Abstract
Sirtuins, a family of histone deacetylases, have a fiercely debated role in regulating lifespan. Contrasting recent observations, we here find that overexpression of sir-2.1, the orthologue of mammalian SirT1, does extend C. elegans lifespan. Sirtuins mandatorily convert NAD+ into nicotinamide (NAM). We here find that NAM and its metabolite, 1-methylnicotinamide (MNA), extend C. elegans lifespan, even in the absence of sir-2.1. We identify anmt-1 to encode a C. elegans orthologue of nicotinamide-N-methyltransferase (NNMT), the enzyme that methylates NAM to generate MNA. Disruption and overexpression of anmt-1 have opposing effects on lifespan independent of sirtuins, with loss of anmt-1 fully inhibiting sir-2.1-mediated lifespan extension. MNA serves as a substrate for a newly identified aldehyde oxidase, GAD-3, to generate hydrogen peroxide acting as a mitohormetic ROS signal to promote C. elegans longevity. Taken together, sirtuin-mediated lifespan extension depends on methylation of NAM, providing an unexpected mechanistic role for sirtuins beyond histone deacetylation.
Sirtuins1 have been identified in yeast2,3 to be crucial for lifespan extension in states of glucose restriction4,5, while some, including our own, subsequent publications were unable to confirm these glycolysis-derived effects6,7. Likewise, some authors demonstrate lifespan extension following over-expression of sirtuins in C. elegans8–10 and Drosophila11,12, whereas others found no significant effect on longevity in either of these organisms13. Impaired sirtuin expression has been linked to reduced longevity in mice1,14, and a recent publication demonstrated that over-expression of the sirtuin isoform SIRT6 extends lifespan in male mice15.
Sirtuins are a family of histone deacetylases that mandatorily require nicotinamide adenine dinucleotide (NAD+) as a co-substrate. The latter is derived from L-tryptophan de novo, from nicotinic acid (NA) or from nicotinamide (NAM) (a.k.a. niacin or vitamin B3)16, all of which are approved drugs or food supplements. During sirtuin-mediated deacetylation of L-lysine residues, NAD+ is converted into NAM and O-acetyl-ADP-ribose. NAM is either recycled into NAD+ or other NA derivatives employing the NAD+ salvage pathway, or it is methylated by nicotinamide-N-methyltransferase (NNMT, EC 2.1.1.1)17 to 1-methylnicotinamide (MNA) (Fig. 1).
Figure 1. Role of Sirtuins within Metabolism of Nicotinic Acid.
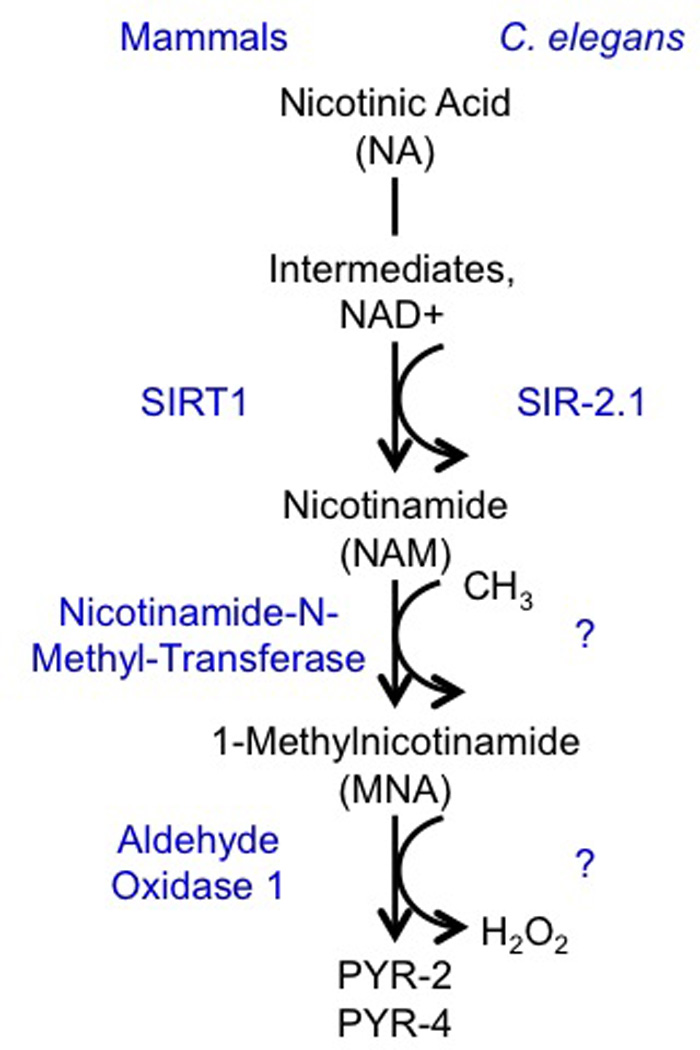
Metabolites are given in black letters, enzymes are given in blue letters.
Published evidence suggests that MNA may promote formation of reactive oxygen species (ROS) by inhibiting complex I of the respiratory chain18. As reviewed elsewhere, increased ROS formation promotes lifespan and metabolic health19.
Given the ROS-inducing capabilities of MNA, we here tested the hypothesis that sirtuins may exert longevity-promoting effects by providing increased amounts of the MNA-precursor NAM, a freely available food supplement, to generate a lifespan-extending ROS signal. We find that sirtuin-mediated lifespan extension fully depends on methylation of NAM to generate a ROS signal that, as with other interventions, is the key cause for longevity.
Results
Effects of NA, NAM, and MNA on C. elegans lifespan
Previously published findings in S. cerevisiae indicate that 5 mM NAM decreases yeast lifespan by impairing activities of the yeast sirtuin SIR2 and pyrazinamidase/nicotinamidase 1 (PNC1)5, while 25 mM NAM decreases nematode lifespan20. We confirm that NAM decreased C. elegans lifespan at 25 mM (Supplementary Results, Supplementary Fig. 1a and Suppl. Table 1 Consistent with a putative role of MNA in the biochemical execution of high-dose NAM supplementation (see above), we find that 1 mM MNA impaired C. elegans lifespan (Suppl Fig. 1b).
We next performed HPLC analysis aiming to quantify the amount of endogenous MNA in unsupplemented adult C. elegans. We observed MNA concentrations in the high nanomolar range (data not shown), though the detection limit of the method prevented us from an exact quantification. We exposed nematodes to 1 µM MNA, and observed an extension of lifespan (Fig. 2a), contrasting the lifespan-reducing effects of MNA at a thousand-fold higher dose described above. The MNA-precursor NAM similarly extended lifespan at the lower dose of 100 µM (Fig. 2b) in contrast to the 250-fold higher dose described above. MNA concentrations tenfold higher or lower than 1 µM showed no detectable effects on lifespan (Suppl. Fig. 1d).
Figure 2. Effects of nicotinic acid (NA), nicotinamide (NAM), and 1-methylnicotinamide (MNA) on C. elegans lifespan in the presence and absence of sir-2.1.
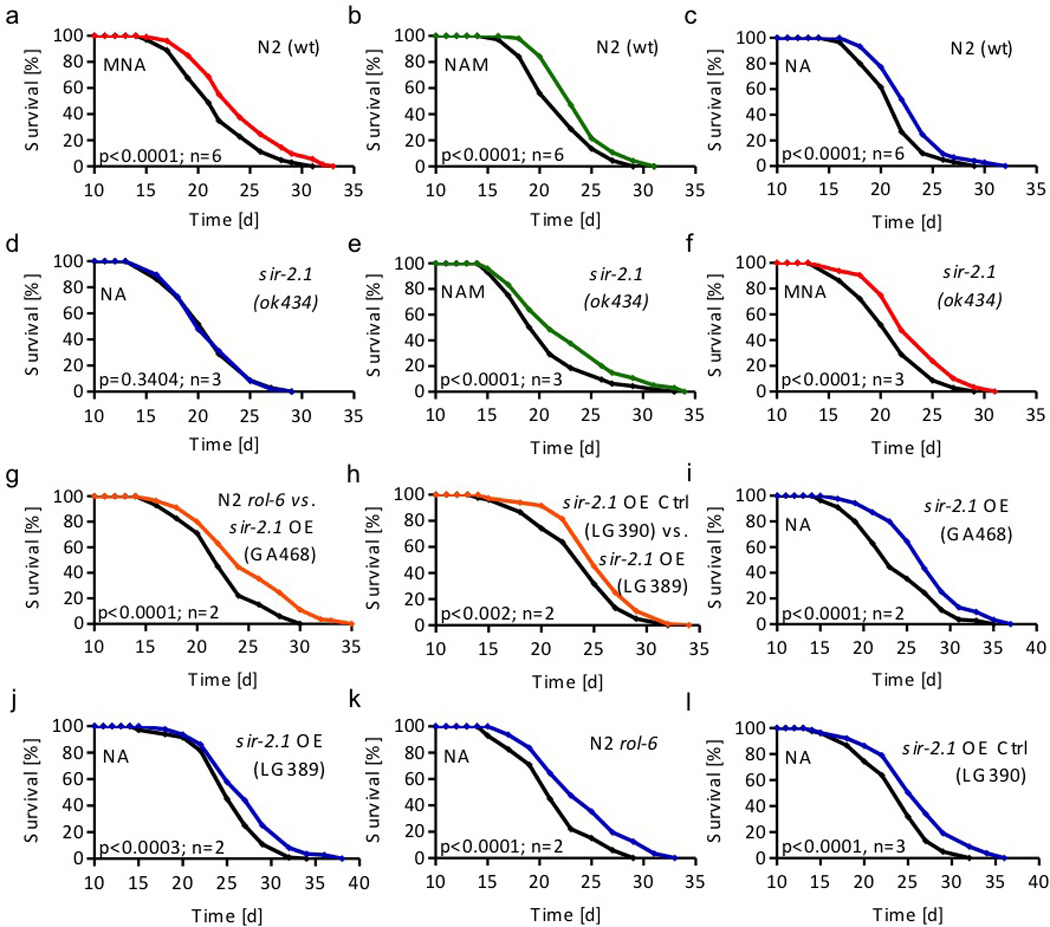
Lifespan analyses of a wild-type (wt) nematodes exposed to 1 µM MNA (red) compared with untreated worms (black); b of wt nematodes exposed to 100 µM NAM (green); c of wt nematodes exposed to 1 mM NA (blue); d of sir-2.1(ok434) nematodes exposed to 1 mM NA; e of sir-2.1(ok434) nematodes exposed to 100 µM NAM; f of sir-2.1(ok434) nematodes exposed to 1 µM MNA; g of sir-2.1 overexpressing nematodes (strain GA468; orange) compared to rol-6 control worms (black); h of sir-2.1 overexpressing nematodes (strain LG389; orange) compared to control worms (strain LG390; black); i of sir-2.1 overexpressing nematodes (strain GA468) exposed to 1 mM NA; j of control LG390 nematodes exposed to 1 mM NA; k of rol-6 control nematodes exposed to 1 mM NA; l of control LG390 nematodes exposed to 1 mM NA. All data were expressed as mean values with n representing the number of independent experiments. Please find further information to statistical analyses in Suppl. Table 1 (also applies to all following C. elegans lifespan assays).
