Abstract
Influenza virus acquires a host-derived lipid envelope during budding, yet a convergent view on the role of host lipid metabolism during infection is lacking. Using a mass spectrometry-based lipidomics approach, we provide a systems-scale perspective on membrane lipid dynamics of infected human lung epithelial cells and purified influenza virions. We reveal enrichment of the minor peroxisome-derived ether-linked phosphatidylcholines relative to bulk ester-linked phosphatidylcholines in virions as a unique pathogenicity-dependent signature for influenza not found in other enveloped viruses. Strikingly, pharmacological and genetic interference with peroxisomal and ether lipid metabolism impaired influenza virus production. Further integration of our lipidomics results with published genomics and proteomics data corroborated altered peroxisomal lipid metabolism as a hallmark of influenza virus infection in vitro and in vivo. Influenza virus may therefore tailor peroxisomal and particularly ether lipid metabolism for efficient replication.
Keywords: ether lipids, sphingolipids, glycerophospholipids, lipid metabolism, biochemistry, systems biology
Influenza viruses hijack host cell machineries for efficient replication and acquire a host-derived lipid envelope during budding. Recent systems-scale studies have primarily addressed the individual roles of genes (1–5) and proteins (6–8) in this process, yet have failed to illustrate how they function together to generate macromolecular precursors for virus production. Host cell lipid metabolism and plasma membrane microdomains are implicated in the biogenesis of virus envelopes. Several studies have dissected the lipid inventory of purified influenza virions (9, 10), whereas others have demonstrated the requirements for de novo fatty acid and sphingolipid biosynthesis and unique cholesterol compositions for virus production at budding sites (11–14).
In addition to the importance of host cell lipid metabolism for the biogenesis of influenza virus envelopes, recent findings suggest a major role for soluble lipid mediators in antiviral responses against influenza virus infection in vivo (15, 16). These soluble lipid mediators originate from membrane glycerophospholipids (GPLs) via phospholipase activity, and (to some extent), are metabolized in peroxisomes. For example, β-oxidation in the peroxisome is crucial for the retroconversion of DHA, the precursor of the lipid mediator protectin D1, which prevents nuclear export of influenza virus RNAs; protectin D1 production is directly inhibited by influenza virus (15). The role of peroxisomes during influenza virus replication is further evident by interaction between influenza virus nonstructural protein 1 (NS1) and multifunctional protein 2 (MFP2/HSD17B4), an antiviral protein essential for peroxisomal β-oxidation (17). Therefore, the collective literature indicates an apparent role for peroxisomes as the initial sites of antiviral signaling (18).
However, a convergent view of the role of lipid metabolic pathways during influenza virus replication, particularly those pathways required in the generation of membranes/envelopes, and their contribution to virus pathogenicity, are lacking. We therefore sought to examine changes in host membrane lipid composition during different stages of infection with influenza virus using analytical biochemistry to explore the requirement of precursors for lipid mediators as well as other peroxisome-derived lipids. While the experimental approach is tried and tested, thus not novel by itself, our detailed and integrative analysis resulted in a comprehensive systems-scale resource of how influenza virus impacts lipid metabolism. Using siRNA knockdown and pharmacological inhibitors, we directly provide a “proof-of-concept” that our resource can be useful to derive novel hypotheses in the emerging field of lipid involvement during virus infections. Using this approach and in combination with meta-data for further support, we reveal novel insights into the role of a class of host membrane components with poorly defined functions, peroxisome-derived ether lipids, during influenza virus replication. Our findings put the peroxisome and its lipid metabolism at the forefront of influenza virus infections in vitro and in vivo.
METHODS
Virus strains, virus production, and purification
Virus stocks were prepared by passaging egg-grown virus strains once in MDCK cells. Virus strains were purified from A549 and MDCK cells as described in the supplementary data.
Lipid extraction of infected cells and purified viruses
A549, Chinese hamster ovary (CHO)-K1, and NRel-4 cells were seeded into 10 cm cell culture dishes 24 h prior to infection. Cells at 80–100% confluence were infected with a 5 ml inoculum of purified influenza virus A/PR/8/34 H1N1 at MOI 5. Virus-infected cells and mock-infected cells were collected at 12, 18, and 24 hours postinfection (hpi) (for A549 cells) or only at 18 hpi (CHO-K1 and NRel-4 cells). Lipid extraction was conducted according to a modified Bligh and Dyer protocol described in the supplementary data.
Quantitative analysis of lipids by HPLC MS/MS
Samples with spiked internal standards were analyzed by ESI-MS. Signal intensities for each lipid species were extracted according to their retention time, normalized to the representative spiked internal standards, and represented as a molar fraction of the total amount of measured lipids. Statistical significance was calculated using an unpaired Student’s t-test (P < 0.05; two-tailed) or a block-design three-way ANOVA corrected by a false discovery rate procedure (for more details, see the supplementary data).
D609 and GW7647 treatment of influenza virus-infected cells
D609 and GW7647 were purchased from Tocris Bioscience (Bristol, UK). A549 cells were infected with influenza virus A/PR/8/34 H1N1 (MOI <1), and serum- and antibiotics-free medium supplemented with D609 (10 µM and 100 µM) or GW7647 (1 µM, 2 µM, and 5 µM) were added at 12 hpi and 1 hpi, respectively. Supernatants and cell lysates were subjected to plaque assay and Western blot at 18 hpi. Cell viability after drug treatment was assessed using Vybrant MTT cell proliferation assay kit (V13154) (Invitrogen®, Life Technologies Co., San Diego, CA) according to the manufacturer’s protocol. For lipid analysis by MS, A549 cells were treated with 10 µM D609 in serum- and antibiotics-free medium for 18 hpi. Significant differences were calculated by Student’s t-test (P < 0.05; two-tailed).
