Abstract
Sphingosine kinases (SphKs), of which there are two isoforms, SphK1 and SphK2, have been implicated in regulation of many important cellular processes. We have developed an assay for monitoring SphK1 and SphK2 activity in real time without the need for organic partitioning of products, radioactive materials, or specialized equipment. The assay conveniently follows SphK-dependent changes in 7-nitro-2-1,3-benzoxadiazol-4-yl (NBD)-labeled sphingosine (Sph) fluorescence and can be easily performed in 384-well plate format with small reaction volumes. We present data showing dose-proportional responses to enzyme, substrate, and inhibitor concentrations. The SphK1 and SphK2 binding affinities for NBD-Sph and the IC50 values of inhibitors determined were consistent with those reported with other methods. Because of the versatility and simplicity of the assay, it should facilitate the routine characterization of inhibitors and SphK mutants and can be readily used for compound library screening in high-throughput format.
Keywords: sphingosine kinase 1, sphingosine kinase 2, NBD-sphingosine, red emission excitation shift, fluorescence sphingosine kinase assay
Sphingosine kinases (SphK), consisting of two isoforms, SphK1 and SphK2, catalyze the formation of sphingosine-1-phosphate (S1P) by transferring the γ-phosphate from ATP to the primary hydroxyl of sphingosine (Sph). S1P is a potent bioactive lipid that can regulate many complex cellular processes, including growth, survival, migration, angiogenesis, and inflammation, to name a few (1–3). Accordingly, abnormal regulation of SphKs that leads to elevated levels of S1P has been implicated in the etiology of cancer, as well as autoimmune and cardiovascular diseases (3). Hence, pharmacologically modulating SphK activity has important therapeutic potential, and technologies that can assist in the discovery of SphK inhibitors are critical tools for their development.
Currently, there are a wide variety of assays that are useful for the characterization of SphK activity in cell extracts or of the recombinant enzymes. However, most lack the capacity to follow SphK activity in real time, precluding the convenient characterization of SphK mutants, competitors, or inhibitors in a high-throughput or “one-pot” approach. The most widely cited method was originally developed by our group and utilizes [γ-32P]ATP and D-erythro-Sph as substrates, and it requires differential organic extraction, TLC separation, and quantification of the radiolabeled S1P product (4). This method is highly accurate but is labor intensive and not high throughput. More specialized variations of the radiolabeling assay have been developed. For example, those that utilize [3H]Sph as a substrate (5) or biotinylated-Sph with [γ-32P]ATP and capture of the radiolabeled phosphorylated product with streptavidin (6), or a method that conveniently avoids organic extraction and TLC separation and quantifies [32P]S1P by scintillation proximity counting (7). However, as with the classic [γ-32P]ATP method, these are not useful for monitoring reactions in real time and also have the drawback of requiring radioactive materials.
Fluorophore-labeled Sphs have also been extensively used as substrates for examining SphK activity (8, 9) and have been shown to be useful surrogates for Sph in the screening of inhibitors. However, these are not high-throughput assays because in order to measure enzymatic activity the phosphorylated products must be isolated by organic fractionation (10) or processed with specialized equipment by capillary electrophoresis (9) or HPLC (11).
A bioluminescence assay has been developed to measure ADP produced when the γ-phosphate from ATP is transferred to Sph (12). However, this is an indirect measure of SphK activity through ATP hydrolysis rates. Among the most accurate methods are those that fractionate and quantify unlabeled reaction products by tandem LC/MS (13, 14). For many researchers, this is not a cost-effective alternative for high-throughput screening of small-molecule libraries as it also requires highly specialized expertise and equipment.
In this report, we present a novel NBD-Sph-based SphK assay that can be used to monitor enzymatic activity in real time. This method is highly sensitive when used with a fluorescence plate reader, can be performed in 384-well format with very small reaction volumes, and can be used to examine the activity of both SphK1 and SphK2. Moreover, assays can also be monitored using absorption at 550 nm in a spectrophotometer or a plate reader, significantly extending and simplifying assay capability by eliminating the need for specialized equipment.
MATERIALS AND METHODS
Materials
The 18-NBD-Sph [omega(7-nitro-2-1,3-benzoxadiazol-4-yl)(2S,3R,4E)-2-aminooctadec-4-ene-1,3-diol] and 18-NBD-S1P were from Avanti Polar Lipids (Alabaster, AL). The (R)-[1-(4-{[3-methyl-5-(phenylsulfonylmethyl)phenoxy]methyl} benzyl) pyrrolidin-2-yl]methanol (PF-543), Escherichia coli Rossetta-2 cells, and pET16b were from EMD-Millipore (Billerica, MA). The n-octyl-β-d-glucopyranoside was from Affymetrix (Santa Clara, CA). All other reagents and chemicals were reagent grade and purchased from Sigma-Aldrich.
