Abstract
Iso-octyl chain-hydroxylated oxysterols were determined in attomoles per 10,000 cells concentrations in 10,000–80,000 cultured pancreatic adenocarcinoma cells, using a sensitive, highly automated nano-LC-ESI-MS-based method. Identified oxysterols included 24S hydroxycholesterol (24S-OHC), 25 hydroxycholesterol (25-OHC), and 27 hydroxycholesterol (27-OHC), while 20S hydroxycholesterol and 22S hydroxycholesterol were not detected. Lower mass limit of quantification was 23 fg (65 amol) for 25-OHC and 27-OHC (100 times lower than our previous method) and 54 fg (135 amol) for 24S-OHC, after derivatization into Girard T hydrazones and online sample cleanup using simplified and robust automatic filtration and filter back flushing solid phase extraction LC/MS/MS. The instrument configuration was easily installed using a commercial nano-LC/MS system. Recoveries in spiked sample were 96, 97, and 77% for 24S-OHC, 25-OHC, and 27-OHC, with within- and between-day repeatabilities of 1–21% and 2–20% relative SD, respectively. The study demonstrates the potential of nano-LC in lipidomics/sterolomics.
Keywords: 24S hydroxycholesterol, 25 hydroxycholesterol, 27 hydroxycholesterol, cholesterol 24-hydroxylase, cholesterol 25-hydroxylase, cholesterol 27-hydroxylase
Oxysterols, which are hydroxylated derivatives of cholesterol, have been implied as activators of the Hedgehog (Hh) signaling pathway through binding to the Smoothened (SMO) receptor (1, 2). The degree of oxysterol activation of SMO is isomer dependent, with iso-octyl chain hydroxylated oxysterols (Fig. 1A) clearly being the most active (3, 4). In addition, the –OH position on the octyl chain is also of great importance for activity [e.g., 25 hydroxycholesterol (25-OHC) is a stronger agonist than 24S hydroxycholesterol (24S-OHC)] (3, 4). Methods are called for to quantify endogenous oxysterol isomers (which are often very low abundance) to fully understand their roles and regulation in Hh signaling (1, 5). In this context, it is advantageous that a method is compatible with small samples (e.g., aggressive tumor cell side populations that are associated with altered signal pathway activity) (6).
Fig. 1.
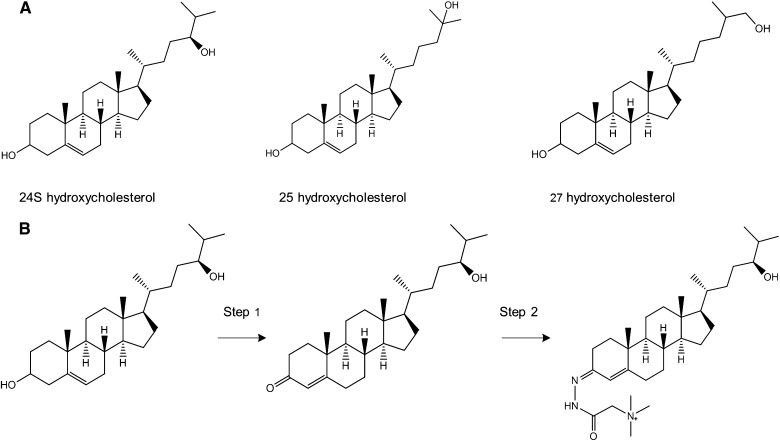
A: The structures of the Hh active oxysterols 24S-OHC, 25-OHC, and 27-OHC. B: Charge tagging of 24S-OHC with Girard T reagent. In step 1, the 3β-hydroxy-5-ene is enzymatically oxidized to 3-oxo-4-ene with cholesterol oxidase (1 h, 37°C). In step 2, the Girard T reagent reacts with the 3-oxo group to form Girard T derivates of oxysterol. See Materials and Methods for more details.
In recent years, LC/MS methods have gained popularity for measuring oxysterols. Due to their neutral nature, oxysterols are not easily ionized with ESI, the most common ionization source in LC/MS. The ionization issue has been solved by, for example, charge tagging of the oxysterols into, for example, picolinyl esters (7), Girard derivates (8–10), and N,N-dimethylglycine esters (11), which enhances sensitivity and selectivity. However, most methods based on the above-mentioned approaches are not optimal for limited cell/tissue samples, as they typically include extensive manual preparation steps and/or analyte-diluting conventional bore LC.
