Abstract
Current challenges exist to widespread clinical implementation of genomic medicine and pharmacogenetics. The University of Florida (UF) Health Personalized Medicine Program (PMP) is a pharmacist-led, multidisciplinary initiative created in 2011 within the UF Clinical Translational Science Institute. Initial efforts focused on pharmacogenetics, with long-term goals to include expansion to disease-risk prediction and disease stratification. Herein we describe the processes for development of the program, the challenges that were encountered and the clinical acceptance by clinicians of the genomic medicine implementation. The initial clinical implementation of the UF PMP began in June 2012 and targeted clopidogrel use and the CYP2C19 genotype in patients undergoing left heart catheterization and percutaneous-coronary intervention (PCI). After 1 year, 1,097 patients undergoing left heart catheterization were genotyped preemptively, and 291 of those underwent subsequent PCI. Genotype results were reported to the medical record for 100% of genotyped patients. Eighty patients who underwent PCI had an actionable genotype, with drug therapy changes implemented in 56 individuals. Average turnaround time from blood draw to genotype result entry in the medical record was 3.5 business days. Seven different third party payors, including Medicare, reimbursed for the test during the first month of billing, with an 85% reimbursement rate for outpatient claims that were submitted in the first month. These data highlight multiple levels of success in clinical implementation of genomic medicine.
Keywords: pharmacogenetics, genomic medicine, implementation, CYP2C19, personalized medicine
INTRODUCTION
In the past decade there have been substantial advances in understanding genetic variability associated with drug response (efficacy and toxicity) and dose requirements [Wang et al., 2011]. In many cases these genetic associations have been widely replicated and the effect sizes are sufficiently large for associations to be predictive in the clinical setting. However, despite these facts and availability of clinical pharmacogenetic tests, there are limited examples of use of pharmacogenetic data to guide drug prescribing in clinical practice. Current challenges to widespread clinical implementation of genomic medicine generally, and pharmacogenetics specifically, include questions about which genetic variants have sufficiently robust evidence to guide clinical decision making, apprehension among busy clinicians about how to order and use such data, few existing genomic clinical decision support (CDS) tools to guide clinicians, lack of standardized processes for storing and using genomic data in clinical care, and limited reimbursement experience with genetic testing, among others.
The University of Florida (UF) Health Personalized Medicine Program (PMP) was established in 2011 as a translational, clinical implementation program within the UF Clinical and Translational Science Institute (CTSI) to address these challenges. Although long-term program goals include incorporation of disease-risk prediction and disease stratification, initial efforts focused on pharmacogenetics due to a broader evidence base, supportive regulatory data in FDA-approved drug labeling, relatively straightforward actions resulting from genetic information in many cases, and fewer perceived (and likely real) concerns about the potential for discrimination based on pharmacogenetic information.
The initial clinical pharmacogenetic implementation within UF PMP was with the antiplatelet agent clopidogrel and CYP2C19 genotype in patients undergoing cardiac catheterization and potentially percutaneous coronary intervention (PCI). Clopidogrel is a prodrug that requires two-step bioactivation, for which CYP2C19 plays a pivotal role. The CYP2C19 gene contains several common loss-of-function polymorphisms and evidence suggests decreased clopidogrel effectiveness in patients carrying a loss-of-function allele [Scott et al., 2013]. The potential for a reduced clopidogrel effect has important clinical implications in the setting of PCI, in which effective dual antiplatelet therapy has demonstrated up to an 85% relative risk reduction compared to aspirin alone [Leon et al., 1998]. In this setting, even a slight decrease in the action of clopidogrel secondary to reduced CYP2C19 bioactivation could have a significant clinical impact [Mega et al., 2010]. The existing data suggest it is in this setting where the clinical benefits of genotype-guided therapy are likely greatest [Johnson et al., 2012b; Scott et al., 2013]. While there are data that question the clinical utility of CYP2C19 genotyping for overall outcome and platelet activity, there are also data that support an increased risk of adverse cardiovascular events and stent thrombosis in individuals undergoing PCI, which led to FDA’s addition of a boxed warning to the clopidogrel product labeling in 2010. Clinical Pharmacogenetics Implementation Consortium guidelines are available to guide use of genetic information with clopidogrel [Scott et al., 2013]. UF PMP chose CYP2C19-clopidogrel as its initial implementation due to the potential for genotype-guided therapy to reduce adverse cardiovascular events, existing regulatory support in the FDA boxed warning, and the potential to impact a large number of patients through a targeted implementation in a defined patient care area involving a relatively small number of physicians. Prior to the launch of the PMP, the interventional cardiologists indicated that they had never ordered a CYP2C19 genetic test, and cited difficulty with ordering, interpreting, and integrating test results into practice as the main barriers that had prevented them from doing so. Therefore, beginning in July 2011, the UF PMP began a stepwise approach for the clopidogrel-CYP2C19 implementation. Steps included (1) synthesis of available evidence; (2) development of a pharmacy and therapeutics (P&T) committee approval process; (3) delivery of genotype results into the electronic health record (EHR) system with associated CDS; and (4) specimen capture into patient care and pathology workflow. In June 2012, the clopidogrel-CYP2C19 implementation was launched. Herein we describe the processes for development of the program, as well as challenges and solutions encountered along the way, and present a summary of data for the first year of using CYP2C19 genotyping data to guide treatment with clopidogrel in post-PCI patients.
MATERIALS AND METHODS
Guiding Principles of the UF PMP
The UF PMP, a pharmacist-led multi-disciplinary clinical implementation initiative, was established with several guiding principles. First, a regulatory body was developed to oversee PMP initiatives to ensure that a clear process for evidence evaluation and program approval existed within the health system. Second, the program launched with a preemptive genotyping focus, using chip-based genotyping. This approach allowed for generation of multiple clinically actionable genotypes that could be immediately used in clinical care or stored in a data repository for future use. The chip-based approach also created the challenges of not only immediately incorporating actionable genotype information into patient care, but also of storing, retrieving, and using genotype data as it becomes clinically actionable in the future. Third, it was decided that program initiatives would be supported through specific informatics CDS within the EHR that provide interpretation and clinical recommendations regarding genetic information. The PMP created a structured process for developing and implementing each initiative.
