Abstract
Currently existing data show that the capability for long-chain PUFA (LC-PUFA) biosynthesis in teleost fish is more diverse than in other vertebrates. Such diversity has been primarily linked to the subfunctionalization that teleostei fatty acyl desaturase (Fads)2 desaturases have undergone during evolution. We previously showed that Chirostoma estor, one of the few representatives of freshwater atherinopsids, had the ability for LC-PUFA biosynthesis from C18 PUFA precursors, in agreement with this species having unusually high contents of DHA. The particular ancestry and pattern of LC-PUFA biosynthesis activity of C. estor make this species an excellent model for study to gain further insight into LC-PUFA biosynthetic abilities among teleosts. The present study aimed to characterize cDNA sequences encoding fatty acyl elongases and desaturases, key genes involved in the LC-PUFA biosynthesis. Results show that C. estor expresses an elongase of very long-chain FA (Elovl)5 elongase and two Fads2 desaturases displaying Δ4 and Δ6/Δ5 specificities, thus allowing us to conclude that these three genes cover all the enzymatic abilities required for LC-PUFA biosynthesis from C18 PUFA. In addition, the specificities of the C. estor Fads2 enabled us to propose potential evolutionary patterns and mechanisms for subfunctionalization of Fads2 among fish lineages.
Keywords: biosynthesis, elongase of very long-chain fatty acids, evolution, fatty acyl desaturase 2, long-chain polyunsaturated fatty acids, teleosts
Biosynthesis of long-chain PUFAs (LC-PUFAs) in vertebrates involves sequential desaturation and elongation of C18 PUFA, linoleic acid (LOA; 18:2n-6), and α-linolenic acid (LNA; 18:3n-3) (1). Synthesis of arachidonic acid (ARA; 20:4n-6) and EPA (20:5n-3) from LOA and LNA, respectively, utilizes the same enzymes and pathways. The predominant pathway involves Δ6 desaturation of LOA or LNA to 18:3n-6/18:4n-3 that are elongated to 20:3n-6/20:4n-3 followed by Δ5 desaturation to ARA/EPA (1), but an alternative pathway with initial elongation of LOA or LNA followed by Δ8 desaturation, an inherent ability of some Δ6 desaturases, may be possible (2). Biosynthesis of DHA (22:6n-3) from EPA can also occur by two pathways. First, the so-called “Sprecher pathway” involves two sequential elongation steps from EPA to 24:5n-3 and a subsequent Δ6 desaturation to 24:6n-3, followed by peroxisomal chain shortening (3). Second, a more direct pathway has been postulated in some marine fish that involves elongation of EPA to docosapentaenoic acid (22:5n-3) followed by Δ4 desaturation to DHA (4, 5).
Dietary PUFAs are essential in fish, although requirements vary with species (6, 7). Generally, C18 PUFAs can satisfy essential FA requirements of freshwater and salmonid species, but most marine fish have a requirement for LC-PUFAs such as EPA and DHA (8). Differing essential FA requirements have been linked to differences in the complement of fatty acyl desaturase (Fads) and elongase of very long-chain FA (Elovl) genes (9–31). Thus, the dependence of many marine fish for dietary LC-PUFAs was caused by deficiency in key enzymatic activities required for their biosynthesis from C18 PUFAs (7, 8). This was hypothesized to be a consequence of marine fish having evolved in an LC-PUFA-rich environment; and thus, there was low evolutionary pressure to retain the ability to desaturate and elongate C18 PUFA. In contrast, lower levels of LC-PUFAs in the food chain meant freshwater species retained the ability to biosynthesize LC-PUFAs from C18 PUFAs (8, 32, 33). This hypothesis was based upon fish that were largely carnivorous in the case of marine species and detritivorous/herbivorous in freshwater species (34). However, trophic level, the position of an organism within the food web, was also investigated, and it was demonstrated that the herbivorous marine fish Siganus canaliculatus (rabbitfish) had the ability to endogenously synthesize LC-PUFAs because two desaturases with Δ4 and Δ6/Δ5 specificities and two elongases (Elovl4 and Elovl5) enabled this species to perform all the enzymatic reactions required in the pathway (4, 28). More recently, other confounding factors, including “trophic ecology” and diadromy, have been proposed (5, 30). Currently, data indicate that the capability for LC-PUFA biosynthesis in teleost fish is more diverse than in other vertebrate groups, and is possibly the result of a combination of factors that interact throughout the evolutionary history of each particular group or species. Such plasticity has been primarily associated with the substrate specificities exhibited by desaturase Fads2, a protein that has subfunctionalized during the evolution of teleosts.
Fads2 has been shown to be the sole Fads-like desaturase existing in teleost genomes, in contrast with other vertebrates that also have another desaturase termed Fads1 (32). Mammalian FADS1 encodes a desaturase with Δ5 activity, whereas FADS2 encodes a Δ6 desaturase (35). Consistently, the majority of teleostei Fads2s are typically Δ6 desaturases (11, 15, 16, 21, 22, 26, 27, 30). Additionally, however, some teleost Fads2s have been functionally characterized as Δ5 (13), Δ4 (4, 5), and bifunctional Δ6/Δ5 (4, 9) desaturases. It has been postulated that the reasons underpinning the diversification of Fads2 specificities observed in certain teleost lineages are the result of adaptations to habitat-specific food-web structures in different environments and over geological timescales (32).
Pike silverside Chirostoma estor (previously Menidia estor) from Lake Pátzcuaro is a highly valued freshwater fish in Mexico (36–39). Although C. estor is a freshwater species, it has a common ancestry with marine atherinopsids (40) and shares many biological and physiological characteristics of marine species (41). Also, unusually for a freshwater fish, tissues of C. estor show high levels of DHA (20–32% of total FAs) and only low levels of EPA (1–3%) in contrast to the FA profile of its zooplankton diet (∼12% DHA, 13% EPA) (42). This suggested that C. estor either selectively accumulates DHA, or has the capacity to convert EPA or other n-3 PUFAs to DHA (7). Interestingly, the activity of the LC-PUFA synthesis pathway in C. estor was very low in freshwater (0 parts per thousand salinity) in comparison to that in higher salinity (5 or 15 parts per thousand), and this was unexpected for a freshwater fish, as they generally show appreciable LC-PUFA synthesis activity (43).
The ancestry of C. estor, pattern of LC-PUFA biosynthesis activity, and effects of salinity, which conflict with the existing paradigm, made this species an ideal candidate for studies investigating the molecular basis of LC-PUFA biosynthesis to gain further understanding of the diversity that these pathways show among teleost lineages. The objective of the present study was the functional characterization of cDNAs encoding desaturases and elongases involved in the LC-PUFA biosynthetic pathways in C. estor. The results indicated that C. estor has all the enzymatic capabilities for endogenous biosynthesis of LC-PUFAs. In addition to the presence of an Elovl5 elongase, C. estor expresses at least two distinct Fads2 enzymes displaying Δ4 and Δ6/Δ5 specificities. The findings on C. estor desaturases provided further insight into the diversification of substrate specificities in teleostei Fads2s and enabled potential evolutionary patterns of subfunctionalized Fads2s among teleost lineages to be proposed.
MATERIALS AND METHODS
Tissue samples
Pike silverside (∼40 g), maintained at the facilities of the Instituto de Investigaciones Agropecuarias y Forestales (UMSNH), Mexico, were anesthetized and euthanized with an overdose of MS-222 (Sigma-Aldrich, Alcobendas, Spain). Tissues including liver, intestine, brain, and muscle were collected in RNAlater (Ambion, Applied Biosystems, Warrington, UK) according to manufacturer’s instructions. Fish maintenance and euthanasia procedures were carried out in compliance with the Official Mexican Standard NOM-062-ZOO-1999 on the technical specifications for the production, care, and use of laboratory animals. This study was also reviewed and approved by the departmental ethics committees of Institute of Aquaculture Torre de la Sal (CSIC, Spain) and Institute of Aquaculture (University of Stirling, UK).
Cloning of fatty acyl desaturases and elongase from C. estor
Total RNA (2 μg) extracted from C. estor tissues (TRI reagent, Sigma-Aldrich) was reverse transcribed using a GoScriptTM reverse transcription system (Promega, Madison, WI) primed with random primers. The primers UNIDF (5′-GGAGAGGAYGCCACGGAGG-3′) and UNIDR (5′-GTCCRCTGAACCAGTCGTTGAA-3′) for fads2 and UNIE5F (5′-CATGGATGGGYCCMAGAGATC-3′) and UNIE5R (5′-GTCTGAATGTAGAAGTTTGAGAAAAG-3′) for elovl5 were used to amplify a partial fragment of these genes from C. estor by PCR with GoTaq® Green Master Mix (Promega) as described previously (30). A mixture of cDNA from liver and brain was used as a template for PCR, which consisted of an initial denaturing step at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 30 s, extension at 72°C for 1 min 30 s, followed by a final extension at 72°C for 5 min. The PCR fragments were purified and sequenced at the DNA Sequencing Service IBMCP-UPV (Valencia, Spain). While one single version of the elovl5-like cDNA sequence was detected, sequencing results of the desaturase-targeted PCR revealed the existence of at least two distinct fads2-like transcripts. Their individual sequences, hereafter referred to as Fads2a and Fads2b, were determined after ligation of the PCR product into pGEM-T Easy vector (Promega) and sequencing as above. The partial sequences within the open reading frame (ORF) of the two desaturases and the elongase were further extended by 5′ and 3′ rapid amplification of cDNA ends (RACE) PCR (FirstChoice® RLM-RACE kit, Ambion) through a two-round (nested) PCR approach as detailed below (see supplementary Table I for primer details).