We have developed a nano-LC-based method enabling quantification of Hh active oxysterols in small samples (10,000–80,000 cells). Features of this method include robust, automatic sample purification, one-vial sample preparation, MS-based monitoring for autoxidation, high chromatographic selectivity, targeted mass spectrometric detection, and a 100-fold increased mass sensitivity compared with our previous efforts (9). The targeting method enabled identification and quantification of Hh active 24S-OHC, 25-OHC, and 27 hydroxycholesterol (27-OHC) in cultured pancreatic adenocarcinoma cells (cell line BxPC-3). The compounds 20S hydroxycholesterol (20S-OHC) and 22S hydroxycholesterol (22S-OHC) were not found to be present above the detection limit in our samples.
MATERIALS AND METHODS
Chemicals and cell lines
All chemicals, reagents, and cells were from commercial suppliers and of analytical grade. See supplementary Section I for details of chemicals, suppliers, and cell culturing.
Preparation of standard solutions, calibration solutions, validation samples, and cell samples
Standard solutions and samples for method development and validation.
Standard solutions containing 13, 27, 54, 81, and 108 pM of each oxysterol, with 30 pmol cholesterol-25,26,27-13C as autoxidation-monitoring standard, were prepared from working solutions (1 nM).
For preparation of spiked validation samples, 10,000 cells were lysed in 300 µl absolute ethanol (Kemetyl, Vestby, Norway) containing 120 pmol cholesterol-25,26,27-13C and spiked with standard solutions to give a concentration of 27, 54, and 108 pM of each oxysterol.
Calibration solutions for quantification.
Calibration solutions were prepared from 1 nM working solutions and covered the concentration range 13–108 pM of each oxysterol. New calibration solutions were made for each assay.
Cell samples.
Cells (10,000–80,000) were lysed in 300 µl absolute ethanol (Kemetyl) containing 120 pmol cholesterol-25,26,27-13C. The samples were stored at –80°C before derivatization of oxysterols into Girard T derivates.
Internal standard.
Aliquots of 50 µl of 1.5 nM internal standard working solution were added to all standard, calibration, and validation solutions and cell samples before evaporation into dryness and derivatization with Girard T as described subsequently.
Charge tagging of oxysterol
Standard and calibration solutions, as well as cell and validation samples, were charge tagged with Girard T reagent as described previously (9) with small changes to adapt to the change in sample size; after evaporation into dryness, the residue was redissolved in 20 µl 2-propanol. Aliquots of 200 µl of 30 µg/ml cholesterol oxidase dissolved in 50 mM phosphate buffer pH 7 were added to all solutions and samples to convert the 3β-hydroxy-5-ene to a 3-oxo-4-ene (Fig. 1B, step 1), performed at 37°C for 1 h using a Grant-Bio PHMT thermoshaker (Grant Instruments, Cambridge, UK) set to 300 rpm. To attach the Girard T reagent (Fig. 1B, step 2), 500 µl of a mixture consisting of 15 mg Girard T reagent, 15 µl glacial acetic acid, and 485 µl methanol was added to the sample. This resulted in a final volume of 720 µl of all solutions. The reaction was completed in the dark at room temperature overnight. Schematic view of the sample preparation is shown in supplementary Section II. Derivatized samples were stored at 4°C and analyzed within a week.
