Abstract
Background
From fundamental studies to industrial processes, synthesis of heterologous protein by micro-organisms is widely employed. The secretion of soluble heterologous proteins in the extracellular medium facilitates their recovery, while their attachment to the cell surface permits the use of the recombinant host cells as protein or peptide supports. One of the key points to carry out heterologous expression is to choose the appropriate host. We propose to enlarge the panel of heterologous secretion hosts by using Streptococcus thermophilus LMD-9. This lactic acid bacterium has a generally recognised as safe status, is widely used in the manufacture of yogurts, fermented milks and cheeses, and is easy to transform by natural competence. This study demonstrates the feasibility of secretion of a heterologous protein anchored to the cell surface by S. thermophilus. For this, we used the cell envelope proteinase (CEP) PrtH of Lactobacillus helveticus CNRZ32 CIRM-BIA 103.
Results
Using S. thermophilus LMD-9 as the background host, three recombinant strains were constructed: i) a negative control corresponding to S. thermophilus PrtS- mutant where the prtS gene encoding its CEP was partially deleted; ii) a PrtH+ mutant expressing the L. helveticus PrtH pro-protein with its own motif (S-layer type) of cell-wall attachment and iii) a PrtH+WANS mutant expressing PrtH pro-protein with the LPXTG anchoring motif from PrtS. The PrtH + and PrtH + WANS genes expression levels were measured by RT-qPCR in the corresponding mutants and compared to that of prtS gene in the strain LMD-9. The expression levels of both fused prtH CEPs genes, regardless of the anchoring motif, reached up-to more than 76% of the wild-type prtS expression level. CEPs were sought and identified on the cell surface of LMD-9 wild-type strain, PrtH+ and PrtH+WANS mutants using shaving technique followed by peptide identification with tandem mass spectrometry, demonstrating that the heterologous secretion and anchoring of a protein of more than 200 kDa was efficient. The anchoring to the cell-wall seems to be more efficient when the LPXTG motif of PrtS was used instead of the S-layer motif of PrtH.
Conclusions
We demonstrated S. thermophilus LMD-9 could heterologously secrete a high molecular weight protein and probably covalently anchor it to the cell-wall.
Keywords: Heterologous expression, Secretion, Cell-wall anchored protein, Streptococcus thermophilus LMD-9, Cell envelope proteinase (CEP), PrtS, PrtH, Lactobacillus helveticus CNRZ32 CIRM-BIA 103
Background
Heterologous protein secretion is being increasingly employed to produce proteins on different living supports. Genetic tools have therefore been developed to present a protein or a peptide specifically on the surface of a microbial vector to study its structure, conformation, activity and/or interactions with its environment. Studies could be conducted in various conditions such as in a culture medium, in dairy matrix, or in vivo, for immune response (vaccination) or physiological effects in gastro intestinal tract [1,2]. In order to secrete heterologous proteins in the extracellular medium or anchor them to the cell-wall, the choice of the host is crucial and a panel of transformable species with their advantages and disadvantages is currently available [1]. The gram negative bacterium Escherichia coli is the most employed for intracellular heterologous expression, while the gram positive bacterium Bacillus subtilis is preferentially chosen for heterologous secretion [1,3]. B. subtilis also displays the advantage to be naturally competent. Indeed, it is possible in a one-step plasmid-free transformation to introduce into the cell and integrate in the chromosome by homologous recombination a foreign DNA. Lactic acid bacteria (LAB) have recently been used as alternative expression hosts to B. subtilis[4]. Among them, Lactococcus lactis presents several advantages, including many tools for genetic modifications, efficient protein secretion capability [4] and only few predicted cell surface proteins [5]. Furthermore, this bacterium possesses only one surface housekeeping protease, HtrA, able to degrade abnormal exported proteins and this protease gene can be inactivated to prevent the degradation of heterologously expressed proteins [4]. Finally, as this species has a generally recognised as safe (GRAS) status and is widely present in dairy products, it could be employed to study proteins behaviour in food (in situ) or in vivo which is not the case for B. subtilis. However, Lc. lactis still lacks the natural competence [6] in contrast to other food grade LAB [7] such as Streptococcus thermophilus.
S. thermophilus has a GRAS status and is widely used for its high acidification ability, in dairy products. This is one of the two dairy starters of yogurt and its recovery in faeces of human volunteers consuming yogurt has definitively established that this bacterium is capable to remain alive during its transit through the digestive tract [8,9]. Further, the production of active β-galactosidase in the second half of the small intestine by the strain FB13 further suggests that S. thermophilus remains metabolically active during its digestive transit. This could explain the improvement of lactose digestion observed in intolerant patients after yogurt consumption [9,10].
Previous works demonstrated that S. thermophilus was able to express intracellular heterologous proteins. Mutants were obtained by electrotransformation and plasmids were used as gene vectors to produce intracellular heterologous proteins. For example, green fluorescent protein [11], Streptomyces cholesterol oxidase [12] and glutamate decarboxylase [13] were heterologously produced in S. thermophilus cytoplasm. The recent discovery of natural competence and its mechanisms in some S. thermophilus strains [14-16] allowed the introduction of heterologous genes in the bacterial cell by natural competence and their chromosomal insertion by double cross-over recombination. Since then, natural competence was used to insert linear DNA fragments corresponding to reporter genes such as the luciferase genes luxAB[17] and the antibiotic resistant genes, i.e. cat for chloramphenicol [17], aphA3 for kanamycin or ery for erythromycin [18] which were intracellularly expressed. Now the challenge is to produce mutants able to export proteins on the cell surface and in this context, S. thermophilus LMD-9 could be a good candidate. Indeed, on the basis of its genome sequence, it is expected that this bacterium would display 2/3 less cell surface proteins than Lc. lactis, which already have few proteins present at the surface [5]. A single cell envelope proteinase (CEP), named PrtS, with high molecular weight is predicted to be covently anchored onto the cell-wall by action of the sortase SrtA [19]. The secretion system exporting this CEP as well as the sortase SrtA could then be good candidates to secrete in the medium heterologous proteins of various molecular weights and to covalently anchor them onto the cell-wall. Further, as in the case of Lc. lactis, the presence of a unique housekeeping protease gene, htrA, is an advantage compared to B. subtilis that possesses multiple housekeeping proteases able to degrade the exported proteins at the surface [3], and htrA of S. thermophilus can be deleted to avoid protein degradation. Finally, S. thermophilus has been detected in human gut by metagenomic sequencing [20], further make possible the use of this bacterium in in vivo studies. Therefore, it could be interesting to further develop genetic tools to produce heterologous proteins on the cell-wall of S. thermophilus LMD-9 strain.
To make the proof of concept that S. thermophilus can be used as a tool for the heterologous secretion of cell-wall anchored proteins, we decided to heterologously express the prtH gene of L. helveticus CNRZ32 in S. thermophilus LMD-9. Indeed, one of the main topics of our laboratories is the study of the activity and specificity of CEPs of lactic acid bacteria, particularly those of S. thermophilus and L. helveticus. The choice of this protease seemed coherent to us knowing that: i) S. thermophilus and L. helveticus CEP genes present similarities in the codon usage; ii) both bacteria are used as co-starters in the manufacturing of Swiss-type cheeses and thus grow in the same environment (milk medium, temperature, pH …), thereby eliminating a potential influence of the environment on the conformation and activity of PrtH; iii) S. thermophilus possesses an active proteolytic system and so anything that seems necessary for the expression and activity of a protease; iv) proteolytic activity would be easy to detect as it is known to confer a growth advantage compared to strains that do not secrete a functional CEP [21,22]. Finally, this experiment may allow us to elucidate the specific role of PrtH in the proteolytic activity of L. helveticus CNRZ32 since four CEP genes were identified in its genome sequence designated as prtH, prtH2, prtH3 and prtH4 and the role of each CEP has not been defined independently yet [23,24].