For desaturase fads2a, first round PCR was performed combining the adaptor-specific 5′RACE outer primer with the gene-specific reverse primer CEFaR1, whose 3′ end sequence contained two nucleotides that differed from the fads2b sequence. A second round PCR with 5′RACE inner primer and CEFaR2 produced a positive band expanding out the putative end of the 5′untranslated region. For the fads2b cDNA sequence, a specific 5′RACE product was not obtained, but the ORF of fads2b was obtained to enable functional characterization as detailed below.
Positive 3′RACE PCR products were obtained using isoform-specific primers and 3′ adaptor primers. First round PCR involved the use of forward primers CEFaF1 (isoform a) and CEFbF1 (isoform b) with the 3′RACE outer primer. First round products were subsequently used as a template for nested PCR with forward primers CEFaF2 (isoform a) and CEFbF2 (isoform b), and with reverse primers consisting of the adaptor primer 3′RACE inner.
A similar approach was followed to obtain the full-length cDNA of C. estor elovl5. The gene-specific primers CEE5R1 and CEE5R2 (5′ RACE) and CEE5F1 and CEE5F2 (3′RACE) used for RACE PCR are listed in supplementary Table I. General RACE PCR conditions consisted of an initial denaturing step at 95°C for 2 min, followed by 32–35 cycles of denaturation at 95°C for 30 s, annealing at 55–60°C for 30 s, extension at 72°C for 2 min 30 s, followed by a final extension at 72°C for 5 min (GoTaq® Green Master Mix, Promega). PCR products were cloned into pGEM-T Easy vector (Promega) and sequenced as above.
Sequence and phylogenetic analyses
The amino acid (aa) sequences of the C. estor desaturases Fads2a and Fads2b, and elongase Elovl5 proteins were compared with those of orthologs from other fish and tetrapods (mammals, amphibians, and birds) and sequence identity scores were obtained using the EMBOSS Needle pairwise sequence alignment tool. For phylogenetic analysis of the C. estor deduced aa sequences of fads2a, fads2b, and elovl5, two trees were constructed using the neighbor-joining method (44), with confidence in the resulting tree branch topology measured by bootstrapping through 10,000 iterations. Desaturase and elongase C. estor sequences were compared with homologous proteins from a variety of vertebrate lineages. The Δ6 desaturase and the PUFA elongase sequences from the oleaginous fungus Mortierella alpina were used as outgroup sequences to construct both rooted trees.
Functional characterization of the C. estor Fads2 and Elovl5 by heterologous expression in yeast
PCR fragments corresponding to the ORF of pike silverside desaturases fads2a and fads2b and elongase elovl5 were amplified from a mixture of cDNA (liver and brain) by PCR using the high fidelity Pfu DNA polymerase (Promega) with primers containing HindIII and SacI restriction sites (underlined in supplementary Table I) as follows. For fads2a, the primer pair CEFVF-CEFaVR was used. For fads2b, the same forward primer CEFVF and the antisense primer CEFbVR were used. While CEFVF was specific for both isoforms, its use in combination with the fads2b-specific primer CEFbVR enabled us to successfully isolate fads2b. Finally, the ORF of the elovl5 was isolated using the primers CEE5VF and CEE5VR (supplementary Table I). PCR consisted of an initial denaturing step at 95°C for 2 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 57°C for 30 s, extension at 72°C for 3 min (desaturase) or 2 min (elongase), followed by a final extension at 72°C for 5 min. The PCR products were subsequently purified (Illustra GFX PCR DNA/gel band purification kit, GE Healthcare, Little Chalfont, UK), digested with the corresponding restriction enzymes (Promega) and ligated into a similarly restricted pYES2 yeast expression vector (Invitrogen, Paisley, UK). The plasmid constructs prepared were designated as pYES2-fads2a, pYES2-fads2b, and pYES2-elovl5.
Yeast competent cells, InvSc1 (Invitrogen), were transformed with the desaturase (pYES2-fads2a, pYES2-fads2b) and elongase (pYES2-elovl5) plasmid constructs and then grown in selective Saccharomyces cerevisiae minimal medium (SCMM)-uracil (10). A single recombinant colony for each enzyme was grown in SCMM-uracil broth and diluted to an optical density (OD)600 of 0.4 in one single Erlenmeyer flask for each potential substrate assayed. The substrate specificities of both Fads2 isoforms were assessed by growing the transgenic yeast in medium supplemented with one of the following substrates: 18:3n-3 or 18:2n-6 for Δ6 desaturation, 20:3n-3 or 20:2n-6 for Δ8 desaturation, 20:4n-3 or 20:3n-6 for Δ5 desaturation, and 22:5n-3 or 22:4n-6 for Δ4 desaturation. The function of C. estor Elovl5 was characterized by growing yeast transformed with pYES2-elovl5 in medium containing one of the following PUFA substrates: 18:3n-3, 18:2n-6, 18:4n-3, 18:3n-6, 20:5n-3, 20:4n-6, 22:5n-3, or 22:4n-6. The FA substrates were added to the yeast cultures at final concentrations of 0.5 mM (C18), 0.75 mM (C20), and 1.0 mM (C22) to compensate for decreased efficiency of uptake with increased chain length (19). Yeast transformed with empty pYES2 were also grown in presence of PUFA substrates as control treatments. After 2 day culture at 30°C, yeast were harvested and washed, and total lipid was extracted by homogenization in chloroform/methanol (2:1, v/v) containing 0.01% butylated hydroxytoluene as antioxidant. Results were confirmed by running the assay with a different transformant colony.
All PUFA substrates, except stearidonic acid (18:4n-3), were from Nu-Chek Prep, Inc. (Elysian, MN). Stearidonic acid and chemicals used to prepare SCMM-uracil were from Sigma-Aldrich, except for the bacteriological agar obtained from Oxoid Ltd. (Hants, UK).
FA analysis of yeast
Total lipids were extracted from yeast samples and fatty acyl methyl esters (FAMEs) were prepared as described previously (9, 30). FAMEs were quantified using a Thermo gas chromatograph (Thermo Trace GC Ultra, Thermo Electron Corporation, Waltham, MA) fitted with an on-column injection system and a flame ionization detector (FID). Additionally, FAMEs were identified using an Agilent 6850 gas chromatograph system coupled to a 5975 series MSD (Agilent Technologies, Santa Clara, CA). The desaturation or elongation conversion efficiencies from exogenously added PUFA substrates were calculated by the proportion of substrate FA converted to desaturated or elongated products as [individual product area/(all products areas + substrate area)] × 100. For the elongase, some of the initial elongation products were further elongated, and thus the accumulated conversions were calculated by summing all elongated products (28). Similarly, the desaturase Fads2b exhibited multifunctional abilities, and thus the conversions on Δ8 substrates (20:3n-3 and 20:2n-6) include stepwise reactions.
Tissue distribution of fads2 and elovl5 transcripts
Expression of the target genes (fads2a, fads2b, and elovl5) was measured by quantitative real-time PCR (qPCR). Total RNA from liver, brain, intestine, and muscle was extracted from three C. estor adults, as described above, and 2 μg of total RNA were reverse transcribed into cDNA (M-MLV reverse transcriptase, Promega) using oligo-dT primer. The qPCR was performed using primers shown in supplementary Table I. Copy numbers of target genes were normalized with copy number of the reference gene ef-1α (GenBank accession number KJ439615). PCR amplicons of each gene were cloned into pGEM-T Easy vector (Promega) that was then linearized, quantified spectrophotometrically (NanoDrop 2000c, Thermo Scientific, Wilmington, DE), and serial-diluted to generate a standard curve of known copy numbers. The qPCR amplifications were carried out in duplicate using a CFX96 BioRad machine (Alcobendas, Spain) in a final volume of 20 μl containing 5 μl diluted (1/20) cDNA, 0.25 μM of each primer, and 4 μl PyroTaq EvaGreen® mix (Cultek Molecular Bioline, Madrid, Spain). Amplifications were carried out with a systematic negative control (no template control) containing no cDNA. The qPCR profiles consisted of an initial activation step at 95°C for 15 min, followed by 40 cycles of 15 s at 95°C, 20 s at the specific primer pair annealing melting temperature (supplementary Table I), and 15 s at 72°C. After the amplification phase, a dissociation curve of 0.5°C increments from 60 to 90°C was performed, enabling confirmation of the amplification of a single product in each reaction. No primer-dimer formation occurred in the no template control. All reactions were carried out in duplicate, a linear standard curve was drawn, and the absolute copy number of the targeted gene in each sample was calculated.
Statistical analysis
Tissue expression (qPCR) results are expressed as mean normalized values (± SE) corresponding to the ratio between the copy numbers of fads2a, fads2b, and elovl5 transcripts and the copy numbers of the reference gene ef-1α. A one-way ANOVA followed by Tukey HSD test (P < 0.05) was performed to compare the expression level among the selected tissues (SPSS, Chicago, IL).