Automatic filtration and filter back flushing/solid phase extraction/nano-LC
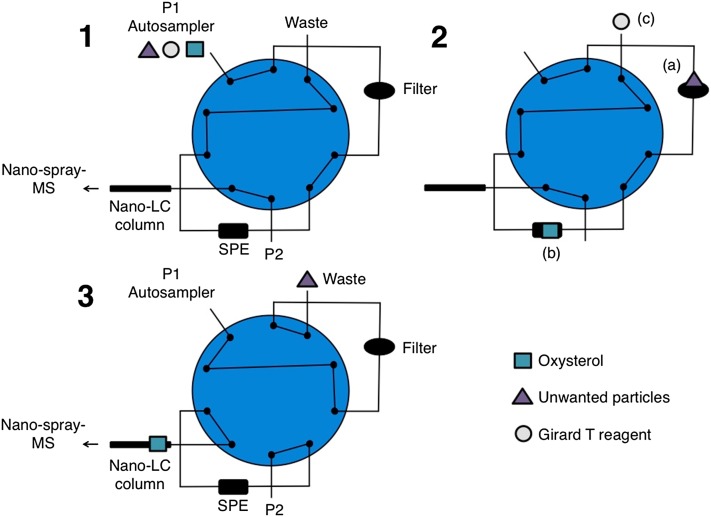
Sample cleanup was performed online using automatic filtration and filter back flushing (AFFL) and online solid phase extraction (SPE), coupled upstream to nano-LC-ESI-Q Exactive MS. With the AFFL setup (Fig. 2 and supplementary Animation I) (12, 13), sample with excess reagent was injected (5 µl) by an Agilent G1377A micro well autosampler (Agilent Technologies, Santa Clara, CA) (Fig. 2, step 1). An Agilent 1100 series pump was used as loading pump and filter back-flush pump [15 µl/min of 0.1% formic acid (aq)]. Cell debris and precipitates/particulate matter from cells were trapped online on a 1 µm Valco (Houston, TX) stainless steel filter (1/16″, 1 µm screen) fitted in a Valco union (1/16″, 0.25 mm bore), while a reversed phase (RP) HotSep Tracy C8 trap column [0.3 mm inner diameter (ID) × 5 mm] from G and T Septech (Ytre Enebakk, Norway) trapped derivatized oxysterols (Fig. 2, step 2). Excess reagent, not sufficiently hydrophobic to be trapped by the RP trap column, was flushed to waste. The valve was subsequently switched after 3.5 min, and the loading pump automatically redirected to back flush the filter unit with 0.1% formic acid (aq) to clean the filter prior to the next injection (Fig. 2, step 3). Simultaneously, an Agilent 1200 series pump eluted the oxysterols from the trap column onto the ACE 3 C18 (0.1 mm ID × 150 mm) RP analytical column (Advanced Chromatography Technologies, Aberdeen, UK) for isocratic separation using a mobile phase consisting of formic acid-water-methanol (0.1:5:95 v/v/v %) (Fig. 2, step 3) at a flow rate of 500 nl/min. Cholesterol residues in the column might accumulate and make a pseudostationary phase (14) and hence disturb the chromatographic separation. To remove cholesterol residues from column, the column was washed with formic acid-water-acetonitrile (ACN) (0.1:5:95 v/v/v %) (40 column volumes) after approximately six injections of cell samples to avoid changing the separation properties of the column. The column switching was performed with a 10-port two-position switching valve from Valco (1/16″, 0.25 mm bore) controlled by the LC pump’s Chemstation software.
Fig. 2.
AFFL using a 10-port two-position switching valve. Sample with excess reagent is injected by autosampler (1). A filter traps all cell debris and particulate matter (2), while Girard T derivates of oxysterols are trapped on the SPE. Excess reagents (more hydrophilic) are flushed into waste. After 3.5 min, the valve is switched automatically (3), and the filter is back flushed by the loading pump simultaneously as the Girard T derivatized oxysterols are eluted from the SPE and onto the nano-LC column for separation. See Materials and Methods for more details and supplementary Animation I. Monitoring of data started when the sample was injected.
MS of oxysterols
MS detection was performed in MS2 mode using a Q Exactive Orbitrap with nanospray flex ion source (Thermo Scientific, Waltham, MA). However, other mass spectrometers (e.g., triple quadrupole) would also be suitable. Ionization was performed in positive mode using a capillary voltage of 2.0 kV. The monitored MS/MS transitions were based on the fragmentation of the Girard T group (see supplementary Section III for illustration of the MS/MS fragmentation) and were 514.44→455.36 (analytes), 520.40→461.36 (internal standard), and 517.40→458.36 (autoxidation monitoring). Collection of data and processing were performed using Xcalibur 2.2 software from Thermo Scientific. More MS instrument parameters (mostly based on recommended settings from the manufacturer) are found in supplementary Section IV.
RESULTS AND DISCUSSION
Our goal was 2-fold: both enabling sensitive, simple, highly automated analysis of small samples (10,000 cell scale) and quantification of 27-carbon iso-octyl hydroxylated oxysterol analytes possibly present in cells. Candidates were 20S-, 22S-, 24S-, 25-, and 27-OHC, which are structural isomers with very similar LC/MS properties.