Influenza virus infection of DHAPAT deficient CHO-K1 cells
CHO-K1 and NRel-4 cells were infected with influenza virus A/PR/8/34 H1N1 (MOI <1) and virus replication was assessed by Western blot and plaque assay after 18 h of infection. Anti-influenza virus matrix protein 2 (M2) and anti-GAPDH antibodies (Santa Cruz, CA) were used to determine virus protein expression. Significant differences were calculated by an unpaired Student’s t-test (P < 0.05; two-tailed).
Knockdown of AGPS and Rab11a by siRNA
Two siRNA constructs targeting alkylglycerone phosphate synthase (AGPS) (Ambion, s16248 and s16249) and Rab11a (Ambion, s16703 and s16704) were used. Full details of the reverse transfection and validation of siRNA constructs is described in the supplementary data.
Catalase assay
A549 cells were infected with influenza virus A/PR/8/34 H1N1 and catalase activity was measured at 18 hpi using the Catalase Assay Kit (Sigma-Aldrich, St. Louis, MO). Significant differences were calculated by an unpaired Student’s t-test (P < 0.05; two-tailed).
Literature mining
To derive a potential model of lipid metabolism in influenza virus-infected cells, we manually incorporated our lipidomics and cell biological data with existing genomics, proteomics, and metabolomics data. Procedures and additional references can be found in the supplementary data.
RESULTS AND DISCUSSION
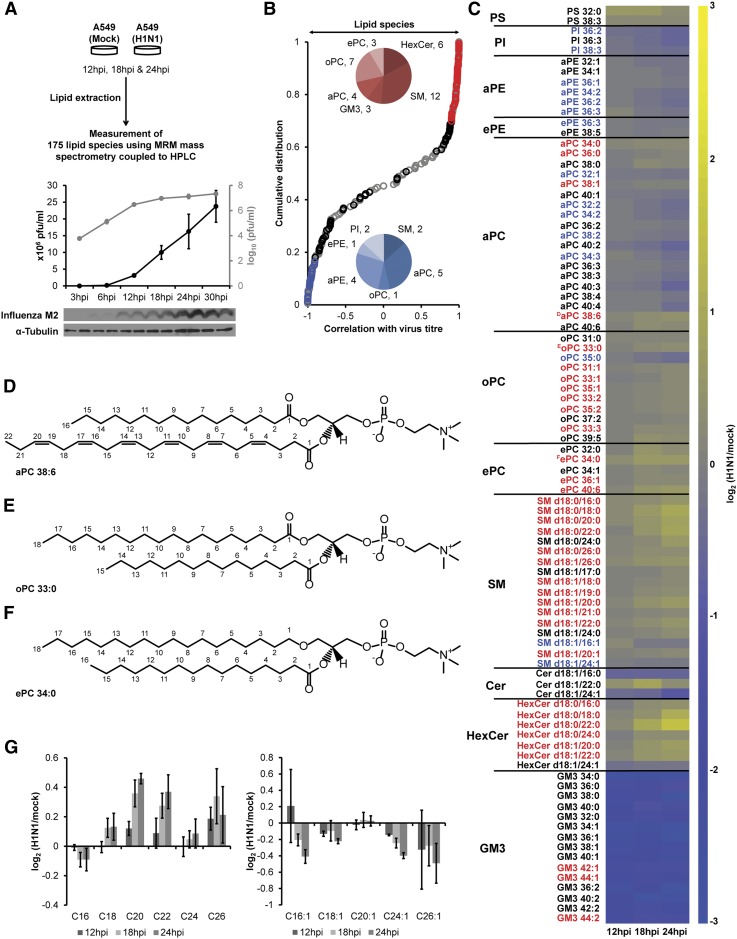
To systematically characterize the temporal changes of host cell membrane lipid composition during influenza virus infection, human lung epithelial (A549) cells were infected with purified influenza virus A/PR/8/34 H1N1 and total cellular lipids were extracted 12, 18, and 24 hpi. A high multiplicity of infection (MOI5) was used to ensure a synchronous, one-round of infection (19). A total of 175 lipid species, representing GPL and sphingolipids (SPLs), two major membrane lipid classes analyzed in this study, were measured using established methodology based on HPLC and ESI-MS and operated in multiple reaction monitoring mode (Fig. 1A, supplementary Table II) (20, 21). The levels of 90 lipid species (i.e., ∼52% of all measured lipids) were significantly altered between H1N1-infected and mock-infected cells (q < 0.006; Fig. 1B, C) at either 18 hpi, 24 hpi, or both. Of these, 35 (Fig. 1B, red pie chart) and 15 (Fig. 1B, blue pie chart) lipid species had correlation coefficients of >0.9 or <−0.9, respectively, with virus titer (Fig. 1B, supplementary Tables I and II). Virus infection lowered the levels of ester-linked GPL, mainly phosphatidylethanolamine and phosphatidylcholine (aPC) species, but increased SPL such as many SM and hexosylceramide (HexCer) species (Fig. 1B, C). These observed changes in lipid species were also reflected by the increase in total amounts of ether-linked PC (ePC), odd chain aPC and SM lipid classes and by the decrease in the total amount of Ganglioside GM3 lipid class across the three independent experiments (supplementary Figs. I, II).
Fig. 1.