RESULTS
C. estor fads2 and elovl5 sequences and phylogenetics
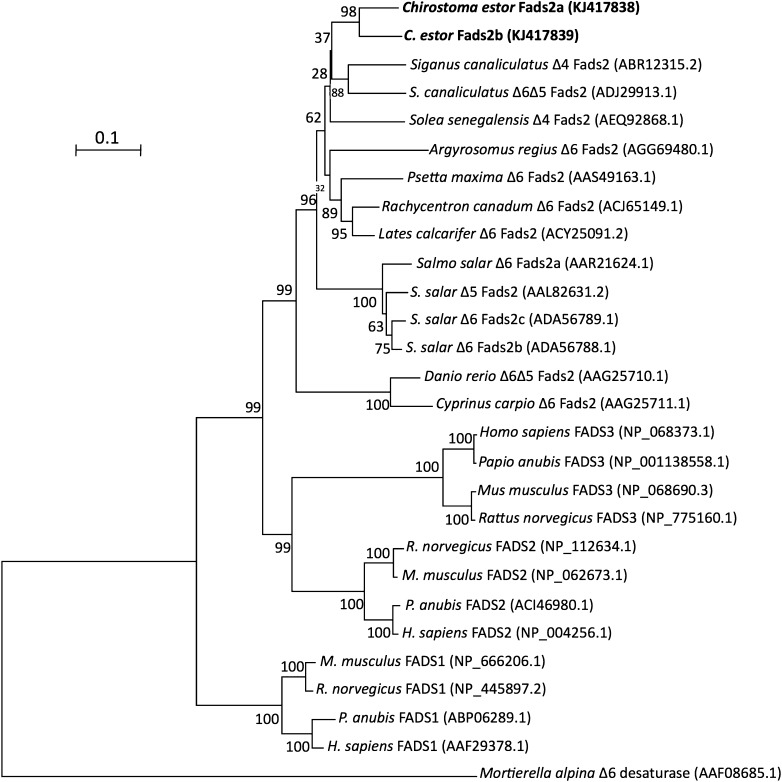
Both pike silverside fads2 desaturases (a and b) have an ORF of 1,326 bp encoding putative proteins of 441 aa that are 88.4% identical to each other. Pairwise aa sequence comparisons of C. estor Fads2 proteins and other homologs from fish (Cyprinus carpio, Siganus canaliculatus, Danio rerio, Solea senegalensis, Argyrosomus regius, Psetta maxima, Lates carcarifer, Rachycentron canadum, and Salmo salar) showed identities ranging from 68.1 to 82.1%, with mammalian FADS2 (Homo sapiens, Mus musculus, and Rattus norvegicus) ranging from 63.3 to 65.5%. Lower identity scores were obtained with other desaturase families from mammals (H. sapiens, M. musculus, and R. norvegicus), namely FADS1 (56.2–58.2% identity) and FADS3 (51.8–55.5%). Sequence comparison of the C. estor Fads2a with the Δ4 desaturase from Thraustochytrium sp. ATCC21685 (45) resulted in 22.7% identity. Analysis of the deduced aa sequence of C. estor clones revealed that they had structural features of fatty acyl desaturases including three histidine boxes (HDFGH, HFQHH, and QIEHH) and a putative cytochrome b5-like domain in the N terminus containing the typical HPGG motif essential for desaturation activity (35). GenBank accession numbers for the newly cloned cDNA sequences are KJ417838 (fads2_a) and KJ417839 (fads2_b).
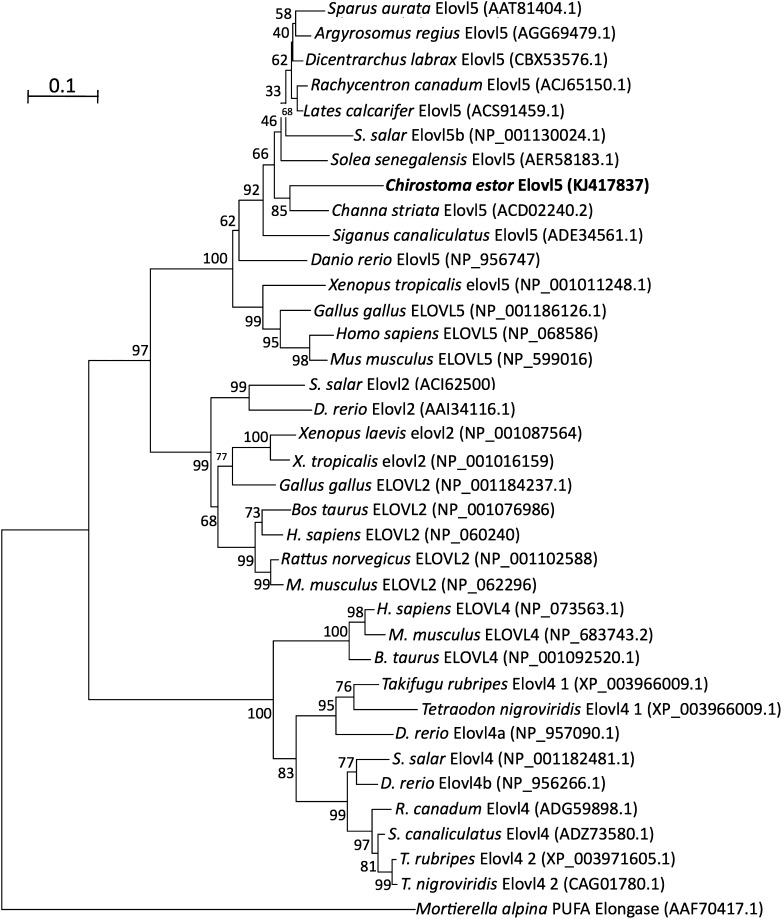
The ORF of the C. estor elovl5 was 885 bp long, encoding a protein of 294 aa. Pairwise comparison of the translated aa sequence showed that the Elovl5 shared 81.1–67.7% identity to Elovl5 proteins from fish (D. rerio, A. regius, C. striata, L. calcarifer, and S. aurata), and 62.5% to that from H. sapiens, 62.7% to that from Xenopus tropicalis, and 64.4% to that of Gallus gallus. Lower identities (38.0–52.3%) were obtained when the C. estor Elovl5 was compared with other elongase families such as Elovl2 and Elovl4 from different vertebrate lineages including teleosts, amphibians, birds, and mammals. The C. estor Elovl5 had a diagnostic histidine box (HVYHH), and lysine (K) and arginine (R) residues at the carboxyl terminus (KKLRVD). The sequence of the elovl5 cDNA from C. estor was deposited in GenBank with accession number KJ417837.
Phylogenetic trees constructed on the basis of aa sequence comparisons of the C. estor cDNAs with homologous proteins from fish and other vertebrate species reflected the identity scores shown above. For desaturases, the C. estor Fads2s clustered with Fads2s from fish and, more distantly, with Fads2s from mammals, birds, and amphibians (Fig. 1). Additionally, Fads1-like proteins, “front-end” desaturases not present in teleost fish (32), grouped separately from the Fads2 cluster that included the C. estor desaturases (Fig. 1). On the other hand, the C. estor Elovl5 grouped together with other teleost and tetrapod (mammals, birds, and amphibians) orthologs, and separately from members of Elovl2 and Elovl4 families from fish and other vertebrates (Fig. 2).
Fig. 1.
Phylogenetic tree comparing the deduced aa sequence of the newly cloned C. estor Fads2-like with Fads1- and Fads2-like desaturases from a variety of vertebrates. The tree was constructed using the neighbor-joining method (44) with MEGA4. The horizontal branch length is proportional to aa substitution per site. The numbers represent the frequencies with which the tree topology presented was replicated after 10,000 bootstrap iterations. All accession numbers are from GenBank database.
Fig. 2.
Phylogenetic tree comparing the deduced aa sequence of the newly cloned C. estor elongase of very long-chain FAs (Elovl) with elongases Elovl2, Elovl4, and Elovl5 from a variety of vertebrates. The tree was constructed using the neighbor-joining method (44) with MEGA4. The horizontal branch length is proportional to aa substitution per site. The numbers represent the frequencies with which the tree topology presented was replicated after 10,000 bootstrap iterations. All accession numbers are from GenBank database.