High sensitivity was achieved by using nano-LC and online sample preparation. Nano-LC columns (50–100 µm ID) allow for compounds to elute in substantially smaller volumes compared with that in more conventional columns (e.g., 1 mm ID or larger). Therefore, analytes enter the ESI-MS in larger concentrations, resulting in enhanced signal. A nanobore column packed with the same stationary phase and particles as used in our previously published microbore LC-based method (9) was used, as the material has good oxysterol separation properties and durability. When replacing our previous microbore LC-ion trap MS methodology with nano-LC coupled with a Q Exactive MS, the mass limit of detection (and quantification) was reduced by a factor of 100 (from 2.5 pg to 23 fg). Separation properties (retention and selectivity) in the nano- and microbore format were similar, as expected.
The oxysterols were charge tagged with Girard T reagent (Fig. 1B, step 2). This established approach was chosen as the entire procedure can be performed in one tube, minimizing sample loss (9). In addition, it is compatible with automated SPE enrichment and cleanup (9).
The Girard T reagent reacts with the keto group as shown in Fig. 1B, step 2. A potential pitfall was that the LC/MS method would not distinguish between species with an already existing keto group and those with keto groups formed during sample preparation. However, analyte analogs with preexisting keto groups were not detected in the analyzed cell samples, investigated by performing Girard T tagging with and without step 1 in Fig. 1B (see supplementary Section V for chromatograms).
The 23 fg mass detection limit of this method corresponds to 13 pM in the vial (720 µl). It is important to notice that the in-vial concentration will be affected by the available number of cells because the resulting total volume is the same for all samples and standard solutions after derivatization. Using more cells per sample will therefore give higher analyte amounts and concentrations, and hence a lower limit of detection per cell. For calculation example, see supplementary Section VI.
A 0.3 mm ID SPE column was coupled online with the nano-LC column. This avoids the overloading effects associated with direct injection of microliter amounts onto nano-LC columns (which can usually only handle nanoliter injections). Such overloading effects may call for extensive column purification routines; in the only previous nano-LC study of oxysterols (to the authors’ knowledge), Karu et al. (10) had to inject a washing solvent between each sample injection to avoid carryover effects. In SPE-nano-LC mode, however, 5 µl injections could easily be SPE trapped, and concentrated analyte bands were subsequently transferred to the LC column in appropriate volumes for satisfactory chromatographic resolution without carryover effects. In addition to allowing rather large injection volumes, online SPE greatly simplifies analysis of our 10,000 cell-scale samples, as excess Girard reagent removal is performed online. This reduces difficult, manual handling of minute samples/fractions, which can be a source of analysis variance (15).
Conventional online SPE-nano-LC can be especially prone to pressure buildups due to the small, relatively easily clogged connections. To avoid this, an AFFL setup was installed upstream to the SPE step. This allows crude samples to be injected directly (here: unfiltered lysates) as remaining cell debris and so forth is trapped on a stainless steel filter and washed out of the system after each sample injection. This reduces off-line sample preparation steps (and hence possible loss of analyte). The AFFL-SPE-nano-LC/MS methodology could handle thousand injections of relatively unprepared samples, representing an unprecedented robustness regarding nano-LC-based instrumentation. Our AFFL-SPE-nano-LC system can be assembled using a commercial SPE-LC system [see (16) for various systems], only requiring replacement of one valve (10 port instead of a 6 port) (13), compared with our original system, which required an additional pump (12).
An additional simplifying aspect of the method was the absence of a (off-line) cholesterol removal step, commonly performed to avoid autoxidation effects that can negatively affect method accuracy. Enzymatically formed side chain-hydroxylated oxysterols (our analytes) are not typically associated with autoxidation; on the other hand, little is known, for example, about the formation of 20S-OHC, as no known enzyme is associated with the formation of this compound. Nonetheless, heavy cholesterol (cholesterol-25,26,27-13C) was added to all samples and calibration solutions. If autoxidation occurred during sample preparation (turning cholesterol into our analytes), heavy cholesterol would be converted to heavy oxysterols. This mass transition was monitored and controlled for each sample with MS. Autoxidation(-like) effects were in fact observed for ∼1 in every 500 samples (these samples were rejected).
