Abstract
It is well-established that over-accumulation of dietary cholesterol in the liver inhibits sterol-regulatory element binding protein (SREBP)-mediated LDL receptor (LDLR) gene transcription leading to a reduced hepatic LDLR mRNA level in hypercholesterolemic animals. However, it is unknown whether elevated cholesterol levels can elicit a cellular response to increase LDLR mRNA turnover to further repress LDLR expression in liver tissue. In the current study, we examined the effect of a high cholesterol diet on the hepatic expression of LDLR mRNA binding proteins in three different animal models and in cultured hepatic cells. Our results demonstrate that high cholesterol feeding specifically elevates the hepatic expression of LDLR mRNA decay promoting factor heterogeneous nuclear ribonucleoprotein (HNRNP)D without affecting expressions of other LDLR mRNA binding proteins in vivo and in vitro. Employing the approach of adenovirus-mediated gene knockdown, we further show that depletion of HNRNPD in the liver results in a marked reduction of serum LDL-cholesterol and a substantial increase in liver LDLR expression in hyperlipidemic mice. Additional studies of gene knockdown in albumin-luciferase-untranslated region (UTR) transgenic mice provide strong evidence supporting the essential role of 3′UTR in HNRNPD-mediated LDLR mRNA degradation in liver tissue. Altogether, this work identifies a novel posttranscriptional regulatory mechanism by which dietary cholesterol inhibits liver LDLR expression via inducing HNRNPD to accelerate LDLR mRNA degradation.
Keywords: low density lipoprotein, posttranscriptional regulation, messenger ribonucleic acid stability, adenylate-uridylate-rich element-binding proteins, heterogeneous nuclear ribonucleoprotein D, hypercholesterolemia
The expression of liver LDL receptor (LDLR) regulates human plasma LDL-cholesterol (LDL-C) homeostasis (1–3). Increased hepatic LDLR expression results in improved clearance of plasma LDL-C through receptor-mediated endocytosis, which is strongly associated with a decreased risk of developing cardiovascular disease in humans (4, 5). Thus far, many studies have demonstrated that intracellular cholesterol levels play a primary role in determination of hepatic LDLR expression levels through a negative feedback mechanism that controls gene transcription mediated by the sterol-regulatory element (SRE) located in LDLR promoter and SRE binding proteins (SREBPs) (6, 7).
In addition to transcriptional regulation, LDLR gene expression is subject to posttranscriptional regulation at the point of mRNA stability (8–11). It is well-known that mRNA stability plays a key role in the control of gene expression both by setting the basal level of gene expression and as a site of regulatory responses (12). In liver cells, when intracellular cholesterol levels rise, LDLR mRNA levels fall substantially along with transcripts of other SREBP target genes (13). While this decrease in LDLR mRNA abundance has been attributed to the transcriptional suppression due to the inhibition of proteolytic processing of SREBP (14, 15), it is currently unknown whether elevated cholesterol levels can also elicit a cellular response to increase LDLR mRNA turnover rate as a rapid means to lower LDLR expression and to further reduce cholesterol uptake from outside of cells.
The stability of LDLR mRNA is modulated through its 3′ untranslated region (UTR) (16). This region harbors four adenylate-uridylate (AU)-rich elements (AREs) (8, 9, 17) that are docking sites for cytoplasmic ARE-binding proteins (ARE-BPs). Some ARE-BPs are mRNA destabilizing inducers, while others can protect the mRNA from endonuclease cleavage (18–20). Our previous studies have identified heterogeneous nuclear ribonucleoprotein (HNRNP)D, HNRNPI, and KH-type splicing regulatory protein (KSRP) as decay-promoting factors that bind to ARE sequences of LDLR 3′UTR and accelerate the mRNA decay in HepG2 cells (21). A recent new study identified human antigen R (HuR) as a stabilizing factor for LDLR mRNA (22). It was shown in HepG2 cells that HuR bound to the proximal ARE site of LDLR 3′UTR and led to an increase in LDLR mRNA half-life. In addition to LDLR mRNA, these ARE-BPs have been shown to regulate the stability of other mRNA targets with diverse cellular functions including inflammation, aging, and cell cycle (23–27). To date, there is no literature report describing the effect of sterols on the expression or activity of these LDLR mRNA binding proteins.
In this current study, we investigated the relationship between dietary cholesterol and LDLR mRNA binding proteins to explore a regulatory role of dietary cholesterol on LDLR mRNA stability. Initially, we examined the expression of LDLR ARE-BPs in liver tissues of mice and hamsters fed a normal diet (ND) or a high cholesterol diet (HCD). We observed that the hepatic expression of LDLR mRNA decay promoting factor HNRNPD was specifically induced by HCD in mice as well as in hamsters, while expressions of HNRNPI, KSRP, and HuR were unaffected by high cholesterol feeding. This in vivo finding was recaptured in subsequent in vitro studies of hepatic cell culture. To understand the impact of elevated HNRNPD levels on LDL-C metabolism, we applied the approach of adenovirus-mediated gene knockdown. Our results demonstrated that blocking HNRNPD-mediated degradation of LDLR mRNA generated profound effects on upregulation of liver LDLR and reduction of serum LDL-C under hypercholesterolemic conditions. This work provides the first evidence suggesting that dietary cholesterol inhibits LDLR gene expression through the dual action mechanisms of transcriptional suppression and acceleration of mRNA decay.
MATERIALS AND METHODS
Animals and diets
C57BL/6J male mice, 8 weeks old, were purchased from Charles River Laboratory and male golden Syrian hamsters, 8–10 weeks old, were purchased from Harlan. Albumin (Alb)-luciferase (Luc)-UTR transgenic mice were generated and bred in the Veterinary Medical Unit of Veterans Affairs Palo Alto Health Care System (28). Two- to three-month-old Alb-Luc-UTR mice (male and female) were used in the diet study. All animals were housed under controlled temperature (72°F) and lighting (12 h light/dark cycle). Animals had free access to autoclaved water and food. For diet studies, C57BL/6J mice and hamsters were fed either a ND or a rodent HCD containing 1.25% cholesterol (Product D12108, Research Diet, Inc.) for 2 weeks. Alb-Luc-UTR mice were fed a ND or a HCD for 4 weeks. Mice were fasted for 4 h and hamsters were fasted overnight before euthanization for serum and liver tissue collections. Livers were immediately removed, cut into small pieces, and stored at −80°C for RNA and protein isolation. All animal procedures and protocols were reviewed and approved by the Institutional Animal Care and Use Committee of the Veterans Affairs Palo Alto Health Care System.
Adenoviral infection in C57BL/6J mice
In one study, 8-week-old male C57BL/6J mice were fed a HCD (n = 10) for 2 weeks. At day 0, mice were fully anesthetized with isoflurane mixed with pure oxygen. Five mice were injected with Ad-shHNRNPD and another five mice were injected with a control adenovirus (Ad-shGFP) at a dose of 6 × 109 pfu/per mouse via retro-orbital sinus in a total volume of 100 μl of sterile PBS. Four hour-fasted serum samples were collected 2 days prior to infection and at days 0, 2, 4, and 7 after infection. At the termination, all animals were euthanized and livers were collected and stored at −80°C for isolation of total RNA and total protein lysates. In another study, 10 ND-fed C57BL/6J mice with similar serum lipid levels were injected with Ad-shHNRNPD (n = 5) or Ad-shGFP (n = 5) at a dose of 6 × 109 pfu/per mouse via retro-orbital sinus in a total volume of 100 μl of sterile PBS. Body weight and food intake were monitored during the treatment. After 7 days of infection, mice were fasted for 4 h before euthanization for serum and liver tissue collections.