Regulatory Body for PMP Oversight
The UF Health Shands Hospital P&T committee was designated as the regulatory body to provide oversight for PMP initiatives. A multidisciplinary PMP subcommittee to the P&T was created to support this function of the P&T committee and provide support to UF PMP.
Genotyping
From program initiation, genetic samples and assays for UF Health PMP have been processed and performed in the UF Health Pathology Laboratories (UFHPL), a College of American Pathologists-accredited Clinical Laboratory Improvements Amendments-licensed (CAP/CLIA) clinical laboratory. Prior to the clopidogrel-CYP2C19 launch, the laboratory used several sources of control materials to validate CYP2C19 assays according to CAP/CLIA quality assurance and quality control requirements (CLIA 42 CFR 493.1253 and CAP GEN 42020–42163) for instrument, assay, and clinical validation on two platforms, Life Technologies (Carlsbad, CA) Quant Studio™ Open Array and GenMark Dx (Carlsbad, CA). Both systems were used during the first year of the program for generation of clinical CYP2C19 genotypes. Genotyping conducted on the Quant Studio™ Open Array utilized a custom genotype array that includes 256 single nucleotide polymorphisms (SNPs). The process for SNP selection and the design and validation of this customized array has been previously described [Johnson et al., 2012a]. The custom genotyping array tests for the *2, *3, *4, *5, *6, *8, *10, and *17 of CYP2C19, with *10 included to ensure proper genotype calling for *2. The GenMark platform tests for the same alleles plus *9 and *13. During the first year of PMP implementation, the cost of CYP2C19 genotyping was covered by the PMP and UFHPL. Beginning in June 2013, UFHPL started generating bills for CYP2C19 genotyping.
Informatics and Clinical Decision Support
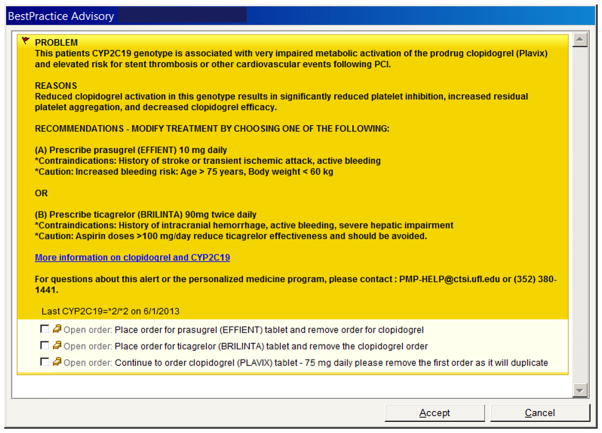
From an informatics perspective, several steps were necessary. First, a new pharmacogenetic test had to be developed as an orderable test within the EHR, which at UF Health is Epic. Then, consistent with a preemptive testing approach, the CYP2C19 pharmacogenetic test panel was added to the standing order set for all patients undergoing left heart catheterization. Translation software was written to convert the SNP genotype data into the star (*) allele nomenclature. A Best Practice Advisory (BPA) was also developed within the EHR that linked a clopidogrel order with an actionable CYP2C19 genotype (Fig. 1). When a clopidogrel order was entered into Epic for a patient in whom an actionable genotype had been determined, the ordering physician was presented with the BPA, from which a drug therapy change could be made by choosing one of the therapy options listed. The ordering physician could also select “cancel” to bypass the BPA, however, we were unable to track each physician’s action when presented with a BPA due to technical limitations of the EHR for generating reports from BPAs.
Figure 1.
Sample Epic Best Practice Advisory alert for clopidogrel-CYP2C19 implementation. BPA alerts are presented to the ordering physician in the EHR when a clopidogrel order is linked with an actionable CYP2C19 genotype. Physicians are provided email and phone contacts for additional help and a weblink to the Pharmacogenomics Knowledgebase (http://www.pharmgkb.org/) for additional information. PCI, percutaneous coronary intervention; PMP, Personalized Medicine Program.
Consent for Genetic Testing and Storage of Genetic Data
During the planning year for the PMP clinical launch, there was substantial discussion among PMP leadership, ethicists, the UF Health Institutional Review Board and clinicians regarding consent for CYP2C19 pharmacogenetic testing. It was concluded that because the CYP2C19 genotyping test was being incorporated as standard of care in the cardiac catheterization laboratory, consent for genotyping would occur during the process for clinical consent for cardiac catheterization and associated care. Therefore, only eight of 256 SNPs on the custom array used to define the CYP2C19 genotype were moved to the EHR as part of standard care in post-PCI patients receiving clopidogrel.
Following program launch, patients were also invited to provide research informed consent to have the remaining 248 of 256 SNPs stored in a data repository (outside the EHR) for: (1) future transfer of genotypes for other genes into the EHR for clinical care once approved within PMP; (2) research purposes within UF CTSI Integrated Data Repository; (3) authorization to store leftover samples in the CTSI biorepository for future research; and (4) permission to be re-contacted for other research studies. Patients not invited for research informed consent, or who declined to consent had the 248 non-CYP2C19 genotype data and their remaining biological sample destroyed.
Collection of the above research informed consent was accomplished electronically, as part of the UF Health pilot for Research Permissions Management System (RPMS), an electronic consenting tool developed at Medical University of South Carolina. This not only provided a mechanism for electronic consent, but also gave laboratory and biorepository personnel electronically accessible status of research consent to know whether the additional genetic data and blood sample should be saved or destroyed.