The chromatographic resolution of our method revealed that, presumably due to generation of syn/anti forms during the derivatization process (7, 17), several of the analytes appeared as two peaks. As these peaks were well resolved, their areas could be combined for quantification. By using methanol as the LC organic modifier instead of ACN, we were able to quantitatively distinguish 24S-OHC and 27-OHC, which coeluted with ACN as modifier. In spiked samples, 27-OHC and 20S-OHC coeluted with methanol as the organic modifier, but it was possible to resolve 20S-OHC from the other oxysterols when using ACN (see supplementary Section VII for chromatograms). In accordance with other recent studies (9, 18, 19), 20S-OHC was not observed in the analyzed cell samples using ACN as mobile phase, even when the sample amount was increased 8-fold. Hence, methanol could be used as the organic modifier for our method for reliable quantification of 27-OHC.
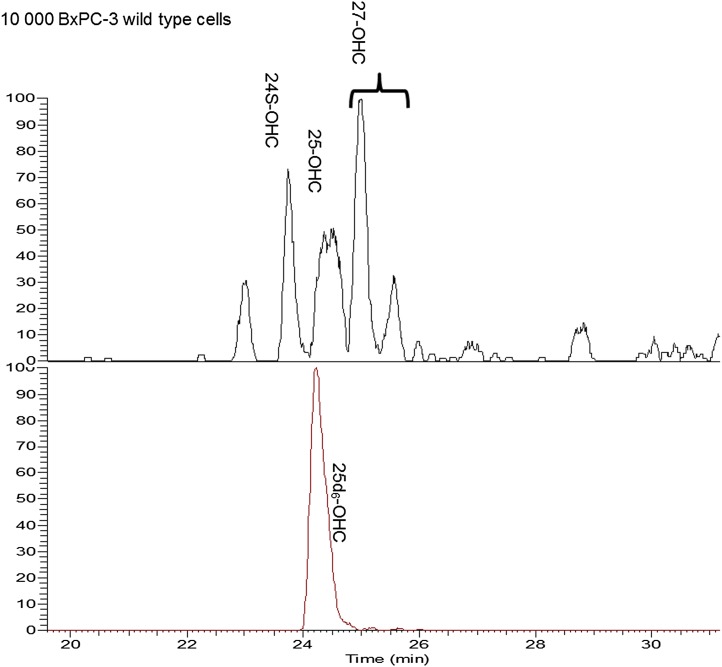
In a pancreatic cell line (BxPC-3) and a derived clonal mutant cell line (Δβcat BxPC-3), the sensitivity of the method allowed detection of 24S-OHC, 25-OHC, and 27-OHC in as little as 10,000 cultured cells (Fig. 3). In contrast, 20S-OHC and 22S-OHC were not detected in the cells even when using larger samples. The compounds 24S-OHC, 25-OHC, and 27-OHC were quantified (38, 19, and 35 fmol/10,000 cells, respectively). In support of these oxysterols being generated in the cells was the within-cell presence of corresponding enzymes responsible for formation of these oxysterols, both at the transcriptional level by RT-PCR and the protein level by Western blot and nano-LC/MS/MS. Also, a consistent and significant upregulation of 27-OHC and its corresponding enzyme (cholesterol 27-oxidase) as function of a cell perturbation (i.e., knockout of β catenin) was observed (for more details, see supplementary Section IX).
Fig. 3.
Extracted ion chromatogram (EIC) from MS/MS analysis of derivatized oxysterols (m/z 514.44→455.36, top) and internal standard (25d6, m/z 520.40→461.40, bottom) in 10,000 BxPC-3 cells.
Validation of the nano-AFFL-SPE-LC/MS/MS method
The concentration limit of detection (cLOD) was defined as the lowest concentration that repeatably produced a chromatographic peak with signal-to-noise ratio >3. The concentration limit of quantification (cLOQ) was defined as the concentration that produced peak areas with a relative SD (RSD) ∼20%. The cLODs were 13 pM derivatized analyte for 24S-OHC, 25-OHC, and 27-OHC, corresponding to 23 fg (65 amol) in 5 µl injected on column. cLOQ was also 13 pM for 25-OHC and 27-OHC, while for 24S-OHC, the cLOD was 27 pM (54 fg, 135 amol injected on column).