Influenza virus infection impacts peroxisomal and sphingolipid metabolism in the host. A: A549 cells were infected with purified influenza virus. 175 lipid species were analyzed by mass spectrometry at 12, 18, and 24 hpi. B: Distribution of Pearson correlation coefficients between 175 lipid species and virus titer (supplementary Tables I, II). Black, red (>0.9) and blue (>−0.9) indicate 90 lipid species altered in influenza virus-infected cells (q < 0.006; supplementary Table II). C: Heatplot showing fold ratios (infected/mock) of 90 lipid species with altered levels upon infection (q < 0.006). Yellow and blue indicate elevated and decreased concentrations, respectively. Lipid species, which correlate with virus titer (B), are indicated by red (>0.9) and blue (<−0.9) fonts. Representative structures to illustrate the differences between ester-linked (D), odd chain (E), and ether-linked (F) PC lipids. Please note that we were able to determine the total carbon fatty acyl composition but did not dissect the exact carbon composition of the two fatty acyl constituents in the measured GPL species. Hence, the two fatty acyl constituents shown in the structures can vary as long as they add up to the respective total carbon fatty acyl compositions. G: Fold ratios of changes in fatty acid chain length composition of Cer, HexCer, and SM lipid species. Results in panels A, B, C, and G are from three independent experiments with three replicates each (n = 9 for each condition). Error bars in G represent ± SDs.
The decrease in the proportion of ganglioside GM3 species likely reflected influenza virus neuraminidase activity (9). The reduced levels of aPC species in influenza virus-infected cells have been previously proposed to be related to impaired aPC biosynthesis as measured by metabolite rates of phospholipid precursors and by global gene and protein expression experiments (8, 22, 23). SREBP1, a major regulator of the one-carbon cycle producing the methyl donor S-adenosylmethionine required for the de novo methylation pathway for aPC biosynthesis, was significantly downregulated in influenza virus-infected cells (22, 24). These changes coincided with the increasing levels of another choline containing lipid, SM, suggesting an important correlation of influenza virus replication with choline lipid metabolism (Fig. 1B,C). Increase in SM and decrease in aPC species could possibly be explained by the activities of inter-related enzyme systems including sphingomyelin synthases (SMS1 and SMS2, which transfer the choline headgroup of PC onto a ceramide backbone to produce SM) and ethanolamine kinase 1, which is downregulated in influenza virus-infected cells (22). However, additional experiments such as quantitative proteomics and enzymatic assays would be required to draw such conclusions. The results presented here provide a good starting point for generation of hypotheses and such future investigations. The upsurge in the proportion of long chain (>38 fatty acyl carbons, >C38) aPC species with polyunsaturated fatty acyls (Fig. 1D), odd chain aPC (Fig. 1E), and ePC (Fig. 1F) suggested altered peroxisomal lipid metabolism in infected cells. Consistent with impaired peroxisomal β-oxidation, we observed an enrichment in C26:0 but a decrease in C24:1 fatty acids in SPL species (Fig. 1G) (25–27); this provided further support for an important role of fatty acyl metabolism during influenza infection (15, 16).
We next tested whether alterations in host membrane lipid levels are detectable also in envelopes harvested from purified virus particles (Fig. 2A). Indeed, aPC, phosphatidylethanolamine, phosphatidylinositol, and GM3 levels were decreased in viruses whereas SM was increased when compared with levels in the producer cell (Fig. 2B, supplementary Table III) (9, 10). To characterize the enrichment of peroxisome-derived ether-linked lipids in influenza virus envelopes and to establish a quantitative correlate between host and virus membrane compositions, we determined the ratios of ePC/aPC in both membrane extracts. To account for the confounding effect of variations in peroxisomal activity among different host cell lines as well as differences in experimental approaches in the published literature, we decided to use the difference between the ePC/aPC ratios of virus particles and uninfected producer cells (ΔVirus-Cell = ePC/aPCVirus−ePC/aPCCell) as a molecular proxy of PC lipid class remodelling to compare a wide variety of different studies (Fig. 2C–F). Envelopes of influenza viruses (circles in Fig. 2C, F) had a significantly higher ePC/aPC ratio than their uninfected producer cells (bars in Fig. 2C, F), whereas other enveloped viruses (including human immunodeficiency virus, murine leukemia virus, vesicular stomatitis virus, dengue virus, and hepatitis C virus) showed equal or lower ePC/aPC ratios (diamonds in Fig. 2D, F, supplementary Table IV). Therefore, peroxisome-dependent remodelling of lipids within the abundant PC class, rather than overall changes to total PC concentration, is specific to influenza virus.
Fig. 2.
Peroxisome-dependent remodelling of PC lipids is specific for influenza virus. A: 159 lipid species from purified A549-grown influenza virions were analyzed by mass spectrometry. Virus purity was assessed by SDS-PAGE and Coomassie blue staining. Virus proteins were identified based on their size. B: Lipid composition of influenza virions (dark gray bars) compared with mock-infected A549 cells at 12 h postinfection (hpi) (light gray bars) (supplementary Table III). Error bars represent ± SDs calculated from three independent experiments (n = 9 for mock-infected A549 cells at 12 hpi) and two independent experiments (n = 6 for purified influenza virions); *P < 0.05, **P < 0.005, and ***P < 0.0005 (unpaired Student’s t-test; two-tailed). C: Ether PC to ester PC (ePC/aPC) ratios of influenza virions (circles) compared with (D) ePC/aPC ratios of other enveloped viruses (diamonds) in relation to uninfected producer cells (bars) (supplementary Table IV); **P < 0.002 (unpaired Student’s t-test; two-tailed). E: Differences in ePC/aPC ratios between P0 and P10 H3N2 strains (supplemetary Fig. III, supplementary Table V). Results are from three independent experiments with two replicates (n = 6); * P < 0.02 (paired Student’s t-test; two-tailed). F: Differences in ePC/aPC ratios in virus versus host membranes (ΔVirus-Cell) of influenza (circles) and other viruses (diamonds) as described for D and E.