Data Collection Metrics and Statistical Analysis
Clopidogrel-CYP2C19 program metrics during year 1 included data collection regarding (a) clinical implementation including test ordering rate, results, and adoption of alternative therapy recommendations; (b) research consent and enrollment; and (c) pathology processes and quality improvement information. Program metrics data were tracked by patient encounter in EHR reports and through daily verification of patient data by PMP staff and/or clinical pharmacists. If a single patient had multiple encounters in the cardiac catheterization laboratory and did not undergo PCI or a pharmacogenetic test, only the first patient encounter was used to track pharmacogenetic test completion. Any patient encounter that resulted in a PCI and/or a pharmacogenetic test was designated as the encounter used to track pharmacogenetic test completion.
The CYP2C19 test adoption rates between the first and last 2 months of implementation were compared in all patients undergoing left heart catheterization and/or PCI using the Pearson χ2 test, and in PCI patients only using a two-sided Fisher’s exact test. All analyses were conducted using SAS version 9.3 (SAS, Cary, NC).
RESULTS
Clinical Implementation of PMP Subcommittee Processes
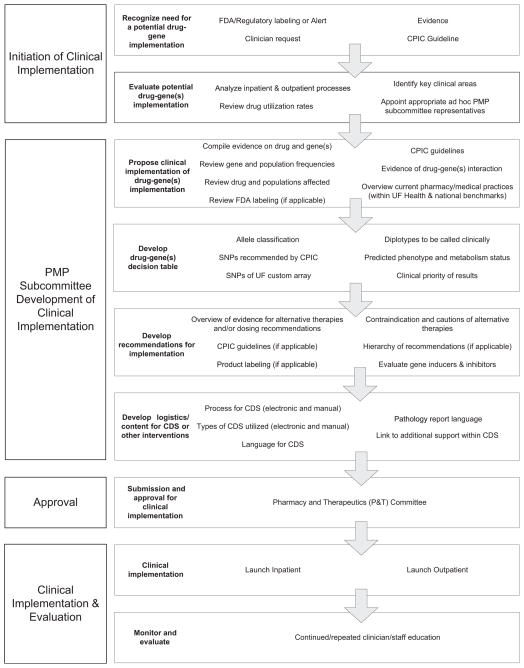
A streamlined process was developed and adopted for moving a pharmacogenetic clinical implementation from concept to reality (Fig. 2). The PMP subcommittee of the P&T committee initially reviewed the evidence to identify potential clinically actionable drug–gene pairs.
Figure 2.
Drug–gene implementation process. CPIC, Clinical Pharmacogenetic Implementation Consortium; UF, University of Florida; PMP, Personalized Medicine Program; SNP, single nucleotide polymorphism; CDS, clinical decision support.
Once clopidogrel-CYP2C19 was selected for this pilot, the committee defined specific genetic polymorphisms and predicted metabolism phenotypes, as appropriate, that would be the basis for pharmacogenetic recommendations. The committee then developed clinical recommendations for use of genetic information, wording of pathology reports and associated CDS alerts, and supportive education materials. These recommendations were presented to the full P&T committee for approval. Once approved, specific genotype information could be reported within a patient’s record in the EHR. The PMP subcommittee then began an ongoing literature evaluation process to identify new evidence that may necessitate a change in pathology reports and CDS wording or other procedures. If such evidence was identified in year 1 or anytime thereafter, any necessary changes would be presented to the full P&T committee for approval after final PMP vetting. Use of a comprehensive process ensured that the clopidogrel-CYP2C19 pilot proceeded through a robust evaluation, with input from thought leaders across the health science center, and subsequent revisions as necessary. Adopting this as standardized procedure within UF PMP provided similar assurances for future implementations.
Clinical Implementation of Clopidogrel-CYP2C19 Clopidogrel
In the case of clopidogrel, evaluation of the available evidence for CYP2C19 resulted in the decision to make clinical recommendations on the polymorphisms that define CYP2C19, including *2, *3, *4, *5, *6, *8, and *17. All but *17 were considered loss of function alleles [Scott et al., 2013]. The published data for *9, *10, and *13 were considered insufficient for clinical use and genotypes that include these alleles were reported as having an unknown predicted phenotype. The resulting phenotype classifications and approved clinical actions for each genotype in post-PCI patients are summarized in Table I. Patients with extensive, ultrarapid, or unknown metabolizer status were recommended to receive usualcare, which was clopidogrel 75 mg daily. Carriers of a *17 allele along with a loss of function allele were considered intermediate metabolizers. Any test results indicative of an “intermediate metabolizer” or “poor metabolizer” phenotype were considered actionable and led to a CDS recommendation for alternative therapy. At program launch, alternative therapy recommendations included (a) prasugrel, (b) ticagrelor, or (c) triple-dose clopidogrel (for intermediate metabolizers only).
TABLE I.
Clinical Recommendations for Patients With Variant CYP2C19 Genotype
| Phenotype | Diplotype results | Clinical recommendation regarding clopidogrel therapya |
|---|---|---|
| Extensive metabolizers | *1/*1 | Usual care |
| Intermediate metabolizers | *1/(*2–*6, *8) (*2–*6, *8)/*17 | Alternative therapy |
| Poor metabolizers | *2/(*2–*6, *8)or any combination with two loss of functional alleles | Alternative therapy |
| Ultrarapid metabolizers | *1/*17, *17/*17 | Usual care |
| Unknown | Any combination containing *10 | Usual care |
Usual care was defined as clopidogrel 75 mg daily. Alternative therapy included either prasugrel 10 mg daily, ticagrelor 90 mg twice daily, or clopidogrel 225 mg daily (triple-dose clopidogrel was removed as an alternative option in March 2013).