Linearity of the method in spiked samples was examined in the range 27–108 pM, and the correlation coefficient (r2) for all linearity curves was >0.99 for all analytes (three concentration points, three spiked samples per concentration point). The slope of the curves corresponded to that in standard solutions (five concentration points, three standard solutions per point). Standard solutions could therefore be used as calibration solutions. Ideally, calibration solutions should be based on cell samples, but this was not possible due to lack of cell matrix without oxysterols. The linear concentration range examined covered the concentration range observed in studied samples.
Repeatability was examined for low (13 pM), medium (54 pM), and high (108 pM) concentration levels. Three individually prepared standard solutions per level were injected over 4 days (total of 12 standard solutions for each concentration level) for all cell-detected analytes. Within-day repeatability at low concentration level was between 2% and 21% RSD for 25-OHC and 27-OHC. The between-day repeatability was between 15% and 20% RSD for 25-OHC and 27-OHC (but somewhat higher for 24S-OHC, 27% RSD) at low concentration level and 2–6% RSD at medium and high levels for all analytes (4 days).
Recovery [or more correctly, the apparent recovery (20)] was examined by comparing samples spiked at three concentration levels with standard solutions of the same concentration levels, low (27 pM), medium (54 pM), and high (108 pM). The recovery was 96, 97, and 77% for 24S-OHC, 25-OHC, and 27-OHC, respectively. See supplementary Section VIII for more details (linearity curves, equations, etc.).
CONCLUSIONS
A highly sensitive, robust, and automated method for quantification of the natural iso-octyl chain hydroxylated oxysterols 24S-OHC, 25-OHC, and 27-OHC was successfully developed and validated and applied to measure oxysterols in small samples (≥10,000 cells). The study illustrates robust and successful use of nanoscale LC in targeted lipidomics/sterolomics, an approach that is usually “reserved” for proteomics. The identification/presence of the endogenous oxysterols in BxPC-3 cells was supported by identification of corresponding enzymes responsible for formation of these oxysterols within the cells. The method will be further used in studying endogenous side chain-hydroxylated oxysterols in the Hh pathway in the context of cancer.
Supplementary Material
Footnotes
Abbreviations:
- ACN
- acetonitrile
- AFFL
- automatic filtration and filter back flushing
- cLOD
- concentration limit of detection
- Hh
- Hedgehog
- ID
- inner diameter
- RP
- reversed phase
- RSD
- relative SD
- SPE
- solid phase extraction
- 20S-OHC
- 20S hydroxycholesterol
- 22S-OHC
- 22S hydroxycholesterol
- 24S-OHC
- 24S hydroxycholesterol
- 25-OHC
- 25 hydroxycholesterol
- 27-OHC
- 27 hydroxycholesterol
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of nine supplementary data sections and one animation.
REFERENCES
- 1.Nachtergaele S., Mydock L. K., Krishnan K., Rammohan J., Schlesinger P. H., Covey D. F., Rohatgi R. 2012. Oxysterols are allosteric activators of the oncoprotein Smoothened. Nat. Chem. Biol. 8: 211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberg-Larsen H., Strand M. F., Krauss S., Wilson S. R. 2014. Metabolites in vertebrate Hedgehog signaling. Biochem. Biophys. Res. Commun. In press. [DOI] [PubMed] [Google Scholar]
- 3.Corcoran R. B., Scott M. P. 2006. Oxysterols stimulate Sonic hedgehog signal transduction and proliferation of medulloblastoma cells. Proc. Natl. Acad. Sci. USA. 103: 8408–8413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dwyer J. R., Sever N., Carlson M., Nelson S. F., Beachy P. A., Parhami F. 2007. Oxysterols are novel activators of the hedgehog signaling pathway in pluripotent mesenchymal cells. J. Biol. Chem. 282: 8959–8968 [DOI] [PubMed] [Google Scholar]