Bioluminescence imaging and adenoviral infection of Alb-Luc-UTR mice
Bioluminescence was detected with the In Vivo Imaging System (IVIS; Xenogen, Alameda, CA) as previously described (28). Alb-Luc-UTR mice received an ip injection of 50 mg/kg D-luciferin 10 min before imaging and were anesthetized with isoflurane during imaging. Photons emitted from living mice were acquired as photons per second/cm2 per steridian (sr) by using Living Image software (Xenogen) and were integrated over 60 s for photon quantification; a region of interest was manually selected and kept constant within all experiments. The signal intensity was converted into photons per s/cm2 per sr.
For adenoviral infection experiments, 1 week prior to the injection, the baseline bioluminescence imaging was obtained from Alb-Luc-UTR mice fed a ND. Ten mice with similar bioluminescent signals were divided into two groups. Five mice in one group were injected with 4 × 109 pfu/per mouse of Ad-shHNRNPD, five mice in the other group were injected with 4 × 109 pfu/per mouse of Ad-shLacZ as the control adenovirus via retro-orbital sinus in a total volume of 100 μl of sterile PBS. Body weight and food intake were monitored during the treatment. After 10 days of infection, bioluminescence imaging was recorded before euthanization of the animals. Bioluminescence was expressed as fold induction over baseline levels. At the termination, animals were euthanized. Livers were harvested for isolation of total RNA, total protein lysates, and Luc activity measurement.
Measurement of serum lipid levels
Serum was isolated at room temperature and stored at −80°C. Standard enzymatic methods were used to determine total cholesterol (TC), LDL-C, and HDL-cholesterol (HDL-C) with commercially available kits purchased from Stanbio Laboratory (Boerne, TX).
RNA isolation and real-time quantitative RT-PCR
Total RNA was isolated from flash-frozen liver tissue using an RNeasy kit (Qiagen, Alameda, CA). Generation of cDNA and real-time PCR were performed as previously described (29). Each liver cDNA sample was run in duplicate. Primer sequences of genes used in quantitative (q)PCR are listed in supplementary Table I. Target mRNA expression in each sample was normalized to the housekeeping gene, GAPDH. The 2−ΔΔCt method was used to calculate relative mRNA expression levels.
Western blot analyses of HNRNPD and LDLR in liver tissues and hepatic cell lines
Approximately 50 mg of frozen liver tissue from individual mice or hamsters was homogenized in 0.3 ml RIPA buffer containing 1 mM PMSF and protease inhibitor cocktail (Roche). After protein quantification, aliquots of homogenates containing 100 μg proteins were used for SDS-PAGE and Western blotting. For hepatic cells, aliquots of cell lysates containing 30 μg proteins were used for SDS-PAGE and Western blotting. HNRNPD p45 isoform was detected with rabbit anti-HNRNPD polyclonal antibody obtained from Novus Biological (NBP1-61684) and p37 isoform was detected by goat polyclonal antibody (sc-22368) from Santa Cruz Biotechnology. LDLR antibody was obtained from BioVision (Mountain View, CA). Mouse anti-human transferrin receptor (TFR) antibody (13-6800) was obtained from Life Technology. Membranes were reprobed with an anti-β-actin antibody (Sigma). Immunoreactive bands of predicted molecular mass were visualized using an ECL plus kit (GE Healthcare Life Sciences, Piscataway, NJ) and quantified with the Alpha View software with normalization by signals of β-actin or signals of TFR.
Cell culture
Human hepatoma HepG2 cells and mouse hepatoma AML12 cells were obtained from ATCC. HepG2 cells were cultured in MEM with 10% FBS. AML12 cells were cultured in a 1:1 mixture of Dulbecco’s modified Eagle’s medium and Ham’s F12 medium with 10% FBS. For cholesterol treatment, cells were seeded in 6-well culture plates and cultured in medium containing 10% lipoprotein-depleted serum (LPDS) overnight. The next day, cells were treated with vehicle DMSO, cholesterol (10 μg/ml cholesterol and 1 μg/ml of 25-hydroxyl cholesterol), or other compounds for 24 h before isolation of total RNA and cell lysates. In each experiment, duplicate wells were used for each condition.
Tissue Luc enzymatic assay
Liver samples of Alb-Luc-UTR mice were homogenized in cell culture lysis buffer (Promega). Protein concentrations of liver homogenates were determined using BCATM protein assay reagent (Pierce). Luc activity from liver homogenate was measured and normalized to tissue protein.
Assay of secreted PCSK9
The secreted proprotein convertase subtilisin/kexin type 9 (PCSK9) in serum of C57BL/6J mice was measured using a mouse PCSK9 ELISA kit obtained from R&D Systems according to the manufacturer’s instructions.
Statistical analysis
All values are expressed as mean ± SEM. To determine statistical significance, one way ANOVA with Dunnett’s post hoc test was carried out using GraphPad Prism 5 software. For two group data analysis, unpaired two-tailed Student’s t-test was applied. P < 0.05 was considered statistically significant.
