-independent HSF1 activation using a combined RNAseq transcriptional profiling and whole cell quantitative proteomics approach. Through these efforts, we demonstrate that stress-independent HSF1 activation does not significantly influence the global proteome not transcriptionally targeted by HSF1. Furthermore, we find that the high levels of HSF1 overexpression employed in previous studies are not required to obtain the beneficial consequences of proteostasis network remodeling. Instead, lower levels of HSF1 activation selectively attenuate intracellular aggregation of misfolding-prone proteins and promote cellular survival in response to stress. Collectively, our results provide an essential framework to evaluate stress-independent HSF1 activation as a therapeutic strategy to correct pathologic imbalances in cellular proteostasis associated with human disease.
Results and Discussion
FKBP·cHSF1 Allows Stress-Independent Activation of the HSF1 Transcriptional Program
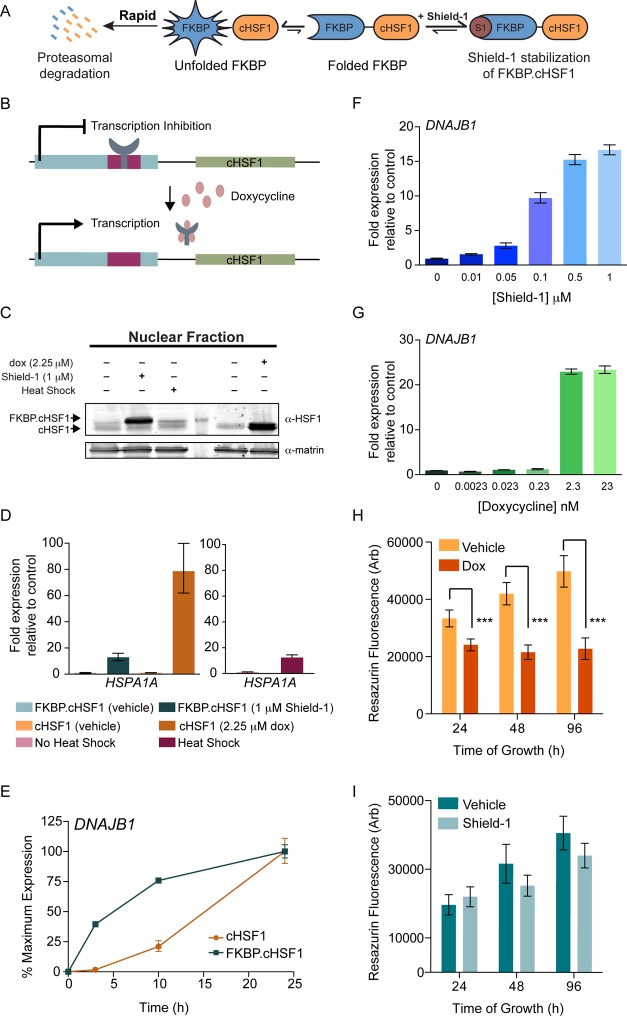
We established a genetically encoded, small-molecule-regulated fusion between a destabilized mutant of FK506 binding protein 12 and constitutively active HSF1 (FKBP·cHSF1; Figure 1A).27 When FKBP·cHSF1 is expressed in mammalian cells, the entire fusion protein is rapidly degraded by the proteasome, repressing HSF1 transcriptional activity. This degradation is prevented by the addition of the FKBP·cHSF1 pharmacologic chaperone Shield-1, which stabilizes the entire protein against degradation and allows cHSF1 transcriptional activity.27
Figure 1.
Shield-1-dependent FKBP·cHSF1 stabilization induces the expression of HSF1 target genes. (A) Illustration showing the posttranslational, Shield-1-mediated regulation of FKBP·cHSF1.27 (B) Illustration showing doxycycline-mediated induction of cHSF1 expression in cells stably expressing the tetracycline-repressor. (C) Immunoblot of the nuclear fraction from HEK293T-REx cells stably expressing FKBP·cHSF1 or dox-regulated cHSF1. Cells were treated for 15 h with vehicle, Shield-1 (1 μM), or dox (2.25 μM), as indicated. As a control, cells stably expressing FKBP·cHSF1 were heat shocked at 42 °C for 1 h and allowed to recover for 2 h prior to lysis. (D) qPCR of HSPA1A in HEK293T-REx cells stably expressing FKBP·cHSF1 or dox-regulated cHSF1. Cells were treated for 15 h with either Shield-1 (1 μM) or dox (2.25 μM), as indicated. HEK293T-Rex cells were also subjected to a 1 h heat shock at 42 °C and a 2 h recovery. Error bars show 95% confidence intervals. (E) qPCR of DNAJB1 in HEK293T-REx cells stably expressing FKBP·cHSF1 (blue) or dox-regulated cHSF1 (orange). Shield-1 (1 μM) or dox (2.25 μM) was added for the indicated time. The expression of DNAJB1 was normalized to the maximal expression observed at 24 h. Error bars show 95% confidence interval. (F) qPCR of DNAJB1 in HEK293T-REx cells stably expressing FKBP·cHSF1 and treated with increasing doses of Shield-1 for 15 h. Error bars show 95% confidence intervals. (G) qPCR of DNAJB1 in HEK293T-REx cells stably expressing dox-regulated cHSF1 and treated with increasing doses of dox for 15 h. Error bars show 95% confidence intervals. (H) Graph showing the growth of HEK293T-Rex cells stably expressing dox-regulated cHSF1. Dox (2.25 μM) was added, as shown, for the indicated time. Error bar shows SEM for n = 4. ***p-value < 0.01. (I) Graph showing the growth of HEK293T-Rex cells stably expressing FKBP·cHSF1. Shield-1 (1 μM) was added for the indicated time.
Here, we further characterize the Shield-1-dependent regulation of FKBP·cHSF1. We employed HEK293T-Rex cells stably expressing FKBP·cHSF1.27 We previously showed that FKBP·cHSF1 affords tight, ligand-dependent control over the expression of HSF1 target genes with no increased expression of these genes in the absence of activating ligand.27 An analogous HEK293T-Rex cell line expressing tet-inducible, unconjugated cHSF1 was prepared for comparison. In these cells, cHSF1 overexpression is induced by the addition of doxycycline (dox) (Figure 1B). These cells show tight, dox-dependent regulation of HSF1 target gene expression (Figure S1A, Supporting Information). We confirmed ligand-dependent regulation of FKBP·cHSF1 and tet-inducible cHSF1 by immunoblotting, showing nuclear HSF1 accumulation upon addition of the appropriate activating ligand (Figure 1C). Similarly, protein levels of the HSF1 target genes increase following Shield-1-dependent FKBP·cHSF1 stabilization or dox-dependent cHSF1 overexpression (Figure S1B,C, Supporting Information). In the absence of activating ligand, the levels of these proteins were identical to controls, further reflecting the tight ligand-dependent control over HSF1 activity in these cells.
qPCR shows ligand-dependent increases in the expression of the HSF1 target gene HSPA1A, which encodes heat shock-inducible HSP70 in each of these cell lines (Figure 1D). The extent of HSPA1A mRNA induction by Shield-1-dependent FKBP·cHSF1 stabilization is nearly identical to that observed in HEK293T-Rex cells subjected to heat shock (42 °C; 1 h) and significantly less than that observed for dox-dependent cHSF1 overexpression (Figure 1D). Similar results were observed for the alternative HSF1 target gene DNAJB1, which encodes a heat shock-inducible HSP40 (DNAJB1) (Figure S1D, Supporting Information). Since the increased expression of HSPA1A and DNAJB1 observed during heat shock reflects levels of gene expression obtainable through endogenous HSF1, these results demonstrate that Shield-1-dependent FKBP·cHSF1 allows for the activation of HSF1 to levels consistent with those obtainable using potential selective small-molecule HSF1 activators.