- 5.Sharpe H. J., de Sauvage F. J. 2012. Signaling: an oxysterol ligand for Smoothened. Nat. Chem. Biol. 8: 139–140 [DOI] [PubMed] [Google Scholar]
- 6.Dembinski J. L., Krauss S. 2009. Characterization and functional analysis of a slow cycling stem cell-like subpopulation in pancreas adenocarcinoma. Clin. Exp. Metastasis. 26: 611–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honda A., Yamashita K., Hara T., Ikegami T., Miyazaki T., Shirai M., Xu G., Numazawa M., Matsuzaki Y. 2009. Highly sensitive quantification of key regulatory oxysterols in biological samples by LC-ESI-MS/MS. J. Lipid Res. 50: 350–357 [DOI] [PubMed] [Google Scholar]
- 8.Griffiths W. J., Hornshaw M., Woffendin G., Baker S. F., Lockhart A., Heidelberger S., Gustafsson M., Sjovall J., Wang Y. 2008. Discovering oxysterols in plasma: a window on the metabolome. J. Proteome Res. 7: 3602–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberg-Larsen H., Strand M. F., Grimsmo A., Olsen P. A., Dembinski J. L., Rise F., Lundanes E., Greibrokk T., Krauss S., Wilson S. R. 2012. High sensitivity measurements of active oxysterols with automated filtration/filter backflush-solid phase extraction-liquid chromatography–mass spectrometry. J. Chromatogr. A. 1255: 291–297 [DOI] [PubMed] [Google Scholar]
- 10.Karu K., Turton J., Wang Y., Griffiths W. J. 2011. Nano-liquid chromatography-tandem mass spectrometry analysis of oxysterols in brain: monitoring of cholesterol autoxidation. Chem. Phys. Lipids. 164: 411–424 [DOI] [PubMed] [Google Scholar]
- 11.Jiang X., Ory D. S., Han X. 2007. Characterization of oxysterols by electrospray ionization tandem mass spectrometry after one-step derivatization with dimethylglycine. Rapid Commun. Mass Spectrom. 21: 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Svendsen K. O., Larsen H. R., Pedersen S. A., Brenna I., Lundanes E., Wilson S. R. 2011. Automatic filtration and filter flush for robust online solid-phase extraction liquid chromatography. J. Sep. Sci. 34: 3020–3022 [DOI] [PubMed] [Google Scholar]
- 13.Røen B. T., Sellevåg S. R., Dybendal K. E., Lundanes E. 2014. Trace determination of primary nerve agent degradation products in aqueous soil extracts by on-line solid phase extraction - liquid chromatography - mass spectrometry using ZrO2 for enrichment. J. Chromatogr. A. 1329: 90–97 [DOI] [PubMed] [Google Scholar]
- 14.Ogden P. B., Coym J. W. 2011. Retention mechanism of a cholesterol-coated C18 stationary phase: van’t Hoff and Linear Solvation Energy Relationships (LSER) approaches. J. Chromatogr. A. 1218: 2936–2943 [DOI] [PubMed] [Google Scholar]
- 15.Hyötyläinen T. 2009. Critical evaluation of sample pretreatment techniques. Anal. Bioanal. Chem. 394: 743–758 [DOI] [PubMed] [Google Scholar]
- 16.Rogeberg M., Malerod H., Roberg-Larsen H., Aass C., Wilson S. R. 2014. On-line solid phase extraction-liquid chromatography, with emphasis on modern bioanalysis and miniaturized systems. J. Pharm. Biomed. Anal. 87: 120–129 [DOI] [PubMed] [Google Scholar]
- 17.Griffiths W. J., Crick P. J., Wang Y. 2013. Methods for oxysterol analysis: past, present and future. Biochem. Pharmacol. 86: 3–14 [DOI] [PubMed] [Google Scholar]
- 18.McDonald J. G., Smith D. D., Stiles A. R., Russell D. W. 2012. A comprehensive method for extraction and quantitative analysis of sterols and secosteroids from human plasma. J. Lipid Res. 53: 1399–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myers B. R., Sever N., Chong Y. C., Kim J., Belani J. D., Rychnovsky S., Bazan J. F., Beachy P. A. 2013. Hedgehog pathway modulation by multiple lipid binding sites on the Smoothened effector of signal response. Dev. Cell. 26: 346–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burns D. T., Danzer K., Townshend A. 2002. Use of the term “recovery” and “apparent recovery” in analytical procedures (IUPAC Recommendations 2002). Pure Appl. Chem. 74: 2201–2205 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.