The PMP subcommittee processes for development of evidence and clinical recommendations and subsequent P&T committee approval were conducted through once- to twice-monthly meetings that occurred over a 5-month period, with testing initiated in June 2012. Subsequently in March 2013, the P&T committee approved removal of the triple-dose clopidogrel option at the request of the PMP subcommittee, due to emerging evidence that reduced confidence in the benefit-to-risk ratio with this option [Price et al., 2011, 2012; Scott et al., 2013].
Once the initial clopidogrel-CYP2C19 recommendations were approved by the P&T committee, changes were made to incorporate this information into the clinical workflow and EHR that are still in place today. For those patients who undergo PCI and have an actionable genotype test result, an alert pops up within the EHR for the ordering physician when the clopidogrel order is placed (Fig. 1). The BPA alert is colored bright yellow in the EHR to designate its importance. Notification is also sent to the cardiovascular clinical pharmacist who works with the cardiologist to ensure drug changes are made for patients discharged prior to the availability of the genotype data. Alternative drug therapy is prescribed as appropriate based on CDS alert recommendations and/or the clinical pharmacist’s follow up and recommendation.
Laboratory Processes
Since launch of the PMP, UFHPL reported the CYP2C19 genotype for 100% of samples received, with an average turnaround time from blood draw to result entry into the medical record of 3.5 business days. Factors influencing sample turnaround time included courier delays in moving samples from the hospital to the off-site laboratory; once-daily batch processing of samples introduced an additional 24-hr delay in some cases if samples were not received in the lab before daily batch run; next-day entry of results in the EHR after sample processing; and the need to process some samples in duplicate for quality assurance purposes. Approximately half of the samples were processed on the Life Technologies Quant Studio™ Open Array, with the remaining samples processed on the GenMark Dx. Technical issues with the Open Array chip manufacturing by Life Technologies necessitated transition to the GenMark Dx system.
Billing for CYP2C19 genotyping began in June 2013, when the Department of Pathology, Immunology and Laboratory Medicine began billing third party payors and the hospital for outpatient and inpatient tests, respectively. During the first month of billing, seven different third party payors (including Medicare) and the hospital reimbursed for the test. Reimbursement was received for 85% of outpatient claims for CYP2C19 genotyping billed in the first month.
Clinical Implementation Data Metrics
From June 25, 2012 through June 25, 2013, a total of 1,097 patients who underwent left heart catheterization were genotyped (Table II), which was 74% of all patients undergoing left heart catheterization. Reasons for not receiving a genotype order and/or test result in the remaining patients included genotype test was ordered but not collected (5%); test was ordered but subsequently cancelled (2%); test was ordered but sample was not received by the laboratory (1%); and opportunity for testing was missed or test was deemed inappropriate by the cardiologist (17%). In the latter category, reasons for not testing included the patient undergoing left heart catheterization for reasons other than clinical suspicion of coronary artery disease (i.e., pre-transplant evaluation), a decision had already been made to use alternative therapy, patient was emergent and test opportunity was missed, the cardiologist did not wish to order the test, or patient refused the test. In most cases, it was deemed that there was very low likelihood of the patient requiring clopidogrel or genotype data for clinical decision making, and the choice to not order a preemptive genotype was appropriate.
TABLE II.
CYP2C19 Test Ordering and Adoption
| Implementation metric | No. of patients (%) | P-Valuea |
|---|---|---|
| Patients undergoing left heart catheterization and/or a PCI (preemptive genotyping) | 1,479 | |
| Number of patients receiving CYP2C19 test in year 1 | 1,097 | |
| Number of tests successfully processed in lab | 1,097 | |
| Year 1 test adoption rate | 1,097/1,479 (74) | |
| First 2 months (July and August 2012) | 113/239 (47) | |
| Last 2 months (May and June 2013) | 208/252 (83) | <0.001 |
| PCI patients only | 291 | |
| Number of patients receiving CYP2C19 test in year 1 | 247 | |
| Number of tests successfully processed in lab | 247 | |
| Year 1 test adoption rate | 247/291 (84) | |
| First 2 months (June and July 2012) | 30/48 (63) | |
| Last 2 months (May and June 2012) | 40/41 (98) | <0.001 |
PCI, percutaneous coronary intervention.
P-Values provided for CYP2C19 test adoption rates between first 2 months of implementation and last 2 months of implementation.
Measures instituted early in year 1 to address missed testing opportunities included (1) automatic selection of the genotype test order on the pre-catheterization order sets; (2) clinical pharmacist monitoring of cardiac catheterization patient population and follow-up with physicians in the event of a missed test; and (3) verbal and written educational initiatives for staff, administration, ordering providers, and phlebotomy such as development of a one-page reference sheet for unit nurses and addition of a reminder to catheterization laboratory tracking sheet for technicians and nurses. These measures resulted in improvement in test order rates in year 1, with 47% (113/239) of patients being genotyped in the first 2 months of the program versus 83% (208/252) in the last 2 months of year 1 (P <0.001). In the subset of patients who subsequently underwent PCI, test order rates increased from 63% (30/48) in the first 2 months to 98% (40/41) in the last 2 months of year 1 (P <0.001).
CYP2C19 drug metabolism phenotype classifications among tested patients are listed in Table III. Of the 1,097 patients genotyped, 38.5% were classified as normal metabolizers, 26.1% were intermediate metabolizers (actionable genotype), 1.7% were poor metabolizers (actionable genotype), 32.9% were ultrarapid metabolizers, and <1% (8/1,097) of patients had a diplotype with an uncharacterized metabolism status (genotype results reported to physician as “unknown”). A total of 27.8% (306/1,097) of all patients genotyped had an actionable result.
TABLE III.