Protein expression and purification
Homo sapiens SphK1 (residues 2–384 with an N-terminal MGHHHHHHHHHHSSG tag) and H. sapiens SphK2 (residues 2–618 with an N-terminal MGHHHHHHHHHHSSGHIEGRQL tag) were cloned into a modified pET16b bacterial expression vector. For expression, transformed E. coli Rossetta-2 cells were grown in Luria broth with appropriate antibiotics at 37°C with shaking until optical density at 600 nm = 0.4, then the temperature was reduced to 18°C. After 3 h at 18°C, expression was induced overnight with 0.1 mM isopropyl β-d-1-thiogalactopyranoside. Cells were harvested by centrifugation and resuspended at a ratio of 10 ml buffer per liter of initial culture in ice-cold buffer: SphK1 resuspension buffer contained 30 mM Tris-HCl [pH 7.4], 300 mM NaCl, 10% glycerol, 5 mM imidazole, 5 mM MgCl2, 5 mM β-mercaptoethanol, and 2.5 mM octyl β-d-glucopyranoside; SphK2 resuspension buffer contained 30 mM Tris-HCl [pH 7.4], 1 M KCl, 10% glycerol, 5 mM imidazole, 5 mM β-mercaptoethanol, and 2.5 mM n-octyl-β-d-glucopyranoside. Cells were disrupted by sonication and centrifuged for 45 min at 25,000 g to remove cell debris, and the supernatants incubated at 4°C with 100 µl Ni-NTA Agarose (Qiagen, Valencia, CA) slurry per liter of initial culture volume (preequilibrated with resuspension buffer described previously) for 2 h with gentle shaking. The resin was extensively washed in batch with resuspension buffer, followed by washing with 2 ml per liter of initial culture volume with resuspension buffer containing 0.5 mM ATP and 5 mM MgCl2. SphKs were eluted with a step gradient containing 20, 30, 50, 60, 70, and 200 mM imidazole. Fractions containing SphK1 were pooled, concentrated, and loaded onto a 16/60 Superdex S75 (GE Healthcare, Pittsburgh, PA) column (120 ml) preequilibrated with 30 mM Tris-HCl [pH 7.4], 300 mM NaCl, 10% glycerol, and 2.5 mM n-octyl-β-d-glucopyranoside, and run at a flow rate of 0.1 ml/min in an AKTA fast protein LC system (GE Healthcare). SphK1 eluted as two peaks, one at a position expected for a monomer and another in the void volume of the column. Fractions corresponding to the monomeric volume were pooled and concentrated to 1 mg/ml (ε = 48,275 M−1 cm−1). SphK2 was similarly purified on 16/60 Superdex S200 column preequilibrated with 30 mM Tris-HCl [7.5], 1 M KCl, 10% glycerol, 3 mM n-octyl-β-d-glucopyranoside, and 1 mM DTT. SphK2 also eluted as two peaks, one in the void volume and another at a volume consistent with monomeric SphK2. Monomeric SphK2 was concentrated to 0.1 mg/ml (ε = 63,660 M−1 cm−1).
NBD-Sph substrate preparation
A stock solution of NBD-Sph in chloroform-methanol (8:2) was dried under nitrogen in a glass tube, solubilized in 5% Triton X-100, and sonicated extensively in a water bath with alternating vigorous vortexing. Concentrated stocks of 3.5 mM NBD-Sph in 5% Triton X-100 were stored frozen at −20°C until use. However, after thawing and prior to use in assays, NBD-Sph solutions were extensively resonicated with alternating vigorous vortexing.
Fluorescence Sph kinase assay
Fluorescence emission was measured with a TECAN Infinite M1000 fluorescence plate reader (Männedorf, Switzerland) at 37°C in 384-well polypropylene plates (Greiner Bio-One, Frickenhausen, Germany). Excitation wavelength was 550 nm, and emission wavelength was set at 584 nm with a 5 nm band-pass. Reactions analyzed using absorption were performed in quartz cuvettes with a Cary 50 Bio UV-visible spectroscopy (UV/Vis) Spectrophotometer (Agilent, Santa Clara, CA), and data were collected either in scanning kinetics mode (400–600 nm) or at 550 nm. Prior to the initiation of reactions, solutions were allowed to equilibrate, as indicated, at either 25°C or 37°C for 10 min. In most cases, assays were initiated with 20× ATP-Mg (20 mM ATP, 200 mM MgCl2, 900 mM Tris-HCl, pH 7.4), after which 384-well plates were shaken or cuvettes were mixed for 15 s for complete mixing. All assays were prepared as master mixes immediately before use in either SphK1 or SphK2 reaction buffer containing enzyme, NBD-Sph, and inhibitors where indicated. SphK1 reaction buffer contained 30 mM Tris-HCl, pH 7.4, 0.05% Triton X-100, 150 mM NaCl, 10% glycerol, 1 mM Na3VO4, 10 mM NaF, and 10 mM β-glycero-phosphate. SphK2 reaction buffer contained 30 mM Tris-HCl, pH 7.4, 0.05% Triton X-100, 200 mM KCl, and 10% glycerol. Due to the poor solubility of NBD-Sph, optimal assay conditions contain 0.05–0.1% Triton X-100. It was important to add NBD-Sph to the master mix last and for the solution to be immediately and extensively vortexed just prior to use. Concentrations of DMSO up to 2% had no appreciable effects on catalytic rates and were used where indicated.