We next compared the rate of ligand-dependent HSF1 transcriptional activation in cells expressing FKBP·cHSF1 or tet-inducible cHSF1. Posttranslational Shield-1-dependent activation of FKBP·cHSF1 results in a rapid increase in DNAJB1 expression, with significant induction observed in <3 h (Figure 1E).27 In contrast, dox-dependent induction of cHSF1 does not significantly increase DNAJB1 mRNA levels following 3 h, although dox-dependent cHSF1 overexpression ultimately results in higher DNAJB1 expression (Figure S1E, Supporting Information). Similar results were observed for the alternative HSF1 target gene HSP90AA1 (Figure S1F, Supporting Information).
We further evaluated the differing capacities for FKBP·cHSF1 and tet-inducible cHSF1 activation to regulate the HSF1 transcriptional program in cells treated with increasing concentrations of the appropriate activating ligand (Figure 1F,G). As reported previously,27 Shield-1-dependent stabilization of FKBP·cHSF1 affords dose-dependent induction of HSF1 target genes such as DNAJB1. Dox-dependent cHSF1 overexpression is not readily dosable, showing increased DNAJB1 expression only once a threshold dox concentration is reached.
The poor dosability of dox-dependent HSF1 activation results in levels of HSF1 activity significantly higher than those observed following heat shock. This is important because it does not realistically represent what selective, small-molecule HSF1 activators could achieve, and very high levels of HSF1 activity could be detrimental to cell viability. Indeed, we found that dox-dependent cHSF1 overexpression reduces cellular viability (Figure 1H). This decrease in cell viability is greater than that observed upon acute heat shock where HSF1 activity is regulated through endogenous stress-dependent signaling (Figure S1G, Supporting Information). In contrast, Shield-1-dependent FKBP·cHSF1 stabilization results in only a mild growth defect (Figure 1I).
Collectively, these results show that Shield-1-dependent FKBP·cHSF1 stabilization enables the rapid, dosable stress-independent activation of HSF1 transcriptional targets to levels similar to those observed by heat shock. Importantly, these results demonstrate our ability to separate the heat-dependent posttranslational regulation of HSF1 from activation of this transcription factor in the absence of high levels of HSF1 transcriptional activity observed using the nondosable, dox-dependent induction of cHSF1. While HSF1 activity during heat shock involves additional levels of transcriptional regulation mediated through stress-dependent processes (e.g., posttranslational modification, chromatin remodeling, and heterooligomerization with other HSF transcription factors3,4), our establishment of FKBP·cHSF1 provides a mechanism to explore alterations in cellular proteostasis that could be induced by stress-independent activation of HSF1 to levels achievable using small-molecule HSF1 activators.
FKBP·cHSF1 Stabilization Activates the HSF1 Transcriptional Program
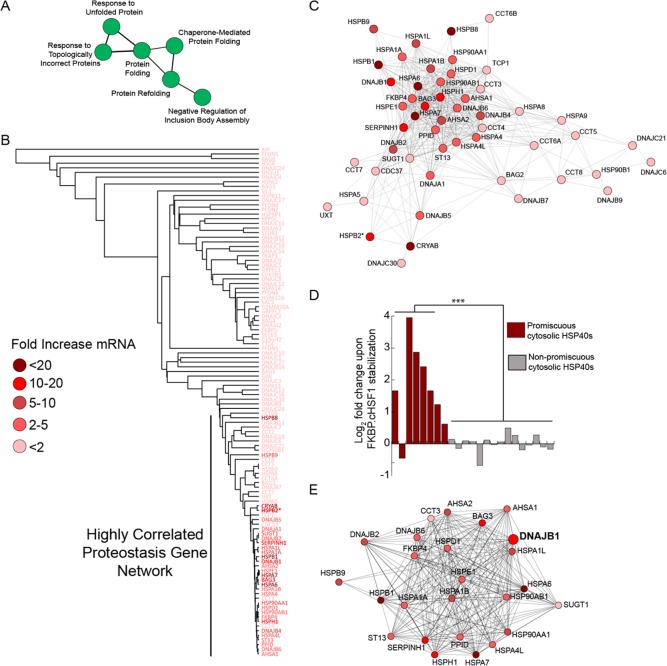
We characterized the impact of stress-independent HSF1 transcriptional activation by Shield-1-dependent FKBP·cHSF1 stabilization on gene expression using RNAseq. An analogous experiment was performed to characterize the impact of dox-dependent cHSF1 overexpression as a control. We observed a total of 397 genes whose expression was altered >2-fold (up- or down-regulated) upon Shield-1-dependent stabilization of FKBP·cHSF1 (FDR < 0.01; Figure 2A and Table S1, Supporting Information). Of these 397 genes, 375 were increased while 22 were decreased. Alternatively, dox-dependent activation of cHSF1 altered the expression of 1122 genes (fold-change >2-fold; FDR < 0.01) (Table S1, Supporting Information). Of these 1122 genes, 906 were increased while 216 were decreased. Genes significantly affected by FKBP·cHSF1 overexpression overlap with those induced by dox-dependent cHSF1 overexpression (96.8% overlap), indicating that the fusion to FKBP does not significantly impact stress-independent HSF1 transcriptional activity (Figure 2A). Genes highly induced by dox-dependent cHSF1 overexpression and Shield-1-dependent FKBP·cHSF1 include many well-known HSF1 target genes induced during heat shock including proteostasis factors such as HSPA1A, HSPA6, DNAJB1, and BAG3(6−8)(Figure S2A, Supporting Information). However, a direct comparison of genes induced by FKBP·cHSF1 versus those induced by heat shock7,28 show that stress-independent HSF1 activation and heat shock have distinct but overlapping transcriptional profiles (Figure S2A, Supporting Information). These differences likely reflect the additional layers of transcriptional regulation involved in the stress-induced heat shock response including chromatin remodeling, HSF1 posttranslational modification, and the involvement of additional transcription factors.3,4,29 This directly demonstrates the importance of using stress-independent HSF1 activation to evaluate the therapeutic potential of HSF1 to influence cellular proteostasis in the context of human disease, as selective small-molecule HSF1 activators will not activate these other stress-dependent transcriptional regulation pathways. Importantly, we did not observe significant increases in transcriptional markers of other stress-responsive signaling pathways such as the unfolded protein response (e.g., BiP,30 Table 1) or the mitochondrial unfolded protein response (e.g., DNAJA3,31 Table 1), showing that stress-independent HSF1 activation does not induce global cellular stress.
Figure 2.