We next determined the lipid composition of two closely related H3N2 influenza virus strains differing in pathogenicity. The parent influenza A strain A/Aichi/2/68 H3N2 (P0) was adapted by ten passages in mice (P10), which ultimately showed higher virulence with enhanced replication fitness because of nonconservative point mutations in hemagglutinin (HA) (Gly218Glu) and NS1 (Asp125Gly) (28). NS1 regulates lipid metabolic genes in a severity-dependent manner (22) and interacts with MFP2/HSD17B4 (see above) (17). The latter mutation has been shown to produce high virus titres with enhanced interferon-β antagonism and to differentially regulate host gene expression (29, 30). We assumed a negligible effect of the mutation in HA (Gly218Glu) on host lipid metabolism because the mutation lies in a region involved in sialic acid linkage recognition, important for entry rather than influenza virus replication within the host cell (28). We showed that the more pathogenic H3N2 strain (P10) exhibited a ∼25% higher ePC/aPC ratio in its envelope than the less pathogenic P0 strain (Fig. 2E, supplementary Fig. III, supplementaryTables IV, V), which was comparable with the variation in ePC/aPC ratios between different influenza viruses (Fig. 2C). Collectively, these results suggest strain-dependent differences with regard to PC class remodelling which might reflect influenza virus pathogenicity and underscore the conserved role of peroxisomes in influenza infection (Fig. 2F).
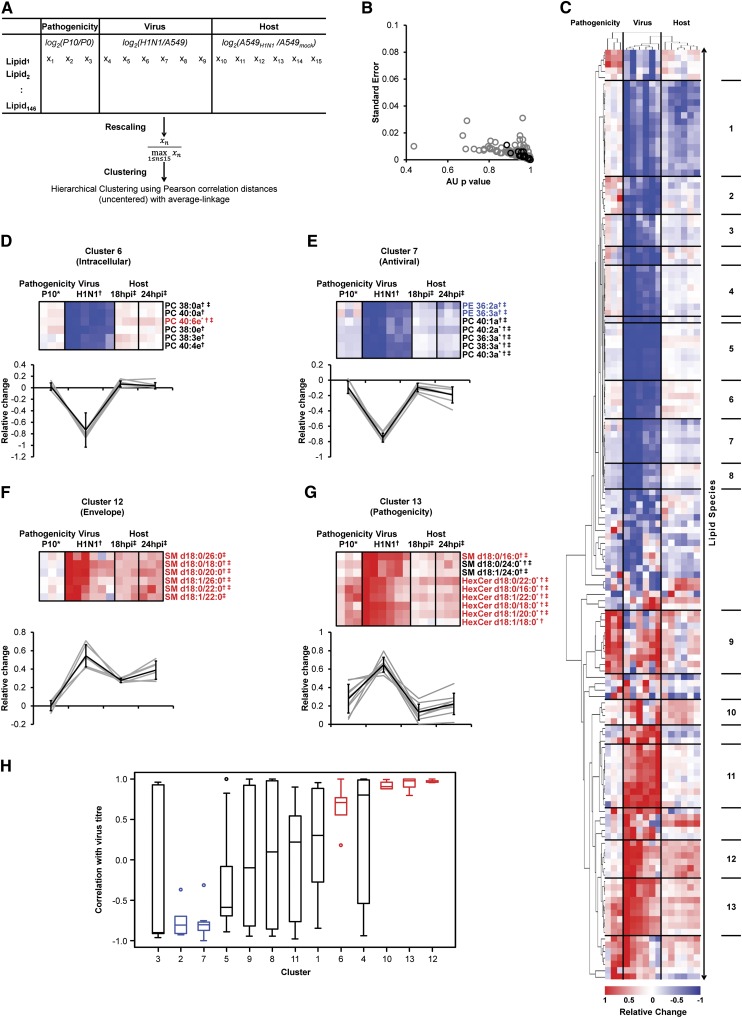
We next sought a systems-scale perspective on the broader impact of influenza virus-induced perturbations of lipid metabolism (Fig. 3). To do so, we used the aforementioned data from i) A549 cell infection (host), ii) purified H1N1 virus (virus), and iii) H3N2 P10 virus (pathogenicity) for unsupervised cluster analysis (Fig. 3A, supplementary Table VI). We assigned 13 clusters (AU p-value >0.9, Fig. 3B) with unique patterns of lipid regulation (Fig. 3C). For instance, lipids with increased concentrations in virus-infected cells but decreased levels in virions are suggestive of intracellular requirements for virus replication as revealed for long chain fatty acid-containing ePC and two aPC species with long but saturated fatty acyls (cluster 6 in Fig. 3C, D). Unsaturated ester-linked PE and PC species were reduced both in virions as well as in infected cells (Fig. 3E), consistent with the downregulation of genes implicated in ester-linked GPL metabolism (22). On the contrary, lipids enriched in both infected cells and virions are indicative of a possible function in virus morphogenesis, as seen for saturated short-chain fatty acid-containing ePC species (cluster 10 in Fig. 3C, supplementary Table VI). Two single clusters highlighted the importance of SPL species for virion formation. While saturated fatty acyls containing SM species were enriched in virus-infected cells as well as virions (Fig. 3F), saturated fatty acyls containing HexCer species were additionally enriched in the more pathogenic virus strain P10 (Fig. 3G). These findings emphasized a general requirement of SM metabolism for virion morphogenesis, which was also reflected by high correlation with virus titer (Figs. 1, 3H) (9, 11, 14). This comparative analysis of lipid levels in purified viruses and infected host cells therefore postulates new hypotheses for the roles of lipids with similar chemistries during different stages of virus replication.
Fig. 3.