RESULTS AND DISCUSSION
SphK-dependent changes in visible spectra of NBD-Sph
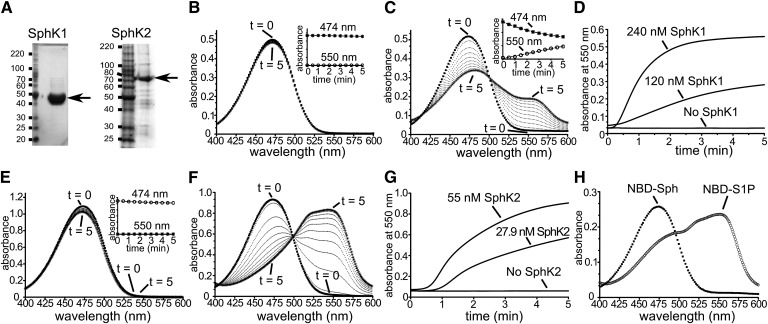
The 15-NBD-Sph, with a shorter sphingoid base, has previously been used for measurements of SphK1 activity (10). During the time course of this assay, samples were collected and sphingolipids extracted with chloroform-methanol (2:1) in alkaline conditions (10). This effectively separated the phosphorylated and nonphosphorylated NBD-labeled Sphs into aqueous and organic phases, respectively. When carrying out this activity assay with recombinant purified SphK1 (Fig. 1A) and 18-NBD-Sph, designated hereafter as NBD-Sph, and prior to organic partitioning, we observed a distinct color change from bright yellow to light red. This led us to examine whether these color changes were quantifiable and SphK dependent. In the presence of SphK1 but lacking ATP (Fig. 1B), or in the absence of both SphK1 and ATP (Fig. 1H), NBD-Sph (30 µM) in 0.05% Triton X-100 had an absorption spectrum with a λmax around 474 nm that was stable and consistent with the known properties of the NBD fluorophore. However, upon addition of SphK1 and ATP, the 474 nm absorbance rapidly decayed and was accompanied by a subsequent rise of a new chromogenic peak around 550 nm (Fig. 1C). Decay of the 474 nm peak was also accompanied by a transient λmax shift of the 474 nm peak to 480 nm (Fig. 1C). Importantly, the rate of increase in absorbance at 550 nm was proportional to the concentration of SphK1 (Fig. 1D), and in control reactions no changes were observed in the absence of either SphK1 or ATP. Similarly, assays with purified recombinant SphK2 (Fig. 1A) showed that NBD-Sph absorbance at 474 nm was stable in SphK2 reaction buffer containing SphK2 but lacking ATP (Fig. 1E), but decay at 474 nm was observed with a rise in absorbance at 550 nm when ATP was added (Fig. 1F). The rate of increase in absorbance at 550 nm was also proportional to SphK2 concentration (Fig. 1G). Importantly, in contrast to NBD-Sph, NBD-S1P in reaction buffer (Fig. 1H) had spectroscopic properties similar to the new chromogenic peaks observed in Fig. 1C and , with a λmax around 550 nm and a shift of the 474 nm peak to 480 nm. However, when similar SphK assays were carried out with the short chain 14-NBD-Sph (15), no changes in absorption were observed as those seen with 18-NBD-Sph (data not shown).
Fig. 1.
SphK1- and SphK2-dependent changes in NBD-Sph visible absorption spectra. A: SDS-PAGE analysis of purified recombinant SphK1 and SphK2 stained with Coomassie brilliant blue G250. B, E: Absorption spectra of NBD-Sph (30 µM) in SphK1 buffer containing 0.05% Triton X-100 and 100 nM SphK1 but lacking ATP (B), or NBD-Sph (60 µM) in SphK2 buffer containing 0.07% Triton X-100, 200 mM KCl, and 100 nM SphK2 but lacking ATP (E). Spectra were recorded every 30 s for 5 min. Insets: Absorbances at 474 nm and 550 nm are unchanged. C, F: Time-resolved scanning absorption spectra of reaction mixtures in B or E after addition of 1 mM ATP. D, G: Time-resolved changes in absorbance at 550 nm of SphK1 (D) or SphK2 (G) assay mixtures containing the indicated concentrations of SphK1 or SphK2, respectively. H: Absorption spectra of NBD-Sph (12.5 µM) and NBD-S1P (12.5 µM) in assay buffer containing 0.05% Triton X-100.