Results
Strategy of the mutant constructions for the heterologous expression of PrtH on the cell-wall of S. thermophilus LMD-9
prtS of S. thermophilus and prtH of L. helveticus both encode high molecular weight proteins (above 180 kDa) secreted on the cell-wall and belonging to the subtilisin-like serine proteinase family. They display a similar organization, which can be separated into five regions important for our genetic constructions (Figure 1A and B): i) the promoter and the ribosome binding site (RBS); ii) the export signal with the signal sequence (S) which addresses the protein to the secretory system (Sec system); iii) the different CEP domains: pro-peptide domain (PP), catalytic domain (PR), A domain (A), B domain (B) only for prtH and helix domain (H); iv) the CEP fixation part consisting of a cell-wall spacer domain (W) and an anchor domain (AN) with a LPXTG anchoring motif for prtS, and of a cell-wall spacer domain (W) with a S-layer domain attachment at the C-terminal end for prtH; v) a stop codon at the end of the last domain followed by a transcription terminator [25,26]. Three main parameters should be considered to achieve PrtH secretion and anchoring: the signal sequence, the transcription level of the CEP and the anchoring motif. As S. thermophilus LMD-9 expresses an active CEP on its cell-wall, we have chosen to use the expression/secretion pathway of PrtS to express and secrete the CEP of L. helveticus. Thus, our strategy to secrete PrtH was to replace the pro-protein sequence of prtS of S. thermophilus LMD-9 with the pro-protein sequence of prtH of L. helveticus CNRZ32 (from the PP to the W domains), maintaining the promoter, RBS, and signal sequence of prtS (Figure 2A and B).
Figure 1.
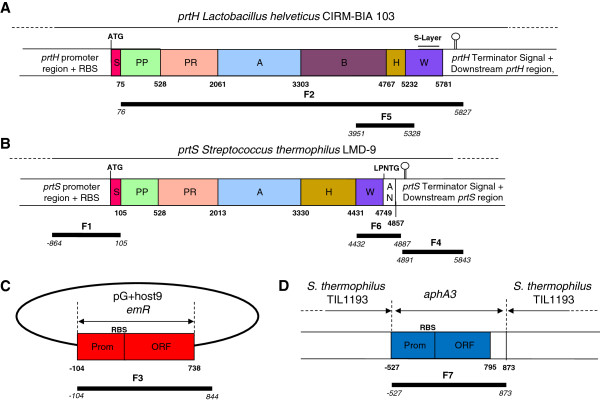
Schematic representation of DNA matrix and fragments amplified for mutant constructions. Black bars (F1 to F7) represent the fragments amplified from prtS, prtH, emR and aphA3 genes used to produce mutants expressing a PrtH fusion protein. Numbers refer to the last nucleotide of a domain (bold characters) or fragment limits (italic characters), starting from the adenosine nucleotide of the start codon of the ORF. The proteinase genes prtH(A) of Lactobacillus helveticus CNRZ32 CIRM-BIA 103 [this study, [28] and prtS(B) of Streptococcus thermophilus LMD-9 [this study, [25] are represented with their domains: upstream regions contain promoter and ribosome binding site (RBS), followed by S (signal sequence domain), PP (propeptide domain), PR (catalytic domain), A (A domain), B (B domain) in prtH, H (helix domain), W (cell-wall spacer domain) containing in prtH a 303 nucleotides S-layer attachment domain (small bar) and AN (anchor domain) in prtS with the LPNTG anchoring site. The downstream regions with the prtH and prtS terminator signals are also represented [22,28]. The erythromycin resistance gene (emR) is presented in the pG+host9 plasmid (C) with its promoter (Prom) and its ORF [48]. The kanamycin resistance gene (aphA3) inserted in S. thermophilus LMD-9 TIL1193 chromosome (D) is presented with its promoter (Prom) and its ORF [18].
Figure 2.
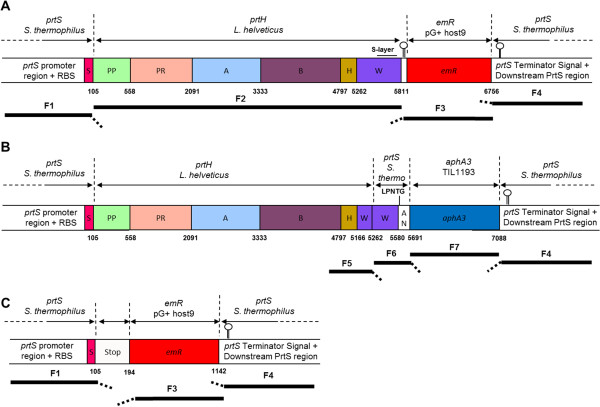
Schematic representation of the fragments used for mutant constructions and resulting mutant loci. Mutant constructions were performed by fusing different fragments from prtS, prtH, emR and aphA3 genes named F1 to F7 (black bars) to produce a PrtH fusion protein. Fragments tails (dashed thick bars) are homologous to the beginning of adjacent fragment in the construction. Numbers in bold characters refer to the last nucleotide of a domain. Nucleotides are numbered starting from the adenosine nucleotide of the start codon of the ORF. A) PrtH+ construction was inserted in S. thermophilus LMD-9 to express L. helveticus CNRZ32 CIRM-BIA 103 CEP (PrtH), instead of its own CEP (PrtS). In prtS chromosome locus, prtS promoter, prtS RBS and prtS S domain were fused to prtH gene composed of PP (propeptide domain), PR (catalytic domain), A (A domain), B (B domain), H (helix domain), W (cell-wall spacer domain) with a 303 nucleotides (nt) S-layer attachment domain (small bar) and a downstream region with the prtH terminator signal. An erythromycin resistance gene (emR) from pG+host9 [48] was inserted between prtH and the downstream region of prtS. B) PrtH+WANS construction replaced the last 450 nt, containing S-layer attachment of prtH W domain and emR gene from PrtH+ mutant, by prtS W and AN domains and aphA3 (kanamycin resistance gene) from S. thermophilus TIL1193 strain [18]. The secreted fusion PrtH+WANS proteinase has been designed to be covalently anchored to the host peptidoglycan thanks to the PrtS LPNTG motif. The PrtS W domain was expecting to bring the fused proteinase above the cell-wall of S. thermophilus. C) PrtS- sequence was obtained from a clone selected among PrtH+ potential mutants where the F2 fragment (prtH ORF) was not inserted. A 89 nt sequence composed by the two F2 primers (with 2 mutations) replaced F2 fragment in the construction.
Regarding the signal sequence, the use of the PRED TAT tool [27] revealed that both CEPs PrtS and PrtH have a signal peptide which addresses them to the Sec system. In gram positive bacteria , this general secretion pathway seems to imply the presence of a peculiar amino acid sequence which follows the signal peptide and could be recognised by the secretory system as previously demonstrated in Lc. lactis[2]. As both PrtS and PrtH are predicted to be secreted by the Sec system, this peculiar sequence should be functional in both CEPs. Therefore, we assumed that this peculiar sequence, located at the beginning of the PP domain of PrtH, would be functional in the Sec system of S. thermophilus. So only the S domain of PrtS was systematically conserved in our protein constructions to preserve the entire PP domain of PrtH in S. thermophilus.
Concerning the anchoring, we decided to test two different cell-wall anchoring systems: either the one from PrtH (S-layer type) or that from PrtS (LPXTG). Indeed, the S-layer motif used by L. helveticus may be not adapted to the cell-wall of S. thermophilus and PrtH may not correctly interact with the cell-wall. Thus, to maximise the anchoring of the proteinase, we replaced its S-layer domain by the W and AN domains of PrtS.
Constructions of PrtH+, PrtH+WANS and PrtS- mutants
In order to heterologously express PrtH on the cell-wall of S. thermophilus LMD-9, we constructed two mutants with two different anchoring systems (Figure 2). The mutant constructions were based on the design of an overlapping PCR (OL PCR) fragment to join the different gene parts, the natural transformation of S. thermophilus with the OL PCR resulting fragment and its integration into the chromosome by double cross-over recombination. The first mutant, named PrtH+, was expected to secrete PrtH which exhibited at the C-terminal of its W domain a S-layer anchoring motif, thought to anchor PrtH to the L. helveticus cell-wall [24,28] (Figure 2A). The 8622 base pair (bp) construction (F1 to F4) allowed the insertion of a 6718 bp foreign DNA by double cross-over recombination, replacing a large part of the prtS locus in S. thermophilus LMD-9 chromosome. As expected, the heterologous fused gene comprised the promoter, the RBS and the Signal sequence (S domain) of prtS, the prtH ORF spanning from the PP domain to 46 nucleotides (nt) downstream the stop codon, an erythromycin resistance gene and the downstream region of prtS.