Life cycle-dependent clusters of lipids revealed by comparative analysis of host and viral lipid profiles. A: Hierarchical clustering was performed on 146 lipid species describing pathogenicity-related differences in lipid composition, influenza virus-enriched lipids, and alterations in host cell lipid metabolism. Values were rescaled to make different data sets comparable for cluster analysis (see Methods for details). B: Approximately unbiased (AU) p-values and standard errors (bootstrap resampling; n = 10,000) of tree splits (gray circles) and 13 assigned clusters (black circles). C: Dendrogram indicating increased (red) and decreased (blue) levels of individual lipid species (supplementary Table VI). D–G: Average changes in lipid species (gray lines) and average relative trends (black line ± SDs) for intracellular (D), antiviral (E), envelope-enriched (F) and pathogenicity-dependent (G) clusters. Correlations of individual lipid species with virus titer is indicated by red (>0.9) and blue (<0.9) fonts, respectively; * altered in a pathogenicity-dependent fashion; † enriched in H1N1; ‡ alterations in infected cells as determined in Figs. 1 and 2. H: Correlation of the 13 clusters with virus titer. Medians are depicted with Q1, Q3, and 1.5*inter-quartile range (IQR). Robust correlations are colored in blue (negative) and red (positive).
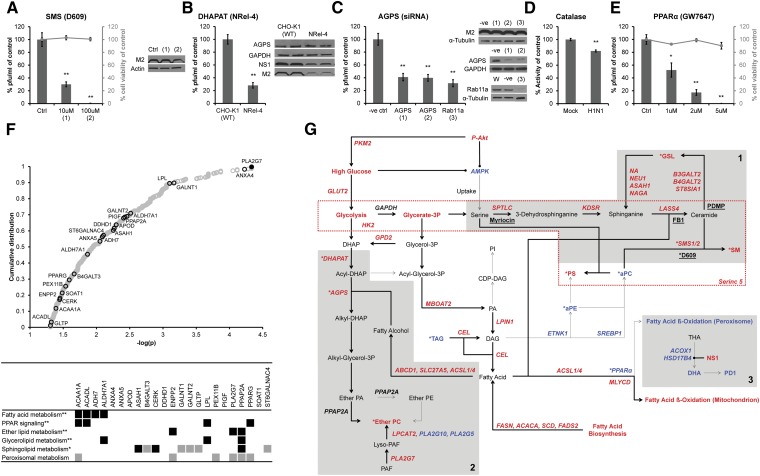
We next evaluated the contribution of ether lipid and SPL metabolism during the late stages of influenza virus replication using a combination of pharmacological inhibitors and genetic silencing. We first treated A549 cells with the SMS inhibitor D609, which inhibits an SM biosynthesis salvage pathway rather than de novo biosynthesis (31) (Fig. 4A, supplementary Fig. IV). D609 was added at 12 hpi and virus titres were reduced in a dose-dependent manner at 18 hpi without affecting virus protein expression and cell viability (Fig. 4A). This result strengthens the previously identified requirement of intact SM biosynthesis for influenza virus production (Fig. 1) (14), but more specifically, implies importance of the salvage pathway.
Fig. 4.
Functional requirements of ether- and sphingolipid metabolism. A: The sphingomyelin synthase (SMS) inhibitor D609 impaired late stages of virus replication (supplementary Fig. IV). Results are shown from three independent experiments with four replicates each (n = 12 for all conditions). B: Impaired virus replication in ether lipid-deficient CHO (NRel-4) cells compared with wild-type cells (CHO-K1) (supplementary Fig. V). Results are from three independent experiments with four replicates each (n = 12 for CHO-K1 and NRel-4 cells). C: Knockdown of AGPS (1 and 2 denote different probes) reduced virus production similar to Rab11a depletion (3) (supplementary Fig. VI). Results are from at least three independent experiments with two to four replicates each (n = 19 for negative control (-ve); n = 16 for AGPS (1); n = 19 for AGPS (2); n = 8 for Rab11a). D: Catalase activity was reduced 20% in virus-infected cells (n = 6 from two independent experiments with three replicates each). E: Induction of peroxisomal β-oxidation by a PPARα agonist (GW7647) inhibited virus production. Results are from three independent experiments with four to six replicates each. [n = 14 for DMSO control (Ctrl), GW7647 (1 µM) and GW7647 (2 µM)] and from one experiment with six replicates [n = 6 for GW7647 (5 µM)]. For A–E, virus titer was determined by plaque assay and represented as % of control ± SEMs; *P < 0.005 and **P < 0.0005 (unpaired Student’s t-test; two-tailed). Expression of proteins was determined by Western blot (one representative blot is shown), using antibodies against AGPS, Rab11a, GAPDH, and α-tubulin (positive controls), and influenza virus NS1 and M2. F: Distribution of host (mouse) susceptibility factors to influenza virus infection (39): lipid-associated genes (black circles), independently identified genes (40) (filled black circles), such as PLA2G7, the most significantly associated factor; remaining genes (open gray circles). Lipid-associated genes were manually annotated to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (gray squares) or by Database for Annotation, Visualization and Integrated Discovery (DAVID) (black squares; * P < 0.05 (EASE score, a modified Fisher Exact P-value) and ** P < 0.05 [EASE score; Benjamini corrected)]. G: Proposed lipid metabolism in influenza virus-infected cells based on combined lipidomics data (this study) with genomics and proteomics results (supplementary Table VII). Genes, proteins, and metabolites are depicted in bold red (proviral or increased), bold blue (antiviral or decreased) and bold black (pro- or antiviral activity not determined) fonts. Fluxes proposed to be increased (bold arrows) or decreased (dashed arrows), inhibitory interactions (round ended bold lines), chemical inhibitors (underlined bold black) and the membrane scaffold of Serinc5 (dashed red box) are also shown. Gray boxes indicate sphingolipid (1), ether lipid (2) and peroxisomal β-oxidation (3) metabolic pathways; *identified in this study.