Functional characterization in yeast
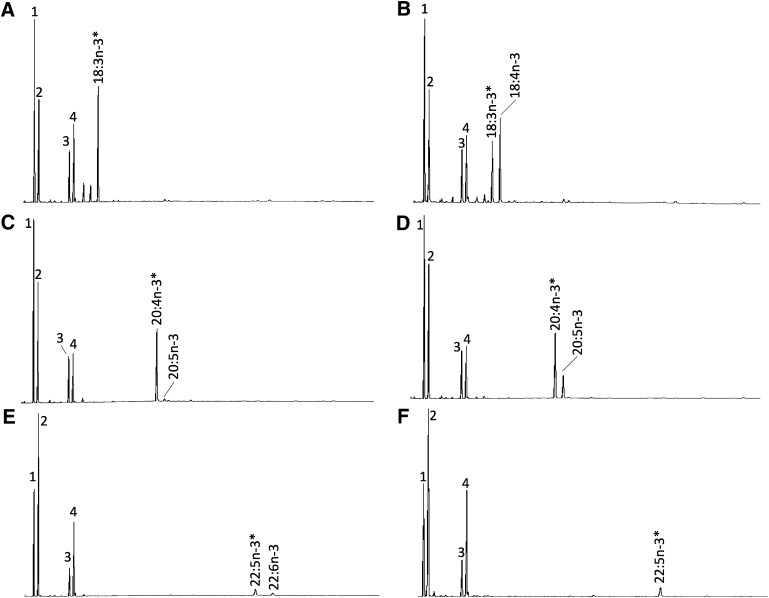
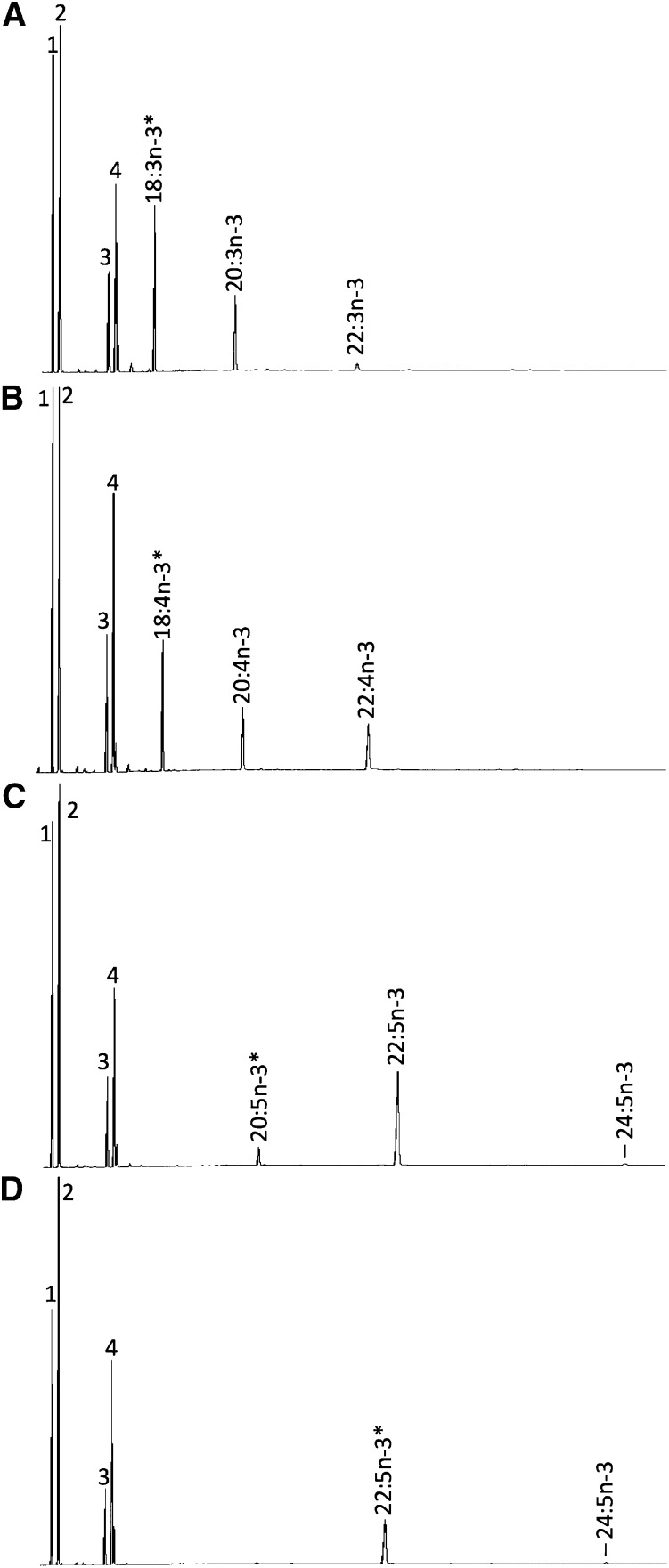
The ORF of C. estor desaturases Fads2a and Fads2b, along with the elongase Elovl5, were functionally characterized in yeast. Control treatments, consisting of yeast transformed with empty pYES2 vector, indicated there was no endogenous activity, as all exogenously added PUFA substrates remained unmodified. Hence, the FA profile of control yeast was characterized by the main endogenous FAs of S. cerevisiae, namely 16:0, 16:1 isomers (16:1n-9 and 16:1n-7), 18:0, and 18:1 isomers (18:1n-9 and 18:1n-7) (22), plus the exogenously added PUFAs (data not shown). This indicated that wild-type yeast do not express activities toward PUFA substrates, in agreement with earlier observations (9, 10). Yeast expressing the C. estor desaturases and elongases showed additional peaks. Yeast transformed with pYES2-fads2a and grown in the presence of 22:5n-3 and 22:4n-6 showed additional peaks corresponding to 22:6n-3 and 22:5n-3, respectively, with identities confirmed by GC-MS (Fig. 3). The conversions of these desaturation reactions are shown in Table 1. These results confirmed that Fads2a from C. estor had Δ4 desaturase activity. Moreover, the C. estor Fads2a exhibited some Δ5 activity on 20:4n-3 that was desaturated to 20:5n-3, although no Δ5 desaturation was detected toward 20:3n-6 (Table 1).
Fig. 3.
Functional characterization of the C. estor Fads2a and Fads2b desaturases in yeast (S. cerevisiae). The FA profiles of yeast transformed with pYES2-fadsa (A, C, E) and pYES2-fadsb (B, D, F) were determined after they were grown in the presence of FA substrates (*) 18:3n-3 (A, B), 20:4n-3 (C, D), 22:5n-3 (E, F). Peaks 1–4 in all panels are the main endogenous FAs of S. cerevisiae, namely 16:0 (peak 1), 16:1 isomers (peak 2), 18:0 (peak 3), and 18:1 isomers (peak 4). Additionally, peaks derived from exogenously added substrates and desaturated products are indicated accordingly. Vertical axis, FID response; horizontal axis, retention time.
TABLE 1.
Functional characterization of the pike silverside (C. estor) desaturases Fads2a and Fads2b in S. cerevisiae
| Conversion (%) | ||||
| FA Substrate | FA Product | Fads2a | Fads2b | Activity |
| 18:3n-3 | 18:4n-3 | 0 | 56.5 | Δ6 |
| 18:2n-6 | 18:3n-6 | 0 | 25.2 | Δ6 |
| 20:3n-3 | 20:4n-3 | 0 | 12.2a | Δ8 |
| 20:2n-6 | 20:3n-6 | 0 | 9.9a | Δ8 |
| 20:4n-3 | 20:5n-3 | 3.3 | 25.8 | Δ5 |
| 20:3n-6 | 20:4n-6 | 0 | 11.6 | Δ5 |
| 22:5n-3 | 22:6n-3 | 28.9 | 0 | Δ4 |
| 22:4n-6 | 22:5n-6 | 10.3 | 0 | Δ4 |
Yeast S. cerevisiae were transformed with pYES2-fadsa and pYES2-fadsb and grown in the presence of Δ6 (18:3n-3 and 18:2n-6), Δ8 (20:3n-3 and 20:2n-6), Δ5 (20:4n-3 and 20:3n-6), and Δ4 (22:5n-3 and 22:4n-6) FA substrates. Conversions were calculated according to the formula [individual product area/(all products areas + substrate area)] × 100.
Conversions of Δ8 substrates (20:3n-3 and 20:2n-6) by Fads2b include stepwise reactions due to multifunctional desaturation abilities. Thus, the conversions of the C. estor Fads2b on 20:3n-3 and 20:2n-6 include the Δ8 desaturation toward 20:4n-3 and 20:3n-6, respectively, and their subsequent Δ5 desaturations to 20:5n-3 and 20:4n-6, respectively.
In spite of its homology with the Fads2a aa sequence, the C. estor Fads2b showed a remarkably different pattern of FA substrate specificity. Whereas no Δ4 desaturation was detected, Fads2b conferred on yeast the ability to desaturate both Δ6 substrates 18:3n-3 and 18:2n-6 to 18:4n-3 and 18:3n-6, respectively, and Δ5 substrates 20:4n-3 and 20:3n-6 to 20:5n-3 and 20:4n-6, respectively (Fig. 3; Table 1), which indicated the C. estor Fads2b had dual Δ6Δ5 activity. Conversions obtained from the yeast assays suggested that both C. estor Fads2 enzymes more efficiently desaturated n-3 than n-6 PUFAs when each homologous pair of substrates for Δ6 desaturase (18:3n-3 and 18:2n-6), Δ5 desaturase (20:4n-3 and 20:3n-6), and Δ4 desaturase (22:5n-3 and 22:4n-6) were compared (Table 1).