Nicotinic acid (NA) is converted to nicotinic acid mononucleotide (NAMN), then to NA adenine dinucleotide (NAAD), and finally to NAD+ via the NAD salvage pathway16. Sirtuins then generate NAM from NAD+. We tested the effect of NA on C. elegans lifespan, and observed longevity-extending effects at a 1 mM NA (Fig. 2c). Consistent with this, supplementation with large amounts of the highly unstable NAD+ has been previously shown to extend nematodal lifespan21. Taken together, these findings indicate that NA and its metabolites, NAM and MNA, uniformly extend C. elegans lifespan at physiologically relevant concentrations.
Supplementation of NAD+ extends C. elegans lifespan in a fashion that depends on SIR-2.121, the nematodal orthologue of SirT18. We exposed sir-2.1-deficient nematodes to 1 mM NA as above and found the lifespan-extending effects of NA to be abolished (Fig. 2d). This suggests that the conversion of NA/NAD+ to NAM is essential for the effects of NA on lifespan extension (Fig. 1c), also consistent with the findings of Hashimoto and colleagues21.
Surprisingly, supplementation of sir-2.1-deficient nematodes with NAM (100 µM) or MNA (1 µM) still extended lifespan (Figs. 2e and f). These findings, and especially those on MNA in sir-2.1-deficient worms (Fig. 2f), are in conflict with the assumption that sirtuins exert their lifespan-extending effects via increased histone deacetylation activity only. These findings rather suggest that a crucial role of sirtuins in lifespan-extension is the conversion of NA/NAD+ into NAM, to ultimately promote formation of MNA, whereas the roles of deacetylation processes has not been evaluated in the current study.
Overexpression of sir-2.1 in nematodes has been shown to extend lifespan more than a decade ago8. A recent publication13 using sir-2.1 transgenic worms derived from the initial study8, however then backcrossing them for several generations to avoid unspecific effects, was unable to confirm these results, i.e. no extension of lifespan was observed13. These backcrossed nematodes have been named GA468. Independently, another backcross named LG389 was performed, still showing lifespan extension9 supporting the initial results8 and opposing the findings in GA46813.
We have repeated these experiments and find that both strains, i.e. GA468 and LG389 are long-lived in comparison to the respective controls, N2 rol-6 and LG390 (Fig. 2g,h). However, supplementation of both sir-2.1-overexpressing strains with NA did not extend lifespan further (Fig. 2i,j) than NA did in controls (Fig. 2a, k and l), indicating that overexpression of sir-2.1 and NA supplementation share a functional denominator.
The methyltransferase ANMT-1 controls longevity
The findings outlined above suggest that conversion of NAM to MNA is an essential step for the extension of lifespan by NA and NAM. In mammals, methylation of NAM is executed by N-methyl-N-transferase (NNMT, EC 2.1.1.1)17. We identified a putative orthologue in C. elegans, now named amine-N-methyltransferase-1 (ANMT-1), by orthology search. We exposed nematodes deficient for this enzyme, now named anmt-1(gk457), to the previously applied concentrations of NA, NAM, and MNA. We found that impairment of anmt-1 abolished the effects of NA and NAM, while the product of ANMT-1, MNA, is still capable of extending lifespan (Figs. 3a to c). These findings indicate that formation of MNA is required to extend lifespan in states of supplementation with NA or NAM, and shows that ANMT-1 acts as a methyltransferase to form MNA from NAM.
Figure 3. Disruption and overexpression of nicotinamide-N-methyltransferase/ANMT-1 indicate that 1-methylnicotinamide (MNA) is key regulator of longevity in wild-type and sir2.1-overexpressing nematodes.
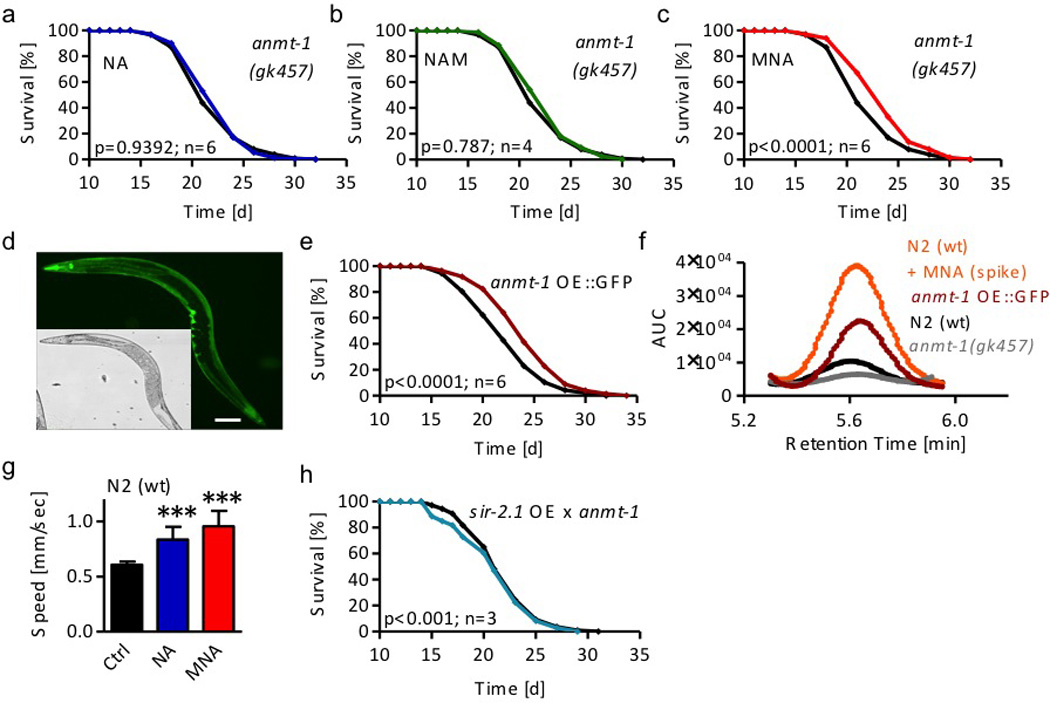
Lifespan analyses a of anmt-1(gk457) nematodes exposed to 1 mM nicotinic acid (NA; blue); b of anmt-1(gk457) nematodes exposed to 100 µM nicotinamide (NAM; green); c of anmt-1(gk457) nematodes exposed to 1 µM MNA (red). d Expression pattern of GFP as a fused surrogate marker of ANMT-1 protein expression in anmt-1 OE::GFP. Scale bar, 100 µm. e Lifespan analyses of anmt-1 OE::GFP (dark red) compared with wt nematodes (black). f HPLC-derived MNA signals in anmt-1(gk457) nematodes (grey), unsupplemented wt worms (black) and MNA-spiked wt extracts (orange) as well as unsupplemented anmt-1 OE::GFP nematodes (dark red). g Average crawling speed of wildtype (wt) nematodes exposed to NA and MNA expressed as mean values with standard deviation of 3 independent experiments and 10 examined nematodes per condition each. h Lifespan analyses of sir-2.1 overexpressing and anmt-1 deficient nematodes (sir-2.1 OE × anmt-1, strain MIR22; turquoise) compared to rol-6 control worms (black).
Conversely, we generated and studied two transgenic strains over-expressing ANMT-1 under the control of its endogenous promoter. Both strains express stably integrated transgenes: one construct was C-terminally fused to a GFP-encoding cDNA (anmt-1 OE::GFP) (Suppl. Fig. 2a), whereas in the other construct, GFP was replaced by a nine-amino acid hemagglutinin tag (anmt-1 OE::HA) (Suppl. Fig. 2b). We confirmed the expression and protein size of both transgenes by immunoblotting (Suppl. Figs. 2c–h). Next, we analyzed the expression pattern in whole anmt-1 OE::GFP nematodes by fluorescence microscopy (Fig. 3d and Suppl. Fig. 2i). We then tested life expectancy in unsupplemented worms. Both constructs showed a similar extension of lifespan in comparison to non-transgenic worms (Figs. 3e and Suppl. Fig. 2j), notably in absence of supplementation with NA or NAM.