To further scrutinise the necessity of ether lipid metabolism during influenza replication, we infected wild-type CHO-K1 and ether lipid-deficient CHO cells (NRel-4) (32) with influenza virus H1N1 A/PR/8/34. NRel-4 cells exhibited lower expression levels of influenza virus proteins NS1 and M2, and a four- to five-fold decrease in virus production when compared with CHO-K1 cells (Fig. 4B). The lipid alterations induced by influenza virus in CHO cells were consistent with the changes in A549 cells (supplementary Fig. V) with the obvious exception of ePC, which cannot be generated in NRel-4 cells because of the reported impairment in their peroxisomal dihydroxyacetone phosphate acyltransferase (DHAPAT) activity. DHAPAT catalyses the first committed step in ether lipid biosynthesis, attaching a fatty acid to dihydroxyacetone phosphate (DHAP) to produce acyl-DHAP.
As acyl-DHAP can also be redirected into TAG biosynthesis (33), and because the two CHO cell variants were not isogenic, we therefore decided to further investigate ether lipid involvement using two siRNA constructs against peroxisomal AGPS. AGPS is the immediate downstream enzyme of DHAPAT in ether lipid biosynthesis, which exchanges the fatty acid of acyl-DHAP with a fatty alcohol. Both probes led to the substantial reduction in enzyme levels (∼70%), as judged by Western blotting (Fig. 4C, supplementary Fig. VI A), to a moderate (<1.5-fold) but significant decrease in ether lipid levels (supplementary Fig. VI B), and, importantly, to a 60% reduction in infectious virus production (Fig. 4C). These results were comparable to siRNA constructs targeting Rab11a, which is required for assembly and budding (34), and demonstrate the functional importance of ether lipid biosynthesis in influenza virus production.
To examine the functional role of the peroxisome, we treated infected cells with a PPARα agonist (GW7647) that induces peroxisomal fatty acyl β-oxidation (35, 36). We hypothesized impaired peroxisomal β-oxidation in influenza virus-infected cells due to the following evidence: 1) catalase activity correlates with peroxisomal β-oxidation and was decreased (Fig. 4D) (37), 2) accumulation of SPL-containing long chain fatty acyls (C26:0) are a molecular marker for impaired peroxisomal β-oxidation (Fig. 1D), and 3) the decrease of C24:1 fatty acyls is linked to reduced acyl-CoA oxidase 1 activity, an enzyme essential for peroxisomal β-oxidation (Fig. 1D) (27). Accordingly, virus production was reduced upon GW7647 treatment of A549 cells without impacting cell viability (Fig. 4E). These findings were consistent with independent studies identifying MFP2/HSD17B4 (see above), acyl-CoA oxidase 1, and carnitine O-octanoyltransferase as antiviral mediators of influenza virus infection (5, 8, 17). They further establish the antiviral role of peroxisomal β-oxidation and PPARα activation and provide a functional explanation for the usage of PPARα agonists as an alternative treatment for influenza virus infection (38).
Finally, we explored the potential in vivo relevance of peroxisomal and SPL metabolism for influenza virus infection through an examination of the reported susceptibility factors in mice (39). We found 23 genes associated with lipid metabolism [5% of total genes measured (39)] that were enriched for peroxisomal and SPL metabolism (Fig. 4F). The platelet-activating factor (PAF) acetylhydrolase, PLA2G7, which exhibits the strongest association, has been independently identified as a host susceptibility factor for influenza infection (40). PLA2G7 hydrolyses PAF, an ePC and activator of platelets and inflammation, producing lyso-PAF, which can be converted to ePC by lyso-PC acyltransferase 2 (Fig. 4G). Consistent with elevated ePC levels in infected cells, influenza virus infection correlated with higher expression levels of PLA2G7 and induced lyso-PC acyltransferase 2 activity in mice (39, 41). These findings provide a link between the metabolic pathways of peroxisomes and lipid meditators involved in inflammation in vivo.
In summary, we present a detailed account of the temporal changes in host cell membrane lipids during influenza virus replication in relation to the composition of virus envelopes and virus pathogenicity. While our study does not directly expose the mechanistic actions of identified lipids in the influenza virus life cycle in detail, the comprehensive systems-scale catalog of lipids reported here is the first of its kind (Fig. 3). The combination and hierarchical clustering of the different datasets provides a powerful framework to derive novel hypotheses in the emerging field of lipid involvement during virus infections. As a result, we present clear evidence that metabolism of ether lipids is functionally important for the production of infectious virions. Further integration of our findings with other published genomics and proteomics data (1, 3–8, 15, 22) led to a systems-scale model of host cell lipid metabolism during influenza virus infection, which will serve as a reference basis for future investigations (Fig. 4G, supplementary Table VII). Based on this analysis, we propose three major lipid metabolic pathways implicated in influenza virus replication: 1) elevated ether lipid and 2) elevated SPL biosynthesis required for influenza virus morphogenesis and 3) decreased peroxisomal β-oxidation associated with intracellular life cycle stages (Fig. 4G). Peroxisomal function is a common metabolic denominator and may therefore represent a key determinant for influenza virus replication. Our detailed analysis represents a major step forward in uncovering that peroxisomes and especially their lipid metabolism are exploited by influenza viruses. Our findings may open entirely new avenues with immediate exploitability for therapeutic interventions against influenza virus infection via peroxisome function.
Supplementary Material
Acknowledgments
The authors thank Sebastien Gagneux, Swiss Tropical and Public Health Institute, and Benhur Lee, University of California, Los Angeles, for scientific discussions and feedback on the manuscript.
Footnotes
Abbreviations:
- aPC
- phosphatidylcholine
- CHO
- Chinese hamster ovary
- DHAP
- dihydroxyacetone phosphate
- DHAPAT
- dihydroxyacetone phosphate acyltransferase
- ePC
- ether-linked PC
- GL
- glycerophospholipid
- HA
- hemagglutinin
- HexCer
- hexosylceramide
- hpi
- hours postinfection
- MFP2/HSD17B4
- multifunctional protein 2
- NS1
- nonstructural protein 1
- PAF
- platelet-activating factor
- PLA2G7
- Phospholipase A2, Group VII (Platelet-Activating Factor Acetylhydrolase, Plasma) SMS, SM synthase
- SPL
- sphingolipid
This work is supported by grants from the National University of Singapore via the Life Sciences Institute (LSI) and the Singapore National Research Foundation under CRP Award No. 2007-04 (to M.R.W.), and the Swiss National Science Foundation (SNSF) Ambizione Award (to X.L.G.).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of five figures, eight tables, and supplementary methods and references.