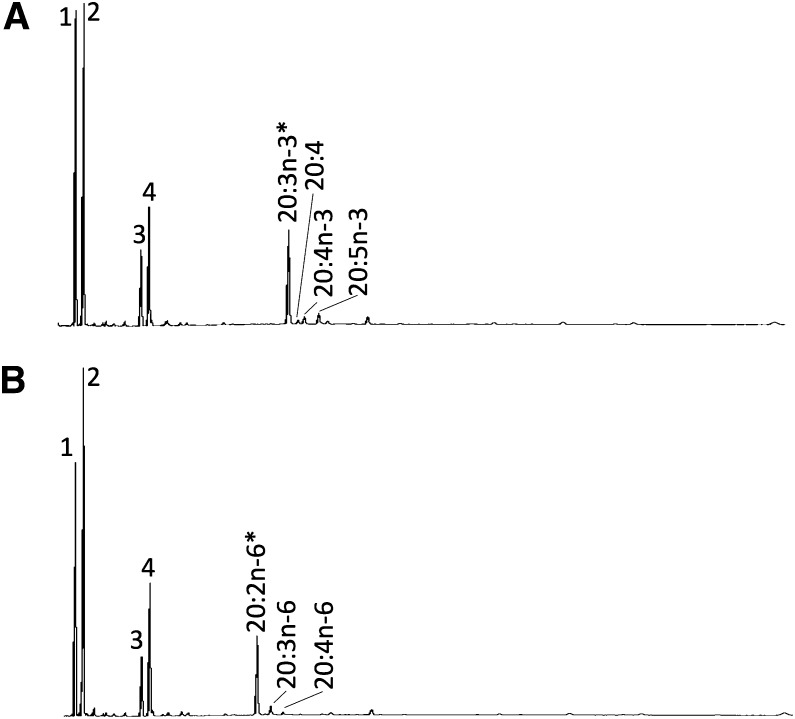
The inherent capability of vertebrate Fads2 enzymes for Δ8 desaturation was investigated in both isoforms. Only Fads2b exhibited the ability to desaturate 20:3n-3 and 20:2n-6 to the corresponding Δ8-desaturated products 20:4n-3 and 20:3n-6, respectively (Table 1). The relative Δ6/Δ8 activity ratio for Fads2b toward n-3 substrates (18:3n-3 vs. 20:3n-3 conversion) was 4.6. Interestingly, the products of Δ8 desaturation, 20:4n-3 and 20:3n-6, were further desaturated to 20:5n-3 and 20:4n-6, respectively, confirming the Δ5 desaturase activity of the enzyme (Fig. 4; Table 1). For 20:3n-3, direct desaturation of 20:3n-3 as Δ6 or Δ5 led to the production of minor peaks identified by GC-MS as non-methylene-interrupted products Δ6,11,14,1720:4 or Δ5,11,14,1720:4 (Fig. 4).
Fig. 4.
Characterization of Δ8 desaturation activities of bifunctional Δ6/Δ5 Fads2b from C. estor. The FA profiles of yeast transformed with pYES2-fadsb were determined after they were grown in the presence of Δ8-desaturase substrates (*), namely 20:3n-3 (A) or 20:2n-6 (B). Peaks 1–4 in all panels are the main endogenous FAs of S. cerevisiae, namely 16:0 (peak 1), 16:1 isomers (peak 2), 18:0 (peak 3), and 18:1 isomers (peak 4). The substrates and their corresponding desaturated products are indicated. The peak indicated as “20:4” is a nonmethylene interrupted FA (Δ6,11,14,1720:4 or Δ5,11,14,1720:4). Vertical axis, FID response; horizontal axis, retention time.
The C. estor elongase exhibited FA substrate specificities consistent with those of Elovl5 enzymes. Thus, transgenic yeast expressing the coding sequence of the pike silverside elovl5 showed activity toward most of the PUFA substrates assayed, with particularly high conversions for most C18 and C20 substrates. Among C18 substrates, high elongations were obtained with 18:3n-3, 18:4n-3, and 18:3n-6, which were converted to the corresponding C20 products 20:3n-3, 20:4n-3, and 20:3n-6, respectively (Table 2). Further elongations to C22 secondary products could be detected in yeast incubated with 18:3n-3, 18:2n-6, and 18:4n-3. For C20 PUFA substrates, almost 90% of total EPA was elongated to 22:5n-3, whereas ARA was elongated to 22:4n-6 to a lower extent (54.8%). C22 PUFA substrates, including 22:5n-3 and 22:4n-6, were only marginally or not converted to longer products (Table 2; Fig. 5). For each pair of homologous substrates considered, conversions obtained from the yeast assays suggested that the C. estor Elovl5 more efficiently elongated n-3 than n-6 PUFAs on a consistent basis (Table 2). Thus, 18:3n-3, 18:4n-3, and 20:5n-3 were elongated to a greater extent than the corresponding n-6 PUFA substrates 18:2n-6, 18:3n-6 and 20:4n-6, respectively. Particularly interesting was the difference in the conversions observed between 18:3n-3 (up to 41.1% converted to longer products) and 18:2n-6 (only 5.5% converted to longer products).
TABLE 2.
Functional characterization of the pike silverside (C. estor) elongase Elovl5 in S. cerevisiae
| FA Substrate | FA Product | Individual Conversion (%) | Accumulated Conversion (%) |
| 18:3n-3 | 20:3n-3 | 35.6 | 41.1 |
| 22:3n-3 | 5.5 | 5.5 | |
| 18:2n-6 | 20:2n-6 | 4.9 | 5.2 |
| 22:2n-6 | 0.3 | 0.3 | |
| 18:4n-3 | 20:4n-3 | 27.2 | 60.2 |
| 22:4n-3 | 33.0 | 33.0 | |
| 18:3n-6 | 20:3n-6 | 27.2 | 27.2 |
| 22:3n-6 | 0.0 | 0.0 | |
| 20:5n-3 | 22:5n-3 | 87.1 | 89.6 |
| 24:5n-3 | 2.5 | 2.5 | |
| 20:4n-6 | 22:4n-6 | 53.8 | 54.8 |
| 24:4n-6 | 1.0 | 1.0 | |
| 22:5n-3 | 24:5n-3 | 1.9 | 1.9 |
| 22:4n-6 | 24:4n-6 | 0.6 | 0.6 |
Individual conversions were calculated according to the formula [individual product area/(all products areas + substrate area)] × 100. Accumulated conversions were computed by summing the individual conversion for each particular product and also those for longer products.
Fig. 5.
Functional characterization of the of the newly cloned C. estor elovl5 in yeast (S. cerevisiae). The FA profiles of yeast transformed with pYES2 containing the coding sequence of elovl5 as an insert were determined after the yeast were grown in the presence of one of the exogenously added FA substrates (*) 18:3n-3 (A), 18:4n-3 (B), 20:5n-3 (C), or 22:5n-3 (D). Peaks 1–4 in all panels are the main endogenous FAs of S. cerevisiae, namely 16:0 (peak 1), 16:1 isomers (peak 2), 18:0 (peak 3), and 18:1 isomers (peak 4). Additionally, peaks derived from exogenously added substrates and elongated products are indicated accordingly in (A)–(D). Vertical axis, FID response; horizontal axis, retention time.
Tissue distribution of fads2 and elovl5 transcripts
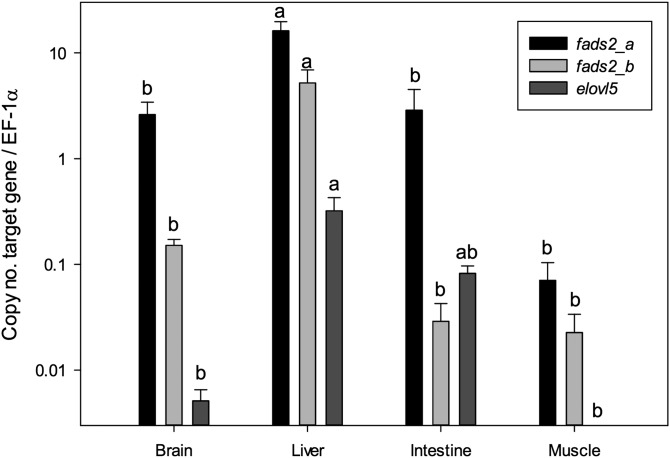
Tissue distribution of the C. estor fads2 (a and b isoforms) and elovl5 mRNA in adult specimens was analyzed by qPCR. Both fads2 transcripts were detected in all four tissues analyzed, with significantly higher expression signals found in liver compared with brain, intestine, and muscle (Fig. 6). With regard to elovl5, liver also showed the highest expression rates, but significant differences could only be established with brain and muscle signals (Fig. 6).
Fig. 6.
Tissue distribution profile of fads2a, fads2b, and elovl5 transcripts in C. estor determined by qPCR. Absolute copy numbers were quantified for each transcript and normalized by copy numbers of elongation factor 1α (EF-1α). Mean values are represented diagrammatically in logarithmic scale. Error bars are standard errors (n = 3). The expressions of a gene in the different tissues studied that share the same letter are not significantly different (one-way ANOVA and Tukey’s tests, P < 0.05).
DISCUSSION
A previous study suggested that C. estor had an active LC-PUFA biosynthesis pathway that enabled this species to endogenously synthesize DHA from PUFA precursors (43). Here, we provide robust data supporting a likely molecular mechanism demonstrating that C. estor expresses genes encoding enzymatic activities that would enable the synthesis of DHA.