To analyze the effects of both ANMT-1 disruption and over-expression on NAM methylation, we compared MNA levels in unsupplemented wild-type worms (Fig. 3f, black) and extracts from such wild-type worms that, after extraction, had been spiked with MNA (Fig. 3f, orange) to confirm the specificity of the signal. Moreover, we analyzed ANMT-1-deficient (Fig. 3f, grey) and overexpressing anmt-1 OE::GFP (Fig. 3f, red) nematodes. These findings indicate that MNA production is strongly reduced in ANMT-1-deficient nematodes, and that ANMT-1-overexpression profoundly increases MNA content, again consistent with ANMT-1 functioning as a methyltransferase and orthologue of NNMT.
Next, we tested whether NA or MNA exert effects on food uptake or locomotion. While no changes in pharyngeal pumping rate, reflecting food uptake, were observed (not shown), both NA and MNA increased the average crawling speed of nematodes (Fig. 3g) insinuating increased fitness.
Lastly, we generated anmt-1-deficient but sir-2.1-overexpressing nematodes, called MIR22, to test whether the absence of the methyltransferase would affect lifespan extension mediated by sir-2.1 overexpression. Unlike the parental GA468 strain (Fig. 2g), anmt-1-deficient MIR22 showed reduced longevity (Fig. 3h), indicating that methylation of NAM exclusively explains lifespan extension due to overexpression of sir-2.1.
MNA is a GAD-3 substrate to form hydrogen peroxide
MNA has been previously suggested to be an inducer of reactive oxygen species (ROS)-production apparently by inhibiting complex I of the mitochondrial respiratory chain18.
We now analyzed whether and how 1 µM MNA induces ROS in C. elegans, as this concentration extends lifespan, as shown above. When we exposed wild-type nematodes to MNA for four hours, we observed an increase in ROS formation as quantified by an intracellular fluorophore MitoTracker Red CM-H2X ROS (Fig. 4a), and the AmplexRed reaction (Fig. 4b) which is specific for formation of hydrogen peroxide. Both approaches indicate that MNA induces formation of ROS and more specifically hydrogen peroxide of exposure.
Figure 4. 1-Methylnicotinamide (MNA) serves as a substrate for aldehyde oxidase/GAD-3 to form hydrogen peroxide.
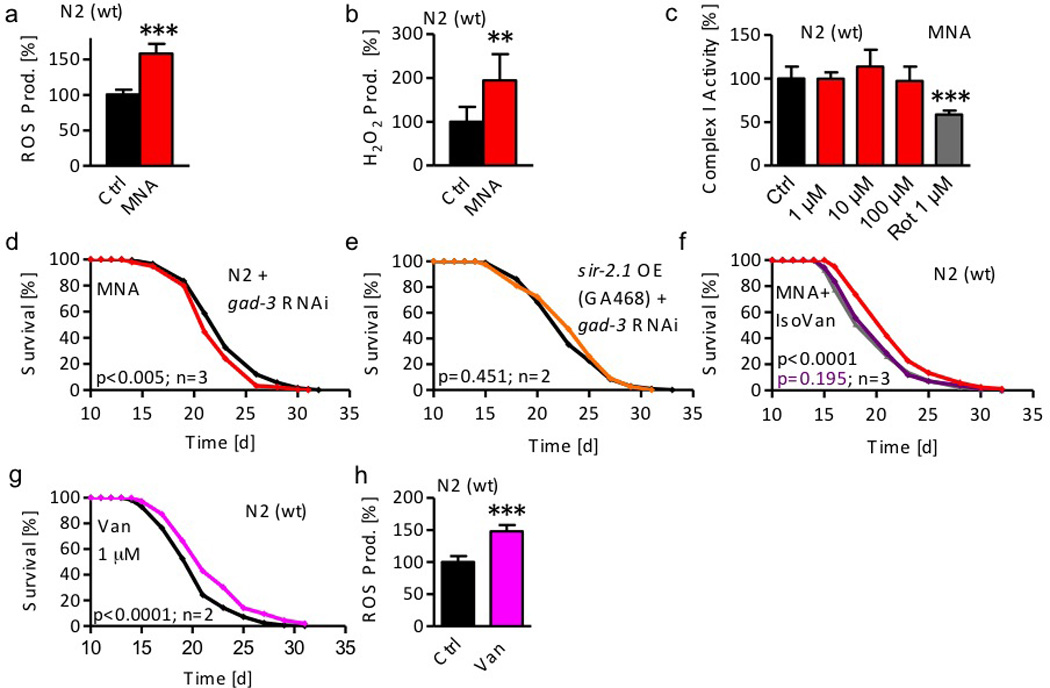
a ROS levels in wild-type (wt) nematodes following exposure to 1 µM MNA for 4 hrs (red bar) compared with untreated nematodes (black bar). b H2O2 production following exposure to 1 µM MNA for 4 hrs. Data represent mean values with standard deviation of at least 2 independent experiments. c Complex I activity after treatment with 1, 10, and 100 µM MNA; the complex I-inhibitor rotenone (1 µM) served as positive control. Data represent mean values with standard deviation of at least 2 independent experiments and n=4 each. Lifespan analyses d of wt nematodes treated with RNAi against gad-3 exposed to 1 µM MNA; e of sir-2.1 overexpressing nematodes (strain GA468) treated with RNAi against gad-3 (orange); f of wt nematodes exposed to MNA (1 µM) in the presence (purple) or absence of IsoVan (red) compared with worms treated with IsoVan only; g of wt nematodes exposed to the AOX1/GAD-3 substrate vanillin (1 µM, pink). h ROS levels in wild-type (wt) nematodes following exposure to 1 µM vanillin for 24 hrs (pink bar) compared with untreated nematodes (black bar). Data represent mean values with standard deviation of 2 independent experiments.
Since MNA has been proposed to inhibit complex I of the mitochondrial respiratory chain18 we next quantified complex I activity in the presence and absence of MNA. While the well-established complex I inhibitor and lifespan-extending22 ROS inducer rotenone served as a positive control, MNA unexpectedly did not affect complex I activity (Fig. 4c) rejecting the hypothesis that MNA generates a ROS signal by inhibiting complex I.
MNA is known to be metabolized by the phase I detoxification enzyme aldehyde oxidase (AOx1, EC 1.2.3.1). Oxidation of MNA by AOx1 forms two metabolites, named 1-methyl-2-pyridone-5-carboxamide (PYR-2) and 1-methyl-4-pyridone-5-carboxamide (PYR-4) raising the possibility that one of these, or both metabolites may execute the MNA-initiated lifespan extension. However, neither PYR-2 nor PYR-4 extended lifespan of C. elegans at concentrations equimolar to the lifespan-extending concentration of MNA (Suppl. Figs. 3a and b).
Unlike aldehyde dehydrogenases (EC 1.2.1.5 and others), AOx1 produces hydrogen peroxide during oxidation of its multiple, mostly xenobiotic substrates as a by-product23. Given the ROS-(Fig. 4a) and specifically hydrogen peroxide-(Fig. 4b) inducing effects of MNA supplementation we next hypothesized that conversion of MNA to PYR-4 and PYR-2 by AOx1 may cause this increase in ROS formation. By homology search we identified the C. elegans GAD-3 protein24 as an AOx1 orthologue. Inhibiting gad-3 expression by RNAi fully abolished the effects of MNA on lifespan (Fig. 4d) indicating that MNA serves as a substrate for GAD-3 to form a hydrogen peroxide-mediated ROS signal that may exert lifespan-extending properties, as further analyzed below. Likewise, when treating sir-2.1-overexpressing GA468 nematodes with RNAi against gad-3, the lifespan-extending effect of sir-2.1 overexpression (Fig. 2g) was abolished (Fig. 4e) suggesting that MNA-mediated formation of hydrogen peroxide by AOX1/GAD-3 explains sirtuins-mediated lifespan-extension.
Independently, we used a specific AOx1-inhibitor, iso-vanillin25, to test whether MNA-mediated lifespan extension depends on GAD-3. While iso-vanillin had no effect on lifespan per se (Suppl. Fig. 3c), co-application of iso-vanillin abolished the effects of MNA on lifespan (Fig. 4f) again indicating that oxidation of MNA by AOx1/GAD-3 is required for lifespan extension.
Lastly, we tested whether an established AOx1/GAD-3 substrate other than MNA may exert lifespan-extending properties in C. elegans. When applying 1 µM of the established AOx1 substrate vanillin26 to C. elegans, we observed an extension of lifespan (Fig. 4g), resembling findings on MNA (Fig. 2a) at the same concentration. Moreover and like MNA, vanillin caused an increase in ROS formation (Fig. 4h). Using a thousand-fold higher dose, and again resembling findings on MNA (Suppl. Fig. 1b), supplementation of worms with 1 mM vanillin shortened lifespan (Suppl. Fig. 1c). These findings indicate that MNA and other substrates of the phase I detoxifying enzyme AOx1/GAD-3 generate hydrogen peroxide and extend lifespan at low doses, whereas high doses are lifespan shortening, reflecting non-linear, i.e. mitohormetic, dose-response characteristics.
MNA induces a transient ROS signal to extend lifespan
We next analyzed whether and how NA or MNA induce ROS in C. elegans at the lifespan-extending concentrations of 1 mM and 1 µM (Figs. 1a and c), respectively, in a time-resolved manner. When we exposed wild-type nematodes to NA (Fig. 5a [relative] and Suppl. Fig. 4a [absolute]) or MNA (Fig. 5b and Suppl. Fig. 4b) at the respective lifespan-extending concentrations for four hours, we observed a transient increase in ROS formation, which was undetectable at 12 or 24 hours after initiation of exposure. Interestingly, at 48 hours and beyond, i.e. in the steady-state, a persistent reduction of ROS levels was observed following exposure to either compound (Figs. 5a and b). Co-treatment with gad-3 RNAi abolished the increase in ROS production after short-term incubation with the respective compound (Figs. 5a and b) and prevented the reduction of ROS levels in the steady-state (Figs. 5a and b).