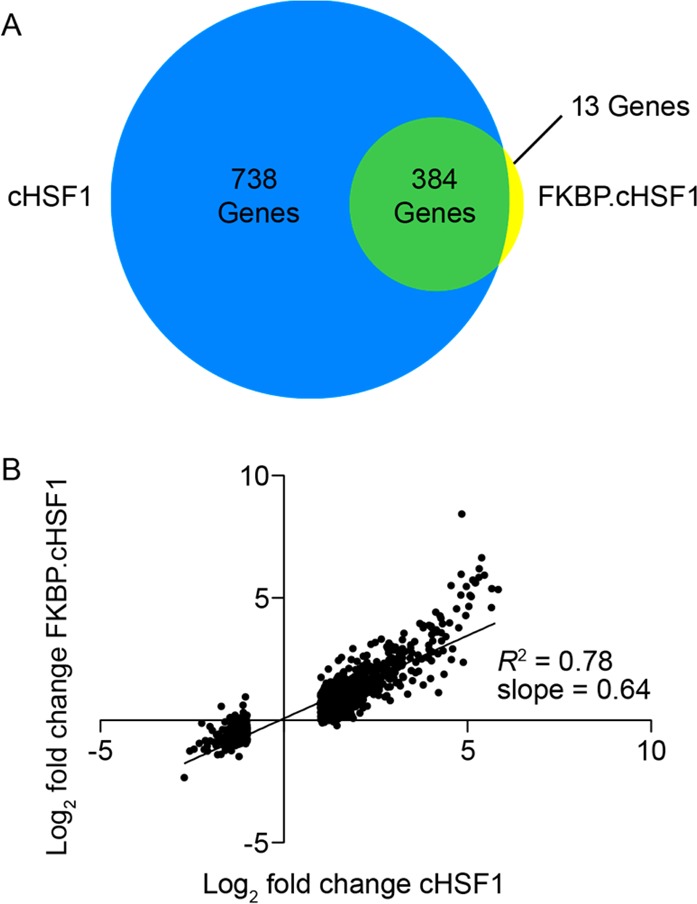
RNAseq transcriptional profiling of stress-independent HSF1 activation. (A) Diagram showing the overlap of genes with >2-fold change (FDR < 0.01) in HEK293T-REx cells following dox-dependent cHSF1 overexpression or Shield-1-dependent FKBP·cHSF1 stabilization. (B) Plot showing the correlation between gene expression in HEK293T-REx cells following Shield-1-dependent FKBP·cHSF1 activation or dox-dependent cHSF1 overexpression. Only genes whose expression is significantly affected (FDR < 0.01) by dox-dependent cHSF1 overexpression are shown.
Table 1. mRNA and Protein Level Alterations for Select Cellular Proteostasis Factors Induced by Stress-Independent HSF1 Activationa.
| fold change
in RNAseq |
fold change in proteomics |
|||
|---|---|---|---|---|
| gene symbol | cHSF1 | FKBP·cHSF1 | cHSF1 | FKBP·cHSF1 |
| Cytosolic Components of ATP-Dependent Chaperoning Pathways | ||||
| HSPA1A | 6.6 | 5.1 | 2.3 | 1.7 |
| HSPA1B | 7.5 | 5.3 | 2.0 | 1.5 |
| HSPA1L | 15.4 | 8.4 | 2.1 | 1.7 |
| HSPA6 | 40.0 | 73.3 | 2.4 | 2.6 |
| DNAJA1 | 3.8 | 3.2 | 1.8 | 1.4 |
| DNAJB1 | 13.0 | 15.5 | 8.5 | 5.4 |
| HSPH1 | 17.9 | 12.2 | 3.8 | 2.3 |
| HSPA4 | 4.4 | 2.3 | 1.8 | 1.4 |
| BAG3 | 19.1 | 19.3 | 7.3 | 3.8 |
| CCT4 | 2.2 | NS | NS | NS |
| HSP90AA1 | 7.2 | 5.1 | 1.7 | 1.5 |
| CHORDC1 | 3.3 | 2.3 | 1.6 | 1.3 |
| AHSA1 | 4.3 | 3.1 | 1.7 | 1.3 |
| Small Heat Shock Proteins | ||||
| HSPB1 | 17.6 | 21.4 | 19.5 | 7.8 |
| HSPB8 | 33.5 | 34.4 | ND | ND |
| HSPB9 | 4.0 | 5.3 | 1.4 | 1.3 |
| CRYAB | 35.5 | 53.1 | ND | ND |
| ER Folding Chaperones | ||||
| HSPA5 (BiP) | NS | NS | NS | NS |
| GRP94 | NS | NS | NS | NS |
| Mitochondrial Folding Chaperones | ||||
| HSPD1 (HSP60) | 4.6 | 4.6 | 1.7 | 1.5 |
| HSPE1 (HSP10) | 4.0 | 3.0 | 1.3 | 1.3 |
| DNAJA3 | NS | NS | NS | NS |
| HSPA9 (mtHSP70) | 1.5 | 1.3 | 1.10 | 1.10 |
NS indicates not significant (FDR < 0.01 in RNAseq; p < 0.1 in proteomics). ND indicates protein not quantified in our TMT-MuDPIT proteomic analysis.
The large number of genes altered >2-fold by dox-dependent cHSF1 overexpression relative to FKBP·cHSF1 activation likely results from the high levels of cHSF1 activation elicited by the tet-inducible system. Consistent with this notion, a plot of the ligand-dependent expression of genes significantly (FDR < 0.01) altered by dox-dependent cHSF1 overexpression versus the expression of the same genes following Shield-1-dependent FKBP·cHSF1 stabilization (Figure 2B) revealed a linear correlation (R2 = 0.78). The slope of this line was 0.64 ± 0.01, reflecting the higher levels of HSF1 activation induced by dox-dependent cHSF1 overexpression. Similar results were observed by qPCR where FKBP·cHSF1-dependent induction of HSF1 target genes including DNAJB1, BAG3, and GRPEL1 were induced ∼0.64-fold relative to dox-dependent cHSF1 overexpression (see Figures S1D and S2B, Supporting Information). These results further show that the FKBP·cHSF1 fusion does not influence HSF1 transcriptional selectivity but results in a more modest induction of HSF1 target genes. Table 1 shows mRNA fold changes for select HSF1 target genes altered by Shield-1-dependent FKBP·cHSF1 stabilization or dox-dependent cHSF1 overexpression.
Stress-Independent HSF1 Activity Increases Cellular Proteostasis Capacity
Functional clustering of the 397 genes whose expression is significantly altered by stress-independent FKBP·cHSF1 activation shows enrichment for cellular proteostasis pathways (Figure 3A and Table S2, Supporting Information). By correlation analysis on the expression of 116 primary cellular proteostasis factors (see Table S3, Supporting Information) following activation of FKBP·cHSF1 or cHSF1, we identify a cluster of cytosolic factors whose expression is affected similarly by stress-independent HSF1 activation (Figure 3B,C and Figure S3A, Supporting Information). This gene network is highly enriched for small heat shock proteins,32 components of the cytosolic HSP70 chaperoning pathway,33,34 and components of the cytosolic HSP90 chaperoning pathway.35,36
Figure 3.