CYP2C19 Pharmacogenetic Test Results and Classification
| Phenotype (metabolism status) | No. of patients (%) n =1,097 patients genotyped | Diplotype |
|---|---|---|
| Extensive metabolizer (normal) | 422 (38.5) | *1/*1 |
| Intermediate metabolizer (impaired) | 287 (26.1) | |
| 218 (19.9) | *1/*2 | |
| 61 (5.6) | *2/*17 | |
| 3 (0.3) | *1/*8 | |
| 1 (0.1) | *4/*17 | |
| 1 (0.1) | *8/*17 | |
| 1 (0.1) | *1/*3 | |
| 1 (0.1) | *1/*4 | |
| 1 (0.1) | *1/*6 | |
| None detected | *1/*5, *3/*17, *5/*17, 6/*17 | |
| Poor metabolizer (very impaired) | 19 (1.7) | |
| 17 (1.5) | *2/*2 | |
| 2 (0.2) | *2/*3 | |
| None detected | *2/*4, *2/*5, *2/*6, *2/*8, *3/*3, *3/*4, *3/*5, *3/*6, *3/*8, *4/*4, *4/*5, *4/*6, *4/*8, *5/*5, *5/*6, *5/*8, *6/*6, *6/*8, *8/*8 | |
| Ultrarapid metabolizer (enhanced) | 361 (32.9) | |
| 303(27.6) | *1/*17 | |
| 58 (5.3) | *17/*17 | |
| Unknown (uncharacterized) | 8 (0.8) | |
| 2 (0.2) | *1/*10 | |
| 1 (0.1) | *2/*10 | |
| 3 (0.1) | *1/*9 | |
| 1 (0.1) | *13/*17 | |
| 1 (0.1) | *4/*17/*17 |
Of patients who were genotyped, 291 individuals underwent subsequent PCI. Among these, 80 patients had an actionable genotype result. A total of 92.5% (74/80) of these were classified as intermediate metabolizers and 7.5% (6/80) as poor metabolizers (Fig. 3). Because use of clopidogrel is standard of care in post-PCI patients at UF Health, drug therapy changes were defined as use of alternative therapy in patients with an actionable genotype. Seventy percent of patients with an actionable genotype (56/80) were changed to alternative therapy. Among poor metabolizers, 100% (6/6) were switched to a new therapy, whereas 68% (50/74) of intermediate metabolizers were changed. Prasugrel was the most common alternative therapy choice, selected in 80% of patients with a change in therapy. Among the 24 patients not switched, reasons documented in the EHR for not making a drug therapy change included a contraindication to alternative therapy (42%), patient was deemed stable on current therapy (21%), or reason unknown/patient was lost to follow-up (37%). Based on feedback during year 1, the higher cost of prasugrel or ticagrelor compared to generic clopidogrel, influenced the choice to not change therapy in a large portion of these cases, although this was not documented in the medical record.
Figure 3.
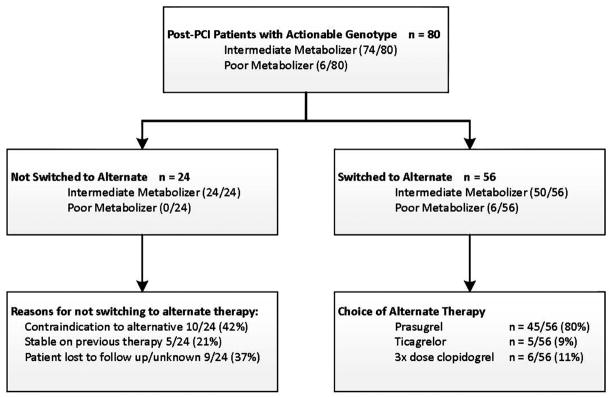
Clinical metrics in PCI patients with actionable genotypes. PCI, percutaneous coronary intervention; 3×, three times; diplotype classifications for intermediate and poor metabolizers provided in Table I.
The number needed to genotype (NNG) was defined as the number of PCI patients who would need to undergo genotyping to have a recommended change in antiplatelet therapy based on genotype. The NNG was 3.64 (291/80); approximately four PCI patients would need to be genotyped to result in one recommended change in therapy.
In nearly all cases, genotype results were reported at the time of or after PCI; rarely, the genotype was available prior to the first antiplatelet order, since antiplatelet therapy is initiated prior to the cardiac catheterization. The exception was a small percentage of patients who had multiple cardiac catheterizations, and thus genotype data were available for procedures that occurred after the genotype data were determined. In all but four patients, a change to alternative therapy was made after the genotype information became available. Four patients with an actionable genotype were placed on an alternate agent from the start of therapy; it is unclear if the decision to stay with that alternate therapy long term was influenced by the genotype data in these patients. In instances for which the genotype was reported after patient discharge, the clinical pharmacist followed up with the physician and patient post-discharge to facilitate a therapy change.
Research Consent and Storage of Genetic Data
Several months after program initiation, patients whose genotypes were determined on the Life Technologies Quant Studio™ Open Array using the custom chip were eligible to participate in the research arm of this project. Not all patients had the opportunity to provide research informed consent, based on timing of their cardiac catheterization procedure and/or patients’ short duration of stay in the cardiac catheterization laboratory or hospital. Approximately 10% of those invited to consent for research declined to participate. The most common reasons given for declining consent were not feeling well, and generally not being interested in research. Few declined consent because they did not want their genetic information stored. Among those consenting, and as shown in Table IV, nearly all patients agreed to be notified if a new pharmacogenetic test result was reported into their medical record, and wanted to be notified if a pharmacogenetic finding was later linked to disease risk, thus making it an incidental finding. The electronic RPMS system for consenting patients on an iPad was highly effective, with minimal documented downtimes with RPMS that led to only 3.1% (11/357) of patients needing to be consented on paper due to technical difficulties with RPMS.
TABLE IV.