Figure 5. 1-Methylnicotinamide (MNA) induces a transient ROS signal which is crucial for C. elegans lifespan extension.
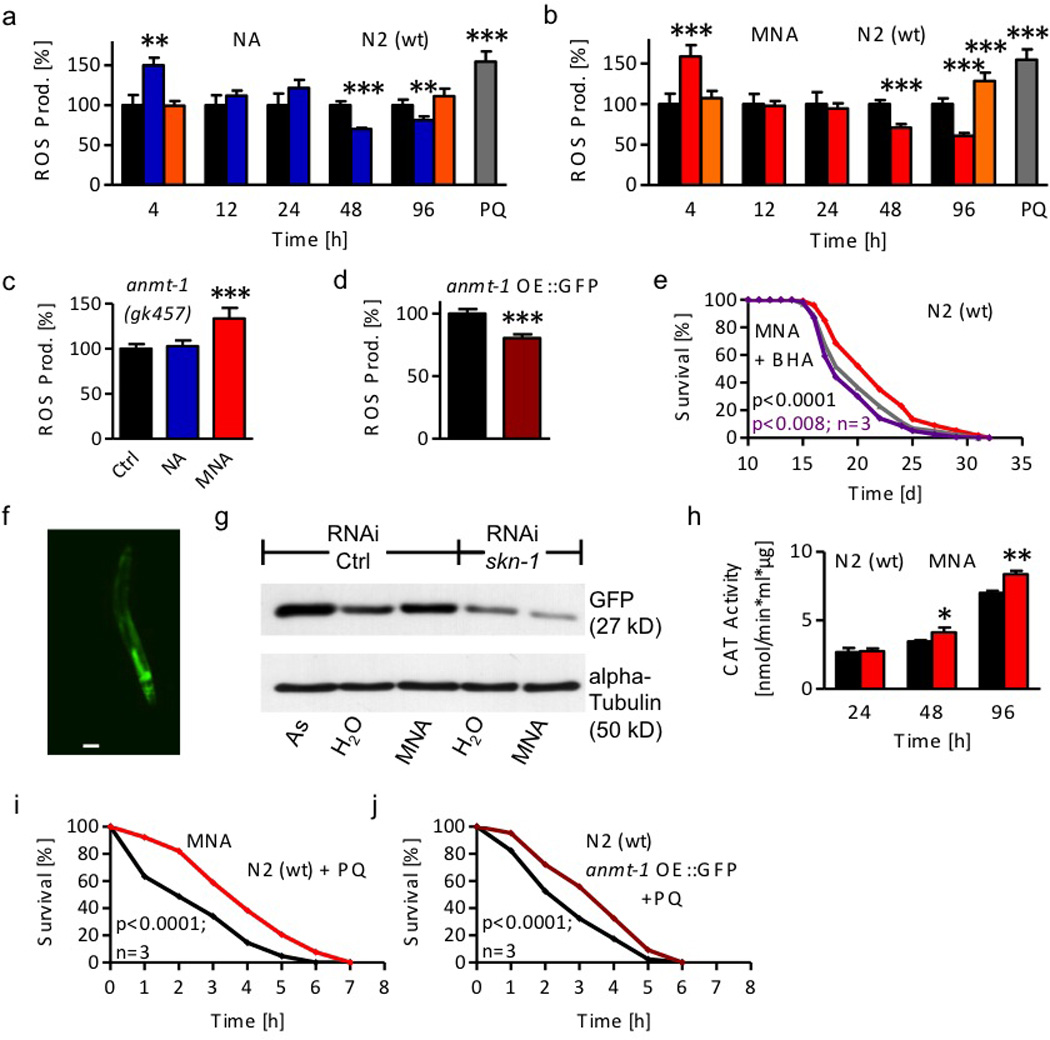
a ROS levels following 1 mM nicotinic acid (NA; blue bars) exposure and co-treatment with gad-3 RNAi (orange bars) compared with untreated wild-type (wt) nematodes (black bars) at different time points. 1 hr paraquat treatment (PQ, grey bar) acts as positive control. b ROS levels following 1 µM MNA (red bars) exposure at different time points. c ROS levels following 4 hr NA and MNA exposure of anmt-1(gk457) nematodes. d Constitutive ROS levels of wt and anmt-1 OE::GFP nematodes (dark red). e Lifespan analyses of wt nematodes exposed to MNA in the presence (purple) or absence of the antioxidant BHA (red) compared with BHA-treated worms (grey). f Fluorescent microphotograph (enlargement 10-fold) of gst-4::GFP nematodes after 48hr MNA treatment. Scale bar, 100 µm. g Western blot against GFP resembling GST-4 promoter activation in gst-4::GFP nematodes in the presence or absence of skn-1 RNAi after 48 hr MNA treatment compared with untreated nematodes. 48 hr arsenite (As) treatment acts as positive control. h Activity of catalase (CAT) in wt nematodes exposed to 1 µM MNA (red bars). Data represent mean values with standard deviation of at least 2 independent experiments. i Survival of wt nematodes in liquid medium containing 50 mM paraquat after 7 days MNA exposure in comparison with untreated nematodes. j Survival of anmt-1 OE::GFP nematodes in comparison with wt nematodes. Data were expressed as mean values of 2 independent experiments and 50 examined nematodes/condition each.
Moreover, the high concentrations of NAM, MNA and vanillin that were shown to reduce lifespan (Suppl. Fig. 1a–c) did not, unlike low concentrations, cause a decrease in ROS levels in the steady state, but rather increased ROS levels after exposure of four days (Suppl. Fig. 4c–e). Moreover, microphotographs of nematodes loaded with the ROS-sensitive fluorophore indicated an increase of ROS formation in worms treated with low doses of either NA (Suppl. Fig. 4g) or MNA (Suppl. Fig. 4h) compared to untreated nematodes (Suppl. Fig. 4f). Paraquat, a known ROS generator, was used as positive control (Suppl. Fig. 4i).
We additionally quantified ROS formation by an independent method based on AmplexRed fluorescence that is proportional to the formation of hydrogen peroxide. These data similarly show that both NA and MNA induce formation of hydrogen peroxide four hours after initiation of exposure (Suppl. Fig. 4j).
We tested whether impairment of anmt-1 would affect ROS levels after exposure to NA and MNA, respectively. In congruence with the findings of NA and MNA on lifespan in anmt-1-deficient worms, we found that NA is incapable of inducing ROS in deficient nematodes, while MNA still induces ROS (Fig. 5c). Since both NA as well as MNA caused a reduction of ROS levels in the steady state, we questioned whether constitutively active over-expression of anmt-1 would similarly reduce ROS levels. We found that unsupplemented anmt-1 OE::GFP nematodes have indeed reduced ROS levels (Fig. 5d).
We exposed nematodes to antioxidants as ROS scavengers following exposure to NA or MNA. As previously shown27, the scavengers butylated hydroxyl-anisole (BHA) (Suppl. Fig. 4k) and N-acetyl-cysteine (NAC) (Suppl. Fig. 4m) had no effect on C. elegans lifespan per se. However, the lifespan-extending capabilities of NA and MNA were completely abolished in the presence of BHA and NAC (Fig. 5e and Suppl. Figs. 4l, n,o).
SKN-1/NRF-2 mediates ROS-induced lifespan extension
To identify the molecular mechanism responsible for transducing the ROS signal into extended longevity, we performed next-generation sequencing (RNAseq) of RNA samples extracted from nematodes that were treated with lifespan-extending doses of either NA or MNA in comparison to unsupplemented worms. By applying two independent statistical methods we identified 1447 differentially expressed genes (DEGs) following MNA treatment (Suppl. Fig. 5a and Suppl. Data Set 1), while NA treatment generates 1098 DEGs (Suppl. Fig. 5b and Suppl. Data Set 2). In part to confirm the RNA sequencing data, we performed quantitative real-time PCR analyses of two of these genes, glutathione-S-transferase 4 (gst-4) and a member of the cytochrome P450 family, cyp-29A2. Both analyses fully confirmed the results obtained with RNAseq (Suppl. Fig. 5c and Suppl. Data Sets 1 and 2).
To test whether NA and MNA share a common mechanistic denominator, we analyzed the number of genes similarly regulated by both interventions. We found that 42 per cent of the DEGs up-regulated by NA are also induced by MNA as well as 79 per cent of the down-regulated DEGs (Suppl. Figs. 5d and e), suggesting that a significant portion of genes is similarly regulated by both compounds. We analyzed MNA-dependent DEGs regarding their biological function, and found that genes which are involved into functional categories like secondary metabolism, detoxification and stress response were over-represented (Suppl. Table 2). In particular, we identified a number of specifically regulated DEGs in regards to oxidative stress response that include a large number of glutathione-S-transferase isoforms, as exemplified in Suppl. Fig. 5f.
We applied in silico methods to identify putative promoter elements that respond to those transcription factors known to promote stress response, namely DAF-16/FOXO and SKN-1/NRF-2. We found that half of the DEGs up-regulated following MNA treatment carry putative DNA binding domains for DAF-16 (Suppl. Fig. 5g), while 73 percent of these DEGs are positive for putative SKN-1 DNA binding elements (Suppl. Fig. 5h).