RESULTS
Upregulation of hepatic HNRNPD expression by HCD in mice and hamsters
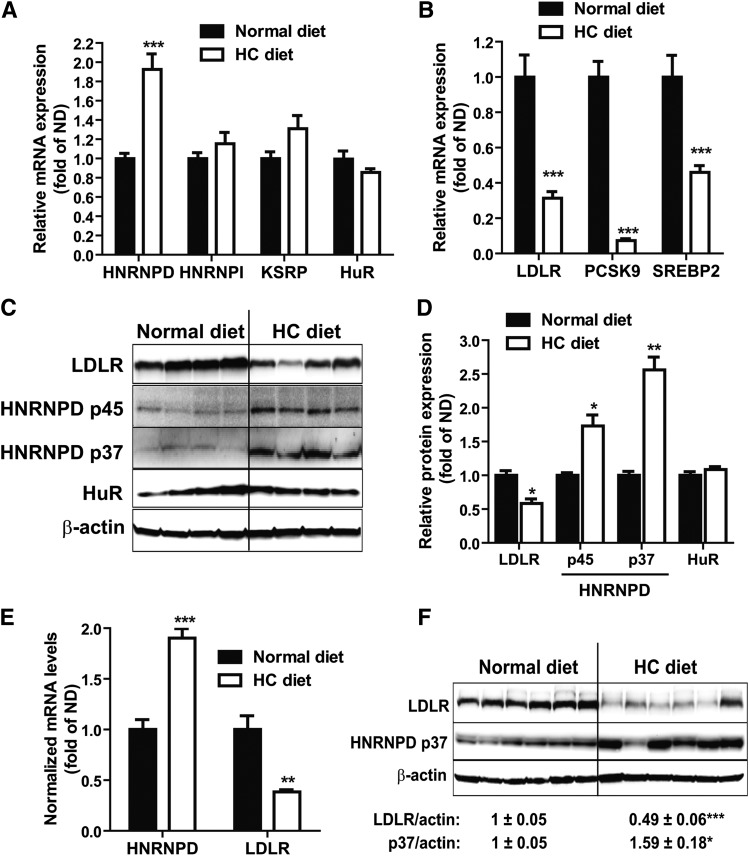
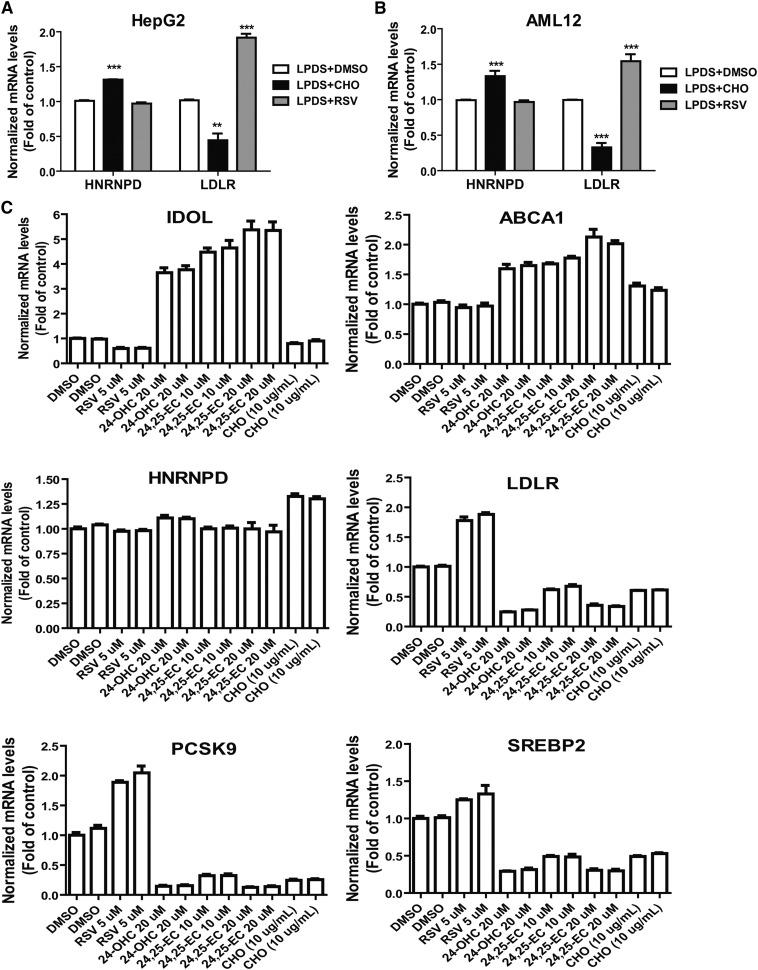
Feeding mice a HCD for 4 weeks increased serum cholesterol levels by ∼2-fold from 122 to 240 mg/dl and serum LDL-C by 5-fold from 36 to 181 mg/dl, as compared with mice fed a ND (supplementary Fig. IA). To determine whether a HCD could alter hepatic expressions of LDLR mRNA binding proteins, we examined mRNA levels of HNRNPD, HNRNPI, KSRP, and HuR in liver samples of HCD mice and compared them to ND mice. Figure 1A shows that a HCD increased HNRNPD mRNA levels by 1.9-fold (P < 0.001) without affecting HNRNPI, KSRP, or HuR mRNA expressions. In contrast and as expected, mRNA levels of LDLR and other SREBP2 target genes, including PCSK9 and SREBP2, were markedly decreased by HCD feeding (Fig. 1B).
Fig. 1.
HCD increases HNRNPD mRNA and protein levels in animal liver. A, B: Mice fed a HCD (n = 10) or a ND (n = 7) for 4 weeks were euthanized after 4 h fasting. Hepatic gene expression was measured by real-time PCR (A, B). ***P < 0.001 compared with the ND group. C: Individual mouse liver protein extracts were prepared and four lysate samples per group were randomly chosen for Western blotting. D: Expression levels of HNRNPD isoforms, HuR and LDLR, were quantified with the Alpha View software with normalization by signals of β-actin. Values are mean ± SEM of four samples per group. *P < 0.05, **P < 0.01 compared with the ND group. E, F: Male golden Syrian hamsters were fed a HCD (n = 6) or a ND (n = 6) for 2 weeks before euthanization. Hepatic mRNA and protein expressions were determined as described in (A–D).
HNRNPD protein has four isoforms, p37, p40, p42, and p45, generated by alterative precursor mRNA splicing (30–32). In mouse liver tissue and mouse primary hepatocytes, only the p37 and p45 isoforms could be detected (28). Utilizing specific antibodies, we examined HNRNPD, HuR, and LDLR protein levels in liver tissues of the ND and HCD groups. The results demonstrated that HCD feeding increased both HNRNPD p45 and p37 isoforms to 1.7- and 2.5-fold, respectively, of control and reduced LDLR protein abundance to 50% of control (Fig. 1C, D). Consistent with the gene expression analysis, protein levels of HuR did not differ between diet groups.
Next, we questioned whether the elevated expression of HNRNPD upon HCD feeding could be detected in other animal models. To this end, we fed hamsters a HCD or a ND for 2 weeks and examined HNRNPD mRNA and protein expressions in liver samples. The serum TC was increased 4-fold in hamsters fed a HCD as compared with hamsters fed a ND (supplementary Fig. IB). A HCD increased the HNRNPD mRNA level approximately 2-fold and reduced the LDLR mRNA level by 2.6-fold as compared with a ND (Fig. 1E). Western blot analysis further corroborated the induction of HNRNPD p37 protein expression in hamster liver by HCD feeding (Fig. 1F). The p45 isoform could not be detected in hamster liver. As observed in the mouse diet study, we did not detect changes in mRNA or protein levels of HuR in hamster livers upon HCD feeding (supplementary Fig. II). Altogether, these results demonstrate, for the first time, that animals responded to a cholesterol-enriched diet by increasing HNRNPD expression in liver tissues where LDLR expressions were reduced.
Depletion of HNRNPD in hypercholesterolemic mice reduced serum LDL-C through upregulation of liver LDLR
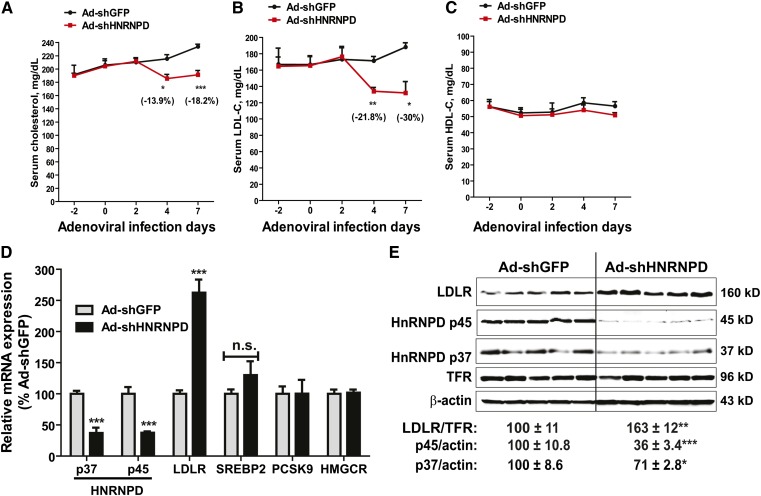
In our previous in vitro studies, we had generated an adenovirus (Ad-shHNRNPD) to express a shRNA that targets all isoforms of mouse and human HNRNPD. Transduction of Ad-shHNRNPD in mouse primary hepatocytes was shown to reduce HNRNPD mRNA and protein levels dose-dependently without affecting mRNA levels of other ARE-BPs (HNRNP I, KSRP) or SREBP2-target genes (HMGCR, PCSK9) (28). Thus, it is a valuable tool to assess the in vivo role of HNRNPD in cholesterol-mediated suppression of liver LDLR expression. Ad-shHNRNPD or control virus (Ad-shGFP) was injected into C57BL/6J mice that were fed a HCD for 2 weeks. We measured the serum lipid levels before, during, and after 7 days of viral injection. While on a HCD, serum cholesterol levels continued rising during the treatment in control mice (Ad-shGFP), a significant drop in serum TC level was observed after 4 days of injection in Ad-shHNRNPD-infected mice (−13.9%, P < 0.05 compared with control mice), and serum TC further declined to a level which was 18.2% (P < 0.001) lower than control mice after 7 days of injection (Fig. 2A). The time-dependent reduction in serum LDL-C levels by Ad-shHNRNPD infection in HCD mice was even more pronounced, with ∼22% lowering after 4 days and 30% lowering after 7 days, as compared with control mice (Fig. 2B). Serum HDL-C levels were not affected by HNRNPD depletion throughout the treatment duration (Fig. 2C).