Research Enrollment and Genetic Data Storage
| Patients approached for research enrollment | 394 |
| Patients declined to participate | 37 (9%) |
| Patients consented for research enrollment | 357 (91%) |
| Electronic consent (RPMS) | 346 |
| Paper consent | 11 |
| Patients consenting to additional research questions | |
| Notification of new pharmacogenetic test result reported in EHR | 328/357 (92%) |
| Notification of incidental pharmacogenetic findings that are later learned to affect disease risk | 342/357 (96%) |
| Re-contact for other research | 253/357 (71%) |
| Electronic consent (RPMS) performance | |
| Number of patients experiencing technical issues with RPMS consent | 5 |
| Number of documented RPMS consent downtimes | 3 (avg 3.5 hr each) |
| Storage of genetic data | |
| Number of DNA samples stored in biorepository | 341/357 (96%) |
| Number of patients with genetic data stored in IDR | 322/357 (90%) |
RPMS, Research Permissions Management System; EHR, electronic health record; IDR, Integrated Data Repository.
DISCUSSION
The UF PMP pilot genomic medicine implementation provides evidence that use of genomic medicine can be successfully incorporated into clinical care by reducing barriers to use of such data. Moreover, it demonstrates that a multidisciplinary team-based approach is critical for implementation and development.
Successful program elements included creating an easily orderable and locally run clinical laboratory test, reporting summarized genotype and phenotype and associated simple interpretations into the EHR for direct use by clinicians, along with CDS within the EHR that notifies clinical pharmacy support to facilitate drug changes and guides clinicians during future patient encounters. Prior to launch of this program, the interventional cardiologists had never ordered a CYP2C19 genetic test, but reduction of barriers and educational approaches of the PMP resulted in these same physicians obtaining a test result for nearly 1,100 patients in the first year of the program.
Similar to other early implementation projects, our experience supports the need for strong institutional commitment to program implementation and adoption within leadership and across multiple disciplines and support staff [Crews et al., 2012; Pulley et al., 2012; Farrugia and Weinshilboum, 2013; Hamilton et al., 2013]. This need has been strongly expressed among early implementers, even with highly targeted drug–gene implementations such as clopidogrel-CYP2C19 [Pulley et al., 2012]. As individual clinical initiatives expand to include larger patient populations and additional drug–gene pairs, we feel it will be essential for programs to actively continue to seek and maintain widespread provider and administrative commitment for long-term success.
Our experiences and lessons learned also echo others in reinforcing the need for a dedicated cadre of clinical experts to evaluate new clinical implementations, review and analyze the evidence base in an ongoing manner, and provide input on target populations, clinical recommendations, and pathology report and CDS wording, among other roles [Pulley et al., 2012; Farrugia and Weinshilboum, 2013]. Within our model, the PMP subcommittee provides this input to the P&T committee and PMP leadership. In addition to the tangible output of this group in areas described above, we have also seen other benefits from the ongoing dialogue it facilitates. For example, inclusion of key clinicians and/or staff from targeted patient care areas as ad hoc subcommittee members during implementation planning allowed these individuals to give valuable insight to the committee, while also increasing awareness and understanding of PMP initiatives. As clinical pharmacogenetics initiatives expand, it will be important for emerging programs to continue to identify strategies for systematically translating evidence review processes and findings to other clinicians and institutions that may be unable to support a comprehensive internal process.
At the outset, it was anticipated that informatics issues would present the greatest challenge. However this was not the case, in part because the informatics team worked within the existing Lab Information System (LIS) and EHR infrastructure. In contrast, some of the biggest barriers were issues we had spent less time addressing. A major lesson learned was the need to educate phlebotomists and floor nurses about the program, as they were the ones who were often doing the blood sample collection. Despite the test being a clinically ordered laboratory test, we encountered multiple problems with sample collection early in the program because those drawing the blood samples were unfamiliar with the test. Additionally, there were unanticipated sample transfer issues that prolonged the test turnaround time. Moving forward with new implementations we are much more proactive in educating all relevant clinical support personnel, and understanding workflow and sample flow in the clinical area to avoid some of these issues.
The first year of the program also made clear that electronic CDS alone is insufficient for ensuring improved patient care based on genetic information. Specifically, in some cases the genotype data became available after the patient had been discharged from the hospital, as most patients are hospitalized less than 24 hr after a PCI. This meant that the CDS, which required the genotype result and an EHR order for clopidogrel, was not triggered on these patients because they had already been discharged. As a result, the clinical pharmacist receiving alerts and following up on patients with actionable genotypes was an essential element to the high success rate in patients having therapy changed based on the genetic information. It is clear that without this clinical pharmacist support, based on patient flow issues within interventional cardiology, many fewer patients would have had their therapy changed.
It is also clear that some of the early successes of the program related to the fact that research funds and support from the UF Pathology Laboratories covered the cost of the genotyping for much of the first year. Once billing for the CYP2C19 genotype was initiated, a decision was made by the interventional cardiologists to transition to ordering the test only in patients undergoing a successful PCI. This strategy to select the patients most likely to benefit from genotyping was a logical one as it would lead to a lower absolute cost of genotyping. However, as genomic medicine initiatives transition from grant- or institutional-supported enterprises to full clinical programs, balancing the financial implications, especially reimbursement for preemptive testing, against the potential future benefits of using genotype data in clinical care will remain important challenges.
In our experience, a model of preemptive genotyping for a single gene is not realistic or feasible in a real-world clinical implementation. As we moved from research funding for genetic testing to clinical billing it became clear that we had to move away from a preemptive model and test only those with a PCI. In these patients, the NNG to identify a patient where an alternative treatment would be recommended was 3.64 (291/80). In comparison, Reese et al. [2012] calculated the NNG to avoid the occurrence of one cardiovascular event based on genotype-guided drug selection as compared to either clopidogrel for all patients or prasugrel for all patients, using case probabilities from the Trial to Assess Improvement in Therapeutic Outcomes by Optimizing Platelet Inhibition with Prasugrel-Thrombolysis in Myocardial Infarction (TRITON-TIMI) 38 studies. The resultant NNGs to prevent one excess cardiovascular event were 23 (genotype-guided therapy compared to clopidogrel for all) and 30 (genotype-guided therapy compared to prasugrel for all). In other words, 23 and 30 patients, respectively, would need to be genotyped to avoid one excess cardiovascular event as compared to clopidogrel- and prasugrel-only strategies [Mega et al., 2009a,b; Reese et al., 2012]. To address cost-effectiveness challenges within our program in year 1, we engaged the UF Bureau of Economics and Business Research to conduct a benefit-cost analysis, which included event and treatment costs and preliminary program data. The resulting analysis and its conclusions were useful in demonstrating the short- and long-term cost impacts of the program to hospital leadership.