Since the findings presented above suggest that both DAF-16- as well as SKN-1-signaling may be involved in sensing the effects of MNA and hence transiently increased ROS formation, we exposed nematodes deficient for daf-16 to MNA or NA. Consistent with the findings from the RNA analyses (Suppl. Fig. 5g), no extension of lifespan was observed in daf-16-deficient worms by MNA (Suppl. Fig. 6a) or NA (Suppl. Fig. 6b), indicating that genes that are induced by DAF-16 contribute to the effects of MNA on lifespan.
Similarly, we tested both NA and MNA on nematodes that are deficient for all three isoforms of skn-1, the strain skn-1(zu135). Consistent with the findings in daf-16-deficient nematodes, lack of skn-1 similarly abolished the lifespan-extending capabilities of MNA, and also NA (Suppl. Figs. 6c,d). This indicates that genes which are induced by SKN-1 contribute to the effects of NA and MNA on lifespan. We studied skn-1(zu135) nematodes that had been reconstituted for a mainly gut-specific isoform of skn-1, named LG357 skn-1;geIs1028. When exposing these reconstituted nematodes to NA or MNA, both compounds were again capable of inducing lifespan (Suppl. Figs. 6e,f), suggesting that intestinal SKN-1-signaling is sufficient to compensate for globally impaired SKN-1-signaling. It should be noted that the SKN-1 orthologue NRF-2 is known to strongly promote expression of AOx129, i.e. the orthologue of GAD-3, shown above to be responsible for MNA-dependent production of hydrogen peroxide.
We next analyzed a well-established SKN-1 target gene, the above mentioned gst-4, by employing a gst-4 reporter strain where the corresponding promoter is coupled to a GFP cDNA30. We observed increased fluorescence following MNA treatment (Fig. 5f) compared to untreated nematodes (Suppl. Fig. 6g), indicating that expression of gst-4 is induced by the compound, consistent with the RNA expression data above. We also quantified GFP protein expression as a surrogate marker for gst-4 expression in the reporter worms30. The known gst-inducer arsenite, as well as MNA, cause a strong induction of gst-4 in comparison to the solvent. Pre-treatment of the reporter worms with RNAi against skn-1 severely diminished the signal, and abolished the induction of gst-4 by MNA (Fig. 5g, Suppl. Fig. 6h and i).
Independently, we quantified activities of SKN-1-dependent ROS-defense enzymes, superoxide dismutase (SOD) and catalase (CAT), following exposure to MNA. Consistent with the fact that aldehyde oxidases, in particular GAD-3, produce hydrogen peroxide rather than other ROS species from their substrates, including MNA (Fig. 4b), we did not detect any induction of SOD activity by MNA (Suppl. Fig. 6j). By contrast, CAT was induced by MNA (Fig. 5h), consistent with the fact that CAT is the predominant enzyme in C. elegans to detoxify hydrogen peroxide. This indicates that the previously shown reduction of ROS levels in the steady state following NA and MNA exposure (Figs. 5a and b), as well as in unsupplemented anmt-1 OE::GFP nematodes (Fig. 5d), likely are a consequence of increased CAT activity following the corresponding intervention. Moreover, these findings strongly suggest that exposure to MNA increases stress resistance, and in particular resistance to oxidative stress. To test this, we exposed worms treated with NA or MNA to a lethal concentration of paraquat, a known ROS inducer. We observed increased survival following NA (Suppl. Fig. 6k) and MNA (Fig. 5i) exposure, indicating increased stress resistance. Consistently, unsupplemented anmt-1 OE::GFP nematodes also show increased resistance to paraquat (Fig. 5j).
Discussion
Sirtuins, a family of histone deacetylases, have a fiercely debated role in regulating lifespan of different species presumably by altering acetylation patterns to promote longevity. We here provide a distinct mechanism by suggesting that sirtuins have a relevant role in promoting production of MNA to extend lifespan (Fig. 6).
Figure 6. An acetylation-independent mechanism for sirtuin function in extending lifespan.
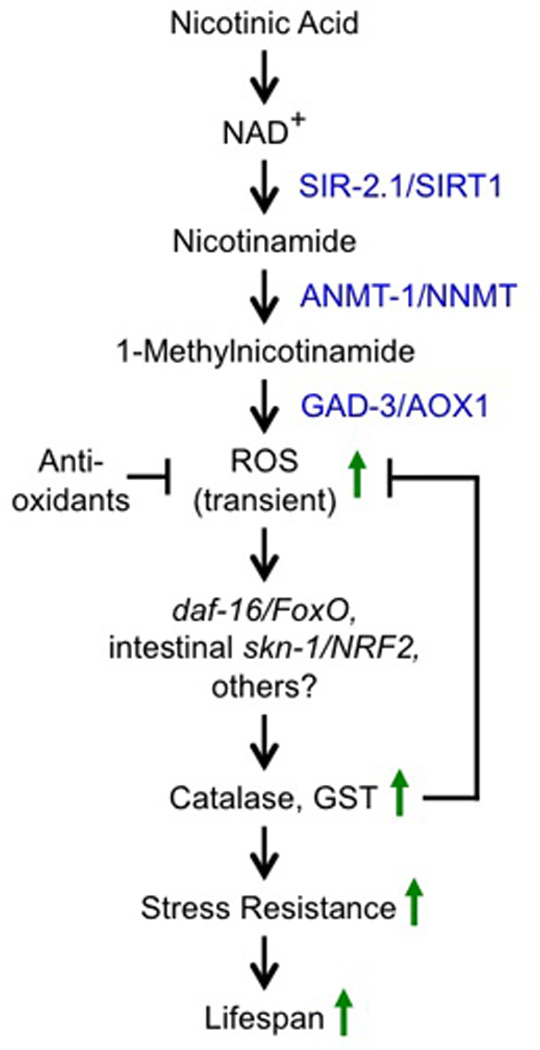
Turnover of the sirtuin cofactor NAD+ to NA and subsequent irreversible methylation to MNA results in generation of hydrogen peroxide by GAD-3 and a downstream mitohormetic response yielding increased stress resistance.
The findings are based on the widely underappreciated fact that sirtuins use NAD+ as an essential co-factor during the deacetylation process, forming the metabolite NAM which becomes methylated by ANMT-1, as established in the current study, to form MNA. The latter serves as a substrate for the newly identified C. elegans AOx1 orthologue, GAD-3, to form hydrogen peroxide which initiates an orchestrated transcriptional response to transiently increased oxidative stress, while it should be noted that additional factors, i.e. beyond increased stress resistance may contribute to lifespan extension.
While the current data were exclusively obtained in C. elegans, it should be noted that in mammals multiple links between different degrees of acetylation, sirtuin activities and functions of distinct proteins, also including histones, have been described, as reviewed elsewhere1. While it appears possible that such changes in acetylation or acylation status impacts life expectancy in mammals, our current data indicate that the absence of ANMT-1 precludes lifespan extension by SIR-2.1 in nematodes, indicating that methylation of NAM is an essential mechanism linking SIR-2.1 activity to lifespan extension. Whether this also applies to other C. elegans sirtuins remains to be evaluated.
Interestingly, ROS-inducing substrates of GAD-3, including MNA, its precursor NAM, and vanillin, exert non-linear effects on lifespan: while low physiological doses are capable of extending lifespan, high and supra-physiological doses exert the opposite effect, i.e. shorten lifespan, presumably by producing an excessive ROS load. A non-linear, a.k.a. hormetic response to ROS31 therefore now appears to be a common denominator for apparently distinct interventions to extend lifespan, including impaired mitochondrial function32,33, calorie restriction7, physical exercise34, impaired TOR signaling35, impaired insulin/IGF-1-signaling27, and sirtuin activation, as shown here.
Sirtuins have been repeatedly proposed to contribute to increased stress resistance and especially increased ROS defense, as reviewed elsewhere36, notably also in regards to a non-linear, i.e. hormetic fashion37. Further supporting this notion, an additional copy of S. cerevisiae SIR2 extends lifespan whereas expression from a high-copy plasmid is toxic3, for at that time unknown reasons. We mechanistically complement these observations solely by supplementing MNA instead of increasing sirtuin activity, providing additional support for the notion that sirtuins exert their lifespan-promoting effects by transiently producing a ROS signal that constitutively induces endogenous ROS defense depending on intestinal SKN-1/NRF-2 and, to a lesser extent, DAF-16/FOXO. Notably, altered FOXO signaling has also been observed in states of sirtuin activation10,38 whereas, to our best knowledge, NRF-2 signaling has not been analyzed in this regard. Transient formation of mitochondrial ROS (mtROS) has been previously linked to the lifespan-extending capabilities of impaired insulin/IGF-1 signaling in a globally SKN-1-dependent fashion27. We here extend this overarching downstream mechanism by demonstrating that the primary formation of hydrogen peroxide acts similar to mitochondrial superoxide formation, while sensing of the supplementation-induced peroxide signal occurs in a intestine-specific fashion.
MNA is currently being examined in clinical trials for various applications, most importantly to lower serum triglycerides39 which contribute to cardio-vascular disease and reduced lifespan in humans40. We would predict that application of exogenous antioxidants would abolish the health-promoting effects of MNA in humans, since the transient ROS signal in C. elegans is essential for the MNA effects, and co-application of antioxidants blocks MNA effects in nematodes. In this regard it is extremely interesting to note that the cholesterol-lowering capacity of the MNA-precursor NA in humans is severely impaired by co-application of antioxidants41, insinuating that NA induces a ROS signal also in humans that contributes to reduced cholesterol levels, strikingly supporting our current mechanistic findings in model organisms (Fig. 6).
Notably, NA has been shown to reduce mortality in humans suffering from cardiovascular disease42 suggesting that methylation of NA metabolites by NNMT as well as MNA supplementation in humans may become versatile approaches to extend human healthspan. Consistently and being known to promote mean lifespan in humans43, physical exercise has been shown to induce both NNMT activity and formation of MNA44.