SphK-dependent changes in NBD-Sph fluorescence emission
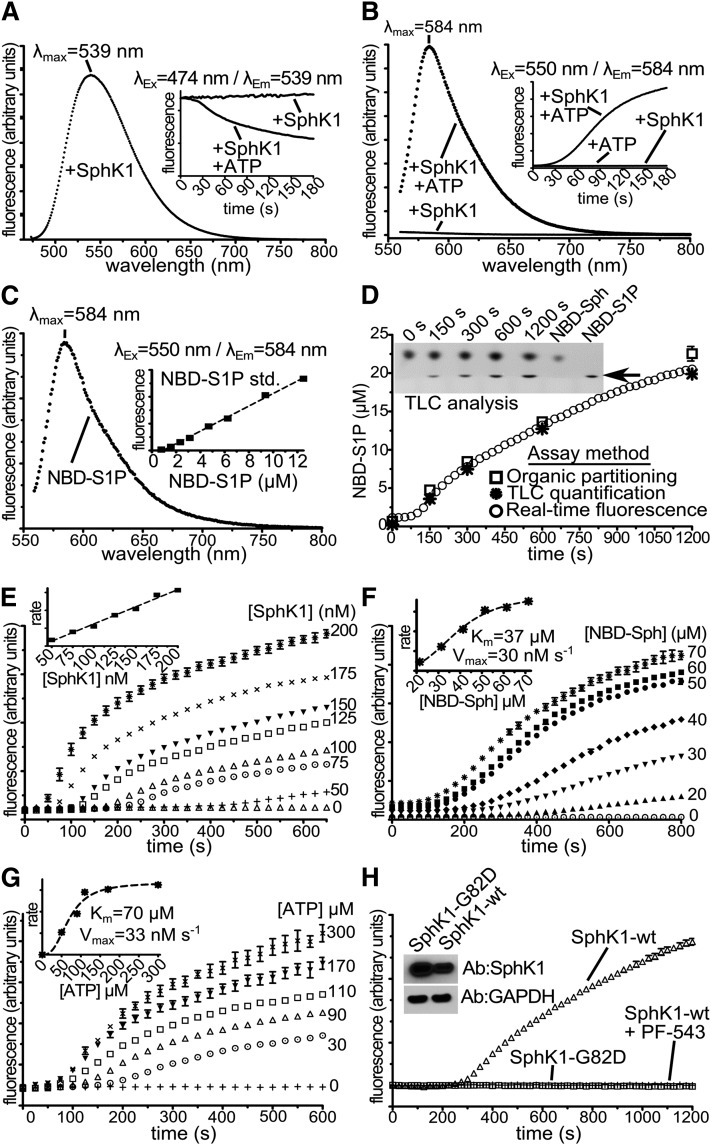
A shift in the wavelength of maximum fluorescence emission of a fluorophore toward higher wavelengths due to a red shift in the excitation wavelength is termed red edge excitation shift (REES). The NBD fluorophore is highly sensitive to its environment, and the REES of NBD-labeled lipids has been used to monitor a variety of biological and biophysical processes in cellular and/or artificial lipid membranes (16, 17). Therefore, we next examined whether the NBD-Sph spectral changes induced by SphK1 or SphK2 activity were accompanied by a corresponding shift in fluorescence emission. To this end, we collected NBD-Sph fluorescence emission spectra upon excitation at 474 nm (Ex474 nm) or 550 nm (Ex550 nm) in the absence or presence of SphKs and/or ATP. As expected, with Ex474 nm, in the presence of SphK1 without ATP, NBD-Sph fluorescence emission spectrum was stable with λmax at 539 nm (Fig. 2A), which rapidly decayed upon addition of ATP (Fig. 2A, inset). Similarly, with Ex550 nm in the presence of SphK1 or SphK2 but lacking ATP, NBD-Sph had little to no emission at 584 nm (Em584 nm) (Figs. 2B and 3A). However, when NBD-Sph was incubated with SphK1 or SphK2 and ATP for 5 min at 25°C, there was a concomitant rise in Ex550/Em584 nm emission (Fig. 2B, inset and Fig. 3A, inset). Importantly, with Ex550 nm, NBD-S1P in reaction buffer (containing 0.05% Triton X-100) had a fluorescence emission spectrum (Fig. 2C) similar to that shown in Figs. 2B and Fig. 3A, with a λmax at 584 nm, and Ex550/Em584 nm emission was proportional to the NBD-S1P concentration over a broad range (Fig. 2C, inset). Thus, NBD-S1P and the fluorogenic species that appears in reactions containing NBD-Sph and SphK have identical spectroscopic properties. Furthermore, TLC analysis (4) of the reactions clearly demonstrates the formation of a product with the same mobility as the NBD-S1P standard (Fig. 2D, inset).