The second mutant, named PrtH+WANS, derived from the first. The anchoring system of the PrtH+ mutant was replaced by the prtS cell-wall spacer and the anchor domains (W and AN domains) of S. thermophilus (Figure 2B and Methods). This was chosen to favour the secretion and the anchoring of the PrtH CEP in the cell-wall of S. thermophilus by covalent binding. Thus, a 4186 bp OL PCR fragment (Figure 2B; F5-F6-F7-F4) replaced by double cross-over recombination the prtH W domain (from the 97th nt of prtH W domain to the end of F3 fragment) by the F6 and F7 fragments.
During the construction of the PrtH+ mutant, one of the erythromycin resistant clones was lacking the prtH gene fragment (F2) (Figure 2C). In this mutant, called PrtS-, the major part of prtS was replaced by an 89 nt sequence and an erythromycin resistance gene, inserted between F1 and F4 fragments as described in the Methods section. This 89 nt sequence was produced during the OL PCR amplification of prtH + fragment by the fusion of F1 and F3 tail fragments. This sequence was composed of the beginning of prtH PP domain directly fused to the 3′ region of F2 (after prtH stop codon). These two fragments had mutations (data not shown) and the resulting 48 aa residues peptide was composed of prtS S domain (MKKKETFSLRKYKIGTVSVLLGAVFLFAGAPSVAA) followed by 13 aa residues (KQQVKASVDSQTK) similar to the N-terminal aa residues of the PrtH PP domain with one mutation changing the first aa residue glutamate (E) of the PP into a lysine residue (K). As PrtS- mutant was deprived of the major part of prtS and no other CEP genes were present in S. thermophilus LMD-9 genome, this mutant was used as a negative control in this study.
For the three constructed mutants, the constructions were checked by sequencing, which also confirmed that no additional or unexpected mutations occurred in the three mutant sequences.
Heterologous gene expressions
The transcription of prtH + and prtH + WANS genes was investigated by real-time quantitative polymerisation chain reaction (RT-qPCR) during the exponential growth of S. thermophilus PrtH+ and PrtH+WANS in milk (Figure 3). This medium was use to maximise the CEP gene expression. Indeed, to grow at high cellular density in milk, which is poor in free amino acids and peptides, S. thermophilus requires an efficient CEP activity to hydrolyse caseins into peptides for nitrogen supply, and this activity has been shown to be maximum during the exponential growth phase of the bacterium [29]. As expected, the prtS, prtH + and prtH + WANS genes were expressed in S. thermophilus LMD-9 wild-type, PrtH+ mutant and PrtH+WANS mutant, respectively. When compared with the expression level of the prtS gene in the wild-type strain, the prtH + and prtH + WANS genes appeared to be approximately 15% and 24% lower, respectively. No CEP gene expression was detected in the PrtS- mutant.
Figure 3.
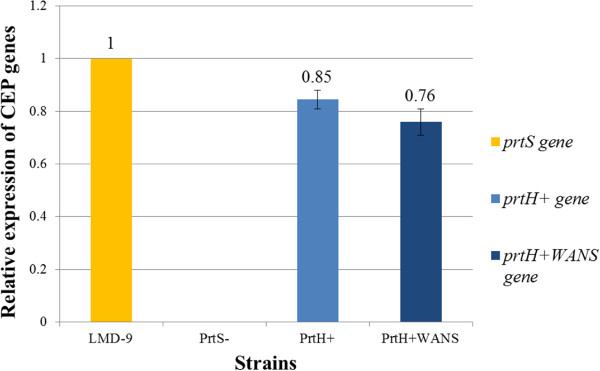
Relative expression of CEP genes in the mutants compared to prtS in S. thermophilus LMD-9. The expression of CEP genes (prtS, prtH+ and prtH+WANS) was detected by RT-qPCR of S. thermophilus wild-type LMD-9 and PrtS-, PrtH+ and PrtH+WANS mutant strains during the exponential growth phase in milk. CEP gene expressions were normalised with respect to the reference gene sigma70/sigma32 for each strain. The normalised expressions of prtH+and prtH+WANS CEP genes were compared to that of the reference prtS gene.
Although CEP genes were expressed in PrtH+ and PrtH+WANS mutants, their growth in skim milk appeared to be very similar to that of the PrtS- mutant. These three mutants grew slightly lower than the wild-type strain LMD-9. Indeed, the three mutants reached an optical density at 480 nm (OD480 nm) of 2 in five hours whereas the wild-type strain reached this OD in less than three hours. Additional activity tests were performed by using β- and αs1- caseins as substrate as described by Sadat-Mekmene et al. [30]. No CEP activity was detected neither on the cell surface nor in the extracellular culture medium of the PrtH+ and PrtH+WANS mutants, whereas activity was detected for both wild-type strains S. thermophilus LMD-9 and L. helveticus CNRZ32 CIRM-BIA 103 (data not shown). However, as the strain CIRM-BIA 103 contains 4 protease genes, the detected activity may be due to one or more proteases and not necessarily to PrtH.
Detection of the heterologous proteins expressed on the bacterial cell surface
To detect the presence of the various proteinases at the surface of S. thermophilus LMD-9 wild-type and mutant strains, a shaving approach was used. This method consists in hydrolysing the cell surface proteins of the living bacteria by trypsin under mild conditions, and in identifying the peptides released from the surface proteins using chromatographic separation coupled on line with tandem mass spectrometry. For each sample, negative controls without trypsin (cf. Methods) were performed. The identified proteins are reported in Additional file 1: shaving results A. The three CEPs PrtS, PrtH+ and PrtH+WANS were identified on the cell-wall of S. thermophilus LMD-9 wild-type strain, PrtH+ and PrtH+WANS mutants, respectively (Table 1 and Additional file 1: shaving results B, C and D). All detected peptides were specific to trypsin hydrolysis (which cuts after lysyl and arginyl amino acid residues), suggesting that these peptides did not result from degradation by another protease. To estimate the coverage of each protein identified by LC-MS/MS experiments, the first step was to calculate the protein abundance index (PAI) which is the number of observed peptides divided by the number of observable peptides per protein. The second step was to calculate the exponentially modified protein abundance index (emPAI) from the PAI values as: emPAI = 10PAI-1. Although shaving is first of all a qualitative method, the emPAI allows a semi quantitative estimation of proteins regarding each sample separately [31,32].
Table 1.
Proteinases identified after cell surface shaving of S. thermophilus strains PrtS - , LMD-9, PrtH + and PrtH + WANS
| |
|
|
PrtS
-
|
LMD-9 |
PrtH
+
|
PrtH
+
WANS |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Description | Prot Id | MW (kDa) | Nb id pept | emPAI | Nb id pept | emPAI | Nb id pept | emPAI | Nb id pept | emPAI |
| Subtilisin-like serine protease (PrtS) |
Q03L35 |
172.9 |
|
|
60 |
112.92 |
|
|
|
|
| PrtH+ |
|
208.6 |
|
|
|
|
40 |
6.86 |
|
|
| PrtH+WANS | 206.0 | 82 | 161.66 | |||||||
emPAI: exponentially modified Protein Abundance Index [31].
Nb id pept: number of identified peptides belonging to identified protein.
Prot Id: Identification sequence from the following databases TrEMBL.
The emPAI of CEPs among identified proteins in the sample was 112.92 for PrtS, 6.86 for PrtH+ and 161.66 for PrtH+WANS (Table 1). PrtS and PrtH+WANS were thus ranked among the most abundant proteins detected in their respective samples (Additional file 1: shaving results A). Those most abundant proteins were also predicted as cell surface proteins. So PrtH+WANS seemed to be present at the cell surface, probably anchored to the cell-wall as it is the case for PrtS. In contrast, PrtH+ appeared to be less abundant among the identified proteins. Identified CEP peptides are presented in Figure 4 and in Additional file 1: shaving results B, C and D. For PrtS, 60 peptides were identified from the PP to H domains, while no peptides corresponded to the S, W or AN domains (Figure 4A and Additional file 1: shaving results B). For PrtH+, 40 peptides were identified from the PP to the W domains except from the H domain, while no peptides corresponded to the S domain (Figure 4A and Additional file 1: shaving results C). Finally, 82 peptides covering the PrtH+WANS sequence from the PP to the PrtS W domains were detected while no peptides originating from the S and AN domains of PrtS or from the H and W domains of PrtH were detected (Figure 4A and Additional file 1: shaving results D). As expected, no CEP was identified on the cell surface of the PrtS- mutant.