However and as a note of caution, it should be mentioned that NNMT activity has been positively associated with the degree of malignancy of various cancers, recently also in regards to epigenetic remodeling of malignant cells45. Our data insinuate that this increase in malignancy is linked to increased stress resistance and hence increased chemotherapy defense of cancer cells following endogenous activation of NNMT by positive selection. Interestingly, this has been experimentally shown elsewhere for such cells46, ideally complementing our findings on increased paraquat resistance in nematodes following increased formation of MNA.
In summary, we have shown that sirtuins contribute to lifespan extension in C. elegans, supported by recently published findings47. Unexpectedly this occurs in an acetylation-independent manner, by instead promoting the metabolic formation of a pro-oxidative substrate, 1-methylnicotinamide (MNA), to induce a phase I - mediated ROS response depending on NNMT/ANMT-1 as well as AOx1/GAD-3.
Online Methods
Chemicals
All chemicals were obtained from Sigma-Aldrich (Munich, Germany) unless stated otherwise. The MNA metabolites PYR-2 and PYR-4 were obtained from TLC Pharmachem (Mississauga, Canada).
Statistical analyses
Data are expressed as means ± SD unless otherwise indicated. Statistical analyses for all data except life-span and stress resistance assays were performed by Student’s t-test (unpaired, two-tailed) or ANOVA after testing for equal distribution of the data and equal variances within the data set. For comparing significant distributions between different groups in the life-span assays and stress resistance assays, statistical calculations were performed using JMP software version 9.0 (SAS Institute Inc., Cary, NC, USA) applying the log-rank test. All other calculations were performed using Excel 2007 and 2010 (Microsoft, Albuquerque, NM, USA). A P-value below 0.05 was considered as statistically significant.
Nematode strains and maintenance
The following C. elegans strains used for this publication were provided by the Caenorhabditis Genetics Center (CGC, Univ. of Minnesota, USA) except for LG389 and LG390 which were kind gifts of Dr. L. Guarente and Dr. M. Viswanathan:
Bristol N2 (wild type), HE1006 rol-6(su1006), EU31 skn-1(zu135), LG357 skn-1(zu135) IV/nT1[qIs51] (IV;V); geIs10 [ges-1p(long)::skn-1c::GFP + rol-6(su1006)], GA468 geIs3[sir-2.1(+) + rol-6(su1006)], CF1038 daf-16(mu86), CL2166 dvIs19[pAF15(gst-4::GFP::NLS)]. Furthermore, MIR14 and MIR16 were obtained by outcrossing strains VC199 sir-2.1(ok434) and VC1061 anmt-1(gk457) respectively four times each to wild type strain Bristol N2. MIR22 was generated by crossing the anmt-1 K.O. strain MIR16 with the sir-2.1 overexpressing strain GA468. Additionally, we used the transgenic strains MIR8 risIs1[anmt-1p::anmt-1::GFP + unc119(+)] and MIR12 risIs2[anmt-1p::anmt-1::HA + unc119(+)] both generated as described below. All newly generated MIR strains (idem sunt MIR8, 12, 14, 16, 22) have been deposited at the CGC under these names.
Compound treatment
Treatment of C. elegans was carried out on NGM agar plates containing the respective compounds. All agar plates were prepared from the same batch of NGM agar, whereas treatment plates were supplemented with the respective compound and control plates with water. After plates were poured and dried for about 30 min, they were sealed and stored at 4°C. Freshly prepared E. coli OP50 were spotted on plates on the previous evening and allowed to dry and grow overnight.
Incubations with compounds started 64 h after synchronization of the population, by washing the synchronized, young adult worms and transferring them to the respective treatment plates using S-buffer.
Treatments with the antioxidants N-acetylcysteine (NAC) and butylated hydroxyanisole (BHA) were performed exactly as previously described27.
Lifespan assays
All lifespan assays were performed at 20°C according to standard protocols and as previously described7. Briefly, a C. elegans population was synchronized as described above at day zero of the lifespan. 64h after egg preparation around 100 nematodes were manually transferred to fresh incubation plates containing the respective compounds. Experiments were conducted in triplicates. For the first 10–14 days, worms were transferred every day and afterwards every second day. Nematodes that show no reaction to gently stimulation were scored as death. Those animals that crawled off the plates or displayed non-natural death particularly due to internal hatching were censored.
RNAi-mediated gene knock-down experiments
For RNAi gene knock-down experiments we applied E. coli HT115 to the worms as previously described7. The clones for skn-1 and gad-3 RNAi were derived from the Ahringer library (Source BioScience, Nottingham, UK) and were sequenced before use. The bacteria were spotted on NGM plates containing additionally 1 mM IPTG, 100 µg/ml Ampicillin and, if required, 12.5 µg/ml tetracycline (all from Applichem, Darmstadt, Germany).
Generation of anmt-1 overexpressing strains
The cDNA of anmt-1 constructs was obtained from GenScript (Piscataway, NJ, USA), modified according to Suppl. Fig. 2, including cloning into a modified pBluescript vector containing the unc-119 rescue gene48. The anmt-1 constructs (Suppl. Fig. 2) were transformed into the unc-119-deficient strain BR584 by bombardment as described48 using a biolistic particle delivery system (PDS-1000/He, Bio-Rad, Hercules, CA, USA).
We obtained a stable insertion for both constructs into the genome, as confirmed by PCR-based offspring analysis over several generations. These strains have been backcrossed five times for the GFP-tagged, and four times for the hemagglutinin-tagged construct. The backcrossed strain MIR8 (risIs1 [anmt-1p::anmt-1::GFP+unc119 (+)]) as well as the strain MIR12 (risIs1 [anmt-1p::anmt-1::HA+unc119(+)]) have been used for experiments.
For genotyping and identification of anmt-1::GFP and anmt-1::HA positive worms we used fluorescence microscopy as described below or conducting single worm PCR analyses (Primer: anmt-1::GFP fwd GGC GAC AAC TGC CGA ACA AGG and rev ACC TTC GGG CAT GGC ACT CTT; anmt-1::HA fwd GCG ACA ACT GCC GAA CAA GGA and rev CAC GGG CGC GAG ATG CTG AA).
Immunoblotting
Nematodes were washed three times with ice-cold S-Buffer and pellets were shock frozen in liquid nitrogen. Frozen pellets were grinded in a nitrogen-chilled mortar and suspended in phosphate buffer containing protease and phosphatase inhibitors (Complete protease inhibitor cocktail [Roche, Penzberg, Germany] and additionally 2 mM sodiumfluoride, 2 mM sodium orthovanadate, 1 mM PMSF, and 2 mM EDTA). Extracts were sonicated three times and centrifuged for 7 min at 12,000 × g. Supernatants were used for protein quantification, and an aliquiot was boiled in Laemmli buffer and applied to SDS-PAGE. Antibodies against the green fluorescent protein (GFP; Anti-GFP; polyclonal rabbit Ab from Cell Signaling, Beverly, MA, USA), hemagglutinin (HA; Anti-HA high affinity monoclonal rat Ab [clone 3F10] from Roche) and α-tubulin (clone DM1A) were used.
Quantification of complex I activity
Activity of complex I of the mitochondrial electron transport chain was determined as described49. Briefly, C. elegans mitochondria were isolated as previously described27 to obtain mitochondrial membranes. NADH was used as complex I substrate, 2,6-dichloroindophenol (DCIP) as electron acceptor. The electrons produced as byproduct during complex I oxidation of NADH reduce the artificial substrate decylubiquinone, which delivers the electrons to DCIP. Reduction of DCIP acts as a surrogate marker for complex I activity and is visible through a color change from blue to colorless that can be measured photometrically at 600 nm. Color change was detected at 20°C every 30 sec for four minutes, than the potentially complex I inhibiting substance was added and color change was again detected for four minutes. Slope ratio after and before substance adding were compared relative to solvent control.
Protein determination
Protein content in nematodes was determined by standard methods as previously described7.
Quantification of ROS using Mitotracker Red CM-H2X ROS
For ROS measurements, MitoTracker Red CM-H2X ROS (Invitrogen, Carlsbad, CA, USA) incubation plates were prepared as following: For each treatment 500 µl heat inactivated OP50 (65°C, 30 min) were mixed with 100 ul MitoTracker Red CM-H2X stock solution (100 µM) and spotted on a large NGM agar plate which was allowed to dry for approx. 20 min. Nematodes were incubated with corresponding compounds, then washed off the plates with S-Basal and allowed to settle by gravitation to remove offspring. Worms were washed two additional times with S-Basal and centrifuged (300g, 30 sec). The worm pellet was transferred to freshly prepared MitoTracker Red CM-H2X solution and incubated for 2 h at 20°C. To remove excess dye from the gut, worms were transferred to NGM agar plates with corresponding compounds or, as a positive control, to plates containing 10 mM paraquat for 1 h at 20°C. Aliquots of 100 µl worm suspension were distributed into 96-well FLUOTRAC™ plate (Greiner Bio-One, Frickenhausen, Germany). Fluorescence intensity was measured in a microplate reader (FLUOstar Optima, BMG Labtech, Offenburg, Germany) using well-scanning mode (ex: 570 nm, em: 610 nm). To normalize fluorescence signal, remaining worm suspension was used for protein determination.