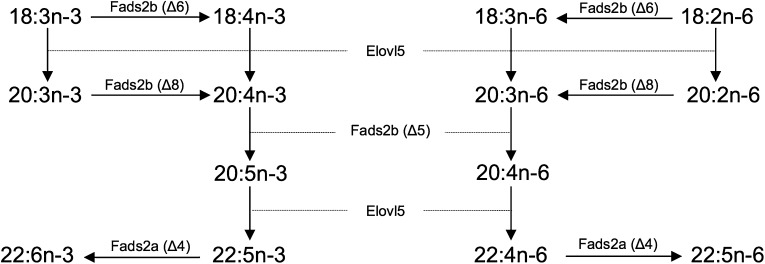
The capabilities exhibited by the two desaturase cDNAs cloned from C. estor (Fads2a and Fads2b) cover the set of desaturation requirements for DHA synthesis from LNA (18:3n-3). Heterologous expression of Fads2b showed this protein was a dual Δ6Δ5 desaturase and, thus, it catalyzed the desaturation of 18:3n-3 to 18:4n-3 (Δ6) and also the Δ5 desaturation required to convert 20:4n-3 to EPA. While the C. estor Fads2a can partly contribute to the Δ5 desaturation leading to EPA biosynthesis as described for Fads2b, the higher conversion activities of Fads2a suggested that its major role in the overall pathway was to catalyze the Δ4 desaturation involved in the direct conversion of 22:5n-3 to DHA. A similar pathway of DHA biosynthesis from EPA was postulated to operate in the rabbitfish, S. canaliculatus (4, 28), and more recently Senegalese sole, S. senegalensis (5). In comparison with the Sprecher pathway, the so-called “Δ4 pathway” is a more direct metabolic route, as it avoids translocation of PUFA intermediates (namely 24:6n-3) between endoplasmic reticulum and peroxisomes, and also the further catabolic step (partial oxidation to DHA) occurring in the latter organelle (3, 46). While we did not test the ability of C. estor desaturases to mediate the Δ6 desaturation of 24:5n-3 to 24:6n-3 required in the Sprecher pathway, we hypothesize that it is not operative in the presence of a more direct and efficient mechanism, such as the Δ4 pathway. In agreement, neither hepatocytes nor enterocytes of C. estor that had been incubated with either [1-14C]18:3n-3 or [1-14C]20:5n-3 showed any recovery of radioactivity in Sprecher pathway intermediates, namely 24:5n-3 to 24:6n-3 (43). Moreover, the functional characterization of the C. estor Elovl5 supports such a hypothesis.
The Elovl5 of C. estor showed virtually no ability to elongate 22:5n-3 to 24:5n-3 as would be required in the Sprecher pathway. This is in agreement with the great majority of fish Elovl5, with the exception of orthologs from rabbitfish (10.6% conversion from 22:5n-3 to 24:5n-3) (28) and, to a lesser extent, cobia (6.6% conversion) (19). Also consistent with Elovl5 from fish, the C. estor Elovl5 showed substantial activity for the elongation of C18 and C20 PUFAs to the corresponding C20 and C22 PUFA products, including the elongations of 18:4n-3 to 20:4n-3 and 20:5n-3 to 22:5n-3, which were catalyzed in yeast at high conversions (60.2 and 89.6%, respectively). Clearly, the C. estor Elovl5 can support all the elongation reactions required for DHA biosynthesis through the Δ4 pathway. Taken together, the functional characterization data for Elovl5, Fads2a, and Fads2b, and the biochemical assays with radiolabeled PUFA substrates (43), predicts a putative pathway of biosynthesis of LC-PUFAs in C. estor from dietary essential C18 PUFAs, 18:3n-3 and 18:2n-6 (Fig. 7). In addition to the DHA biosynthetic pathway described above, two possible pathways for EPA and ARA biosynthesis are shown. First, the “classical”, and likely the most prominent, pathway involving Δ6 desaturation→elongation→Δ5 desaturation is possible through the consecutive action of Fads2b, Elovl5, and Fads2b, respectively. Second, the alternative “Δ8 pathway” proceeds through an initial elongation of dietary essential C18 PUFAs by Elovl5, followed by two consecutive desaturations catalyzed by Fads2b, first as Δ8 and second as Δ5 (Fig. 7). The C. estor Elovl5 was effective in elongating both 18:3n-3 and 18:2n-6 as required in the Δ8 pathway, elongase abilities previously demonstrated in Elovl5 from Southern bluefin tuna (20) and meagre (30). Moreover, the desaturase activities required for the Δ8 pathway, namely Δ8 and Δ5, were demonstrated by Fads2b, but not Fads2a. While the activity of Fads2a as Δ4 desaturase suggested a steric impediment disabling the insertion of new double bonds at the Δ6 or Δ8 positions, the Fads2b desaturase showed the ability to desaturate both 20:3n-3 and 20:2n-6 at the Δ8 carbon.
Fig. 7.
The biosynthesis pathway of long-chain PUFAs from α-linolenic (18:3n-3) and linoleic (18:2n-6) acids in C. estor. Enzymatic activities shown in the scheme are predicted from heterologous expression in S. cerevisiae of the Δ4 and Δ6/Δ5 desaturases (Fads2a and Fads2b, respectively) and the Elovl5-like elongase characterized in the present study.
The ability of fish Fads2 to catalyze Δ8 desaturation has been regarded as a characteristic primarily of marine species, and thus relatively low values of Δ6/Δ8 ratio; i.e., the ratio of the conversions toward 18:3n-3 and 20:3n-3 are shown by Fads2 from species of marine origin (2). The Δ6/Δ8 of the C. estor Fads2b was 4.6, slightly above the range of Δ6/Δ8 ratios of marine fish desaturases (1.8–4.2), but well below those of desaturases from salmonid/freshwater species (12.0–91.2) (2). Moreover, the activities of the LC-PUFA biosynthetic pathways measured in hepatocyte and enterocyte primary cultures from C. estor (40) were generally lower than those obtained in Atlantic salmon (47), but higher than those obtained in the marine teleost Atlantic cod (Gadus morhua) (15). Interestingly, the tissue distribution of the desaturases and elongases generally reflected a “freshwater fish” pattern for C. estor. Thus, liver had the highest mRNA levels for both desaturases and the elongase, and appeared as a major metabolic site for LC-PUFA biosynthesis, over intestine, brain, and muscle. Consistently with the mRNA tissue distribution data, previous studies highlighted the unusually high contents of DHA in C. estor liver, brain, and muscle, and also other tissues including gonads and adipose tissues (38). Similar tissue distribution patterns were observed in freshwater/salmonid species, including Atlantic salmon (14, 18) and zebrafish (17), in which liver and intestine exhibited the highest expression signals of fads2 and elovl2 and elovl5 elongase genes. Marine fish species, including Asian sea bass, Atlantic cod, and cobia, had the highest expression levels of LC-PUFA biosynthesis genes in brain (15, 19, 21). The relatively low Δ6/Δ8 ratio of Fads2b, which was typical of marine species on one hand, and the higher expression of desaturase/elongase mRNA in liver, which was typical of freshwater species on the other, probably reflected the particular evolutionary history of C. estor. Therefore, while C. estor shares some “marine” features with most of its atherinopsid counterparts, it also reflects some characteristics typically found in freshwater species (42).
The substrate specificities of C. estor desaturases described here are not entirely unique among fish Fads2 enzymes and, as mentioned above, similar substrate specificities (Δ4 and Δ6Δ5) were found in two desaturases isolated from rabbitfish (4). Furthermore, a Δ6Δ5 Fads2 and a Δ4 Fads2 were previously identified from zebrafish (9) and Senegalese sole (5), respectively, and a monofunctional Δ5 Fads2 was also cloned from Atlantic salmon (13). Provided the Gnathostomata (jawed fish) ancestral Fads2 had Δ6 desaturase activity (32), as for mammalian FADS2s (35), the expansion of Fads2s in teleosts has been accompanied by subfunctionalization in the enzyme derived from independent mutations in the primary aa sequence (32). Although a recent study identified a single aa residue as determining the differential ability for 22:5n-3 elongation between ELOVL2 and ELOVL5 elongases in rat (48), identification of specific domains/residues controlling the functionality of desaturases has been elusive and studies are restricted to nonvertebrate enzymes (49–51).
However, the above said, the existence of three Δ6Δ5 desaturases and three Δ4 desaturases distributed in four distinct species, allows us to explore potential evolutionary scenarios for teleostei Fads2 subfunctionalization. The recently revised tree of life of bony fish (52), based on 369 families, allows us to investigate whether the diversity of substrate specificities of C. estor desaturases can be related in an evolutionary context along with those of other desaturases with Δ4 and Δ6Δ5 activities, previously described from S. canaliculatus (4), S. senegalensis (5), and D. rerio (9). The three species possessing a Δ4 desaturase (S. canaliculatus, S. senegalensis, and C. estor) belong to three different clades including Percomorpharia (S. canaliculatus), Carangimorphariae (S. senegalensis), and Ovalentariae (C. estor), all sharing a common ancestor (∼115 Ma). It is tempting to speculate that Fads2 enzymes in fish with Δ4 activity are restricted to these three groups, albeit these groups contain >200 families and so the activity is likely further restricted to specific families, or even individual species within the groups. Thus, other species within these three groups, including A. regius, T. thynnus, L. calcarifer, D. labrax, S. aurata, R. canadum (Percomorpharia), Psetta maxima (Carangimorphariae), and Oreochromis niloticus (Ovalentariae), have Fads2 enzymes functionally characterized as Δ6 desaturases (11, 16, 19, 21, 26, 27, 29, 30). However, investigation of desaturases from representatives of other phylogenetic branches is required to confirm this hypothesis.
Establishing a potential pattern of distribution for Fads2 enzymes in teleost fish lineages showing Δ6Δ5 activity is more speculative, as the three species displaying these activities (D. rerio, S. canaliculatus, and C. estor) are more distantly related. While the groups of C. estor and S. canaliculatus are relatively close and thus the presence of Δ6Δ5 Fads2s might have a similar restricted evolutionary context as described for Δ4 desaturases, the existence of a dual Δ6Δ5 desaturase in D. rerio, belonging to the more ancient cyprinid lineage, does not follow common evolutionary patterns, although it is possible that convergent evolution may have occurred. Assuming the ancient Fads2 was a Δ6 desaturase (32), subfunctionalization of some Fads2 in distantly related lineages (cyprinids vs. atherinopsids and siganids) led to the acquisition of Δ5 desaturase activity, possibly through discrete mutations at the catalytic site. According to the proposed time-calibrated tree, the emergence of the dual Δ6Δ5 in cyprinids occurred ∼100 Ma ago (52), as the desaturase from common carp (also cyprinid) does not show dual activity.