Figure 4.

Schematic location of peptides identified from proteinase sequences after cell surface shaving. Cell surface proteins of S. thermophilus LMD-9, PrtH+, PrtH+WANS strains were hydrolysed by trypsin and peptides were identified by chromatography coupled on line with ESI quadrupole-orbitrap tandem mass spectrometry. A. Peptides corresponding to CEPs were aligned with PrtS, PrtH+ and PrtH+WANS protein sequences. Peptide sequences are represented with blue bars under the corresponding domains of CEPs: S, PP, PR, A, B, W and AN. B. Peptides identified (blue bars) around the cleavage site between PP and PR domains of S. thermophilus LMD-9 PrtS CEP. Amino acids and their position from the S domain of PrtS are indicated.
Discussion
Although tools required for the expression of heterologous proteins in LAB have been recently developed [4], B. subtilis and E. coli remain the mostly employed bacteria [1,3]. However, the expression of proteins on the bacterial surface remains a challenge regardless the bacterial host because the final structure of the secreted protein is influenced by various parameters. For example, pH, temperature or ionic environment can impair the final conformation and subsequent function of the heterologous protein. Moreover, the cell-wall composition (peptidoglycan, S-layer, …), which differed from one species to another [33,34], should influence the protein environment. Thus, enlarging the host panel to express heterologous proteins on the cell surface offers new choices between various secretion or cell-wall anchoring systems. LAB host can also be used as vectors in various milk derived products. To the best of our knowledge, this study reports for the first time the secretion to the cell-wall of a high molecular weight heterologous protein (above 200 kDa), i.e. the CEP PrtH of L. helveticus. Two mutants were constructed by natural transformation to secrete instead of the CEP PrtS of S. thermophilus LMD-9 wild-type strain, a PrtH (with its own S-layer attachment motif) and a PrtH+WANS protein (where the PrtH anchoring system was replaced by the PrtS one).
CEPs identification and location by shaving
To determine whether proteinases were really present on the cell surface, even at low level, we chose to shave the surface of the live cells of S. thermophilus wild-type and mutant strains by trypsin, a proteinase of well-known specificity, widely used in proteomics for protein identification by gel or gel-free based methods. This leads to the release of peptides from the cell surface proteins. This study presents, to the best of our knowledge, the first use of the shaving method on a S. thermophilus strain. The results have been very successful, not only because the shaving allowed us proving that the heterologous proteins were actually expressed but it also allowed detecting other proteins on the cell surface of the mutants and the wild-type strain. Forty-three proteins were thus identified. Searching their subcellular locations either by using the Uniprot data bank or by similarity to subcellular located proteins [35] revealed 17 proteins defined as cell surface (CS), or membrane-located (M) proteins, 22 cytoplasmic proteins and four uncharacterised proteins (Additional file 1: shaving results A). Only two cytoplasmic proteins were identified in all samples: the elongation factor Tu and the ATP-binding subunit of Clp protease and DnaK/DnaJ chaperones (Additional file 1: shaving results A). To estimate the abundance of each protein in each sample compared to all detected proteins, we calculated the protein content in percent using the following formula: ((emPAI of CS + M)/total emPAI)×100 [31]. Thus extracellular proteins (CS + M) represented 99.97%, 99.92%, 98.82% and 99.92% of all detected proteins for PrtS- mutant, LMD-9, PrtH+ mutant and PrtH+WANS mutant, respectively.
The identification of few intracellular proteins, at a low level, is intrinsic to shaving [36] and can be explained either by a slight cell lysis during shaving leading to the release of intracellular proteins [37] or to the presence of moonlighting multitask proteins such as enolase [38].
Regarding the CEPs, PrtS, PrtH+ and PrtH+WANS were identified in shaving supernatants of S. thermophilus LMD-9 wild-type strain, PrtH+ mutant and PrtH+WANS mutant, respectively, while no CEP was identified for PrtS- mutant strain (Table 1). As no signal peptide was identified, this suggests that the three CEPs were excreted through the Sec system, and consequently located on the cell-wall of S. thermophilus. So, if the first amino acids following the signal sequence are crucial for the translocation by the sec system (see “Strategy of the mutant constructions” in the Results part), we showed that the peculiar sequence present in PrtH of L. helveticus is functional in S. thermophilus. The wild-type strain S. thermophilus LMD-9 secretes PrtS CEP covalently anchored to the cell-wall [39]. This protein was highly abundant as shown by the high value of the emPAI (Table 1 and Additional file 1: shaving results A). This was also the case for PrtH+WANS, but not for PrtH+. This suggests that PrtH+WANS is probably covalently anchored to the cell-wall of S. thermophilus LMD-9, by the action of the sortase SrtA which recognizes the LPXTG motif, as it is the case for PrtS. Thus, the anchored system could be crucial for having highly abundant protein at the cell surface and it appears that the S-layer motif arising from L. helveticus strain seemed less adapted than the LPXTG motif to attach proteins to the cell-wall of S. thermophilus. Indeed, the emPAI of PrtH+ don’t rank it among the most abundant proteins detected in its sample, contrary to PrtH+WANS (Additional file 1: shaving results A). This difference is also encountered with the number of identified peptides as 40 peptides were identified for CEP of PrtH+ mutant against 84 peptides for PrtH+WANS mutant (Figure 4A and Additional file 1: shaving results C and D). The lower number of PrtH+ identified peptides could be explained by a lower interaction of PrtH+ with the cell-wall leading to its higher release into the extracellular medium. Indeed, the absence of S-layer proteins surrounding the cell of S. thermophilus could have impaired the interaction between the W domain of PrtH and the peptidoglycan [33,34]. Actually, Hu et al. [40] showed that an excess of fusion green fluorescent protein (GFP) harbouring a S-layer motif incubated one hour with S. thermophilus and Lc. lactis cells was able to be fixed on their surface but at low level compared to other LAB expressing S-layer proteins on their cell-wall.
Regarding the PrtS maturation, peptide LSVADETTAITNQEEAKPQ, corresponding to the C-terminal extremity of the PP domain, and peptide NIDSNTIITVPK to the N-terminal end of the PR domain were detected, which does not correspond to the theoretical cleavage sites for trypsin (Figure 4B). They could result from a cleavage between the last residue (Q) of the PP domain and the first residue (N) of the PR domain that occurs during the maturation process of PrtS, as shown by Fernandez-Espla et al. [25] and Chang et al. [41]. The concomitant presence of the complete sequence of the peptide LSVADETTAITNQEEAKPQNIDSNTIITVPK suggested that some PrtS molecules were also in their immature form, as stated by Chang et al. [41]. As both mature and immature forms were present at the cell surface, the maturation step could occur after the CEP anchoring.
CEP activity issues
The activity of PrtH+ and PrtH+WANS was neither detected at the cell surface of S. thermophilus nor in the medium. Moreover, cell-wall heterologous CEPs secreted in S. thermophilus mutants were not active on β and αs1- caseins in contrast to the cells of wild-type strains of L. helveticus[30] and S. thermophilus LMD-9 (results not shown). This lack of activity could explain the difference of growth rate observed in milk between mutants and the wild-type strain. One hypothesis to explain the inactivity of the PrtH is the presence of a mutation in the gene sequence. Indeed, in this study, the strain L. helveticus CNRZ32 CIRM-BIA 103 was used (Accession: PRJEB1537; Taxonomy ID: 1226332) and the analysis of its prtH gene sequence revealed an unexpected difference compared to that of L. helveticus CNRZ32 prtH sequence previously published [GenBank: AF133727.1; GI: 5758038] although both strains derived from the same initial strain of the CNRZ collection. This difference consisted of a 83 aa residues imperfect duplication which overlaps the end of the H domain and the beginning of the W domain. This insertion, which extends twice the helix domain H, may change the conformation of the protein, the way it can be included in the cell wall or the environment around the catalytic domain PR. Previous works showed that the strains of L. helveticus CIRM-BIA 103 and CNRZ32 contain four protease genes, and showed a similar specific activity of the cell envelope proteinases [30], but it is not possible to know whether all CEPs are really active or only some of them.