Amplex Red-based quantification of hydrogen peroxide
Worms were removed from plates with 50 mM sodium-phosphate buffer pH 7.4, washed twice and transferred into an upright plexiglas cylinder (1.5 ml volume) with continuous stirring at low speed (100 rpm) at 20°C. Firstly determination of fluorescence was done without horse radish peroxidase (HRP) only in the presence of 1 µM Amplex Red (Invitrogen, Carlsbad, CA, USA) to detect possible unspecific increase in fluorescence (which was not observed). Next, 0.01 U/ml HRP was added and changes of fluorescence were recorded with a fluorescence detector (LF402 ProLine, IOM, Berlin, Germany) for at least 15 minutes at excitation and emission wavelengths of 571 nm and 585 nm, respectively. Immediately afterwards, worms were removed and collected for protein determination to normalize fluorescence values.
Detection of MNA using high-pressure liquid chromatography (HPLC)
For detection and quantification of MNA we modified a previously described protocol50. Worms were incubated and harvested as described above and stored at −80 °C. The worms were grinded in liquid nitrogen adding 200 µl of 100 µM HCl, sonicated and centrifuged for 5 min at 12,000 × g. A 10 µl aliquot of the supernatant was taken for protein determination. Supernatant was transferred to a new reaction tube and 50 µl of 20% trichloric acid (Applichem) was added, mixed and centrifuged at 12000 × g for 10 min at room temperature. 200 µl of the supernatant was added to 100 µl of acetophenone (100 µM in abs. Ethanol; Roth), mixed for 10 s and stored on ice for 10 min. Thereafter, 200 µl sodium hydroxide (6 M) was added, mixed and incubated for 3 h in ice water. Afterwards, we added 100 µl formic acid, mixed and incubated the solution for 25 min in ice water. Finally, we boiled the samples for 6 min. For HPLC (Jasco PU-1580, 3-line degaser DG-1580-53, ternary gradient unit LG-1580-02, autosampler AS-1555-10, fluorescence detector FP-920; all from Jasco, Easton, MD, USA) we used the following settings: injection volume: 100 µl samples, 20 µl standard; column: Kromasil 100, C18, 250 × 4 mm, 5 µm (Kromasil, Eka Chemicals, Separation Products, Bohus, Sweden); buffer A: 0.01 M sodium heptanesulfonate (Applichem); 0.5 % triethylamine (Merck) with 85% ortho-phosphoric acid (pH 3.2), 22 % acetonitrile; buffer B : acetonitrile/H2 O 4:1; flow rate: 1.0 ml/min; gradient: adding buffer B at 10 min running time with successive increase reaching 100 % at 15min and successive decrease at 25 min and 0% after 30 min; temperature: 20 °C; detection: excitation wave length: 366 nm, emission wave length: 418 nm.
Locomotion assay
Single worm movements within a liquid system were recorded using a digital CCD camera (Moticam 2300, Motic, St. Ingbert, Germany) coupled microscope (SMZ 168, Motic, St. Ingbert, Germany) with a subsequent analysis using the program DanioTrack (Loligo Systems, Tjele, Denmark). Worms (n=10–15) at the age of seven days of adult age were transferred from agar plates to S-buffer and immediately afterwards a 20 second video sequence was recorded. During the subsequent video analysis, the DanioTrack software subtracted the background and determined the centre of gravity of all object pixels in contrast with the background. Finally, the moving distance of this worm gravity centre was tracked and calculated. Thus, this locomotion assay can be considered as a quantitative analysis of the maximum movement capacity of a single worm.
Superoxide dismutase and catalase activity assays
Antioxidant enzyme activities (SOD, CAT) in nematodes were determined by standard photometric assays as previously described27.
gst-4 reporter strain
The transcriptional activity of the SKN-1-target gst-4 was determined using the reporter strain CL216630. Worms were pre-incubated for 48 h with and without MNA, maintained on E.coli HT115 with L4440 control RNAi or skn-1-RNAi, respectively. Immunoblotting was performed as described above.
Fluorescence microscopy
Worms were treated with MitoTracker Red CM-H2X ROS exactly as described above. Individual worms were placed on agarose pads and paralyzed with 1 mg/ml tetramisole. Worms were examined under a fluorescence microscope (Axiovert 100, Zeiss, Oberkochen, Germany) using a specific filter set (BP546/12, FT580, LP590, all nm) and pictures were taken with a digital camera (Moticam 2300).
GFP signals of gst-4 reporter worms were examined under the before-mentioned fluorescence microscope using a specific filter set (BP450-490, FT510, LP520, all nm). The GFP fluorescence pictures of anmt-1 OE::GFP strain MIR8 were taken with a Leica DMI6000B unit using excitation wavelength 470/40 nm, emission wavelength 525/50 nm and a Andor iXon EM CCD camera.
Paraquat stress assay
Resistance to lethal oxidative stress derived from the ROS inducer paraquat was determined using stress assay in liquid medium. Pre-incubated (7 d) worms were transferred to a 96-well plate (flat bottom) with S-Basal containing 50 mM paraquat (Acros Organics, Geel, Belgium). Stress resistance was determined by tapping the plate and counting dead and alive worms. We used 6 worms per well and 8 wells per condition, i.e. 48 worms in total per experiment.
Extraction of RNA
Total RNA was isolated using QIAzol (Qiagen, Hilden, Germany) based on the phenol/chloroform extraction method. Afterwards the RNA was quantified photometrically with a NanoDrop 1000 (PeqLab, Erlangen, Germany) and stored at −80 °C until use.
Real-Time PCR
Reverse transcription and quantitative real-time PCR was carried out using the GoTaq 2-Step RT-qPCR System (Promega, Madison, WI, USA) and a gene detection kit (PrimerDesign Ltd., Southampton, UK) according to the manufacturer’s instructions on LightCycler 480 system (Roche, Mannheim, Germany). Data were normalized to cdc-42 and analyzed using the Δ/Δ-CT method. Primer sequences used for gst-4 are fwd TCG GTC AGT CAA TGT CTA TCA C and rev CGG AAA AAG AAT ATG AAA TCT CTG TAT, for cyp29A2 fwd AAC TGA AAC AAA TAA AGG AGA TGA CTA and rev GAC TTT TCC ATC GTT CAC CAT ATC, and for cdc-42 (housekeeping gene) fwd TAT AGC ACA ACC AAC TAC ATC CC and rev AAA AGG AAA GAA GAA AAA CAA ACT GG.
Next-generation sequencing (RNAseq)
Total RNA was inspected for degradation using Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). For library preparation an amount of 2 µg of total RNA per sample was processed using Illumina’s TruSeq™ RNA Sample Prep Kit (Illumina; San Diego; CA, USA) following the manufacturer’s instruction. Each library includes its own index sequencing to allow multiplexing. The libraries were sequenced using v3 sequencing chemistry and a HiSeq2000 (Illumina, San Diego, CA, USA) in a single read approach with 50 cycles resulting in reads with length of 50 nucleotides. Libraries were sequenced in a multiplex manner pooling four libraries per lane. Sequencing ends up with around 30–40 mio reads per sample. Sequence data were extracted in FastQ format and used for mapping approach.
The deep sequencing data obtained and discussed in the current publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE49662.
Bioinformatics of RNA expression data
All reads were mapped against the C. elegans genome (WS220) using the TopHat tool51. Only unique mapable reads were regarded. For counting the reads per gene (raw counts) the Python package HTSeq (http://www.huber.embl.de/users/anders/HTSeq/doc/overview.html) was used in mode ‘union’ together with the Ensembl gene annotation.
Raw counts for the genes were analyzed using the R Statistical Computing Environment and the Bioconductor packages DESeq52 and edgeR53. Both packages provide statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting p-values were adjusted using the Benjamini and Hochberg's approach for controlling the false discovery rate (FDR). If both FDR values (by DESeq and edgeR) were smaller than p=0.05, genes were assigned as differentially expressed.
FunCat analyses
For analyses of functional annotations of C. elegans genes we used FungiFun as previously described54.
Promoter analyses
The search for SKN-1 and DAF-16 transcription factor binding sites (TFBS) was done within the proximal promoter region of all identified differential expressed genes 1.5 kb upstream of each predicted start codon. Therefore, a FASTA file containing all promoter regions of the corresponding differential expressed genes was created using WormMart. Next, the remaining sequence file was scanned for one or more matches to the position-specific scoring matrix (PSSM) of skn-1 and daf-16 using the matrix scan function of the pattern-matching program RSAT (regulatory sequence analysis tools). The PSSM contain the nucleotide frequency at each position within the binding sites and were obtained for the database Transfac and experimentally verified as described55,56. The threshold P-value, which indicates the risk of false positive predictions, was set to 0.0005.
Supplementary Material
Acknowledgments
Most C. elegans strains used in this work were provided by the Caenorhabditis Genetics Center (Univ. of Minnesota, USA), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). The strains LG389 and LG390 were a kind gift of Dr. L. Guarente and Dr. M. Viswanathan. The excellent technical assistance of Ivonne Heinze, Beate Laube, Annett Müller, Susann Richter and Waltraud Scheiding, as well as the excellent secretarial assistance of Mandy Schalowski are gratefully acknowledged. This work is in part (Suppl. Fig. 5 only) supported by the research program of the Jena Centre for Systems Biology of Ageing (JenAge) funded by the German Ministry for Education and Research (Bundesministerium für Bildung und Forschung; support code BMBF 0315581). D.A.S. is supported by grants from the NIH/NIA, the United Mitochondrial Disease Foundation, and the Glenn Medical Foundation. Funding for this project was denied by the German Research Association (Deutsche Forschungsgemeinschaft, DFG), grant application number RI 1976/3-1.
Footnotes
Conflict of interests: D.S. is a consultant and inventor on patents licensed to GlaxoSmithKline, PA, a company developing sirtuin-based medicines.