Fig. 2.
SphK1 activity-dependent changes in NBD-Sph fluorescence emission. A–C: λEx = 474 nm (A) or λEx = 550 nm (B, C) fluorescence emission spectra of NBD-Sph (A, B) (30 µM) or NBD-S1P (C) (25 µM) at 25°C in SphK1 reaction buffer containing 0.05% Triton X-100, 100 nM SphK1 (A, B), without or with ATP as indicated (A, B). Insets: Time-resolved fluorescence emission with λEx = 474 nm/λEm = 539 nm (A) or λEx = 550 nm/λEm = 584 nm (B) in the absence or presence of 1 mM ATP as indicated, or λEx = 550 nm/λEm = 584 nm (C) emission of NBD-S1P standards (n = 3). D: Parallel reactions at 37°C with 50 µM NBD-Sph, 150 nM SphK1, 1 mM ATP, and 0.1% Triton X-100 were monitored by TLC (4), organic partitioning (10), or with time-resolved fluorescence emission (λEx = 550 nm/λEm= 584 nm), and turnover determined with a NBD-S1P standard curve. n = 3. Initial rates for λEx = 550 nm/λEm= 584 nm emission were calculated from 125 to 400 s. E–G: Time-resolved fluorescence emission at 37°C with λEx = 550 nm/λEm= 584 nm of reactions containing 0.1% Triton X-100, 1 mM ATP (E, F), 50 µM NBD-Sph (E, G), 150 nM SphK1 (F, G), and increasing SphK1 (E), NBD-Sph (F), or ATP (G) concentrations. n = 3 (E); n = 4 (F); n = 3 (G). Insets: Initial rates versus [SphK1] (E), initial rates versus [NBD-Sph] (F), and initial rate versus [ATP] (G) plots. H: λEx = 550 nm/λEm= 584 nm time-resolved fluorescence emission at 37°C of reactions containing 0.1% Triton X-100, 1 mM ATP, 1.2 µg of lysates from HEK293T cells overexpressing SphK1-WT with or without 250 nM PF-543 as indicated, or catalytically inactive SphK1-G82D. Inset: Immunoblots of extracts probed with the indicated antibodies. Experiments in D–H were performed in 384-well 20 µl format in a fluorescence plate reader.
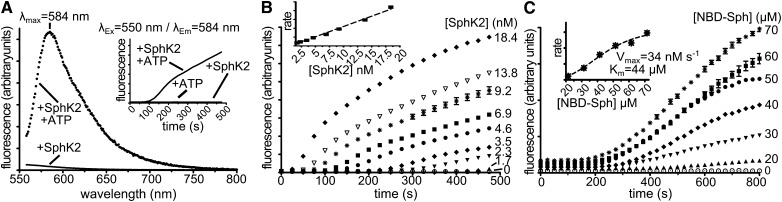
Fig. 3.
SphK2 activity-dependent changes in NBD-Sph fluorescence emission. A: λEx = 550 nm fluorescence emission spectra of NBD-Sph (75 µM) at 25°C in SphK2 reaction buffer containing 0.07% Triton X-100, 200 mM KCl, and 13.8 nM SphK2, but no ATP. Inset: Time-resolved fluorescence emission with λEx = 550 nm and λEm = 583 nm in the absence or presence of ATP or SphK2 as indicated. B: Time-resolved fluorescence emission of NBD-Sph (75 µM) at 37°C in SphK2 reaction buffer containing 0.07% Triton X-100, 200 mM KCl, 1 mM ATP, and increasing SphK2 concentrations (n = 3). Inset: Initial rates versus [SphK2] plot. C: Time-resolved fluorescence emission at 37°C of solutions containing 0.1% Triton X-100, 200 mM KCl, 1 mM ATP, 6.9 nM SphK2, and increasing NBD-Sph concentrations. n = 3. Inset: Initial rates versus [NBD-Sph] plot. Experiments in B and C were performed in 384-well 20 µl format in a fluorescence plate reader. Fluorescence emission was measured with λEx = 550 nm and λEm= 583 nm.