Stress-independent HSF1 activation remodels cytosolic proteostasis pathways. (A) Image showing the functional clustering of GO annotations identified in genes showing a >2-fold change (FDR < 0.01) in HEK293T-REx cells following Shield-1-dependent FKBP·cHSF1 stabilization. See Table S2 (Supporting Information) for additional GO annotations identified in this analysis. (B) Dendrogram of primary proteostasis factors (see Table S3, Supporting Information) whose expression is highly correlated following stress-independent HSF1 activation. The color indicates the fold change in mRNA levels observed for specific highly correlated proteostasis factors following Shield-1-dependent FKBP·cHSF1 stabilization, as indicated by the legend. * indicates a gene only identified in cHSF1 overexpression RNAseq. (C) Cluster analysis of primary proteostasis factors whose expression is highly correlated following stress-independent HSF1 activation. The color indicates the fold change in mRNA levels observed for specific highly correlated proteostasis factors following Shield-1-dependent FKBP·cHSF1 stabilization, as indicated for B. A full correlation of the proteostasis factors described in Table S3 (Supporting Information) is shown in Figure S2A (Supporting Information). (D) Bar graph showing the log2 fold change mRNA of promiscuous (red) and nonpromiscuous (gray) client binding cytosolic HSP40 cochaperones in HEK293T-REx cells following Shield-1-dependent FKBP·cHSF1 stabilization. ***p-value < 0.005. See Table S3 (Supporting Information) for the identity of the specific HSP40s used in this analysis. (E) Graph showing the genes whose expression is highly correlated with DNAJB1 following HSF1 activation. The color indicates the fold-change in mRNA levels induced by FKBP·cHSF1 stabilization.
Increases in cytosolic proteostasis capacity induced by stress-independent FKBP·cHSF1 activation are best illustrated by the selective increase in cytosolic HSP40s that bind misfolded, aggregation-prone proteins. HSP40s function as cochaperones in the HSP70 chaperoning pathway through the recruitment of misfolded client proteins to HSP70 for ATP-dependent chaperoning and/or promoting HSP70 ATPase activity.34,33 HSP40s can be classified into four categories defined by their binding to substrates: (1) promiscuous client binding, (2) selective client binding, (3) no client binding, and (4) unknown client binding.34 Stress-independent HSF1 activation selectively induces the expression of promiscuous cytosolic HSP40s (e.g., DNAJB1 and DNAJA1) but not HSP40s that demonstrate selective or no client binding (e.g., DNAJC10 and DNAJC11) (Figure 3D). Since promiscuous HSP40s are the cochaperones most responsible for recruiting misfolded, aggregation-prone proteins to ATP-dependent HSP70 chaperoning,34,37,38 selective remodeling of the HSP40 cochaperone pool will result in the increased flux of misfolded, aggregation-prone clients to HSP70s for ATP-dependent chaperoning to the exclusion of more selective, pathway-specific HSP70 activities.
This expression of promiscuous HSP40s induced by stress-independent HSF1 activation is highly correlated with other proteostasis factors that cooperatively prevent intracellular protein aggregation. We observe strong correlations between the promiscuous client binding HSP40 DNAJB1 and other components of the cytosolic HSP70 chaperoning pathway including cytosolic HSP70s (e.g., HSPA1A, HSPA1B, and HSPA6) and cytosolic nucleotide exchange factors (e.g., HSPH1 and HSPA4L) (Figure 3E). Furthermore, the increased expression of DNAJB1 correlates with the increased expression of small heat shock proteins (e.g., HSPB1 and HSPB9), potentially reflecting functional cooperation between small heat shock proteins and the HSP70 chaperoning pathways for attenuating intracellular protein aggregation through stress-independent HSF1 activation.19 Similar correlations are observed for other promiscuous cytosolic HSP40 cochaperones (Figure S3B,C, Supporting Information).
Quantitative Proteomics Reveals Remodeling of Cytosolic Proteostasis Pathways Induced by Stress-Independent HSF1 Activation
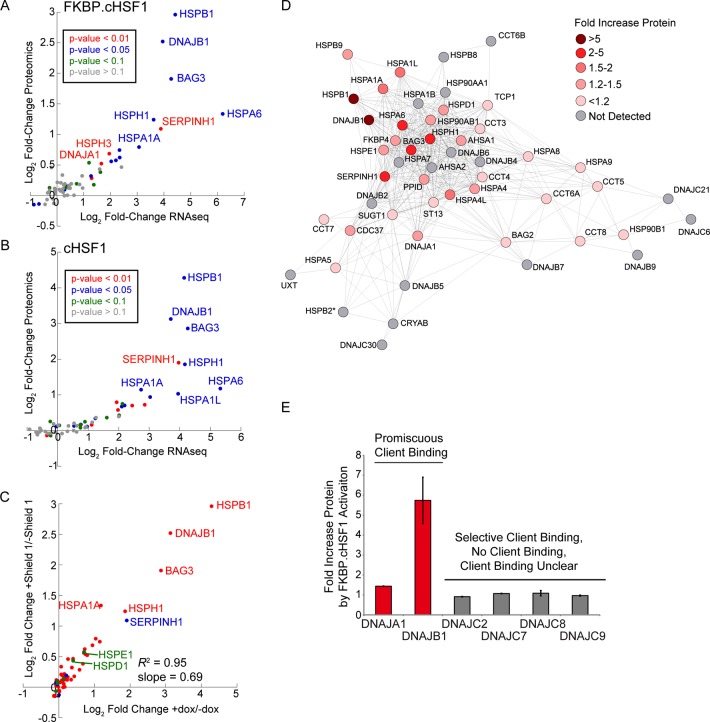
We quantified proteome-level remodeling of cytosolic proteostasis pathways induced by stress-independent HSF1 activation using tandem mass tag (TMT)-multidimensional protein identification technology (MuDPIT) quantitative proteomics.39−41 We performed TMT-MuDPIT proteomics in triplicate on whole cellular proteomes isolated from HEK293T-Rex cells in the presence or absence of Shield-1-dependent FKBP·cHSF1 stabilization or dox-dependent cHSF1 overexpression. The conditions used for this TMT-MuDPIT analysis were identical to those employed in our RNAseq analysis (see Supporting Information for details). In each TMT-MuDPIT experiment, we quantified intracellular levels of >2400 mammalian proteins (Table S4, Supporting Information). Select TMT-MuDPIT data for proteostasis proteins are reported in Table 1.
Proteostasis factors whose mRNA levels are significantly increased by Shield-1-dependent FKBP·cHSF1 stabilization also have increased protein levels (Figure 4A). Identical results were observed for dox-dependent cHSF1 overexpression (Figure 4B). Despite the qualitative correlation between gene expression and protein levels, our results show that mRNA fold changes are generally greater than those observed at the protein level. This observation confirms the importance of combining transcriptional and proteomic profiling of cHSF1 or FKBP·cHSF1 activation to obtain an accurate picture of HSF1-dependent remodeling of cellular proteostasis pathways.30
Figure 4.
TMT-MuDPIT shows proteome level remodeling of cytosolic proteostasis pathways induced by stress-independent HSF1 activation. (A) Plot showing the log2 fold change in proteomics vs the log2 fold change in RNAseq for primary proteostasis factors (see Table S3, Supporting Information) in HEK293T-REx cells following Shield-1-dependent FKBP·cHSF1 stabilization. (B) Plot showing the log2 fold change in proteomics vs the log2 fold change in RNAseq for primary proteostasis factors (see Table S3, Supporting Information) in HEK293T-REx cells following dox-dependent cHSF1 overexpression. (C) Plot showing the Shield-1-dependent FKBP·cHSF1 stabilization vs log2 fold change protein levels for proteostasis factors following dox-dependent cHSF1 overexpression. The proteins are colored by intracellular localization: cytosolic/nuclear (red), blue (ER), green (mitochondrial), and membrane-associated (orange). (D) Illustration showing the protein-level remodeling of the highly correlated cellular proteostasis network shown in Figure 3C. (E) Graph showing the fold increase in protein levels of cytosolic HSP40 proteins identified in our TMT-MuDPIT analyses. HSP40 proteins are categorized based on their client binding; promiscuous HSP40s are shown in red, and nonpromiscuous HSP40s are shown in gray.