Contribution to work: K.S. and J.M. designed, performed, and evaluated all experiments with the following exceptions. M.G. and M.P. performed next generation sequencing analysis of mRNA, whereas sample provision, RNA extraction and quality control were done by K.S. Bioinformatical evaluation was done by S.P., R.G., I.H., and St.S. Promoter analysis and gene classification was done by K.S. and J.M. A.S. and T.H. helped with strain constructions. S.W., D.K., A.P., M.B., S.S., K.Z., N.L.P., Y.K., D.A.S. and H.Y.C. were involved in the study design, sample contribution, and contributed several assays. The entire work was designed and supervised by M.R. The manuscript was written by K.S., J.M. and M.R. All authors discussed and commented on the manuscript.
References
- 1.Nogueiras R, et al. Sirtuins: physiological modulators of metabolism. Physiol Rev. 2012;92:1479–1514. doi: 10.1152/physrev.00022.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sinclair DA, Mills K, Guarente L. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 3.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin SJ, et al. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 5.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulz TJ, et al. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 9.Viswanathan M, Guarente L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature. 2011;477:E1–E2. doi: 10.1038/nature10440. [DOI] [PubMed] [Google Scholar]
- 10.Rizki G, et al. The evolutionarily conserved longevity determinants HCF-1 and SIR-2.1/SIRT1 collaborate to regulate DAF-16/FOXO. PLoS Genet. 2011;7:e1002235. doi: 10.1371/journal.pgen.1002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banerjee KK, et al. dSir2 in the adult fat body, but not in muscles, regulates life span in a diet-dependent manner. Cell Rep. 2012;2:1485–1491. doi: 10.1016/j.celrep.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 13.Burnett C, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boily G, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanfi Y, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 16.Gossmann TI, et al. NAD(+) biosynthesis and salvage - a phylogenetic perspective. FEBS J. 2012;279:3355–3363. doi: 10.1111/j.1742-4658.2012.08559.x. [DOI] [PubMed] [Google Scholar]
- 17.Cantoni GL. Methylation of nicotinamide with soluble enzyme system from rat liver. J Biol Chem. 1951;189:203–216. [PubMed] [Google Scholar]
- 18.Fukushima T, et al. Radical formation site of cerebral complex I and Parkinson's disease. J Neurosci Res. 1995;42:385–390. doi: 10.1002/jnr.490420313. [DOI] [PubMed] [Google Scholar]
- 19.Ristow M, Zarse K. How increased oxidative stress promotes longevity and metabolic health: The concept of mitochondrial hormesis (mitohormesis) Exp Gerontol. 2010;45:410–418. doi: 10.1016/j.exger.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 20.van der Horst A, Schavemaker JM, Pellis-van Berkel W, Burgering BM. The Caenorhabditis elegans nicotinamidase PNC-1 enhances survival. Mech Ageing Dev. 2007;128:346–349. doi: 10.1016/j.mad.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto T, Horikawa M, Nomura T, Sakamoto K. Nicotinamide adenine dinucleotide extends the lifespan of Caenorhabditis elegans mediated by sir-2.1 and daf-16. Biogerontology. 2010;11:31–43. doi: 10.1007/s10522-009-9225-3. [DOI] [PubMed] [Google Scholar]
- 22.Schmeisser S, et al. Neuronal ROS signaling rather than AMPK/Sirtuin-mediated energy sensing links dietary restriction to lifespan extension. Molecular Metabolism. 2013;2:92–102. doi: 10.1016/j.molmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kundu TK, Hille R, Velayutham M, Zweier JL. Characterization of superoxide production from aldehyde oxidase: an important source of oxidants in biological tissues. Arch Biochem Biophys. 2007;460:113–121. doi: 10.1016/j.abb.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawyer JM, et al. Overcoming redundancy: an RNAi enhancer screen for morphogenesis genes in Caenorhabditis elegans. Genetics. 2011;188:549–564. doi: 10.1534/genetics.111.129486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panoutsopoulos GI, Beedham C. Enzymatic oxidation of phthalazine with guinea pig liver aldehyde oxidase and liver slices: inhibition by isovanillin. Acta Biochim Pol. 2004;51:943–951. [PubMed] [Google Scholar]
- 26.Crawford Dl, Sutherland JB, Pometto AL, III, Miller JM. Production of an aromatic aldehyde oxidase by Streptomyces viridosporus. Archives of Microbiology. 1982;131:351–355. [Google Scholar]
- 27.Zarse K, et al. Impaired insulin/IGF1-signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 2012;15:451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 29.Maeda K, et al. Aldehyde oxidase 1 gene is regulated by Nrf2 pathway. Gene. 2012;505:374–378. doi: 10.1016/j.gene.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 30.Link CD, Johnson CJ. Reporter transgenes for study of oxidant stress in Caenorhabditis elegans. Methods Enzymol. 2002;353:497–505. doi: 10.1016/s0076-6879(02)53072-x. [DOI] [PubMed] [Google Scholar]
- 31.Van Raamsdonk JM, Hekimi S. Superoxide dismutase is dispensable for normal animal lifespan. Proc Natl Acad Sci U S A. 2012;109:5785–5790. doi: 10.1073/pnas.1116158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dillin A, et al. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 33.Yang W, Hekimi S. Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans. Aging Cell. 2010;9:433–447. doi: 10.1111/j.1474-9726.2010.00571.x. [DOI] [PubMed] [Google Scholar]
- 34.Ristow M, et al. Antioxidants prevent health-promoting effects of physical exercise in humans. Proc Nat Acad Sci. 2009;106:8665–8670. doi: 10.1073/pnas.0903485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan Y, Schroeder EA, Ocampo A, Barrientos A, Shadel GS. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab. 2011;13:668–678. doi: 10.1016/j.cmet.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Webster BR, Lu Z, Sack MN, Scott I. The role of sirtuins in modulating redox stressors. Free Radic Biol Med. 2012;52:281–290. doi: 10.1016/j.freeradbiomed.2011.10.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merksamer PI, et al. The sirtuins, oxidative stress and aging: an emerging link. Aging (Albany NY) 2013 doi: 10.18632/aging.100544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 39.Gebicki J, Chlopicki S. Method for treating hypertriglyceridemia, dyslipidemia and hyperchlosterolemia with a 1-methylnicotinamide salt. US Patent US 7,935,717 B2. 2011;10 [Google Scholar]
- 40.Nordestgaard BG, Benn M, Schnohr P, Tybjaerg-Hansen A. Triglycerides and risk of myocardial infarction, ischemic heart disease, and death in men and women. JAMA. 2007;298:299–308. doi: 10.1001/jama.298.3.299. [DOI] [PubMed] [Google Scholar]
- 41.Brown BG, et al. Simvastatin and niacin, antioxidant vitamins, or the combination for the prevention of coronary disease. N Engl J Med. 2001;345:1583–1592. doi: 10.1056/NEJMoa011090. [DOI] [PubMed] [Google Scholar]
- 42.Canner PL, et al. Fifteen year mortality in Coronary Drug Project patients: long-term benefit with niacin. J Am Coll Cardiol. 1986;8:1245–1255. doi: 10.1016/s0735-1097(86)80293-5. [DOI] [PubMed] [Google Scholar]
- 43.Warburton DE, Nicol CW, Bredin SS. Health benefits of physical activity: the evidence. Can Med Ass J (CMAJ) 2006;174:801–809. doi: 10.1503/cmaj.051351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chlopicki S, et al. Single bout of endurance exercise increases NNMT activity in the liver and MNA concentration in plasma. Pharmacol Rep. 2012;64:369–376. doi: 10.1016/s1734-1140(12)70777-6. [DOI] [PubMed] [Google Scholar]
- 45.Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol. 2013;9:300–306. doi: 10.1038/nchembio.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parsons RB, et al. The expression of nicotinamide N-methyltransferase increases ATP synthesis and protects SH-SY5Y neuroblastoma cells against the toxicity of Complex I inhibitors. Biochem J. 2011;436:145–155. doi: 10.1042/BJ20101685. [DOI] [PubMed] [Google Scholar]
- 47.Mouchiroud L, et al. The NAD(+)/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
References to Online Methods
- 48.Segref A, Hoppe T. Analysis of ubiquitin-dependent proteolysis in Caenorhabditis elegans. Methods Mol Biol. 2012;832:531–544. doi: 10.1007/978-1-61779-474-2_38. [DOI] [PubMed] [Google Scholar]
- 49.Janssen AJ, et al. Spectrophotometric assay for complex I of the respiratory chain in tissue samples and cultured fibroblasts. Clin Chem. 2007;53:729–734. doi: 10.1373/clinchem.2006.078873. [DOI] [PubMed] [Google Scholar]
- 50.Musfeld C, Biollaz J, Belaz N, Kesselring UW, Decosterd LA. Validation of an HPLC method for the determination of urinary and plasma levels of N1-methylnicotinamide, an endogenous marker of renal cationic transport and plasma flow. J Pharm Biomed Anal. 2001;24:391–404. doi: 10.1016/s0731-7085(00)00425-8. [DOI] [PubMed] [Google Scholar]
- 51.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Priebe S, Linde J, Albrecht D, Guthke R, Brakhage AA. FungiFun: a web-based application for functional categorization of fungal genes and proteins. Fungal Genet Biol. 2011;48:353–358. doi: 10.1016/j.fgb.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 55.Wingender E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief Bioinform. 2008;9:326–332. doi: 10.1093/bib/bbn016. [DOI] [PubMed] [Google Scholar]
- 56.Blackwell TK, Bowerman B, Priess JR, Weintraub H. Formation of a monomeric DNA binding domain by Skn-1 bZIP and homeodomain elements. Science. 1994;266:621–628. doi: 10.1126/science.7939715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