A second hypothesis is that the heterologous PrtH could be immature. This was observed for PrtH+WANS mutant, with the identification of the peptide VYYANDSSADNMANVSTVWNNYK (Additional file 1: shaving results D) which overlaps the PP and PR domains of PrtH+WANS CEP. In the case of L. helveticus, the maturation process could involve another actor to obtain a functional CEP. L. helveticus CNRZ32 has two maturation proteins in the chromosome belonging to the peptidylprolyl cis/trans isomerase family [23], named PrtM [UniProtKB/TrEMBL: A4UAD9] and PrtM2 [UniProtKB/TrEMBL: A4UAE0]. According to Broadbent et al.[23], PrtM could be required for PrtH maturation whereas PrtM2 should have a role in maturation of the other L. helveticus CEPs. Even if the maturation mechanism on S. thermophilus cell-wall is still not well established, this species does not seem to secrete maturase [5]. Maturation of PrtS could be achieved through an automaturation process or other factors not yet identified. One potential experiment to promote maturation of PrtH on the cell surface could be to introduce the prtM gene from L. helveticus CNRZ32 in PrtH+WANS mutant chromosome. However, this experiment requires multiple mutant constructions to expect to obtain active heterologous proteinases. Indeed, the expression level required to efficiently act on PrtH and the location and anchoring of PrtM on the bacterium surface are still ambiguous.
Finally, the lack of activity could be due to the chosen host and/or to the selected heterologous protein. Indeed, we cannot exclude that the microenvironment of the cell-wall (presence or absence of S-layer proteins) may influence the correct folding of the enzyme. This latter problem is also encountered in the other hosts commonly used for heterologous secretion. Furthermore, although PrtS activity on the cell-wall of S. thermophilus was demonstrated [29,41,42], the quantity of secreted proteins by this pathway is still unknown in S. thermophilus LMD-9 and heterologous CEPs as well as PrtS may be displayed at a too low level on the cell surface to detect the activity of heterologous CEPs. Anyway, despite the lack of activity, our tool actually allows secretion and anchoring of proteins of high molecular weight at the cell surface of S. thermophilus.
Conclusion
This study demonstrated that S. thermophilus LMD-9 strain is able to secrete and probably anchor heterologous proteins on its cell-wall and gives a new tool to enlarge the panel of heterologous secretion hosts. This S. thermophilus strain is a powerful tool regarding its ability for natural competence that allows bypassing plasmid construction to produce recombinant proteins. Moreover, its food grade status added to its wide industrial use and its ability to survive in gastrointestinal tract make S. thermophilus LMD-9 a good candidate to release recombinant proteins in a dairy matrix or directly in gastrointestinal tract. At this stage, our tool requires selection genes to be constructed which could constitute a negative point in most applications. However, we can imagine to dispense with the use of a selection gene for further constructions, thanks to the highly transformation rate of S. thermophilus LMD-9 [14,15,17].
Methods
Bacterial strains and growth conditions
Strains used in this work were Lactobacillus helveticus CNRZ32 CIRM-BIA 103, Streptococcus thermophilus LMD-9 [43], S. thermophilus LMD-9 TIL1193 [18] and three S. thermophilus LMD-9 mutants: PrtH+, PrtS- and PrtH+WANS (this study). They were cultivated at 42°C, in MRS (Difco) for L. helveticus and in M17 supplemented with 2% of lactose (LM17) [44] for S. thermophilus strains. For natural transformation experiments, LM17 and reconstituted chemically defined medium (CDM) supplemented with 2% of lactose (LCDM) [45] were used. A 10% reconstituted skim milk was used for strain conservation and precultures of S. thermophilus strains. When necessary, antibiotics were added to the media: erythromycin (Sigma) at 5 μg.mL-1 (for cultures of PrtH+ and PrtS- mutants) or kanamycin (Sigma) at 1 mg.mL-1 (for culture of PrtH+WANS mutant). The absorbance at 480 nm of skim milk cultures was measured after 1:10 dilution in a clarification solution of EDTA 0.2% (pH 12).
DNA extraction and PCR
General molecular biology techniques, DNA extraction and PCR amplifications were achieved according to Green and Sambrook [46], and/or supplier’s recommendations. PCR fragments were obtained using the Taq DNA polymerase or the Phusion high fidelity DNA polymerase (Fermentas, Saint Rémy-lès-Chevreuse, France) in a Mastercycler proS (Eppendorf). All primers used in this work are listed in Table 2 and in Additional file 2: sequencing primers and Additional file 3: qPCR primers. They were designed by using Primer3Plus [47] and purchased from Eurogentec (Seraing, Belgium). To produce overlapping regions between adjacent fragments (see above), a short nucleotide sequence homologous to the beginning of the adjacent fragment was added at the 5′ end of one of the primer used to amplify the desired fragment (in italics in Table 2).
Table 2.
Primers and resulting fragments used in overlap extension PCR for construction of PrtH + and PrtH + WANS mutants
| Fragment | Primer | Sequence (5′ – 3′) | Length (bp) | DNA matrix (strain) | Product size (kbp) | Hyb temp (°C) | Overlap product size (kbp) | Overlap hyb temp (°C) |
|---|---|---|---|---|---|---|---|---|
| PrtH+ |
|
|
|
|
|
|
|
|
| F1 |
Style 1-1* |
ACAAATTCATGCCGTTCATAAG |
22 |
gDNA (LMD-9) |
0.986 |
59.5 |
8.622 |
55 |
| PrtSssUp(H)_R |
GCCTTAACTTGTTGTTCTGCAGCTACCGATGGTGC |
35 |
||||||
| F2 |
PrtHpp_F |
GAACAACAAGTTAAGGCTAGTGTTGACAGCCAAACAAAAAC |
41 |
gDNA (CNRZ32 CIRM-BIA 103) |
5.752 |
62 |
||
| PrtHterm_R |
AAAAGAGTAATGATCCTTCTCATTACTCTTTCATTATATGTAAATGATTATTTTAC |
56 |
||||||
| F3 |
EmpG9(H)_F |
GAGAAGGATCATTACTCTTTTCAAACTTAAGAGTGTGTTGATAGTGC |
47 |
pG+host 9 |
0.969 |
59.5 |
||
| EmpG9_R |
GGACCTCTTTAGCTCCTTGG |
20 |
||||||
| F4 |
PrtSDn(Em)_F |
CCAAGGAGCTAAAGAGGTCCATAATAAAACCGCTTAATCATTGTG |
45 |
gDNA (LMD-9) |
0.973 |
49 |
||
| PrtSDn_R |
CGTCTATCAATCTTGTATTTTCTTG |
25 |
||||||
| PrtH+WANS |
|
|
|
|
|
|
|
|
| F5 |
UpH_F |
CGGTATCAAGTGGGGTACTCG |
21 |
gDNA (PrtH+) |
1.395 |
69 |
4.186 | 55 |
| UpH(WANS)_R |
CTTGCTTGGCTTGCAGAAACAGGAGCTGCAACTTGGTTATC |
41 |
||||||
| F6 |
WANS_F |
TCTGCAAGCCAAGCAAG |
17 |
gDNA (LMD-9) |
0.473 |
58 |
||
| WANS(aphA3)_R |
CTCAAATGGTTCGCTGGTTAGCAAACTTGTGATAAAGC |
38 |
||||||
| F7 |
aphA3_F** |
CCAGCGAACCATTTGAG |
17 |
gDNA (TIL1193) |
1.4 |
54 |
||
| aphA3_R** |
GTTGCGGATGTACTTCAG |
18 |
||||||
| F4 |
PrtSDn(aphA3)_F |
CTGAAGTACATCCGCAACATAATAAAACCGCTTAATCATTGTG |
43 |
gDNA (LMD-9) |
0.971 |
61.7 |
||
| PrtSDn_R | CGTCTATCAATCTTGTATTTTCTTG | 25 | ||||||
Primers Style 1-1, PrtSDn_R, and UpH_F correspond to those used for overlapping PCR reaction and italicised nucleotides correspond to homologous tails.
The name, sequence, length, DNA matrix, product size (including the added tails), hybridization temperature (hyb temp), overlap product size and overlap hybridisation temperature (hyb temp) are indicated.
bp (base pairs); kbp (kilobase pairs) and gDNA (genomic DNA) * [42], ** [18].