Fig. 2.
Strong effects of HNRNPD depletion on serum cholesterol levels and hepatic LDLR expression levels of hypercholesterolemic mice. A–C: Ten C57BL/6J male mice were fed a HCD for 2 weeks. Five mice were injected with 6 × 109 pfu/mouse Ad-shHNRNPD; another five mice were injected with 6 × 109 pfu/mouse Ad-shGFP. Four hour-fasted serum samples were collected 2 days prior to injection and at days 0, 2, 4, and 7 after infection. Serum samples were analyzed for TC (A), LDL-C (B), and HDL-C (C). *P < 0.05, **P < 0.01, and ***P < 0.001 compared with the Ad-shGFP group at the same time points. D: Hepatic mRNA levels of HNRNPD, LDLR, SREBP2, PCSK9, and HMGCR were assessed by qRT-PCR using specific primers. After normalization with GAPDH mRNA levels, the relative levels are presented, and the results are the mean ± SEM of five animals per group with duplicate measurement of each cDNA sample. ***P < 0.001 compared with the Ad-shGFP group. n.s., not significant. E: Western blotting with antibodies to HNRNPD p37, HNRNPD p45, and LDLR was conducted by analyzing individual liver homogenates of each group (n = 5 per group). Membranes were reprobed with anti-TFR or anti-β-actin antibody. The protein abundances of HNRNP D p37/p45 were quantified using the Alpha View software with normalization by signals of β-actin. The protein abundances of LDLR were quantified using the Alpha View software with normalization by signals of TFR. Values are the mean ± SEM of five samples per group. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with the Ad-shGFP group.
Utilizing real-time qRT-PCR, we analyzed mRNA levels of HNRNPD and LDLR in all liver samples. We applied isoform-specific primers to separately measure p37 and p45 mRNA levels. In addition, we measured mRNA levels of a few SREBP2 target genes, including PCSK9, HMGCR, and SREBP2. Figure 2D shows that in Ad-shHNRNPD-infected mice, mRNA levels of p37 and p45 isoforms were both significantly reduced to approximately 40% of that in control mice (P < 0.001), which was associated with a marked increase in the LDLR mRNA level to 260% of control (P < 0.001). In contrast to LDLR, HRNPD depletion did not affect mRNA levels of other SREBP2 target genes that were measured.
Western blot analysis of individual liver homogenates further demonstrated that the p45 protein level in Ad-shHNRNPD-infected mice was reduced to 36% of control and p37 protein was lowered to 71% of control by the expression of HNRNPD shRNA. This reduction of HNRNPD protein abundance was associated with an increase of 163% of control in the hepatic LDLR protein level (P < 0.01) (Fig. 2E). Measurements of body weight and food intake throughout the treatments did not detect significant changes associated with Ad-shHNRNPD injection (supplementary Fig. IIIA, B), thereby ruling out the influence of food intake as a causing factor for reduced serum cholesterol levels. In addition, we measured PCSK9 serum levels. As we expected, knockdown of HNRNPD expression in liver had no impact on serum PCSK9 concentration (supplementary Fig. IIIC). Taken together, these results provided the first in vivo evidence demonstrating a key role of HNRNPD in modulation of hepatic LDLR expression under hypercholesterolemic conditions.
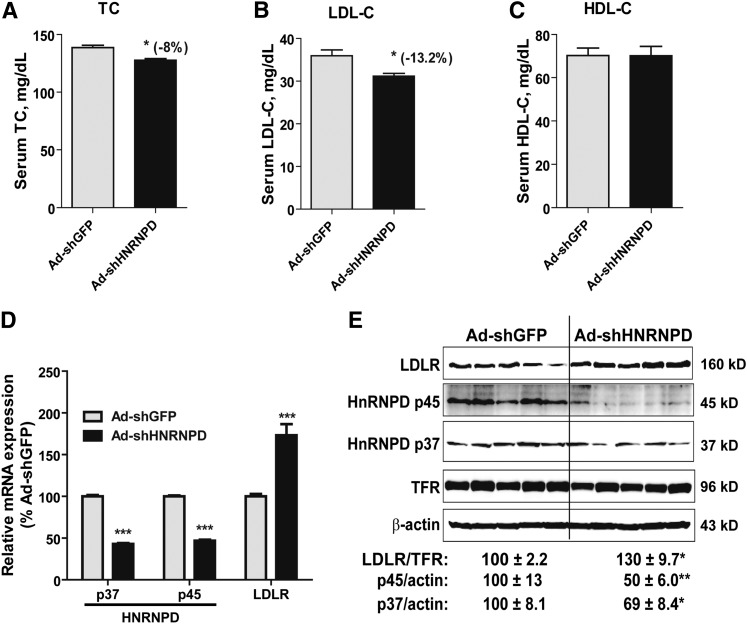
Next, we examined the effect of Ad-shHRNPD injection in normolipidemic mice. Ad-shHNRNPD injection at the same dose into mice fed a ND resulted in a relatively modest decrease in TC (−8%, P < 0.05; Fig. 3A) and LDL-C (−13%, P < 0.05; Fig. 3B) compared with the control mice infected with Ad-shGFP. Again, HDL-C levels were not affected by Ad-shHNRNPD injection (Fig. 3C).
Fig. 3.
HNRNPD depletion had modest effects on serum cholesterol levels and hepatic LDLR expression levels in mice fed a ND. A–C: Ten C57BL/6J male mice were fed a ND for 2 weeks. Five mice were injected with 6 × 109 pfu/mouse Ad-shHNRNPD; another five mice were injected with 6 × 109 pfu/mouse Ad-shGFP. Four hour-fasted serum samples were collected after 7 days of infection. Serum samples were analyzed for TC (A), LDL-C (B), and HDL-C (C). *P < 0.05 compared with the Ad-shGFP group at the same time points. D: Hepatic mRNA levels of HNRNPD p37/p45 and LDLR were assessed by qRT-PCR using specific primers. After normalization with GAPDH mRNA levels, the relative levels are presented, and the results are the mean ± SEM of four to five animals per group with duplicate measurement of each cDNA sample. ***P < 0.001 compared with the Ad-shGFP group. E: Western blotting with antibodies to HNRNPD p37, HNRNPD p45, and LDLR was conducted as described in Fig. 2E. Values are the mean ± SEM of five samples per group. *P < 0.05, **P < 0.01 compared with the Ad-shGFP group.