We also experienced some unanticipated challenges with the genotyping array selected for this program. The Life Technologies Open Array platform has historically been used in the research laboratory setting and was utilized in this program as the platform for a Laboratory Developed Test. The adaptation of research instruments and tests in a clinical laboratory presented certain challenges. As an example, the provision from Life Technologies of frequent software updates, which in a research laboratory might be positive, requires regulatory revalidation of the assay each time such a change is made in a clinical laboratory. Better understanding among manufacturers of the CAP/CLIA laboratory requirements as manufacturers migrate their products from purely research usage into the clinical laboratory setting will be important to the long term success of genomic medicine. Additionally, there were unexpected changes in the manufacturing of the array that required us to move away from the Open Array product in the later months of year 1. We continue to work to solve these technical challenges, and recognize that such roadblocks may occur for any early adopters of genomic medicine.
In summary, we launched a genomic medicine program focused in pharmacogenetics, with the initial pilot targeting clopidogrel-CYP2C19. During the first year, over 1,000 genetic test results were reported into the medical record and 56 post-PCI patients had their therapy changed as a result of the genetic information. The data clearly show increased rates of adoption over the first year, and by the final 2 months of year 1, nearly all PCI patients had a CYP2C19 genetic test ordered. While certain expected and unexpected challenges were encountered, the program was deemed a success and additional pharmacogenetic implementations are planned for the coming year.
Acknowledgments
Grant sponsor: National Institutes of Health; Grant numbers: UL1 TR000064; U01 HL105198; U01 GM074492; U01 HG007269.
This work was supported in part by grants for the National Institutes of Health to J.A.J. and D.N.R., including UL1 TR000064, U01 HL105198, U01 GM074492, and U01 HG007269. We also acknowledge and thank the University of Florida and UF Health Shands Hospital for in-kind support of the Personalized Medicine Program and the large number of faculty and staff who made the pilot genomic medicine implementation a success.
Biographies
Kristin W. Weitzel, PharmD is Associate Director, UF Health Personalized Medicine Program and Clinical Associate Professor, Pharmacotherapy and Translational Research. Her research and practice interests include clinical implementation of pharmacogenetics and educational innovation.
Amanda R. Elsey, M.H.A. is Program Manager, UF Health Personalized Medicine Program.
Taimour Y. Langaee, M.S.P.H., Ph.D. is Research Associate Professor and Graduate Coordinator, Pharmacotherapy and Translational Research and Director of the Genotyping Laboratory, Center for Pharmacogenomics and Director of the CTSI Genotyping Core. His research interests are pharmacogenomics in cardiovascular disease; pharmacogenetics of dichloroacetate; clinical, genetic, and microbial interactions in inflammatory bowel disease; and microbial resistance to antibiotics.
Benjamin Burkley, M.S. is Scientific Research Manager, Center for Pharmacogenomics. His research focuses on metabolic and cardiovascular pharmacogenomics.
David R. Nessl, P.M.P., M.B.A., is Chief Technology Officer, Department of Pathology, UF College of Medicine. His primary interest is applying project management to building high-tech businesses.
Aniwaa Owusu Obeng, PharmD is a member of the Charles Bronfman Institute for Personalized Medicine and Assistant Professor of Medicine, General Internal Medicine at the Icahn School of Medicine at Mount Sinai. She also serves as Clinical Pharmacogenomics Coordinator in the Department of Pharmacy at The Mount Sinai Medical Center. At the time of implementation and data collection, Dr. Obeng was a pharmacogenomics resident in the Center for Pharmacogenomics and UF Health.
Benjamin J. Staley, PharmD is a Clinical Pharmacist, UF Health and adjunct Clinical Assistant Professor in the Department of Pharmacotherapy and Translational Research, College of Pharmacy. He specializes in quality improvement, patient safety and quality initiatives, and use of information technology to improve clinical processes.
Hui-Jia Dong, Ph.D., H.C.L.D. (A.B.B.) is Technical Director, UF Health Pathology Laboratories. Her work primarily focuses on implementing new technologies in molecular laboratory diagnostics, new test development, and laboratory management.
Robert W. Allan, M.D. is Medical Director, UF Health Pathology Laboratories and Associate Professor of Genitourinary Pathology, Hematopathology, and Surgical Pathology, UF College of Medicine. His research interests are principally in translational research for prostate, bladder, and kidneys.
J. Felix Liu, Ph.D. is Director, Clinical Translational Science Information Technology Program and Courtesy Research Assistant Professor, Department of Pathology, Immunology and Laboratory Medicine, UF College of Medicine.
Rhonda Cooper-DeHoff, PharmD, M.S. is Associate Professor, Departments of Pharmacotherapy and Translational Research and Pharmaceutics, College of Pharmacy and Division of Cardiovascular Medicine, College of Medicine. She is Associate Director of the Center for Pharmacogenomics. Her research interests include hypertension, metabolic syndrome, diabetes, and pharmacogenomics.
R. David Anderson, M.D. is Associate Professor of Medicine, Director of Interventional Cardiology, and Director of the Cardiac Catheterization Laboratory, College of Medicine and UF Health.
Michael Conlon, Ph.D. is Director of Biomedical Informatics, Associate Director, and Chief Operating Officer of the UF CTSI.