In summary, the present study demonstrates that C. estor expresses desaturase and elongase genes encoding all the enzymatic activities required for the biosynthesis of DHA from the C18 precursor LNA. While the C. estor Elovl5 accounted for all the elongation steps, two distinct Fads2-like desaturase enzymes displaying Δ4 and Δ6Δ5 specificities operate along the pathway. More importantly, the uncommon substrate specificities of Fads2s from C. estor and other species like D. rerio, S. canliculatus, and S. senegalensis enabled us to propose potential evolutionary scenarios that explain the distribution of such subfunctionalized Fads2s among fish lineages.
Supplementary Material
Footnotes
Abbreviations:
- aa
- amino acid
- ARA
- arachidonic acid (20:4n-6)
- Elovl
- elongase of very long-chain FA
- Fads
- fatty acyl desaturase
- FAME
- fatty acyl methyl ester
- FID
- flame ionization detector
- LC-PUFA
- long-chain PUFA
- LNA
- α-linolenic acid (18:3n-3)
- LOA
- linoleic acid (18:2n-6)
- OD
- optical density
- ORF
- open reading frame
- qPCR
- quantitative real-time PCR
- RACE
- rapid amplification of cDNA ends
- SCMM
- Saccharomyces cerevisiae minimal medium
This study and O.M. were supported by a Marie Curie Reintegration Grant within the 7th European Community Framework Programme (PERG08-GA-2010-276916, LONGFA). Additional funding was obtained from CONACYT, Mexico (INSAM FOINS 102/2012) and from the Ministry of Science and Innovation (Spanish Government) through the OCTOPHYS Project (AGL-2010-22120-C03-02). The authors have no conflicts of interest related to this work.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one table.
REFERENCES
- 1.Cook H. W., McMaster R. C. R. 2004. Fatty acid desaturation and chain elongation in eukaryotes. In Biochemistry of Lipids, Lipoproteins and Membranes. D.E. Vance and J. E. Vance, editors. Elsevier, Amsterdam. 181–204. [Google Scholar]
- 2.Monroig Ó., Li Y., Tocher D. R. 2011. Delta-8 desaturation activity varies among fatty acyl desaturases of teleost fish: high activity in delta-6 desaturases of marine species. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 159: 206–213 [DOI] [PubMed] [Google Scholar]
- 3.Sprecher H. 2000. Metabolism of highly unsaturated n-3 and n-6 fatty acids. Biochim. Biophys. Acta. 1486: 219–231 [DOI] [PubMed] [Google Scholar]
- 4.Li Y., Monroig Ó., Zhang L., Wang S., Zheng X., Dick J. R., You C., Tocher D. R. 2010. Vertebrate fatty acyl desaturase with Δ4 activity. Proc. Natl. Acad. Sci. USA. 107: 16840–16845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morais S., Castanheira F., Martínez-Rubio L., Conceição L. E. C., Tocher D. R. 2012. Long-chain polyunsaturated fatty acid synthesis in a marine vertebrate: ontogenetic and nutritional regulation of a fatty acyl desaturase with Δ4 activity. Biochim. Biophys. Acta. 1821: 660–671 [DOI] [PubMed] [Google Scholar]
- 6.Bell M. V., Tocher D. R. 2009. Biosynthesis of polyunsaturated fatty acids in aquatic ecosystems: general pathways and new directions. In Lipids in Aquatic Ecosystems. M.T. Arts, M. Brett, and M. Kainz, editors. Springer-Verlag, New York, NY. 211–236. [Google Scholar]
- 7.Tocher D. R. 2003. Metabolism and functions of lipids and fatty acids in teleost fish. Rev. Fish. Sci. 11: 107–184 [Google Scholar]
- 8.Tocher D. R. 2010. Fatty acid requirements in ontogeny of marine and freshwater fish. Aquacult. Res. 41: 717–732 [Google Scholar]
- 9.Hastings N., Agaba M., Tocher D. R., Leaver M. J., Dick J. R., Sargent J. R., Teale A. J. 2001. A vertebrate fatty acid desaturase with Δ5 and Δ6 activities. Proc. Natl. Acad. Sci. USA. 98: 14304–14309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agaba M., Tocher D. R., Dickson C., Dick J. R., Teale A. J. 2004. Zebrafish cDNA encoding a multifunctional enzyme involved in the elongation of polyunsaturated, monounsaturated and saturated fatty acids. Mar. Biotechnol. (NY). 6: 251–261 [DOI] [PubMed] [Google Scholar]
- 11.Zheng X., Seiliez I., Hastings N., Tocher D. R., Panserat S., Dickson C. A., Bergot P., Teale A. J. 2004. Characterization and comparison of fatty acyl Δ6 desaturase cDNAs from freshwater and marine teleost fish species. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 139: 269–279 [DOI] [PubMed] [Google Scholar]
- 12.Agaba M. K., Tocher D. R., Dickson C. A., Zheng X., Dick J. R., Teale A. J. 2005. Cloning and functional characterisation of polyunsaturated fatty acid elongases from marine and freshwater teleost fish. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 142: 342–352 [DOI] [PubMed] [Google Scholar]
- 13.Hastings N., Agaba M. K., Tocher D. R., Zheng X., Dickson C. A., Dick J. R., Teale A. J. 2004. Molecular cloning and functional characterization of fatty acyl desaturase and elongase cDNAs involved in the production of eicosapentaenoic and docosahexaenoic acids from α-linolenic acid in Atlantic salmon (Salmo salar). Mar. Biotechnol. (NY). 6: 463–474 [DOI] [PubMed] [Google Scholar]
- 14.Zheng X., Tocher D. R., Dickson C. A., Dick J. R., Bell J. G., Teale A. J. 2005. Highly unsaturated fatty acid synthesis in vertebrates: new insights with the cloning and characterization of a delta6 desaturase of Atlantic salmon. Lipids. 40: 13–24 [DOI] [PubMed] [Google Scholar]
- 15.Tocher D. R., Zheng X., Schlechtriem C., Hasting N., Dick J. R., Teale A. J. 2006. Highly unsaturated fatty acid synthesis in marine fish: cloning, functional characterization, and nutritional regulation of fatty acyl Δ6 desaturase of Atlantic cod (Gadus morhua L.). Lipids. 41: 1003–1016 [DOI] [PubMed] [Google Scholar]
- 16.González-Rovira A., Mourente G., Zheng X., Tocher D. R., Pendón C. 2009. Molecular and functional characterization and expression analysis of a Δ6 fatty acyl desaturase cDNA of European sea bass (Dicentrarchus labrax L.). Aquaculture. 298: 90–100 [Google Scholar]
- 17.Monroig Ó., Rotllant J., Sánchez E., Cerdá-Reverter J. M., Tocher D. R. 2009. Expression of long-chain polyunsaturated fatty acid (LC-PUFA) biosynthesis genes during zebrafish Danio rerio early embryogenesis. Biochim. Biophys. Acta. 1791: 1093–1101 [DOI] [PubMed] [Google Scholar]
- 18.Morais S., Monroig Ó., Zheng X., Leaver M. J., Tocher D. R. 2009. Highly unsaturated fatty acid synthesis in Atlantic salmon: characterization of Elovl5- and Elovl2-like elongases. Mar. Biotechnol. (NY). 11: 627–639 [DOI] [PubMed] [Google Scholar]
- 19.Zheng X., Ding Z., Xu Y., Monroig Ó., Morais S., Tocher D. R. 2009. Physiological roles of fatty acyl desaturases and elongases in marine fish: characterisation of cDNAs of fatty acyl Δ6 desaturase and Elovl5 elongase of cobia (Rachycentron canadum). Aquaculture. 290: 122–131 [Google Scholar]
- 20.Gregory M. K., See V. H., Gibson R. A., Schuller K. A. 2010. Cloning and functional characterisation of a fatty acyl elongase from southern bluefin tuna (Thunnus maccoyii). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 155: 178–185 [DOI] [PubMed] [Google Scholar]
- 21.Mohd-Yusof N. Y., Monroig Ó., Mohd-Adnan A., Wan K. L., Tocher D. R. 2010. Investigation of highly unsaturated fatty acid metabolism in the Asian sea bass, Lates calcarifer. Fish Physiol. Biochem. 36: 827–843 [DOI] [PubMed] [Google Scholar]
- 22.Monroig Ó., Zheng X., Morais S., Leaver M. J., Taggart J. B., Tocher D. R. 2010. Multiple genes for functional Δ6 fatty acyl desaturases (Fad) in Atlantic salmon (Salmo salar L.): Gene and cDNA characterization, functional expression, tissue distribution and nutritional regulation. Biochim. Biophys. Acta. 1801: 1072–1081 [DOI] [PubMed] [Google Scholar]
- 23.Monroig Ó., Rotllant J., Cerdá-Reverter J. M., Dick J. R., Figueras A., Tocher D. R. 2010. Expression and role of Elovl4 elongases in biosynthesis of very long-chain fatty acids during zebrafish Danio rerio early embryonic development. Biochim. Biophys. Acta. 1801: 1145–1154 [DOI] [PubMed] [Google Scholar]