For DNA constructions, each PCR fragment was first amplified with Phusion high fidelity DNA polymerase with the minimum of DNA matrix i.e. enough to have 500 to 1000 matrix copy per PCR reaction. PCR were performed according to polymerase supplier’s recommendations. Fragments were then purified using high pure PCR product purification kit (Roche Applied Science, Meylan, France) and eluted in elution buffer as recommended by Fontaine et al. [17]. For both constructions produced by OL PCR, the four purified fragments were pooled in equal amount and PCR-amplified, in a final volume of 20 μL, with the forward primer of the 5′ end fragment and the reverse primer of the 3′ end fragment (Table 2). For sequencing, PCR fragments were amplified with the Taq polymerase and sequencing primers (Additional file 2: sequencing primers) and shipped to the company Beckman Coulter Genomics (Essex, U.K.).
Genetic material and mutant constructions
Four DNA matrices were used for mutant constructions: the prtH gene [locus_tag: LHCIRMBIA103_00753; NCBI Accession Number: PRJEB1537; Taxonomy ID: 1226332] of L. helveticus CNRZ32 CIRM-BIA 103 (Figure 1A), the prtS gene [locus tag: STER_0846; NCBI Reference Sequence: YP_820283.1; GI: 116627664] from S. thermophilus LMD-9 (Figure 1B), the erythromycin resistance gene (Figure 1C) from pG+host9 plasmid DNA [48] and the kanamycin resistance gene (Figure 1D) from S. thermophilus LMD-9 TIL1193 genomic DNA (gDNA) [18].
Two constructions were obtained using OL PCR. Primers used for OL PCR are described in Table 2. The prtH + 8622 bp construction was produced by assembling 4 fragments in this order: F1, F2, F3 and F4 (Figure 2). For F1 and F4 respectively, a 969 bp sequence composed of the promoter, RBS, and S domain of prtS and the 953 bp downstream region of prtS fragment starting 33 nt after prtS stop codon were amplified from S. thermophilus LMD-9 gDNA (Figure 1B). For F2, a 5752 bp region of the prtH open reading frame (ORF) sequence, from PP to 46 nt after stop codon was amplified from L. helveticus CNRZ32 CIRM-BIA 103 gDNA (Figure 1A). For F3, a 948 bp sequence (Figure 1C) containing an erythromycin resistance gene was amplified from pG+host9 plasmid DNA [48].
The prtH + WANS 4186 bp construction results from the assembly of 4 fragments F5, F6, F7 and F4 (Figure 2B). For F5, a 1378 bp region composed of the last 817 nt of B domain, the H domain and the first 96 nt of W domain of prtH excluding the S-layer sequence was amplified from L. helveticus CNRZ32 CIRM-BIA 103 gDNA (Figure 1A). For F6, a 456 bp sequence with W and AN domains of prtS plus 30 nt after prtS stop codon was amplified from S. thermophilus LMD-9 gDNA (Figure 1B). For F7, a 1400 bp sequence of kanamycin resistance gene (Figure 1D) was amplified from S. thermophilus LMD-9 TIL1193 gDNA [18]. The F4 region was the same as the one used for PrtH+ construction.
Natural transformation
Linear DNA fragments obtained by OL PCR were introduced in S. thermophilus LMD-9 by natural transformation as described by Gardan et al.[18]. Briefly, S. thermophilus LMD-9 was grown at 42°C in chemically defined medium (CDM) during 5–6 hours, until OD600 nm = 1.8-2 (exponential growth phase). Culture was then diluted in CDM in order to obtain OD600 nm = 0.05 and incubated at 42°C about 1 hour, until OD600 nm reached 0.2. Then, 100 μL of culture was mixed with 3 μL of OL PCR product and incubated 30 min at 42°C. After the adding of 900 μL of LM17, the mixture was re-incubated 40 min at 42°C. Cells were then concentrated 10 times by centrifugation (5 min, 3900 g) and resuspension with their own supernatant, and spread on LM17 agar plates supplemented with the appropriate antibiotic.
The prtH + OL PCR fragment was introduced into S. thermophilus LMD-9 to obtain a PrtH+ mutant. Likewise, the prtH + WANS OL PCR fragment was introduced in the PrtH+ mutant to obtain the PrtH+WANS mutant. Integration of OL PCR fragments at the targeted locus is achieved through the presence of 2 fragments of approximately 1000 bp homologous to the targeted locus and surrounding the foreign DNA. These homologous regions were included into the F1 or F5, and F4 fragments (Figures 1 and 2).
RNA extraction, reverse transcription and quantitative real-time PCR
S. thermophilus LMD-9, PrtS-, PrtH+ and PrtH+WANS were grown in skim milk up-to OD480 nm = 1–2 (exponential growth phase), before milk coagulation. Milk caseins were removed according Chopard et al. [49]: at 6 mL of skim milk culture were added 795 μL of saline solution (NaCl 0.85% (wt/v); sodium glycerophosphate 0.5% (wt/v); tween 80 0.1% (v/v); pH 7) and 195 μL of trisodium citrate solution 1 M. The cell pellets were harvested by centrifugation (15 min, 3900 g at 4°C) and a second washing with the same solutions was done. The following step was performed using the RNA extract Kit Aurum (Bio-Rad, Marnes La Coquette, France) and cell lysis step was modified as follows: to improve cell lysis, an ultrasonic bath (Bioruptor, Diagenode, Liège, Belgium) (15 s 4 times at maximal power) was performed twice. An additional mechanic milling with 250 mg of Ø 0.1 – 0.25 mm glass beads (Fisher Scientific, Illkirch, France) was also performed in 2 mL microtubes by vortexing first 60 s then twice 30 s, with at least 2 min stay on ice between each milling. The supernatant was collected after centrifugation at 12,000 g for 1.5 min at room temperature.
After nucleic acids extraction, two DNAse steps were performed: a first step with DNAse I (Bio-Rad) followed by a phenol-chloroform extraction [46] and a second step with RNase-free DNase I (Ambion, Courtaboeuf, France). The last step was repeated until PCR control with sigma70 primers was negative after 30 PCR cycles [42]. Total RNA concentrations were determined by measuring the absorbance at 260 nm using a spectrophotometer Nanodrop-1000 (Thermo Scientific, Illkirch, France). Complementary DNAs (cDNA) were synthesized from 1 μg of RNA by using Moloney murine leukemia virus reverse transcriptase (Invitrogen, Saint Aubin, France) according to the manufacturer’s instructions.
Quantitative PCR (qPCR) reactions were performed on CFX96 Touch™ Real-Time PCR Detection System (Biorad) following manufacturer’s instructions. qPCR primers were designed with Primer3Plus software [47] to amplify approximately 130 bp fragments of: sigma70/sigma32 (reference gene), prtS (positive control) and prtH (Additional file 3: qPCR primers). Serial dilutions of each cDNA sample (LMD-9, PrtH+ and PrtH+WANS) were carried out to check efficiency of each primer pair and obtain standard curves (Ct = f(log initial cDNA concentration)). Relative quantities were obtained after comparing to the standard curves and normalized using the following formula: R = (relative quantity of gene of interest)/(relative quantity of reference gene). Ratios obtained for prtH gene in PrtH+ and PrtH+WANS mutants were then compared to that of prtS gene in LMD-9. Quantification was carried out twice independently and the mean and SEM (Standard Error of Mean) were determined.
Surface tryptic digestion of live cells (shaving)
S. thermophilus LMD-9 wild-type and the three mutants were grown at 42°C in 50 mL of 10% reconstituted skim milk to OD480 nm = 2 (exponential growth phase). Cells were washed first in tri-sodium citrate solution 0.25 M (Carlo Erba, Val de Reuil, France) and centrifuged at 500 g, 10 min at 4°C. The supernatants containing bacterial cells were then centrifuged at 8,000 g, 10 min at 4°C and washed at least three times with 100 mM Tris–HCl, pH 7.5 containing 150 mM NaCl to remove the residual caseins. Washed cells were concentrated to OD650 nm = 30 and suspended in PBS containing 5 mM DL-Dithiothreitol (Sigma-Aldrich, St Quentin Fallavier, France) solution, adjusted to pH 8.5. Five μg of sequenced grade trypsin (Promega, Charbonnières, France) were added to 500 μL of the concentrated cell solution. In parallel, negative controls were carried out by adding the trypsin buffer without the enzyme. Cells were incubated 1 hour at 37°C with shaking (180 rpm). Supernatants were harvested by centrifugation at 10,000 g, 10 min at room temperature and filtrated through a 0.45 μm filter (Millex PVDF, 13 mm, Millipore, Molsheim, France). Supernatants were incubated overnight at 37°C with shaking (100 rpm) in presence of 1 μg of trypsin. The reaction was stopped by adding 15 μL TriFluoroacetic Acid (TFA) 10% (v/v) (Sigma Aldrich) and samples were stored at -20°C before mass spectrometry analysis.