We observe a strong correlation between alterations in protein levels of cellular proteostasis factors induced by Shield-1-dependent FKBP·cHSF1 stabilization or dox-dependent cHSF1 overexpression (R2 = 0.95) (Figure 4C). The slope of this correlation is 0.69 ± 0.02, which is nearly identical to that observed for a similar comparison of mRNA increases afforded by FKBP·cHSF1 and cHSF1 activation (Figure 2B), further demonstrating that Shield-1-dependent FKBP·cHSF1 activation does not significantly affect stress-independent, HSF1-mediated proteostasis remodeling but offers more subtle remodeling of proteostasis pathways as compared to dox-dependent cHSF1 overexpression.
Mapping alterations in cytosolic proteostasis factors induced by Shield-1-dependent FKBP·cHSF1 stabilization at the protein level on the gene network identified in our bioinformatics analysis (see Figure 3C) confirm that a central core of cytosolic proteostasis factors are altered by stress-independent HSF1 activation (Figure 4D). In particular, core pathways that attenuate pathologic protein aggregation are remodeled, including small heat shock proteins and components of cytosolic HSP70/HSP90 chaperoning pathways. Furthermore, our proteomic analysis confirms that stress-independent FKBP·cHSF1 activation increases cytosolic HSP40 cochaperones that have promiscuous client binding but not those that have selective or no client binding (Figure 4E). Collectively, these results substantiate the stress-independent, HSF1-mediated remodeling of cytosolic proteostasis pathways observed by transcriptional profiling and provide a framework to identify proteostasis network components and pathways accessible through stress-independent HSF1 activation that can be targeted to alleviate pathologic intracellular accumulation of misfolded proteins involved in human disease.
Stress-Independent HSF1 Activation Does Not Globally Disrupt the Cellular Proteome
A potential consequence of cytosolic proteostasis remodeling induced by stress-independent HSF1 activation is nontranscriptional alterations in the stability of cellular proteins. These nontranscriptional alterations in protein stability would be reflected by altered steady-state concentrations of proteins whose expression is not affected by stress-independent HSF1 activation. We plotted the −log p-value vs log2 fold-change of protein of alterations in intracellular protein levels induced by FKBP·cHSF1 activation to identify such nontranscriptional alterations in protein levels (Figure 5A). This analysis identified 24 proteins whose intracellular levels significantly increased (fold change >1.4 fold; p-value < 0.1). Alternatively, we observed no proteins whose intracellular levels decreased (fold-change >1.4; p-value < 0.1). Importantly, these proteins were transcriptionally induced by FKBP·cHSF1 (Figure 5B). Similar results were observed for dox-dependent cHSF1 overexpression (Figure 5C,D). While we cannot rule out alterations in intracellular levels of proteins not identified in our MuDPIT analyses, these results suggest that stress-independent HSF1 activation does not globally remodel the composition of the cellular proteome in HEK293T-Rex cells. These results indicate that stress-independent HSF1 activation may provide a therapeutic approach to selectively reduce intracellular protein aggregation without significantly impacting cellular proteome function.
Figure 5.
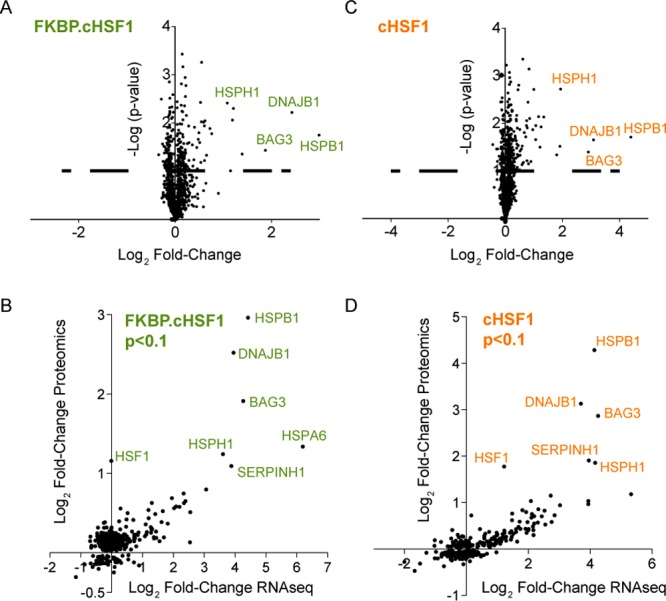
HSF1 activation does not globally disrupt the composition of the cellular proteome. (A) Plot of −log p-value vs log2 fold change (Shield-1/vehicle) for proteins identified in our TMT-MuDPIT analysis of HEK293T-REx cells stably expressing FKBP·cHSF1 ± Shield-1 (1 μM; 15 h). (B) Plot showing log2 fold change proteomics (Shield-1/vehicle) vs log2 fold change RNAseq (Shield-1/vehicle) for proteins identified in HEK293T-REx cells stably expressing FKBP·cHSF1. Only proteins showing a p-value < 0.1 in the TMT-MuDPIT proteomic analysis are shown. (C) Plot of −log p-value vs log2 fold change (dox/vehicle) for proteins identified in our TMT-MuDPIT analysis of HEK293T-REx cells stably expressing dox-regulated cHSF1 and treated with or without dox (2.25 μM; 15 h). (D) Plot showing log2 fold change proteomics (dox/vehicle) vs log2 fold change RNAseq (dox/vehicle) for proteins identified in HEK293T-REx cells stably expressing FKBP·cHSF1. Only proteins showing a p-value < 0.1 in the TMT-MuDPIT proteomic analysis are shown.