REFERENCES
- 1.Brass A. L., Huang I. C., Benita Y., John S. P., Krishnan M. N., Feeley E. M., Ryan B. J., Weyer J. L., van der Weyden L., Fikrig E., et al. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 139: 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hao L., Sakurai A., Watanabe T., Sorensen E., Nidom C. A., Newton M. A., Ahlquist P., Kawaoka Y. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 454: 890–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karlas A., Machuy N., Shin Y., Pleissner K. P., Artarini A., Heuer D., Becker D., Khalil H., Ogilvie L. A., Hess S., et al. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 463: 818–822 [DOI] [PubMed] [Google Scholar]
- 4.König R., Stertz S., Zhou Y., Inoue A., Hoffmann H. H., Bhattacharyya S., Alamares J. G., Tscherne D. M., Ortigoza M. B., Liang Y., et al. 2010. Human host factors required for influenza virus replication. Nature. 463: 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shapira S. D., Gat-Viks I., Shum B. O., Dricot A., de Grace M. M., Wu L., Gupta P. B., Hao T., Silver S. J., Root D. E., et al. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 139: 1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coombs K. M., Berard A., Xu W., Krokhin O., Meng X., Cortens J. P., Kobasa D., Wilkins J., Brown E. G. 2010. Quantitative proteomic analyses of influenza virus-infected cultured human lung cells. J. Virol. 84: 10888–10906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dove B. K., Surtees R., Bean T. J., Munday D., Wise H. M., Digard P., Carroll M. W., Ajuh P., Barr J. N., Hiscox J. A. 2012. A quantitative proteomic analysis of lung epithelial (A549) cells infected with 2009 pandemic influenza A virus using stable isotope labelling with amino acids in cell culture. Proteomics. 12: 1431–1436 [DOI] [PubMed] [Google Scholar]
- 8.Kroeker A. L., Ezzati P., Halayko A. J., Coombs K. 2012. The response of primary human airway epithelial cells to Influenza infection: a quantitative proteomic study. J. Proteome Res. 11: 4132–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerl M. J., Sampaio J. L., Urban S., Kalvodova L., Verbavatz J. M., Binnington B., Lindemann D., Lingwood C. A., Shevchenko A., Schroeder C., et al. 2012. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J. Cell Biol. 196: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polozov I. V., Bezrukov L., Gawrisch K., Zimmerberg J. 2008. Progressive ordering with decreasing temperature of the phospholipids of influenza virus. Nat. Chem. Biol. 4: 248–255 [DOI] [PubMed] [Google Scholar]
- 11.Hidari K. I., Suzuki Y., Suzuki T. 2006. Suppression of the biosynthesis of cellular sphingolipids results in the inhibition of the maturation of influenza virus particles in MDCK cells. Biol. Pharm. Bull. 29: 1575–1579 [DOI] [PubMed] [Google Scholar]
- 12.Munger J., Bennett B. D., Parikh A., Feng X. J., McArdle J., Rabitz H. A., Shenk T., Rabinowitz J. D. 2008. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat. Biotechnol. 26: 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossman J. S., Jing X., Leser G. P., Lamb R. A. 2010. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 142: 902–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tafesse F. G., Sanyal S., Ashour J., Guimaraes C. P., Hermansson M., Somerharju P., Ploegh H. L. 2013. Intact sphingomyelin biosynthetic pathway is essential for intracellular transport of influenza virus glycoproteins. Proc. Natl. Acad. Sci. USA. 110: 6406–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morita M., Kuba K., Ichikawa A., Nakayama M., Katahira J., Iwamoto R., Watanebe T., Sakabe S., Daidoji T., Nakamura S., et al. 2013. The lipid mediator protectin D1 inhibits influenza virus replication and improves severe influenza. Cell. 153: 112–125. [DOI] [PubMed] [Google Scholar]
- 16.Tam V. C., Quehenberger O., Oshansky C. M., Suen R., Armando A. M., Treuting P. M., Thomas P. G., Dennis E. A., Aderem A. 2013. Lipidomic profiling of influenza infection identifies mediators that induce and resolve inflammation. Cell. 154: 213–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolff T., O’Neill R. E., Palese P. 1996. Interaction cloning of NS1-I, a human protein that binds to the nonstructural NS1 proteins of influenza A and B viruses. J. Virol. 70: 5363–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixit E., Boulant S., Zhang Y., Lee A. S., Odendall C., Shum B., Hacohen N., Chen Z. J., Whelan S. P., Fransen M., et al. 2010. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 141: 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritter J. B., Wahl A. S., Freund S., Genzel Y., Reichl U. 2010. Metabolic effects of influenza virus infection in cultured animal cells: Intra- and extracellular metabolite profiling. BMC Syst. Biol. 4: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan R., Uchil P. D., Jin J., Shui G., Ott D. E., Mothes W., Wenk M. R. 2008. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J. Virol. 82: 11228–11238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shui G., Stebbins J. W., Lam B. D., Cheong W. F., Lam S. M., Gregoire F., Kusonoki J., Wenk M. R. 2011. Comparative plasma lipidome between human and cynomolgus monkey: are plasma polar lipids good biomarkers for diabetic monkeys? PLoS ONE. 6: e19731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Billharz R., Zeng H., Proll S. C., Korth M. J., Lederer S., Albrecht R., Goodman A. G., Rosenzweig E., Tumpey T. M., Garcia-Sastre A., et al. 2009. The NS1 protein of the 1918 pandemic influenza virus blocks host interferon and lipid metabolism pathways. J. Virol. 83: 10557–10570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caric-Lazar M., Schwarz R. T., Scholtissek C. 1978. Influence of the infection with lipid-containing viruses on the metabolism and pools of phospholipid precursors in animal cells. Eur. J. Biochem. 91: 351–361 [DOI] [PubMed] [Google Scholar]