Tandem mass spectrometry
Mass spectrometry (MS) experiments were performed using a nanoRSLC Dionex U3000 system fitted to a Q Exactive mass spectrometer (Thermo Scientific, San Jose, USA) equipped with a nanoelectrospray ion source. A preliminary sample concentration step was performed on a nanotrap PepMap 100 (C18, 3 μm, 75 μm Inner Diameter (ID) × 20 mm Length (L)) (Dionex, Amsterdam, Netherlands). Separation was performed on a reverse-phase column PepMap RSLC C18 3 μm, 100 Å (75 μm ID, 150 mm L) (Dionex, Amsterdam, Netherlands) at 35°C, using solvent A (2% (v/v) acetonitrile, 0.08% (v/v) formic acid and 0.01% (v/v) TFA in deionized water) and solvent B (95% (v/v) acetonitrile, 0.08% (v/v) formic acid and 0.01% (v/v) TFA in deionized water). 5-60% of solvent B in 46 min and 60-80% in 1 min was applied as separation gradient at a flow rate of 0.3 μL/min. Eluted peptides were directly electrosprayed into the mass spectrometer operated in positive mode and a voltage of 2 kV with the help of a Proxeon Nanospray Flex ion source (Thermo Scientific, San Jose, USA). Spectra were recorded in full MS mode and selected in a mass range 300–2000 m/z for MS spectra with a resolution of 70,000 at m/z 200. For each scan, the ten more intense ions were selected for fragmentation. MS/MS spectra were recorded with a resolution of 17,500 at m/z 200 and the parent ion was subsequently excluded of the analysis during 15 s. The instrument was externally calibrated according to the supplier’s procedure.
To identify peptides, all data (MS and MS/MS) were submitted to X! Tandem using the X! Tandem pipeline developed by PAPPSO (Plateforme d’Analyse Protéomique de Paris Sud-Ouest (PAPPSO), INRA, Jouy-en-Josas, France, http://pappso.inra.fr).
The search was performed against a database composed of the taxonomy Bacilli from http://www.uniprot.org (Taxon identifier: 91061) to which was added the deduced sequences of the two proteins of PrtH+ and PrtH+WANS. Database search parameters were specified as follows: trypsin cleavage was used and the peptide mass tolerance was set to 10 ppm for MS and 0.02 Da for MS/MS. Oxidation of methionine was selected as a variable modification. Semi-tryptic peptides were allowed during the “refinement” process of X!tandem. For each peptide identified, a minimum score corresponding to an e-value below 0.05 was considered as a prerequisite for peptide validation.
The identified proteins were conserved when at least three specific peptides were identified.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
XL carried out the experiments and contributed to the redaction of the manuscript. VG supervised shaving experiments and results analysis and contributed to the redaction. VBB provided technical assistance for mass spectrometry. JJ provided experience and knowledge for mass spectrometry. SL initiated this work and corrected the manuscript. AD initiated and supervised this work. MG supervised the project and contributed to the redaction of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Proteins identified after cell surface shaving of S. thermophilus strains PrtS - , LMD-9, PrtH + and PrtH + WANS.
Sequencing primers.
Primers used for qPCR experiments.
Contributor Information
Xavier Lecomte, Email: lecomtexa@gmail.com.
Valérie Gagnaire, Email: valerie.gagnaire@rennes.inra.fr.
Valérie Briard-Bion, Email: valerie.briard@rennes.inra.fr.
Julien Jardin, Email: julien.jardin@rennes.inra.fr.
Sylvie Lortal, Email: sylvie.lortal@rennes.inra.fr.
Annie Dary, Email: annie.dary@univ-lorraine.fr.
Magali Genay, Email: magali.genay@univ-lorraine.fr.
Acknowledgements
We are grateful to Dr. Rozenn GARDAN for her advices in CDM and natural transformation methods, Dr. Wessam GALIA for his help in qPCR data analysis and Dr. Gwenaël JAN for his help in shaving method. We also thank Dr. Zeeshan HAFEEZ for his language corrections.
L. helveticus CNRZ32 CIRM-BIA 103 was kindly provided by the Centre International de Ressources Microbiennes-Bactéries d’Intérêt Alimentaire, INRA, Rennes, France.
Xavier LECOMTE is the recipient of a Ph.D. fellowship from the “Ministère de l’Enseignement Supérieur et de la Recherche”. This work was financially supported by the Région Lorraine and the Région Bretagne.
References
- Bernaudat F, Frelet-Barrand A, Pochon N, Dementin S, Hivin P, Boutigny S, Rioux JB, Salvi D, Seigneurin-Berny D, Richaud P, Joyard J, Pignol D, Sabaty M, Desnos T, Pebay-Peyroula E, Darrouzet E, Vernet T, Rolland N. Heterologous expression of membrane proteins: choosing the appropriate host. PLoS One. 2011;6(12):e29191. doi: 10.1371/journal.pone.0029191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Loir Y, Azevedo V, Oliveira SC, Freitas DA, Miyoshi A, Bermudez-Humaran LG, Nouaille S, Ribeiro LA, Leclercq S, Gabriel JE, Guimaraes VD, Oliveira MN, Charlier C, Gautier M, Langella P. Protein secretion in Lactococcus lactis: an efficient way to increase the overall heterologous protein production. Microb Cell Fact. 2005;4:2. doi: 10.1186/1475-2859-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westers L, Westers H, Quax WJ. Bacillus subtilis as cell factory for pharmaceutical proteins: a biotechnological approach to optimize the host organism. Biochim Biophys Acta-Mol Cell Res. 2004;1694:299–310. doi: 10.1016/j.bbamcr.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Morello E, Bermudez-Humaran LG, Llull D, Sole V, Miraglio N, Langella P, Poquet I. Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. J Mol Microbiol Biotechnol. 2008;14:48–58. doi: 10.1159/000106082. [DOI] [PubMed] [Google Scholar]
- Zhou MM, Theunissen D, Wels M, Siezen RJ. LAB-Secretome: a genome-scale comparative analysis of the predicted extracellular and surface-associated proteins of Lactic Acid Bacteria. BMC Genomics. 2010;11:651. doi: 10.1186/1471-2164-11-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wydau S, Dervyn R, Anba J, Ehrlich SD, Maguin E. Conservation of key elements of natural competence in Lactococcus lactis ssp. FEMS Microbiol Lett. 2006;257:32–42. doi: 10.1111/j.1574-6968.2006.00141.x. [DOI] [PubMed] [Google Scholar]
- Johnsborg O, Eldholm V, Havarstein LS. Natural genetic transformation: prevalence, mechanisms and function. Res Microbiol. 2007;158:767–778. doi: 10.1016/j.resmic.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Elli M, Callegari ML, Ferrari S, Bessi E, Cattivelli D, Soldi S, Morelli L, Feuillerat NG, Antoine JM. Survival of yogurt bacteria in the human gut. Appl Environ Microbiol. 2006;72:5113–5117. doi: 10.1128/AEM.02950-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mater DDG, Bretigny L, Firmesse O, Flores MJ, Mogenet A, Bresson JL, Corthier G. Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus survive gastrointestinal transit of healthy volunteers consuming yogurt. FEMS Microbiol Lett. 2005;250:185–187. doi: 10.1016/j.femsle.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Guarner F, Perdigon G, Corthier G, Salminen S, Koletzko B, Morelli L. Should yoghurt cultures be considered probiotic? Br J Nutr. 2005;93:783–786. doi: 10.1079/bjn20051428. [DOI] [PubMed] [Google Scholar]
- Solaiman DKY, Somkuti GA. Construction of a green-fluorescent protein-based, insertion-inactivation shuttle vector for lactic acid bacteria and Escherichia coli. Biotechnol Lett. 1997;19:1175–1179. [Google Scholar]
- Somkuti GA, Solaiman DKY, Johnson TL, Steinberg DH. Transfer and expression of A streptomyces cholesterol oxidase gene in Streptococcus thermophilus. Biotechnol Appl Biochem. 1991;13:238–245. [PubMed] [Google Scholar]
- Renye JA, Somkuti GA. Vector-mediated chromosomal integration of the glutamate decarboxylase gene in Streptococcus thermophilus. Biotechnol Lett. 2012;34:549–555. doi: 10.1007/s10529-011-0802-6. [DOI] [PubMed] [Google Scholar]