Fluorescence SphK assays in 384-well plate format
The ability to perform enzymatic assays quickly, quantifiably, and in high throughput is critical to accurately screen large inhibitor libraries and hence accelerate drug discovery. Moreover, fluorescence is much more sensitive than UV/Vis absorption, which allows reaction volumes and reagents to be decreased. Because of the greater signal-to-noise ratio, we conducted experiments to validate SphK activity assays in high-throughput 384-well format with Ex550/Em584 nm fluorescence emission. Reactions in a total volume of 20 µl were initiated by the addition of 1 µl of 20× ATP-Mg. Ex550/Em584 nm fluorescence emission progress curves were recorded in the presence of increasing amounts of SphK1 (Fig. 2E) and SphK2 (Fig. 3B). In control reactions without ATP, no changes in Ex550/Em584 nm fluorescence emission were observed over the time course of reactions. Increasing amounts of SphK1 (Fig. 2E) or SphK2 (Fig. 3B) showed higher rates and maximum fluorescence emission signal at 584 nm. Initial rates estimated from these experiments showed linearity over a wide range of SphK1 (Fig. 2E, inset) and SphK2 (Fig. 3B, inset) concentrations. There was a short lag in the appearance of Ex550/Em584 nm emission, and initial rates were fit to the linear portion of progress curves. Because the lag was inversely proportional to enzyme concentration, it is likely due to the requirement of sufficient formation of NBD-S1P for the red shift to be detectable. Rates of NBD-S1P formation quantified by Ex550/Em584 nm emission normalized with internal NBD-S1P standards were identical to those determined by TLC (4) or by organic partitioning of NBD-Sph and NBD-S1P (10) (Fig. 2D). Moreover, kcat calculated from Ex550/Em584 nm emission (0.16 s−1) was in good agreement with the previously reported value for 15-NBD-Sph (0.4 s−1) (10).
We next performed experiments that examined fluorescence emission rates at fixed enzyme/ATP concentration and increasing amounts of NBD-Sph. Progress curves with SphK1 (Fig. 2F) or SphK2 (Fig. 3C) showed proportional increases in fluorescence at concentrations below 50 µM, which plateaued at higher concentrations. Data fitting analysis of initial rates estimated apparent Km values of 38 µM for SphK1 and 44 µM for SphK2, which are consistent with the reported value for SphK1 with 15-NBD-Sph (10). Both SphK1 (Fig. 2G) and SphK2 (data not shown) also showed dose-dependent rate increases as a function of ATP concentration. The apparent calculated Km for ATP with SphK1 was 70 µM (Fig. 2G, inset), which is in excellent agreement with previous reports (18).
It was of interest to examine whether this assay could be used to measure endogenous basal or overexpressed SphK activity in mammalian cells. The assay can readily detect NBD-S1P formation in lysates from cells overexpressing mammalian SphK1, but as expected, no activity was detected in lysates from cells overexpressing mammalian catalytically inactive SphK1 (Fig. 2H). However, the signal is too low to accurately measure basal SphK activity. Importantly, overexpressed SphK1 activity was strongly inhibited by PF-543 (19), showing specificity to a SphK1 catalyzed reaction (Fig. 2H).
Fluorescence assays in 384-well plate format for SphK inhibitor studies
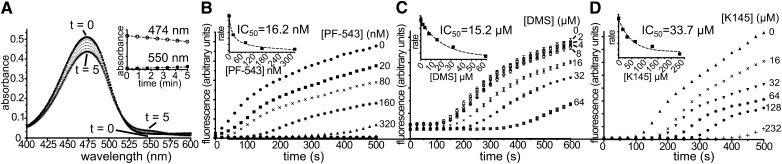
It was next important to determine whether the REES fluorescence SphK assay could be used to screen for small-molecule inhibitors of SphK1 or SphK2. As a first approach, we examined SphK1 activity in the presence of the potent SphK1 inhibitor PF-543 (250 nM) (19), which almost completely suppressed the ATP/SphK1-dependent appearance of the 550 nm peak (Fig. 4A). To further evaluate the dynamic range of this assay, we compared the SphK1 inhibitors PF-543 and N,N-dimethylsphingosine (DMS), which have IC50 values in the nanomolar and micromolar ranges, respectively. In assays with SphK1, PF-543 showed strong inhibition (Fig. 4B), and data analysis (Fig. 4B, inset) revealed an IC50 value of 16 nM. A previous study using 1 µM FITC-Sph as a substrate reported an IC50 of 3.6 nM (19). SphK1 inhibition kinetics with DMS revealed an IC50 of 15.2 µM (Fig. 4C), which is in excellent agreement with previous reports (20). We also examined the effect of the SphK2-specific inhibitor K145 (21). As can be seen in Fig. 4D, K145 suppressed SphK2 activity with an IC50 of 33.7 µM, which is slightly higher than the Ki value determined with the conventional radioactive assay (21). Taken together, these results suggest that our new fluorescence method is suitable for rapid screening of SphK1 and SphK2 inhibitors.
Fig. 4.