Analysis of hepatic gene expression by real-time qRT-PCR showed that the substantial reductions of HNRNPD isoforms in ND mice increased LDLR mRNA levels to 173% of control (Fig. 3D). In addition to LDLR and HNRNPD, we assessed mRNA levels of other LDLR ARE-BPs (HNRNPI, KSRP, and HuR) and genes involved in the cholesterol biosynthetic pathway (PCSK9, HMG-CoA reductase, and SREBP2) in all liver samples. The results showed that none of these mRNA levels were altered by Ad-shHNRNPD infection (data not shown).
Western blot analysis further demonstrated that LDLR protein levels were increased to 130% of control by Ad-shHNRNPD injection while HNRNPD p45 and p37 isoforms were reduced to 50 and 69% of control in normolipidemic mice, respectively (Fig. 3E). Collectively, these data demonstrated that hepatic depletion of HNRNPD produced a modest yet significant impact on LDLR expression and on LDLR-mediated removal of serum cholesterol under normolipidemic conditions.
In addition, we compared mRNA and protein levels of HNRNPD p37/45 isoforms in liver samples of control mice (Ad-shGFP) fed a ND or HCD (supplementary Fig. IVA, B). Again, we detected significant increases in HNRNPD mRNA and protein levels in HCD-fed C57BL/6J mice. Utilizing qRT-PCR, we further analyzed mRNA levels of five HNRNPD targets (30) including cyclin B2, cyclin D1, IL-1β, IL-6, and TNFα in these liver samples (supplementary Fig. IVC). Consistent with an increased HNRNPD expression by HCD, mRNA levels of four out of five HNRNPD targeted transcripts were significantly reduced in liver samples of HCD mice compared with ND-fed mice.
Finally, by Western blotting we directly compared liver LDLR protein levels in all four groups of mice, including control mice (Ad-shGFP) fed a ND or HCD and Ad-shHNRNPD-infected mice under both diets. Results showed that a cholesterol diet lowered LDLR by 25% (P < 0.001) as compared with the ND group in Ad-shGFP-infected mice. Depletion of HNRNPD resulted in a 136% (P < 0.001) increase in ND-fed livers and a 119% (P < 0.01) increase in HCD-fed livers as compared with LDLR protein levels in control mice fed a ND (supplementary Fig. V). Again, the increase in LDLR abundance by HNRNPD depletion was higher when compared with HCD control groups (159%). These data suggest that in this mouse model, depletion of HNRNPD can overcome dietary cholesterol-mediated repression of hepatic LDLR.
Collectively, the results of protein and mRNA analyses from ND and HCD mice clearly demonstrate that cholesterol feeding elevates the LDLR mRNA degrading protein HNRNPD, and that eliminating HNRNPD in cholesterol-loaded liver produced stronger effects in restoring LDLR mRNA and protein levels, which translated into a substantially improved removal of serum LDL-C in hypercholesterolemic mice.
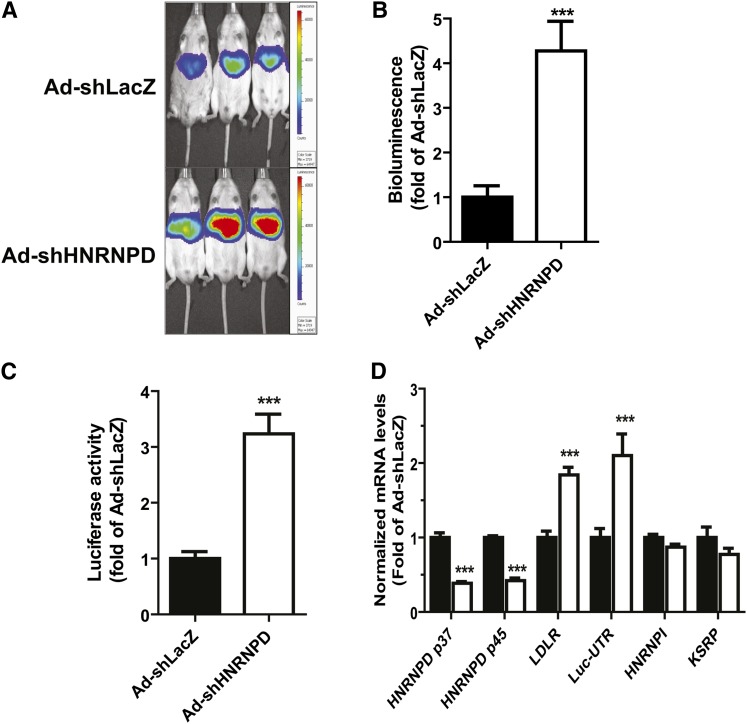
In vivo knockdown of HNRNPD elevated liver LDLR expression through mRNA stabilization in Alb-Luc-UTR mice
To further investigate the underlying mechanism of Ad-shHNRNPD-mediated increase in hepatic LDLR expression, we utilized transgenic mice (Alb-Luc-UTR) that express Luc-LDLR3′UTR reporter gene (Luc-UTR) under the control of liver-specific Alb promoter (28). Alb-Luc-UTR is a unique in vivo model to study the function of 3′UTR in mediating LDLR mRNA decay in liver tissue. We injected Alb-Luc-UTR mice with either Ad-shHNRNPD or with a control adenovirus, Ad-shLacZ, while all mice were on a ND. We conducted live bioluminescence imaging before and after 10 days of viral infections on all mice. Figure 4A shows representatives of bioluminescent imaging of Ad-shHNRNPD- and Ad-shLacZ-infected mice. Figure 4B is the summarized imaging results (mean ± SEM) showing that Ad-shHNRNPD infection increased bioluminescence signal 4.2-fold over the control virus (P < 0.001). We further measured ex vivo Luc activity of individual liver samples and observed a 3.2-fold higher Luc activity in Ad-shHNRNPD-infected livers as compared with control (Fig. 4C). Utilizing real-time qRT-PCR, we analyzed liver mRNA levels of HNRNPD p45/p37, Luc-UTR transgene, and endogenous LDLR mRNA (Fig. 4D). Similar to C57BL/6J mice, expression of HNRNPD shRNA in the liver of Alb-Luc-UTR mice resulted in marked reductions of p45 and p37 mRNA levels with corresponding increases of Luc-UTR mRNA level (2.1-fold of control) and mouse endogenous LDLR mRNA level (1.8-fold of control). As observed in C57BL/6J mice, hepatic mRNA levels of other mRNA binding proteins (HNRNPI, KSRP) were not affected by Ad-shHNRNPD injection.
Fig. 4.
The effect of Ad-shHNRNPD injection on hepatic gene expression of Alb-Luc-UTR mice. Alb-Luc-UTR mice (n = 5 per group) were injected with 4 × 109 pfu/mouse Ad-shHNRNPD or control adenovirus (Ad-shLacZ). Bioluminescence imaging was recorded before viral injection and was recorded after 10 days of infection before euthanizing the animals. The results are the mean ± SEM of five animals per group, ***P < 0.001. A: Representative bioluminescent emissions before termination. B: Bioluminescence was quantified from each mouse and expressed as fold induction over the baseline values. The fold induction in control mice is expressed as one. C: Luc activity from liver homogenate was measured and normalized to tissue protein and relative activity was presented. D: Hepatic gene expression was measured by real-time PCR.