- Blomqvist T, Steinmoen H, Havarstein LS. Natural genetic transformation: a novel tool for efficient genetic engineering of the dairy bacterium Streptococcus thermophilus. Appl Environ Microbiol. 2006;72:6751–6756. doi: 10.1128/AEM.01156-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine L, Dandoy D, Boutry C, Delplace B, De Frahan MH, Fremaux C, Horvath P, Boyaval P, Hols P. Development of a versatile procedure based on natural transformation for marker-free targeted genetic modification in Streptococcus thermophilus. Appl Environ Microbiol. 2010;76:7870–7877. doi: 10.1128/AEM.01671-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine L, Goffin P, Dubout H, Delplace B, Baulard A, Lecat-Guillet N, Chambellon E, Gardan R, Hols P. Mechanism of competence activation by the ComRS signalling system in streptococci. Mol Microbiol. 2013;87:1113–1132. doi: 10.1111/mmi.12157. [DOI] [PubMed] [Google Scholar]
- Fontaine L, Boutry C, De Frahan MH, Delplace B, Fremaux C, Horvath P, Boyaval P, Hols P. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J Bacteriol. 2010;192:1444–1454. doi: 10.1128/JB.01251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardan R, Besset C, Guillot A, Gitton C, Monnet V. The oligopeptide transport system is essential for the development of natural competence in Streptococcus thermophilus Strain LMD-9. J Bacteriol. 2009;191:4647–4655. doi: 10.1128/JB.00257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh YJ, Goin C, O’Flaherty S, Altermann E, Hutkins R. Specialized adaptation of a lactic acid bacterium to the milk environment: the comparative genomics of Streptococcus thermophilus LMD-9. Microb Cell Fact. 2011;10(Suppl 1):S22. doi: 10.1186/1475-2859-10-S1-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin JJ, Li RQ, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li JH, Xu JM, Li SC, Li DF, Cao JJ, Wang B, Liang HQ, Zheng HS, Xie YL, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:U59–U70. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandoy D, Fremaux C, De Frahan MH, Horvath P, Boyaval P, Hols P, Fontaine L. The fast milk acidifying phenotype of Streptococcus thermophilus can be acquired by natural transformation of the genomic island encoding the cell-envelope proteinase PrtS. Microb Cell Fact. 2011;10(Suppl 1):S21. doi: 10.1186/1475-2859-10-S1-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadat-Mekmene L, Genay M, Atlan D, Lortal S, Gagnaire V. Original features of cell-envelope proteinases of Lactobacillus helveticus. A review. Int J Food Microbiol. 2011;146:1–13. doi: 10.1016/j.ijfoodmicro.2011.01.039. [DOI] [PubMed] [Google Scholar]
- Broadbent JR, Cai H, Larsen RL, Hughes JE, Welker DL, De Carvalho VG, Tompkins TA, Ardo Y, Vogensen F, De Lorentiis A, Gatti M, Neviani E, Steele JL. Genetic diversity in proteolytic enzymes and amino acid metabolism among Lactobacillus helveticus strains. J Dairy Sci. 2011;94:4313–4328. doi: 10.3168/jds.2010-4068. [DOI] [PubMed] [Google Scholar]
- Genay M, Sadat L, Gagnaire V, Lortal S. prtH2, not prtH, is the ubiquitous cell-wall proteinase gene in Lactobacillus helveticus. Appl Environ Microbiol. 2009;75:3238–3249. doi: 10.1128/AEM.02395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Espla MD, Garault P, Monnet V, Rul F. Streptococcus thermophilus cell wall-anchored proteinase: release, purification and biochemical and genetic characterization. Appl Environ Microbiol. 2000;66:4772–4778. doi: 10.1128/aem.66.11.4772-4778.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen RJ. Multi-domain, cell-envelope proteinases of lactic acid bacteria. Antonie Van Leeuwenhoek. 1999;76:139–155. [PubMed] [Google Scholar]
- Bagos PG, Nikolaou EP, Liakopoulos TD, Tsirigos KD. Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics. 2010;26:2811–2817. doi: 10.1093/bioinformatics/btq530. [DOI] [PubMed] [Google Scholar]
- Pederson JA, Mileski GJ, Weimer BC, Steele JL. Genetic characterization of a cell envelope-associated proteinase from Lactobacillus helveticus CNRZ32. J Bacteriol. 1999;181:4592–4597. doi: 10.1128/jb.181.15.4592-4597.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbal S, Hemme D, Renault P. Characterization of a cell envelope-associated proteinase activity from Streptococcus thermophilus H-strains. Appl Environ Microbiol. 1993;59:177–182. doi: 10.1128/aem.59.1.177-182.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadat-Mekmene L, Jardin J, Corre C, Mollé D, Richoux R, Delage MM, Lortal S, Gagnaire V. Simultaneous presence of PrtH and PrtH2 proteinases in Lactobacillus helveticus Strains improves breakdown of the pure alphas1-casein. Appl Environ Microbiol. 2011;77:179–186. doi: 10.1128/AEM.01466-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human splicesome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcour J, Ferain T, Deghorain M, Palumbo E, Hols P. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie Van Leeuwenhoek. 1999;76:159–184. [PubMed] [Google Scholar]
- Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST plus: architecture and applications. BMC Bioinforma. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis N, Larsen MR, Cordwell SJ. Improved accuracy of cell surface shaving proteomics in Staphylococcus aureus using a false-positive control. Proteomics. 2010;10:2037–2049. doi: 10.1002/pmic.200900564. [DOI] [PubMed] [Google Scholar]
- Olaya-Abril A, Gomez-Gascon L, Jimenez-Munguia I, Obando I, Rodriguez-Ortega MJ. Another turn of the screw in shaving Gram-positive bacteria: Optimization of proteomics surface protein identification in Streptococcus pneumoniae. J Proteomics. 2012;75:3733–3746. doi: 10.1016/j.jprot.2012.04.037. [DOI] [PubMed] [Google Scholar]
- Henderson B, Martin A. Bacterial virulence in the moonlight: Multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect Immun. 2011;79:3476–3491. doi: 10.1128/IAI.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu SM, Kong J, Sun ZL, Han LL, Kong WT, Yang P. Heterologous protein display on the cell surface of lactic acid bacteria mediated by the s-layer protein. Microb Cell Fact. 2011;10:86. doi: 10.1186/1475-2859-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang OK, Perrin C, Galia W, Saulnier F, Miclo L, Roux E, Driou A, Humbert G, Dary A. Release of the cell-envelope protease PrtS in the growth medium of Streptococcus thermophilus 4 F44. Int Dairy J. 2012;23:91–98. [Google Scholar]
- Galia W, Perrin C, Genay M, Dary A. Variability and molecular typing of Streptococcus thermophilus strains displaying different proteolytic and acidifying properties. Int Dairy J. 2009;19:89–95. [Google Scholar]
- Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee JH, Diaz-Muniz I, Dosti B, Smeianov V, Wechter W, Barabote R. et al. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A. 2006;103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi BE, Sandine WE. Improved medium for lactic Streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letort C, Juillard V. Development of a minimal chemically-defined medium for the exponential growth of Streptococcus thermophilus. J Appl Microbiol. 2001;91:1023–1029. doi: 10.1046/j.1365-2672.2001.01469.x. [DOI] [PubMed] [Google Scholar]
- Green MR, Sambrook J. Molecular Cloning: A Laboratory Manual. N. Y.: Cold Spring Harbor Laboratory Press; 2012. [Google Scholar]
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguin E, Prevost H, Ehrlich SD, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopard MA, Schmitt M, Perrerad E, Chamba JF. Qualitative aspect of proteolytic activity of thermophilic lactobacilli using in Swiss cheeses. Lait. 2001;81:183–194. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteins identified after cell surface shaving of S. thermophilus strains PrtS - , LMD-9, PrtH + and PrtH + WANS.
Sequencing primers.
Primers used for qPCR experiments.