Fluorescence assay for SphK1 and SphK2 inhibitors. A: Time-resolved absorption spectra of NBD-Sph (30 µM) in SphK1 buffer containing 0.05% Triton X-100, 1 mM ATP, 2% DMSO, and 100 nM SphK1 in the presence of 250 nM PF-543 at 25°C. Spectra were recorded every 30 s for 5 min. B, C: Time-resolved fluorescence emission at 37°C of 50 µM NBD-Sph in SphK1 reaction buffer containing 0.05% Triton X-100, 1 mM ATP, 2% DMSO, 150 nM SphK1, and increasing concentrations of PF-543 (B) or DMS (C). n = 3. Insets: Initial rates versus [PF-543] plot (B) and initial rates versus [DMS] plot (C). D: Time-resolved fluorescence emission of 75 µM NBD-Sph in SphK2 reaction buffer containing 0.07% Triton X-100, 200 mM KCl, 1.2% DMSO, 1 mM ATP, 6.9 nM SphK2, and increasing concentrations of K145. Inset: Initial rates versus [K145] plot. Experiments in B–D were performed in 384-well 20 µl format in a fluorescence plate reader. Fluorescence emission was measured with λEx = 550 nm and λEm= 583 nm.
CONCLUSIONS
In summary, we have developed a simple, convenient, and accurate real-time assay that can be used to rapidly determine SphK1 and SphK2 activity using NBD-Sph as a substrate. Our assay exploits a spectral REES in the environmentally sensitive properties of the NBD fluor on NBD-Sph compared with NBD-S1P, which was completely dependent on SphK catalyzed phosphorylation of NBD-Sph to NBD-S1P. Our results suggest that NBD-S1P tends to associate in Triton X-100 micelles and that interactions between NBD groups on NBD-S1P are likely responsible for the red shift, similar to that observed with NBD-cholesterol in artificial membranes, where increasing concentrations show REES spectral transitions (16). Our assay does not require radioactive materials or isolation of products and is sufficiently sensitive for high-throughput screening of chemical libraries and characterization of lead compounds. In addition, it should be an excellent tool to quickly examine the kinetic properties of SphK mutants with minimal labor and as a potential tool for studying pre-steady state properties and the mechanism of substrate binding of these enzymes (22, 23), which remain poorly characterized. As SphK activity has been implicated in the etiology of many types of aggressive cancers, as well as autoimmune and cardiovascular diseases, this is an important step forward in the development of pharmacological agents that can target these important lipid kinases.
Acknowledgments
The authors thank Drs. William Barton and Jessica Bell for their invaluable assistance with equipment and discussions, and Drs. Kai Liu and Shijun Zhang for providing 14-NBD-Sph.
Footnotes
Abbreviations:
- DMS
- N,N-dimethylsphingosine
- NBD
- 7-nitro-2-1,3-benzoxadiazol-4-yl
- PF-543
- (R)-[1-(4-{[3-methyl-5-(phenylsulfonylmethyl)phenoxy]methyl}benzyl) pyrrolidin-2-yl]methanol
- REES
- red edge excitation shift
- S1P
- sphingosine-1-phosphate
- Sph
- sphingosine
- SphK
- sphingosine kinase
This work was supported by National Institutes of Health Grants R37GM043880 (S.S.), R01CA61774 (S.S.), and 5T32HL094290 (S.L.).
REFERENCES
- 1.Pyne N. J., Pyne S. 2010. Sphingosine 1-phosphate and cancer. Nat. Rev. Cancer. 10: 489–503 [DOI] [PubMed] [Google Scholar]
- 2.Cyster J. G., Schwab S. R. 2012. Sphingosine-1-phosphate and lymphocyte egress from lymphoid organs. Annu. Rev. Immunol. 30: 69–94 [DOI] [PubMed] [Google Scholar]
- 3.Kunkel G. T., Maceyka M., Milstien S., Spiegel S. 2013. Targeting the sphingosine-1-phosphate axis in cancer, inflammation and beyond. Nat. Rev. Drug Discov. 12: 688–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olivera A., Barlow K. D., Spiegel S. 2000. Assaying sphingosine kinase activity. Methods Enzymol. 311: 215–223 [DOI] [PubMed] [Google Scholar]
- 5.Vessey D. A., Kelley M., Karliner J. S. 2005. A rapid radioassay for sphingosine kinase. Anal. Biochem. 337: 136–142 [DOI] [PubMed] [Google Scholar]
- 6.Roberts J. L., Moretti P. A., Darrow A. L., Derian C. K., Vadas M. A., Pitson S. M. 2004. An assay for sphingosine kinase activity using biotinylated sphingosine and streptavidin-coated membranes. Anal. Biochem. 331: 122–129 [DOI] [PubMed] [Google Scholar]