Upregulation of HNRNPD mRNA levels in hepatic cell lines by cholesterol
In an attempt to recapture the inducing effect of cholesterol on HNRNPD expression under in vitro conditions, HepG2 cells and mouse AML12 cells were cultured in medium containing LPDS overnight and were then treated with cholesterol or rosuvastatin (RSV) for 24 h. Gene expression analysis by qRT-PCR shows that cholesterol supplementation increased HNRNPD mRNA levels to 131% (P < 0.001) of control in HepG2 cells (Fig. 5A) and elevated HNRNPD mRNA levels to 133% (P < 0.001) of control in AML12 cells (Fig. 5B) while RSV had no effect on HNRNPD expression in both cell lines. In contrast to HNRNPD, LDLR mRNA levels were suppressed by cholesterol treatment and were strongly elevated by RSV. These data suggest that HNRNPD gene expression is not regulated by the SREBP pathway but could be a downstream target of the liver X receptor (LXR) signaling pathway that regulates many genes involved in cholesterol metabolism in liver tissue (33).
Fig. 5.
Induction of HNRNPD mRNA expression by cholesterol in cultured hepatic cell lines. HepG2 cells (A) or AML12 cells (B) were seeded in 6-well culture plates and cultured in medium supplemented with 10% LPDS overnight. The next day, DMSO (as vehicle), cholesterol (10 μg/ml cholesterol + 1 μg/ml 25-hydroxycholesterol), or 5 μM RSV was added to the cells for 24 h prior to isolation of total RNA. Hepatic mRNA levels of HNRNPD, LDLR, and GAPDH were assessed by qRT-PCR using specific primers with triplicate measurement of each cDNA sample. After normalization with GAPDH mRNA levels, the relative levels are presented. The data shown are the summarized results of two separate experiments in which duplicate wells were used in each condition. **P < 0.01, ***P < 0.001 compared with control. CHO, cholesterol. C: HepG2 cells cultured overnight in 10% LPDS medium were treated with RSV, 24-OHC, 24,25-EC, or CHO at the indicated concentrations for 24 h. Gene expression analysis was conducted as in (A) and (B). Duplicate wells were used for each treatment condition.
It is well-known that certain oxysterols, including 24S-hydroxycholestrol (24-OHC) and 24,25-epoxycholesterol (24,25-EC), are endogenous LXR ligands (34). Activation of LXR by oxysterols positively regulates LXR target genes and inhibits SREBP processing. Thus, we treated HepG2 cells cultured in LPDS medium with 24-OHC (20 μM) or 24,25-EC (10 and 20 μM) for 24 h. In this set of experiments, we included RSV and cholesterol as positive controls. Figure 5C shows that mRNA levels of inducible degrader of the LDLR (Idol) and ABCA1 were markedly elevated in cells treated with 24-OHC and 24,25-EC. Surprisingly, HNRNPD mRNA levels were unchanged by 24,25-EC at both 10 and 20 μM concentrations and were only elevated approximately 10% over control by 24-OHC. As we expected, mRNA levels of SREBP2 target genes, including LDLR, PCSK9, and SREBP2, were strongly repressed by oxysterols. Similar to the finding in HepG2 cells, we observed no substantial changes in HNRNPD mRNA levels in AML12 cells after 24 h treatment with 24-OHC or 24,25-EC, despite huge increases in Idol and ABCA1 mRNA levels (supplementary Fig. VI). Altogether, these results show that in cultured hepatic cells, HNRNPD mRNA expression was modestly and consistently elevated by cholesterol, but it did not respond to activators of LXR or SREBP. The molecular mechanisms by which cholesterol affects HNRNPD expression remain to be investigated.
DISCUSSION
It has been well-established that cholesterol feeding reduces LDLR mRNA and mRNAs encoding multiple enzymes in the cholesterol biosynthetic pathway in the livers of mice (15) and hamsters (13, 14). Over-accumulation of dietary cholesterol in the liver prevents the proteolytic activation of SREBPs and leads to a decline in the expression of SREBP target genes, including LDLR and SREBP2 itself. For decades, it has been thought that this transcriptional suppression is the sole mechanism accounting for reduced LDLR mRNA levels. The results from this current study demonstrate, for the first time, that dietary cholesterol reduces liver LDLR mRNA through dual action mechanisms of transcriptional suppression and acceleration of mRNA decay mediated by HNRNPD, the LDLR mRNA decay promoting ARE binding protein.
We began this study by exploring the relationship between cholesterol feeding and cellular abundance of LDLR mRNA binding proteins. From our previous in vitro studies, HNRNPD, along with two other LDLR ARE-BPs (HNRNPI and KSRP), were demonstrated to act as mRNA decay-promoting factors (21). Knocking down each of these proteins in HepG2 cells resulted in elevations of LDLR mRNA. Besides these negative regulators, a recent study reported that HuR, a ubiquitously expressed ARE binding protein, was capable of binding the proximal ARE site of LDLR mRNA 3′UTR and prolonging the mRNA half-life (22). Thus, initially, we examined the mRNA expression of these known negative and positive regulators of LDLR mRNA stability in livers of animals fed a normal chow diet containing little dietary cholesterol or fed a cholesterol-enriched rodent diet. These investigations led us to discover that feeding animals a HCD, specifically and uniformly elevated the expression of HNRNPD in liver tissue.
The significant induction of HNRNPD mRNA and protein by a HCD in both mice and hamsters suggested that this mRNA decay-promoting protein might contribute to the downregulation of LDLR mRNA levels in cholesterol-loaded livers.
Adenovirus Ad-shHNRNPD expresses a shRNA that targets all isoforms of mouse and human HNRNPD. Our previous in vitro studies have demonstrated that transduction of Ad-shHNRNPD in mouse primary hepatocytes, as well as in HepG2 cells, substantially reduced HNRNPD cellular levels with corresponding increases in LDLR mRNA and protein levels (28). Thus, in the present study with this valuable tool in hand, we wanted to address two important questions:1) whether knockdown of this decay-promoting protein would increase liver LDLR expression and affect serum LDL-C levels; and 2) in what physiological conditions changes in LDLR mRNA stability would generate the most impact on serum LDL-C levels.
We administered equal amounts of Ad-shHNRNPD or control adenovirus (Ad-shGFP) to C57BL/6J mice fed a HCD or a ND. Our results clearly showed that compared with normolipidemic mice, reduction of hepatic HNRNPD protein in hyperlipidemic mice produced greater effects on elevation of liver LDLR mRNA levels (2.6-fold in HCD mice vs. 1.7-fold in ND mice) and on lowering serum TC (−18.2% vs. −8%) and LDL-C (−30% vs. −13.2%). These new findings suggest that when the intracellular cholesterol level is elevated due to HCD and the SREBP pathway is repressed, stabilizing LDLR mRNA by blocking HNRNPD-mediated mRNA decay might be a viable means to increase hepatic LDLR activity to remove excessive LDL-C from the circulation. This could be clinically important.