- 24.Walker A. K., Jacobs R. L., Watts J. L., Rottiers V., Jiang K., Finnegan D. M., Shioda T., Hansen M., Yang F., Niebergall L. J., et al. 2011. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell. 147: 840–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettus B. J., Baes M., Busman M., Hannun Y. A., Van Veldhoven P. P. 2004. Mass spectrometric analysis of ceramide perturbations in brain and fibroblasts of mice and human patients with peroxisomal disorders. Rapid communications in mass spectrometry. Rapid Commun. Mass Spectrom. 18: 1569–1574 [DOI] [PubMed] [Google Scholar]
- 26.Sargent J. R., Coupland K., Wilson R. 1994. Nervonic acid and demyelinating disease. Med. Hypotheses. 42: 237–242 [DOI] [PubMed] [Google Scholar]
- 27.Vluggens A., Andreoletti P., Viswakarma N., Jia Y., Matsumoto K., Kulik W., Khan M., Huang J., Guo D., Yu S., Sarkar J., Singh I., Rao M. S., Wanders R. J., Reddy J. K., Cherkaoui-Malki M. 2010. Reversal of mouse Acyl-CoA oxidase 1 (ACOX1) null phenotype by human ACOX1b isoform [corrected]. Lab. Invest . 90: 696–708 [DOI] [PubMed] [Google Scholar]
- 28.Narasaraju T., Sim M. K., Ng H. H., Phoon M. C., Shanker N., Lal S. K., Chow V. T. 2009. Adaptation of human influenza H3N2 virus in a mouse pneumonitis model: insights into viral virulence, tissue tropism and host pathogenesis. Microbes Infect . 11: 2–11 [DOI] [PubMed] [Google Scholar]
- 29.Forbes N. E., Ping J., Dankar S. K., Jia J. J., Selman M., Keleta L., Zhou Y., Brown E. G. 2012. Multifunctional adaptive NS1 mutations are selected upon human influenza virus evolution in the mouse. PLoS ONE. 7: e31839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forbes N., Selman M., Pelchat M., Jia J. J., Stintzi A., Brown E. G. 2013. Identification of adaptive mutations in the influenza a virus non-structural 1 gene that increase cytoplasmic localization and differentially regulate host gene expression. PLoS ONE. 8: e84673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adibhatla R. M., Hatcher J. F., Gusain A. 2012. Tricyclodecan-9-yl-xanthogenate (D609) mechanism of actions: a mini-review of literature. Neurochem. Res. 37: 671–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagan N., Hajra A. K., Larkins L. K., Lazarow P., Purdue P. E., Rizzo W. B., Zoeller R. A. 1998. Isolation of a Chinese hamster fibroblast variant defective in dihydroxyacetonephosphate acyltransferase activity and plasmalogen biosynthesis: use of a novel two-step selection protocol. Biochem. J. 332: 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hajra A. K., Larkins L. K., Das A. K., Hemati N., Erickson R. L., MacDougald O. A. 2000. Induction of the peroxisomal glycerolipid-synthesizing enzymes during differentiation of 3T3–L1 adipocytes. Role in triacylglycerol synthesis. J. Biol. Chem. 275: 9441–9446 [DOI] [PubMed] [Google Scholar]
- 34.Bruce E. A., Digard P., Stuart A. D. 2010. The Rab11 pathway is required for influenza A virus budding and filament formation. J. Virol. 84: 5848–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown P. J., Stuart L. W., Hurley K. P., Lewis M. C., Winegar D. A., Wilson J. G., Wilkison W. O., Ittoop O. R., Willson T. M. 2001. Identification of a subtype selective human PPARalpha agonist through parallel-array synthesis. Bioorg. Med. Chem. Lett. 11: 1225–1227 [DOI] [PubMed] [Google Scholar]
- 36.Reddy J. K., Hashimoto T. 2001. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu. Rev. Nutr. 21: 193–230 [DOI] [PubMed] [Google Scholar]
- 37.Périchon R., Bourre J. M. 1995. Peroxisomal beta-oxidation activity and catalase activity during development and aging in mouse liver. Biochimie. 77: 288–293 [DOI] [PubMed] [Google Scholar]
- 38.Fedson D. S. 2008. Confronting an influenza pandemic with inexpensive generic agents: can it be done? Lancet Infect. Dis. 8: 571–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boivin G. A., Pothlichet J., Skamene E., Brown E. G., Loredo-Osti J. C., Sladek R., Vidal S. M. 2012. Mapping of clinical and expression quantitative trait loci in a sex-dependent effect of host susceptibility to mouse-adapted influenza H3N2/HK/1/68. J. Immunol. 188: 3949–3960 [DOI] [PubMed] [Google Scholar]
- 40.Nedelko T., Kollmus H., Klawonn F., Spijker S., Lu L., Hessman M., Alberts R., Williams R. W., Schughart K. 2012. Distinct gene loci control the host response to influenza H1N1 virus infection in a time-dependent manner. BMC Genomics. 13: 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia C. C., Russo R. C., Guabiraba R., Fagundes C. T., Polidoro R. B., Tavares L. P., Salgado A. P., Cassali G. D., Sousa L. P., Machado A. V., et al. 2010. Platelet-activating factor receptor plays a role in lung injury and death caused by Influenza A in mice. PLoS Pathog. 6: e1001171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.