- 7.Kharel Y., Mathews T. P., Kennedy A. J., Houck J. D., Macdonald T. L., Lynch K. R. 2011. A rapid assay for assessment of sphingosine kinase inhibitors and substrates. Anal. Biochem. 411: 230–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ettmayer P., Billich A., Baumruker T., Mechtcheriakova D., Schmid H., Nussbaumer P. 2004. Fluorescence-labeled sphingosines as substrates of sphingosine kinases 1 and 2. Bioorg. Med. Chem. Lett. 14: 1555–1558 [DOI] [PubMed] [Google Scholar]
- 9.Yangyuoru P. M., Otieno A. C., Mwongela S. M. 2011. Determination of sphingosine kinase 2 activity using fluorescent sphingosine by capillary electrophoresis. Electrophoresis. 32: 1742–1749 [DOI] [PubMed] [Google Scholar]
- 10.Billich A., Ettmayer P. 2004. Fluorescence-based assay of sphingosine kinases. Anal. Biochem. 326: 114–119 [DOI] [PubMed] [Google Scholar]
- 11.Jin Y. X., Yoo H. S., Kihara A., Choi C. H., Oh S., Moon D. C., Igarashi Y., Lee Y. M. 2006. Sphingosine kinase assay system with fluorescent detection in high performance liquid chromatography. Arch. Pharm. Res. 29: 1049–1054 [DOI] [PubMed] [Google Scholar]
- 12.Vidugiriene J., Zegzouti H., Goueli S. A. 2009. Evaluating the utility of a bioluminescent ADP-detecting assay for lipid kinases. Assay Drug Dev. Technol. 7: 585–597 [DOI] [PubMed] [Google Scholar]
- 13.Highkin M. K., Yates M. P., Nemirovskiy O. V., Lamarr W. A., Munie G. E., Rains J. W., Masferrer J. L., Nagiec M. M. 2011. High-throughput screening assay for sphingosine kinase inhibitors in whole blood using RapidFire(R) mass spectrometry. J. Biomol. Screen. 16: 272–277 [DOI] [PubMed] [Google Scholar]
- 14.Jin Y. X., Shi L. H., Yoo H. S., Lee Y. M., Kihara A., Igarashi Y., So H. Y., Yim Y. H. 2008. A sphingosine kinase activity assay using direct infusion electrospray ionization tandem mass spectrometry. Anal. Biochem. 380: 35–40 [DOI] [PubMed] [Google Scholar]
- 15.Peters C., Billich A., Ghobrial M., Hogenauer K., Ullrich T., Nussbaumer P. 2007. Synthesis of borondipyrromethene (BODIPY)-labeled sphingosine derivatives by cross-metathesis reaction. J. Org. Chem. 72: 1842–1845 [DOI] [PubMed] [Google Scholar]
- 16.Pucadyil T. J., Mukherjee S., Chattopadhyay A. 2007. Organization and dynamics of NBD-labeled lipids in membranes analyzed by fluorescence recovery after photobleaching. J. Phys. Chem. B. 111: 1975–1983 [DOI] [PubMed] [Google Scholar]
- 17.Haldar S., Chattopadhyay A. 2013. Application of NBD-labeled lipids in membrane and cell biology. In Fluorescent Methods to Study Biological Membranes. Vol. 13. Y. Mély and G. Duportail, editors. Springer, Berlin. 37–50. [Google Scholar]
- 18.Olivera A., Kohama T., Tu Z., Milstien S., Spiegel S. 1998. Purification and characterization of rat kidney sphingosine kinase. J. Biol. Chem. 273: 12576–12583 [DOI] [PubMed] [Google Scholar]
- 19.Schnute M. E., McReynolds M. D., Kasten T., Yates M., Jerome G., Rains J. W., Hall T., Chrencik J., Kraus M., Cronin C. N., et al. 2012. Modulation of cellular S1P levels with a novel, potent and specific inhibitor of sphingosine kinase-1. Biochem. J. 444: 79–88 [DOI] [PubMed] [Google Scholar]
- 20.Edsall L. C., Van Brocklyn J. R., Cuvillier O., Kleuser B., Spiegel S. 1998. N,N-Dimethylsphingosine is a potent competitive inhibitor of sphingosine kinase but not of protein kinase C: modulation of cellular levels of sphingosine 1-phosphate and ceramide. Biochemistry. 37: 12892–12898 [DOI] [PubMed] [Google Scholar]
- 21.Liu K., Guo T. L., Hait N. C., Allegood J., Parikh H. I., Xu W., Kellogg G. E., Grant S., Spiegel S., Zhang S. 2013. Biological characterization of 3-(2-amino-ethyl)-5-[3-(4-butoxyl-phenyl)-propylidene]-thiazolidine-2,4-dione (K145) as a selective sphingosine kinase-2 inhibitor and anticancer agent. PLoS ONE. 8: e56471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lima S., Spiegel S. 2013. Sphingosine kinase: a closer look at last. Structure. 21: 690–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z., Min X., Xiao S. H., Johnstone S., Romanow W., Meininger D., Xu H., Liu J., Dai J., An S., et al. 2013. Molecular basis of sphingosine kinase 1 substrate recognition and catalysis. Structure. 21: 798–809 [DOI] [PubMed] [Google Scholar]