The function of HNRNPD in mRNA turnover has been extensively studied. To date, most studies that characterize HNRNPD-mediated mRNA decay were conducted in a cell culture system (31, 32) or a cell free system (35, 36). Because our results showed the strong inverse correlation of HNRNPD cellular abundance with LDLR mRNA levels in Ad-shHNRNPD-infected C57BL/6J mice, it was crucial to demonstrate that the effect of HNRNPD was indeed mediated through LDLR3′UTR without the involvement of LDLR gene transcription in vivo. Our Alb-Luc-UTR transgenic mouse is the first in vivo model to exclusively study the function of 3′UTR in liver tissue in living mice. Driven by the liver-specific Alb promoter, the Luc-UTR is exclusively expressed in liver tissue. By infecting the transgenic mice with Ad-shHNRNPD virus or the control adenovirus, we showed that adenovirus-mediated depletion of HNRNPD strongly increased hepatic bioluminescent signals, Luc activity, and Luc-UTR mRNA expression along with elevating endogenous LDLR mRNA levels in these transgenic mice. These in vivo data clearly validated the action mechanism of Ad-shHNRNPD via reducing HNRNPD-mediated LDLR mRNA degradation through 3′UTR.
In this present study, we attempted to examine the cholesterol induction of HNRNPD in cultured hepatoma cells of mouse and human. We consistently detected elevations of HNRNPD mRNA levels in these hepatic cell lines, but the increases were relatively modest as compared with in vivo data. We have also treated hepatic cells with oxysterols that are strong activators of LXR or RSV to activate SREBP2. None of these treatments altered HNRNPD mRNA expression in cultured hepatic cells. How dietary cholesterol induces HNRNPD expression in liver tissue could be complex, and further in depth investigations are required to delineate the underlying mechanisms.
HNRNPD has been shown to regulate the stability or translational efficiency of diverse mRNA targets (30). Many of its target mRNAs encode proteins involved in the cell cycle, apoptosis, or inflammation. In the past, studies of HNRNPD have been focused on its target interaction and activity, little is known about how HNRNPD is regulated under normal physiological conditions or in disease situations. The results of our current studies not only demonstrate the key role of HNRNPD in regulation of lipid metabolism by targeting LDLR, but also unravel its regulation by dietary cholesterol in liver tissue.
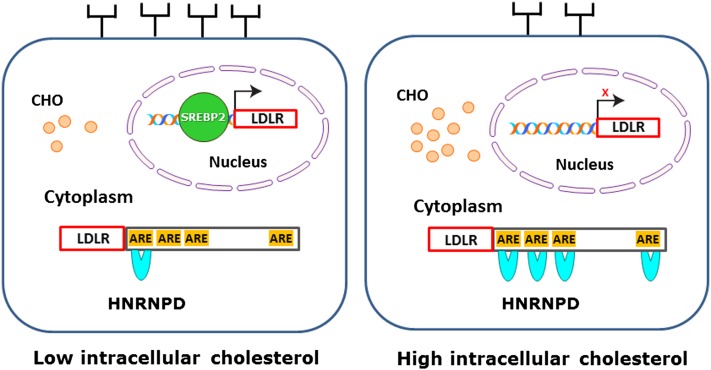
While the exact molecular mechanisms underlying the inducing effect of cholesterol feeding on hepatic HNRNPD expression in mice and hamsters await further investigations, results from the current study allow us to propose a model (Fig. 6) in which accumulation of intracellular cholesterols in liver reduce LDLR mRNA and subsequent protein amounts on the surface of hepatocytes by preventing the binding of SREBP2 to SRE1 of the LDLR promoter and by increasing the interaction of HNRNPD with ARE motifs of the LDLR3′UTR. This unique two-tier regulation at the transcriptional and posttranscriptional levels likely represents a natural response of liver tissue to cholesterol-enriched diets to reduce LDLR-mediated cholesterol uptake from circulation in order to maintain cellular cholesterol homeostasis.
Fig. 6.
A model for the two-tier regulation of hepatocyte LDLR expression by intracellular cholesterol (CHO). When the intracellular cholesterol level is low, mature SREBP2 is translocated to nucleus where it binds the SRE-1 site of the LDLR promoter and stimulates gene transcription, while ARE sites in 3′UTR are minimally occupied due to the low cellular abundance of HNRNPD, leading to mRNA stabilization. These dual effects result in more LDLR proteins reaching the hepatocyte surface. Conversely, when dietary cholesterol accumulates in the liver, the increased expression of HNRNPD in cytoplasm accelerates LDLR mRNA degradation by binding to multiple ARE sites in 3’UTR, and in nucleus SRE-1 site of LDLR promoter is not occupied by SREBP2, leading to a blockage of LDLR gene transcription. The fall in transcription rate combined with enhanced decay of mRNA ensures a reduced amount of LDLR reaching to the hepatocyte surface.
Supplementary Material
Footnotes
Abbreviations:
- Alb
- albumin
- ARE
- adenylate-uridylate-rich element
- ARE-BP
- adenylate-uridylate-rich element-binding protein
- AU
- adenylate-uridylate
- 24
- 25-EC, 24,25-epoxycholesterol
- HCD
- high cholesterol diet
- HDL-C
- HDL-cholesterol
- HNRNP
- heterogeneous nuclear ribonucleoprotein
- HuR
- human antigen R
- KSRP
- KH-type splicing regulatory protein
- LDL-C
- LDL-cholesterol
- LDLR
- LDL receptor
- LPDS
- lipoprotein-depleted serum
- Luc
- luciferase
- Luc-UTR
- luciferase-LDL receptor 3′ untranslated region
- LXR
- liver X receptor
- ND
- normal diet
- 24-OHC
- 24S-hydroxycholesterol
- PCSK9
- proprotein convertase subtilisin/kexin type 9
- RSV
- rosuvastatin
- SRE
- sterol-regulatory element
- SREBP
- sterol-regulatory element binding protein
- TFR
- transferrin receptor
- UTR
- untranslated region
This study was supported by the Department of Veterans Affairs (Office of Research and Development, Medical Research Service) and by grants 1R01AT002543-01A1 and 1R01AT006336-01A1 from the National Center of Complementary and Alternative Medicine. The authors declare no financial conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of six figures and one table.