- 24.Carmona-Antoñanzas G., Monroig Ó., Dick J. R., Davie A., Tocher D. R. 2011. Biosynthesis of very long-chain fatty acids (C>24) in Atlantic salmon: cloning, functional characterisation, and tissue distribution of an Elovl4 elongase. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 159: 122–129 [DOI] [PubMed] [Google Scholar]
- 25.Monroig Ó., Webb K., Ibarra-Castro L., Holt G. J., Tocher D. R. 2011. Biosynthesis of long-chain polyunsaturated fatty acids in marine fish: characterisation of an Elovl4-like elongase from cobia Rachycentron canadum and activation of the pathway during early life stages. Aquaculture. 312: 145–153 [Google Scholar]
- 26.Morais S., Mourente G., Ortega A., Tocher J. A., Tocher D. R. 2011. Expression of fatty acyl desaturase and elongase genes, and evolution of DHA:EPA ratio during development of unfed larvae of Atlantic bluefin tuna (Thunnus thynnus L.). Aquaculture. 313: 129–139 [Google Scholar]
- 27.Santigosa E., Geay F., Tonon T., Le Delliou H., Kuhl H., Reinhardt R., Corcos L., Cahu C., Zambonino-Infante J. L., Mazurais D. 2011. Cloning, tissue expression analysis, and functional characterization of two Δ6-desaturase variants of sea bass (Dicentrarchus labrax L.). Mar. Biotechnol. (NY). 13: 22–31 [DOI] [PubMed] [Google Scholar]
- 28.Monroig Ó., Wang S., Zhang L., You C., Tocher D. R., Li Y. 2012. Elongation of long-chain fatty acids in rabbitfish Siganus canaliculatus: cloning, functional characterisation and tissue distribution of Elovl5- and Elovl4-like elongases. Aquaculture. 350–353: 63–70 [Google Scholar]
- 29.Tanomman S., Ketudat-Cairns M., Jangprai A., Boonanuntanasarn S. 2013. Characterization of fatty acid delta-6 desaturase gene in Nile tilapia and heterogenous expression in Saccharomyces cerevisiae. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 166: 148–156 [DOI] [PubMed] [Google Scholar]
- 30.Monroig Ó., Tocher D. R., Hontoria F., Navarro J. C. 2013. Functional characterisation of a Fads2 fatty acyl desaturase with Δ6/Δ8 activity and an Elovl5 with C16, C18 and C20 elongase activity in the anadromous teleost meagre (Argyrosomus regius). Aquaculture. 412–413: 14–22 [Google Scholar]
- 31.Xie D., Chen F., Lin S., Wang S., You C., Monroig Ó., Tocher D. R., Li Y. 2014. Cloning, functional characterization and nutritional regulation of Δ6 fatty acyl desaturase in the herbivorous euryhaline teleost Scatophagus argus. PLoS ONE. 9: e90200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castro L. F. C., Monroig Ó., Leaver M. J., Wilson J., Cunha I., Tocher D. R. 2012. Functional desaturase Fads1 (Δ5) and Fads2 (Δ6) orthologues evolved before the origin of jawed vertebrates. PLoS ONE. 7: e31950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carmona-Antoñanzas G., Tocher D. R., Taggart J. B., Leaver M. J. 2013. An evolutionary perspective on Elovl5 fatty acid elongase: comparison of Northern pike and duplicated paralogs from Atlantic salmon. BMC Evol. Biol. 13: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tacon A. G. J., Metian M., Turchini G. M., de Silva S. S. 2010. Responsible aquaculture and trophic level implications to global fish supply. Rev. Fish. Sci. 18: 94–105 [Google Scholar]
- 35.Guillou H., Zadravec D., Martin P. G. P., Jacobsson A. 2010. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 49: 186–199 [DOI] [PubMed] [Google Scholar]
- 36.Martínez-Palacios C. A., Ríos-Duran M. G., Fonseca-Madrigal J., Toledo-Cuevas M., Sotelo Lopez A., Ross L. G. 2008. Developments in the nutrition of Menidia estor Jordan 1880. Aquacult. Res. 39: 738–747 [Google Scholar]
- 37.Martínez-Palacios C. A., Barriga-Tovar E., Taylor J. F., Ríos-Duran G., Ross L. G. 2002. Effect of temperature on growth and survival of Chirostoma estor estor, Jordan 1879, monitored using a simple video technique for remote measurement of length and mass of larval and juvenile fishes. Aquaculture. 209: 369–377 [Google Scholar]
- 38.Martínez-Palacios C. A., Racotta I. S., Ríos-Duran M. G., Palacios E., Toledo-Cuevas M., Ross L. G. 2006. Advances in applied research for the culture of Mexican silversides (Chirostoma, Atherinopsidae). Biocell. 30: 137–148 [PubMed] [Google Scholar]
- 39.Martínez-Palacios C. A., Ambriz-Cervantes L., Ríos-Duran M. G., Jauncey K. J., Ross L. G. 2007. Dietary protein requirement of juvenile Mexican silverside (Chirostoma estor estor Jordan 1879), a stomachless zooplanktophagous fish. Aquacult. Nutr. 13: 304–310 [Google Scholar]
- 40.Barbour C. D. 1973. The systematics and evolution of the genus Chirostoma Swainson (Pisces: Atherinidae). Tulane Stud. Zool. Bot. 18: 97–141 [Google Scholar]
- 41.Martínez-Palacios C. A., Comas Morte J., Tello-Ballinas J. A., Toledo-Cuevas M., Ross L. G. 2004. The effects of saline environments on survival and growth of eggs and larvae of Chirostoma estor estor Jordan 1879 (Pisces: Atherinidae). Aquaculture. 238: 509–522 [Google Scholar]
- 42.Martínez-Palacios C. A., Ríos-Duran M. G., Campos-Mendoza A., Toledo-Cuevas M., Aguilar-Valdez M. A., Ross L. G. 2003. Desarrollo tecnológico alcanzado en el cultivo del pez blanco de Pátzcuaro. In Historia y Avances del Cultivo de Pescado Blanco. Avances del Cultivo de Pescado Blanco. P. M. Rojas-Carillo and D. F. Castellanos, editors. Pridsa Impresión, México D. F. 169–190. [Google Scholar]
- 43.Fonseca-Madrigal J., Pineda-Delgado D., Martinez-Palacios C. A., Rodriguez C., Tocher D. R. 2012. Effect of salinity on the biosynthesis of n-3 long-chain polyunsaturated fatty acids in silverside Menidia estor. Fish Physiol. Biochem. 38: 1047–1057 [DOI] [PubMed] [Google Scholar]
- 44.Saitou N., Nei M. 1987. The neighbor-joining method. A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- 45.Qiu X., Hong H. P., Mackenzie S. L. 2001. Identification of a Δ4 fatty acid desaturase from Thraustochytrium sp. involved in biosynthesis of docosahexaenoic acid by heterologous expression in Saccharomyces cerevisiae and Brassica juncea. J. Biol. Chem. 276: 31561–31566 [DOI] [PubMed] [Google Scholar]
- 46.Ferdinandusse S., Denis S., van Roermund C. W. T., Wanders R. J. A., Dacremont G. 2004. Identification of the peroxisomal-oxidation enzymes involved in the degradation of long-chain dicarboxylic acids. J. Lipid Res. 45: 1104–1111 [DOI] [PubMed] [Google Scholar]
- 47.Zheng X., Torstensen B. E., Tocher D. R., Dick J. R., Henderson R. J., Bell J. G. 2005. Environmental and dietary influences on highly unsaturated fatty acid biosynthesis and expression of fatty acyl desaturase and elongase genes in liver of Atlantic salmon (Salmo salar). Biochim. Biophys. Acta. 1734: 13–24 [DOI] [PubMed] [Google Scholar]
- 48.Gregory M. K., Cleland L. F., James M. J. 2013. Molecular basis for differential elongation of omega-3 docosapentaenoic acid by the rat Elovl5 and Elovl2. J. Lipid Res. 54: 2851–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sasata R. J., Reed D. W., Loewen M. C., Covello P. S. 2004. Domain swapping localizes the structural determinants of regioselectivity in membrane-bound fatty acid desaturases of Caenorhabditis elegans. J. Biol. Chem. 279: 39296–39302 [DOI] [PubMed] [Google Scholar]
- 50.Song L-Y., Lu W-X., Hu J., Yin W-B., Chen Y-H., Wang B-L., Wang R. R-C., Hu Z-M. 2013. The role of C-terminal amino acid residues of a Δ6-fatty acid desaturase from blackcurrant. Biochem. Biophys. Res. Commun. 431: 675–679 [DOI] [PubMed] [Google Scholar]
- 51.Kurdrid P., Sirijuntarut M., Subudhi S., Cheevadhanarak S., Hongsthong A. 2008. Truncation mutants highlight a critical role for the N- and C-termini of the Spirulina Δ6 desaturase in determining regioselectivity. Mol. Biotechnol. 38: 203–209 [DOI] [PubMed] [Google Scholar]
- 52.Betancur-R R., Broughton R. E., Wiley E. O., Carpenter K., López J. A., Li C., Holcroft N. I., Arcila D., Sanciangco M., Cureton J. C., II, et al. 2013. The tree of life and a new classification of bony fishes. PLoS Curr. 5 10.1371/currents. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.