Michael J. Clare-Salzler, M.D. is Professor and Chair, UF Department of Immunology, Pathology, and Laboratory Medicine. His research focuses on establishing the cellular, molecular, and genetic basis for the immunopathogenesis of autoimmune endocrine diseases.
David R. Nelson, M.D. is Professor of Medicine, Molecular Genetics, and Microbiology, Director of the CTSI, and Associate Dean for Clinical Research in the Division of Gastroenterology, Hepatology, and Nutrition, College of Medicine. He leads the CTSI and advances clinical/translational research efforts on campus. His personal research interests are immunopathogenesis and treatment of chronic hepatitis C and hepatocellular carcinoma.
Julie A. Johnson, PharmD is Director, UF Health Personalized Medicine Program, Distinguished Professor of Pharmacy and Medicine, and Dean of the College of Pharmacy. Her research focus is pharmacogenomics, with particular focus on cardiovascular drugs, and clinical implementation of pharmacogenomics.
References
- Crews KR, Hicks JK, Pui CH, Evans WE. Pharmacogenomics and individualized medicine: Translating science into practice. Clin Pharmacol Ther. 2012;92:467–475. doi: 10.1038/clpt.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrugia G, Weinshilboum RM. Challenges in implementing genomic medicine: The Mayo Clinic Center for Individualized Medicine. Clin Pharmacol Ther. 2013;94:204–206. doi: 10.1038/clpt.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AB, Oishi S, Yano EM, Gammage CE, Marshall NJ, Scheuner MT. Factors influencing organizational adoption and implementation of clinical genetic services. Genet Med. 2014;16:238–245. doi: 10.1038/gim.2013.101. [DOI] [PubMed] [Google Scholar]
- Johnson JA, Burkley BM, Langaee TY, Clare Salzler MJ, Klein TE, Altman RB. Implementing personalized medicine: Development of a cost-effective customized pharmacogenetics genotyping array. Clin Pharmacol Ther. 2012a;92:437–439. doi: 10.1038/clpt.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JA, Roden DM, Lesko LJ, Ashley E, Klein TE, Shuldiner AR. Clopidogrel: A case for indication-specific pharmacogenetics. Clin Pharmacol Ther. 2012b;91:774–776. doi: 10.1038/clpt.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon MB, Baim DS, Popma JJ, Gordon PC, Cutlip DE, Ho KKL, Giambartolomei A, Diver DJ, Lasorda DM, Williams DO, Pocock SJ, Kuntz RE. A clinical trial comparing three antithrombotic-drug regimens after coronary-artery stenting. N Engl J Med. 1998;339:1665–1671. doi: 10.1056/NEJM199812033392303. [DOI] [PubMed] [Google Scholar]
- Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, Walker JR, Antman EM, Macias W, Braunwald E, Sabatine MS. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009a;360:354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, Walker JR, Antman EM, Macias WL, Braunwald E, Sabatine MS. Cytochrome P450 genetic polymorphisms and the response to prasugrel: Relationship to pharmacokinetic, pharmacodynamic, and clinical outcomes. Circulation. 2009b;119:2553–2560. doi: 10.1161/CIRCULATIONAHA.109.851949. [DOI] [PubMed] [Google Scholar]
- Mega JL, Simon T, Collet J, Anderson JL, Antman EM, Bliden K, Cannon CP, Danchin N, Giusti B, Gurbel P, Horne BD, Hulot J, Kastrati A, Montalescot G, Neumann F, Shen L, Sibbing D, Steg PG, Trenk D, Wiviott SD, Sabatine MS. Reduced-function CYP2C219 genotype and risk of adverse clinical outcomes among patients treated with clopidgrel predominantly for PCI: A meta-analysis. JAMA. 2010;304:1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MJ, Berger PB, Teirstein PS, Tanguay JF, Angiolillo DJ, Spriggs D, Puri S, Robbins M, Garratt KN, Bertrand OF, Stillabower ME, Aragon JR, Kandazari DE, Stinis CT, Lee MS, Manoukian SV, Cannon CP, Schork NJ, Topol EJ GRAVITAS Investigators. Standard- vs high-dose clopidogrel based on platelet function testing after percutaneous coronary intervention: The GRAVITAS randomized trial. JAMA. 2011;305:1097–1105. doi: 10.1001/jama.2011.290. [DOI] [PubMed] [Google Scholar]
- Price MJ, Murray SS, Angiolillo DJ, Lillie E, Smith EN, Tisch RL, Schork NJ, Teirstein PS, Topol EJ GIFT Investigators. . Influence of genetic polymorphisms on the effect of high- and standard-dose clopidogrel after percutaneous intervention: The GIFT (Genotype Information and Functional Testing) study. J Am Coll Cardiol. 2012;59:1928–1937. doi: 10.1016/j.jacc.2011.11.068. [DOI] [PubMed] [Google Scholar]
- Pulley JM, Denny JC, Peterson JF, Bernard GR, Vnencak-Jones CL, Ramirez AH, Delaney JT, Bowton E, Brothers K, Johnson K, Crawford DC, Schildcrout J, Masys DR, Dilks HH, Wilke RA, Clayton EW, Shultz E, Laposata M, McPherson J, Jirjis JN, Roden DM. Operational implementation of prospective genotyping for personalized medicine: The design of the Vanderbilt PREDICT project. Clin Pharmacol Ther. 2012;92:87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese ES, Mullins CD, Beitelshees AL, Onukwugha E. Cost-effectiveness of cytochrome P450 2C19 genotype screening for selection of antiplatelet therapy with clopidogrel or prasugrel. Pharmacotherapy. 2012;32:322–332. doi: 10.1002/PHAR.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SA, Sangkuhl K, Stein CM, Hulot JS, Mega JL, Roden DM, Klein TE, Sabatine MS, Johnson JA, Shuldiner AR. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94:317–323. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Mcleod H, Weinshilbourm RM. Genomics and drug response. N Engl J Med. 2011;364:1143–1153. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]