Stress-Independent FKBP·cHSF1 Activation Attenuates the Toxicity Induced by Cellular Stress
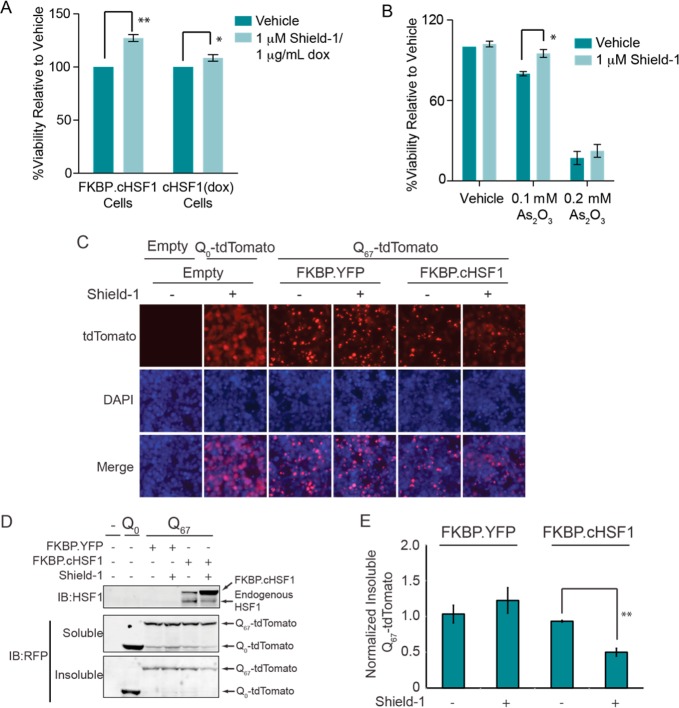
We evaluated the capacity of stress-independent HSF1 transcriptional activity to protect the cytosolic proteostasis environment by monitoring the capacity of FKBP·cHSF1 activation to attenuate cellular toxicity induced by heat stress or the environmental toxin arsenite (As(III)), two cellular insults that induce imbalances in cytosolic proteostasis.42−44 FKBP·cHSF1 activation afforded a small but significant increase in cellular viability when HEK293T-Rex cells are challenged with heat (Figure 6A) or As(III) (Figure 6B), consistent with the capacity of stress-independent HSF1 activation to protect cells against stress.
Figure 6.
Stress-independent FKBP·cHSF1 activation attenuates intracellular aggregation of an aggregation-prone model protein. (A) Plot showing the resazurin fluorescence at 24 h of HEK293T-REx cells overexpressing FKBP·cHSF1 or tet-inducible cHSF1 following heat shock (43 °C for 3 h). Cells were pretreated for 15 h with Shield-1 (1 μM) or dox (2.25 μM), as indicated. Error bar shows SEM for n = 6; **p-value < 0.002 and *p-value < 0.01. (B) Plot showing resazurin fluorescence for HEK293T-Rex cells overexpressing FKBP·cHSF1. Cells were pretreated with 15 h with vehicle or Shield-1 (1 μM) then subjected to a 10 h treatment with the indicated concentration of As(III). Resazurin fluorescence was measured following a 24 h recovery. Error bar shows SEM for n = 6; *p-value < 0.01. (C) Fluorescence image of HEK293T cells cotransfected with plasmids encoding polyQ0-tdTomato, poly-Q67-tdTomato, FKBP·cHSF1, or FKBP·YFP, as indicated. Shield-1 (1 μM) was added 36 h prior to imaging, as indicated. (D) Immunoblot of soluble and insoluble fractions from HEK293T cells cotransfected with a combination of plasmids encoding polyQ0-tdTomato or poly-Q67-tdTomato, and FKBP·cHSF1 or FKBP·YFP, as indicated. Shield-1 (1 μM) was added 36 h prior to lysis. (E) Quantification of insoluble polyQ67-tdTomato from immunoblots as shown in D. ** indicates a p-value < 0.002; n = 3.
We next evaluated the capacity of stress-independent HSF1 activation to attenuate cytosolic aggregation of a model, misfolding-prone protein. Cytosolic protein aggregation is a pathologic hallmark of numerous human diseases.45 Remodeling of the cytosolic proteostasis environment by stress-dependent HSF1 activation or HSF1 overexpression attenuates pathologic intracellular aggregation of destabilized, misfolding-prone proteins such as polyQ.13,46 To explore whether stress-independent FKBP·cHSF1 activation can attenuate pathologic aggregation of destabilized proteins, we transfected FKBP·cHSF1 into HEK293T cells stably expressing a protein fusion between an aggregation-prone polyglutamine sequence consisting of 67 glutamine residues and fluorescent tdTomato (polyQ67-tdTomato). polyQ67-tdTomato efficiently aggregates in these cells shown by the fluorescent puncta observed by fluorescence microscopy (Figure S4, Supporting Information). Cells expressing the nonaggregation-prone polyQ0-tdTomato show disperse fluorescence, reflecting the lack of aggregation. FKBP·cHSF1 activation decreases the population of intracellular polyQ67-tdTomato puncta, indicating decreased intracellular aggregation (Figure 6C). FKBP·cHSF1 activation also attenuates the accumulation of polyQ67-tdTomato into insoluble aggregates (Figure 6D,E). Importantly, these effects cannot be attributed to alterations in intracellular polyQ67-tdTomato, as total protein levels were not significantly affected by FKBP·cHSF1 activation (Figure 6D). The addition of Shield-1 did not influence puncta formation or incorporation of polyQ67-tdTomato into insoluble aggregates in cells transfected with a control plasmid expressing FKBP·YFP (Figure 6C–E). These results show that stress-independent activation of HSF1 to levels achievable with potential small-molecule HSF1 activators is sufficient to increase cytosolic proteostasis capacity and attenuate pathologic protein aggregation.
Concluding Remarks
The holistic activation of stress-responsive signaling pathways has been demonstrated to be more effective in correcting pathological imbalances in proteostasis associated with human disease than the isolated activation of an individual proteostasis network component.29 The control that HSF1 exhibits over cytosolic proteostasis makes it an attractive therapeutic target to attenuate pathologic imbalances in intracellular proteostasis associated with human disease.3,10,47 The comparisons presented herein between FKBP·cHSF1 and tet-inducible cHSF1 validate FKBP·cHSF1 as a highly regulatable genetic approach to induce the stress-independent HSF1 transcriptional program, providing a critical resource to explore the therapeutic potential for stress-independent HSF1 activation to ameliorate pathologic imbalances in intracellular protein aggregation associated with human disease. We show that stress-independent FKBP·cHSF1 activation alters the composition of the cytosolic proteostasis network to promote cellular viability and prevent intracellular aggregation of misfolding-prone proteins. Additionally, we demonstrate the capacity for stress-independent activation of FKBP·cHSF1 to attenuate intracellular protein aggregation and increase cellular viability in response to proteotoxic insult. Furthermore, transcriptional and proteomic profiling of FKBP·cHSF1 activation provides significant insight into the proteome remodeling afforded by stress-independent HSF1 activation and reveals unique, highly coordinated networks of HSF1-regulated genes that promote cytosolic proteostasis maintenance. Our results validate stress-independent HSF1 activation as a strategy to correct pathologic imbalances in cellular proteostasis associated with human disease, providing significant motivation to identify selective small-molecule HSF1 activators.
Methods
Reagents, Plasmids, and Antibodies
Sodium (meta)arsenite was purchased from Sigma-Aldrich Co. Shield-1 was purchased from Clontech. FKBP·cHSF1·pDEST40, FKBP·YFP·pDEST40, and tet-inducible cHSF1·pTREX-Dest30 were prepared as described previously.27 To construct ptdtomato-N1-polyQ67-tdtomato plasmid, the DNA fragment corresponding to the polyQ coding sequence was amplified by PCR using DNA oligonucleotides, 5′- CCGGAATTCGCCGCCACCATGCACCATCACCACCAGCAACAG-3′ and 5′-GGTGGATCCCCGCCCTCCAGTGGGTGGGGAAA-3′ as primers and pEYFP-N1-82Q as the template.48 The PCR product was digested with EcoRI and BamHI, and cloned into EcoRI- and BamHI-digested ptdTomato-N1. The following antibodies were used in this analysis: rabbit polyclonal α-matrin-3 (Bethyl Laboratories), rabbit polyclonal α-HSF1 (Stressgen), and rabbit polyclonal α-RFP.