REFERENCES
- 1.Spady D. K. 1992. Hepatic clearance of plasma low density lipoproteins. Semin. Liver Dis. 12: 373–385 [DOI] [PubMed] [Google Scholar]
- 2.Brown M. S., Goldstein J. L. 1986. A receptor-mediated pathway for cholesterol homeostasis. Science. 232: 34–47 [DOI] [PubMed] [Google Scholar]
- 3.Goldstein J. L., Brown M. S. 1990. Regulation of the mevalonate pathway. Nature. 343: 425–430 [DOI] [PubMed] [Google Scholar]
- 4.Marcoux C., Tremblay M., Fredenrich A., Jacques H., Krimbou L., Nakajima K., Davignon J., Cohn J. S. 1998. Plasma remnant-like particle lipid and apolipoprotein levels in normolipidemic and hyperlipidemic subjects. Atherosclerosis. 139: 161–171 [DOI] [PubMed] [Google Scholar]
- 5.Homma Y. 2004. Predictors of atherosclerosis. J. Atheroscler. Thromb. 11: 265–270 [DOI] [PubMed] [Google Scholar]
- 6.Brown M. S., Goldstein J. L. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of the membrane bound transcription factor. Cell. 89: 331–340 [DOI] [PubMed] [Google Scholar]
- 7.Brown M. S., Goldstein J. L. 1999. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. USA. 96: 11041–11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson G. M., Vasa M. Z., Deeley R. G. 1998. Stabilization and cytoskeletal-association of LDL receptor mRNA are mediated by distinct domains in its 3′ untranslated region. J. Lipid Res. 39: 1025–1032 [PubMed] [Google Scholar]
- 9.Wilson G. M., Roberts E. A., Deeley R. G. 1997. Modulation of LDL receptor mRNA stability by phorbol esters in human liver cell culture models. J. Lipid Res. 38: 437–446 [PubMed] [Google Scholar]
- 10.Goto D., Okimoto T., Ono M., Shimotsu H., Abe K., Tsujita Y., Kuwano M. 1997. Upregulation of low density lipoproteinreceptor by gemfibrozil, a hypolipidemic agent, in human hepatoma cells through stabilization of mRNA transcripts. Arterioscler. Thromb. Vasc. Biol. 17: 2707–2712 [DOI] [PubMed] [Google Scholar]
- 11.Kong W., Wei J., Abidi P., Lin M., Inaba S., Li C., Wang Y., Wang Z., Si S., Pan H., et al. 2004. Berberine is a promising novel cholesterol-lowering drug working through a unique mechanism distinct from statins. Nat. Med. 10: 1344–1351 [DOI] [PubMed] [Google Scholar]
- 12.Parker R., Song H. 2004. The enzyme and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11: 121–127 [DOI] [PubMed] [Google Scholar]
- 13.Sheng Z., Otani H., Brown M. S., Goldenstein J. L. 1995. Independent regulation of sterol regulatory element-binding proteins 1 and 2 in hamster liver. Proc. Natl. Acad. Sci. USA. 92: 935–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimomura I., Bashmakov Y., Shimano H., Horton J. D., Goldstein J. L., Brown M. S. 1997. Cholesterol feeding reduces nuclear forms of sterol regulatory element binding proteins in hamster liver. Proc. Natl. Acad. Sci. USA. 94: 12354–12359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korn B. S., Shimomura I., Bashmakov Y., Hammer R. E., Horton J. D., Goldstein J. L., Brown M. S. 1998. Blunted feedback suppression of SREBP processing by dietary cholesterol in transgenic mice expressing sterol-resistant SCAP(D443N). J. Clin. Invest. 102: 2050–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto T., Davis C. G., Brown M. S., Schneider W. J., Casey M. L., Goldstein J. L., Russell D. W. 1984. The human LDL receptor: A cysteine-rich protein with multiple Alu sequence in its mRNA. Cell. 39: 27–38 [DOI] [PubMed] [Google Scholar]
- 17.Yashiro T., Yokoi Y., Shimizu M., Inoue J., Sato R. 2011. Chenodeoxycholic acid stabilization of LDL receptor mRNA depends on 3′-untranslated region and AU-rich element-binding protein. Biochem. Biophys. Res. Commun. 409: 155–159 [DOI] [PubMed] [Google Scholar]
- 18.Barreau C., Paillard L., Osborne H. S. 2005. Survey and summary AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33: 7138–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang T., Kruys V., Huez G., Gueydan C. 2002. AU-rich element-mediated translational control: complexity and multiple activity of trans-activating factors. Biochem. Soc. Trans. 30: 952–958 [DOI] [PubMed] [Google Scholar]
- 20.Meyer S., Temme C., Wahle E. 2004. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 39: 197–216 [DOI] [PubMed] [Google Scholar]
- 21.Li H., Chen W., Zhou Y., Abidi P., Sharpe O., Robinson W. H., Liu J. 2009. Identification of mRNA-binding proteins that regulate the stability of LDL receptor mRNA through AU-rich elements. J. Lipid Res. 50: 820–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yashiro T., Nanmoku M., Shimizu M., Inoue J., Sato R. 2013. 5-Aminoimidazole-4-carboxamide ribonucleoside stabilizes low density lipoprotein receptor mRNA in hepatocytes via ERK-dependent HuR binding to an AU-rich element. Atherosclerosis. 226: 95–101 [DOI] [PubMed] [Google Scholar]
- 23.Lin S., Wang W., Wilson G. M., Yang X., Brewer G., Holbrook N. J., Gorospe M. 2000. Down-regulation of cyclin D1 expression by prostaglandin A(2) is mediated by enhanced cyclin D1 mRNA turnover. Mol. Cell. Biol. 20: 7903–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pont A. R., Sadri N., Hsiao S. J., Smith S., Schneider R. J. 2012. mRNA decay factor AUF1 maintains normal aging, telomere maintenance, and suppression of senescence by activation of telomerase transcription. Mol. Cell. 47: 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Briata P., Chen C. Y., Giovarelli M., Pasero M., Trabucchi M., Ramos A., Gherzi R. 2011. KSRP, many functions for a single protein. Front. Biosci. (Landmark Ed.) 16: 1787–1796. [DOI] [PubMed] [Google Scholar]
- 26.Ruggiero T., Trabucchi M., Ponassi M., Corte G., Chen C. Y., al-Haj L., Khabar K. S., Briata P., Gherzi R. 2007. Identification of a set of KSRP target transcripts upregulated by PI3K-AKT signaling. BMC Mol. Biol. 8: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raineri I., Wegmueller D., Gross B., Certa U., Moroni C. 2004. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 32: 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh A. B., Li H., Kan C. F., Dong B., Nicolls M. R., Liu J. 2014. The Critical role of mRNA destabilizing protein heterogeneous nuclear ribonucleoprotein D in 3′ untranslated region-mediated decay of low-density lipoprotein receptor mRNA in liver tissue. Arterioscler. Thromb. Vasc. Biol. 34: 8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong B., Kan C. F., Singh A. B., Liu J. 2013. High-fructose diet downregulates long-chain acyl-CoA synthetase 3 expression in liver of hamsters via impairing LXR/RXR signaling pathway. J. Lipid Res. 54: 1241–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gratacós F. M., Brewer G. 2010. The role of AUF1 in regulated mRNA decay. RNA. 1: 457–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loflin P., Chen C-Y. A., Shyu A-B. 1999. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 13: 1884–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barker A., Epis M. R., Porter C. J., Hopkins B. R., Wilce M. C. J., Wilce J. A., Giles K. M., Leedman P. J. 2012. Sequence requirements for RNA binding by HuR and AUF1. J. Biochem. 151: 423–437 [DOI] [PubMed] [Google Scholar]
- 33.Spann N. J., Garmire L. X., McDonald J. G., Myers D. S., Milne S. B., Shibata N., Reichart D., Fox J. N., Shaked I., Heudobler D., et al. 2012. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell. 151: 138–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jakobsson T., Treuter E., Gustafsson J-A., Steffensen K. R. 2012. Liver X receptor biology and pharmacology: new pathways, challenges and opportunities. Trends Pharmacol. Sci. 33: 394–404 [DOI] [PubMed] [Google Scholar]
- 35.Mazan-Mamczarz K., Kuwano Y., Zhan M., White E. J., Martindale J. L., Lai A., Gorospe M. 2009. Identification of a signature motif in target mRNAs of RNA-binding protein AUF1. Nucleic Acids Res. 37: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zucconi B. E., Wilson G. M. 2013. Assembly of functional ribonucleoprotein complexes by AU-rich element RNA-binding protein 1 (AUF1) requires base-dependent and -independent RNA contacts. J. Biol. Chem. 288: 28034–28048 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.