Cell Culture
HEK293T-REx (Invitrogen) cells were cultured in DMEM (CellGro) supplemented with 10% fetal bovine serum (CellGro) and 1% penicillin/streptomycin/glutamine (CellGro). HEK293T-REx cells were cultured in 5 mg/mL blasticidin (InvivoGen) to maintain the tet repressor. HEK293T-REx cells were transiently transfected using calcium phosphate. Stable, clonal HEK293T-REx cell lines were selected from transiently transfected populations using 500 μg/mL G-418 sulfate (Cellgro) for cells expressing pcDNA-DEST40 or pTREx-DEST30 vectors. All stable cell lines were maintained in the appropriate selective antibiotics.
Nuclear Extractions and Western Blot Analyses
HEK293T-REx cells were harvested by scraping, and cell pellets were obtained by centrifugation at 1500 rpm. Cells were lysed in 10 mM Hepes (pH 7.5), 50 mM NaCl, 0.5 M sucrose, 0.1 mM EDTA, 0.5% Triton X-100, and complete EDTA-free protease inhibitors (Roche). Nuclear lysates were prepared as previously described.30 Protein concentrations were normalized by Bradford assays (Bio-Rad). The normalized nuclear lysates were separated by SDS–PAGE and transferred to nitrocellulose membranes. Following blocking and incubation with appropriate primary antibodies, membranes were incubated with 680 and 800 nm fluorophore-labeled secondary antibodies (Li-COR Biosciences) and detected with the Odyssey Infrared Imaging System (Li-COR Biosciences).
Quantitative RT-PCR
The relative mRNA expression levels of target genes were measured using quantitative RT-PCR. The protocol is detailed in the Supporting Information; primers used are listed in Table S3 (Supporting Information).
RNA-seq Analyses
FKBP·cHSF1 or cHSF1(dox) cells in 6-well plates were treated for 12 h with vehicle, 1 μM Shield-1 (for FKBP·cHSF1), or 2.25 μM dox (for cHSF1(dox)) in biological triplicate at 37 °C. Cells were harvested, and RNA was extracted using the RNeasy Mini Kit (Qiagen). Genomic DNA was removed by on-column digestion using the RNase-free DNase Set (Qiagen). Detailed protocols for the RNA-seq analyses are provided in the Supporting Information.
Tandem Mass Tag Liquid Chromatography–Mass Spectrometry Analyses
FKBP·cHSF1 or cHSF1(dox) cells in 6-well plates were treated for 12 h with vehicle, 1 μM Shield-1 (for FKBP·cHSF1), or 2.25 μM dox (for cHSF1(dox)) in biological triplicate at 37 °C. TMT-labeled peptides were prepared from RIPA-lysed cells according to standard protocols and analyzed by MuDPIT mass spectrometry. Detailed protocols for cell lysis and mass spectrometric analyses are provided in the Supporting Information.
Acknowledgments
J.C.G. thanks NHLBI (HL099245) for financial support. T.N. thanks the Uehara Memorial Foundation for a postdoctoral fellowship. M.D.S. thanks the American Cancer Society for a postdoctoral fellowship. R.L.W. thanks the Ellison Medical Foundation, Arlene and Arnold Goldstein, and the National Institutes of Health (AG036634 and AG046495 for funding). R.I.M. and T.N. were supported by the Ellison Medical Foundation, the National Institutes of Health (GM038109, GM081192, AG026647, and NS047331), and the Daniel F. and Ada L. Rice Foundation. We also thank J. Kelly, M. Petrascheck, and the TSRI DNA Core Facility for fruitful discussions and experimental assistance.
Supporting Information Available
Supplemental experimental protocols, complete data tables of RNA-Seq, TMT-MuDPIT, and GO annotation analyses, and supplemental figures supporting the data presented in the main text. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Calamini B.; Morimoto R. I. (2012) Protein homeostasis as a therapeutic target for diseases of protein conformation. Curr. Top. Med. Chem. 12, 2623–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers E. T.; Morimoto R. I.; Dillin A.; Kelly J. W.; Balch W. E. (2009) Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 78, 959–991. [DOI] [PubMed] [Google Scholar]
- Anckar J.; Sistonen L. (2011) Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu. Rev. Biochem. 80, 1089–1115. [DOI] [PubMed] [Google Scholar]
- Akerfelt M.; Morimoto R. I.; Sistonen L. (2010) Heat shock factors: integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11, 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S.; Loew C.; Korner R.; Pinkert S.; Theis M.; Hayer-Hartl M.; Buchholz F.; Hartl F. U. (2014) Interplay of acetyltransferase EP300 and the proteasome system in regulating heat shock transcription factor 1. Cell 156, 975–985. [DOI] [PubMed] [Google Scholar]
- Mendillo M. L.; Santagata S.; Koeva M.; Bell G. W.; Hu R.; Tamimi R. M.; Fraenkel E.; Ince T. A.; Whitesell L.; Lindquist S. (2012) HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 150, 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page T. J.; Sikder D.; Yang L.; Pluta L.; Wolfinger R. D.; Kodadek T.; Thomas R. S. (2006) Genome-wide analysis of human HSF1 signaling reveals a transcriptional program linked to cellular adaptation and survival. Mol. Biosyst. 2, 627–639. [DOI] [PubMed] [Google Scholar]
- Trinklein N. D.; Murray J. I.; Hartman S. J.; Botstein D.; Myers R. M. (2004) The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Mol. Biol. Cell 15, 1254–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Billy E.; Powers M. V.; Smith J. R.; Workman P. (2009) Drugging the heat shock factor 1 pathway: exploitation of the critical cancer cell dependence on the guardian of the proteome. Cell Cycle 8, 3806–3808. [DOI] [PubMed] [Google Scholar]
- Gestwicki J. E.; Garza D. (2012) Protein quality control in neurodegenerative disease. Prog. Mol. Biol. Transl. Sci. 107, 327–353. [DOI] [PubMed] [Google Scholar]
- Whitesell L.; Lindquist S. (2009) Inhibiting the transcription factor HSF1 as an anticancer strategy. Expert Opin. Ther. Targets 13, 469–478. [DOI] [PubMed] [Google Scholar]
- Westerheide S. D.; Morimoto R. I. (2005) Heat shock response modulators as therapeutic tools for diseases of protein conformation. J. Biol. Chem. 280, 33097–33100. [DOI] [PubMed] [Google Scholar]
- Fujimoto M.; Takaki E.; Hayashi T.; Kitaura Y.; Tanaka Y.; Inouye S.; Nakai A. (2005) Active HSF1 significantly suppresses polyglutamine aggregate formation in cellular and mouse models. J. Biol. Chem. 280, 34908–34916. [DOI] [PubMed] [Google Scholar]
- Krobitsch S.; Lindquist S. (2000) Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl. Acad. Sci. U.S.A. 97, 1589–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A.; Ross L.; Nachman I.; Bar-Nun S. (2012) Aggregation of polyQ proteins is increased upon yeast aging and affected by Sir2 and Hsf1: novel quantitative biochemical and microscopic assays. PLoS One 7, e44785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chafekar S. M., and Duennwald M. L. (2012) Impaired heat shock response in cells expressing full-length polyglutamine-expanded huntingtin, PLoS One, 7, DOI: 10.1371/journal.pone.0037929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce A.; Podlutskaya N.; Halloran J. J.; Hussong S. A.; Lin P. Y.; Burbank R.; Hart M. J.; Galvan V. (2013) Over-expression of heat shock factor 1 phenocopies the effect of chronic inhibition of TOR by rapamycin and is sufficient to ameliorate Alzheimer’s-like deficits in mice modeling the disease. J. Neurochem. 124, 880–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper W.; Nijholt D. A.; Hoozemans J. J. (2011) The unfolded protein response and proteostasis in Alzheimer disease: preferential activation of autophagy by endoplasmic reticulum stress. Autophagy 7, 910–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F. U.; Bracher A.; Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332. [DOI] [PubMed] [Google Scholar]
- Cohen E. (2012) Aging, protein aggregation, chaperones, and neurodegenerative disorders: mechanisms of coupling and therapeutic opportunities. Rambam Maimonides Med. J. 3, e0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef D. W.; Turski M. L.; Thiele D. J. (2010) Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoS Biol. 8, e1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamini B.; Silva M. C.; Madoux F.; Hutt D. M.; Khanna S.; Chalfant M. A.; Saldanha S. A.; Hodder P.; Tait B. D.; Garza D.; Balch W. E.; Morimoto R. I. (2012) Small-molecule proteostasis regulators for protein conformational diseases. Nat. Chem. Biol. 8, 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.; Au Q.; Yoon I. S.; Tremblay M. H.; Yip G.; Zhou Y.; Barber J. R.; Ng S. C. (2009) Identification of small-molecule HSF1 amplifiers by high content screening in protection of cells from stress induced injury. Biochem. Biophys. Res. Commun. 390, 925–930. [DOI] [PubMed] [Google Scholar]
- Bersuker K.; Hipp M. S.; Calamini B.; Morimoto R. I.; Kopito R. R. (2013) Heat shock response activation exacerbates inclusion body formation in a cellular model of Huntington disease. J. Biol. Chem. 288, 23633–23638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.; Hu C.; Huang M.; Jiang M.; Lu L.; Tang J. (2013) Heat shock transcription factor 1 attenuates TNFalpha-induced cardiomyocyte death through suppression of NFkappaB pathway. Gene 527, 89–94. [DOI] [PubMed] [Google Scholar]
- Jolly C.; Morimoto R. I. (2000) Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J. Natl. Cancer Inst. 92, 1564–1572. [DOI] [PubMed] [Google Scholar]
- Shoulders M. D.; Ryno L. M.; Cooley C. B.; Kelly J. W.; Wiseman R. L. (2013) Broadly applicable methodology for the rapid and dosable small molecule-mediated regulation of transcription factors in human cells. J. Am. Chem. Soc. 135, 8129–8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R.; Hurt J. A.; Krykbaeva I.; Taipale M.; Lindquist S.; Burge C. B. (2013) Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell 49, 439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabulas R. M.; Raychaudhuri S.; Hayer-Hartl M.; Hartl F. U. (2010) Protein folding in the cytoplasm and the heat shock response. Cold Spring Harbor Perspect. Biol. 2, a004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoulders M. D.; Ryno L. M.; Genereux J. C.; Moresco J. J.; Tu P. G.; Wu C.; Yates J. R. III.; Su A. I.; Kelly J. W.; Wiseman R. L. (2013) Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 3, 1279–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge J. E.; Horibe T.; Hoogenraad N. J. (2007) Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS One 2, e874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga H. H.; Garrido C. (2012) HSPBs: small proteins with big implications in human disease. Int. J. Biochem. Cell Biol. 44, 1706–1710. [DOI] [PubMed] [Google Scholar]
- Hageman J.; van Waarde M. A. W. H.; Zylicz A.; Walerych D.; Kampinga H. H. (2011) The diverse members of the mammalian HSP70 machine show distinct chaperone-like activities. Biochem. J. 435, 127–142. [DOI] [PubMed] [Google Scholar]
- Kampinga H. H.; Craig E. A. (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt W. B.; Morishima Y.; Peng H. M.; Osawa Y. (2010) Proposal for a role of the Hsp90/Hsp70-based chaperone machinery in making triage decisions when proteins undergo oxidative and toxic damage. Exp Biol. Med. (Maywood, NJ, U.S.) 235, 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. C.; Agashe V. R.; Siegers K.; Hartl F. U. (2004) Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5, 781–791. [DOI] [PubMed] [Google Scholar]
- Park S. H.; Kukushkin Y.; Gupta R.; Chen T.; Konagai A.; Hipp M. S.; Hayer-Hartl M.; Hartl F. U. (2013) PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 154, 134–145. [DOI] [PubMed] [Google Scholar]
- Borges J. C.; Seraphim T. V.; Mokry D. Z.; Almeida F. C.; Cyr D. M.; Ramos C. H. (2012) Identification of regions involved in substrate binding and dimer stabilization within the central domains of yeast Hsp40 Sis1. PLoS One 7, e50927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauniyar N.; Gao B.; McClatchy D. B.; Yates J. R. III. (2013) Comparison of protein expression ratios observed by sixplex and duplex TMT labeling method. J. Proteome Res. 12, 1031–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn M. P.; Ulaszek R.; Deciu C.; Schieltz D. M.; Yates J. R. III. (2002) Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Anal. Chem. 74, 1650–1657. [DOI] [PubMed] [Google Scholar]
- Washburn M. P.; Ulaszek R. R.; Yates J. R. III. (2003) Reproducibility of quantitative proteomic analyses of complex biological mixtures by multidimensional protein identification technology. Anal. Chem. 75, 5054–5061. [DOI] [PubMed] [Google Scholar]
- Kim Y. H.; Park E. J.; Han S. T.; Park J. W.; Kwon T. K. (2005) Arsenic trioxide induces Hsp70 expression via reactive oxygen species and JNK pathway in MDA231 cells. Life Sci. 77, 2783–2793. [DOI] [PubMed] [Google Scholar]
- McMillan D. R.; Xiao X.; Shao L.; Graves K.; Benjamin I. J. (1998) Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J. Biol. Chem. 273, 7523–7528. [DOI] [PubMed] [Google Scholar]
- Song X.; Chen Z.; Wu C.; Zhao S. (2010) Abrogating HSP response augments cell death induced by As2O3 in glioma cell lines. Can. J. Neurol. Sci. 37, 504–511. [DOI] [PubMed] [Google Scholar]
- Gidalevitz T.; Ben-Zvi A.; Ho K. H.; Brignull H. R.; Morimoto R. I. (2006) Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311, 1471–1474. [DOI] [PubMed] [Google Scholar]
- Sittler A.; Lurz R.; Lueder G.; Priller J.; Lehrach H.; Hayer-Hartl M. K.; Hartl F. U.; Wanker E. E. (2001) Geldanamycin activates a heat shock response and inhibits huntingtin aggregation in a cell culture model of Huntington’s disease. Hum. Mol. Genet. 10, 1307–1315. [DOI] [PubMed] [Google Scholar]
- Neef D. W.; Jaeger A. M.; Thiele D. J. (2011) Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat. Rev. Drug Discovery 10, 930–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Nollen E. A.; Kitagawa K.; Bindokas V. P.; Morimoto R. I. (2002) Polyglutamine protein aggregates are dynamic. Nat. Cell Biol. 4, 826–831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.