SUMMARY
Ever since eukaryotes subsumed the bacterial ancestor of mitochondria, the nuclear and mitochondrial genomes have had to closely coordinate their activities, as each encode different subunits of the oxidative phosphorylation (OXPHOS) system. Mitochondrial dysfunction is a hallmark of aging, but its causes are debated. We show that, during aging, there is a specific loss of mitochondrial, but not nuclear, encoded OXPHOS subunits. We trace the cause to an alternate PGC-1α/β-independent pathway of nuclear-mitochondrial communication that is induced by a decline in nuclear NAD+ and the accumulation of HIF-1α under normoxic conditions, with parallels to Warburg reprogramming. Deleting SIRT1 accelerates this process, whereas raising NAD+ levels in old mice restores mitochondrial function to that of a young mouse in a SIRT1-dependent manner. Thus, a pseudohypoxic state that disrupts PGC-1α/β-independent nuclear-mitochondrial communication contributes to the decline in mitochondrial function with age, a process that is apparently reversible.
INTRODUCTION
One of the most conserved and robust phenomena in biology is a progressive decline in mitochondrial function with age, leading to a loss of cellular homeostasis and organismal health (Lanza and Nair, 2010; Wallace et al., 2010). There is considerable debate, however, about why mitochondrial homeostasis is disrupted in the first place. The original idea of Harman, that reactive oxygen species (ROS) from mitochondria are a primary cause of aging (Harman, 1972), has been challenged by recent studies of long-lived species and genetically altered animals (Lapointe and Hekimi, 2010).
Though most mitochondrial genes have been transferred to the nuclear genome, 13 subunits of the oxidative phosphorylation (OXPHOS) system remain, demanding functional communication between the nucleus and mitochondria to form stoichiometric OXPHOS complexes. This is mediated in large part by the peroxisome proliferator-activated receptor-γ coactivators α and β (PGC-1α and PGC1-1β), which along with NRF-1 and -2, induce nuclear-encoded proteins, such as TFAM (mitochondrial transcription factor A), that carry out the replication, transcription, and translation of mitochondrial DNA (mtDNA) (Larsson, 2010).
Mammalian sirtuins (SIRT1-7) are a conserved family of NAD+-dependent lysine-modifying acylases that control physiological responses to diet and exercise (Haigis and Sinclair, 2010). The expression of SIRT1, an NAD+-dependent deacetylase, is elevated in a number of tissues following calorie restriction (CR) (Cohen et al., 2004), an intervention that extends lifespan in diverse species. Overexpression or pharmacological activation of SIRT1 reproduces many of the health benefits of CR, including protection from metabolic decline, cardiovascular disease, cancer, and neurodegeneration (Haigis and Sinclair, 2010; Libert and Guarente, 2013). Some of the health benefits of SIRT1 have also been linked to improved mitochondrial function (Baur et al., 2006; Gerhart-Hines et al., 2007; Price et al., 2012; Rodgers et al., 2005). Indeed, increased expression of neuronal SIRT1 extends mouse lifespan (Satoh et al., 2013), though its role in aging in lower organisms has been challenged (Burnett et al., 2011).
A hallmark of cancer is a shift away from OXPHOS toward anaerobic glycolysis that provides cells with sufficient substrates for biomass. This metabolic reprogramming, known as the Warburg effect (Warburg, 1956), is driven by several different pathways, including mTOR, c-Myc, and hypoxia-inducible factor 1 (HIF-1α) (Dang, 2012). Interestingly, SIRT1 increases HIF-1α transcriptional activity (Lim et al., 2010), SIRT3 destabilizes HIF-1α protein (Bell et al., 2011; Finley et al., 2011), and SIRT6 functions as a HIF-1α corepressor (Zhong et al., 2010), raising the possibility that HIF-1α may also be relevant to aging. Consistent with this, in C. elegans, Hif-1 regulates lifespan and the response to CR (Leiser and Kaeberlein, 2010). A role for HIF-1α in mammalian aging, however, has not been explored.
In this study, we provide evidence for a PGC-1α/β-independent pathway of mitochondrial regulation that plays a role in the aging process. Activity of this pathway declines during aging due to changes in nuclear NAD+ levels, causing a pseudo-hypoxia-driven imbalance between nuclear- and mitochondrially encoded OXPHOS subunits—a process that is prevented by CR and is reversed by raising NAD+, with implications for treating age-related diseases, including cancer.
RESULTS
Aging Leads to a Specific Decline in Mitochondrially Encoded Genes
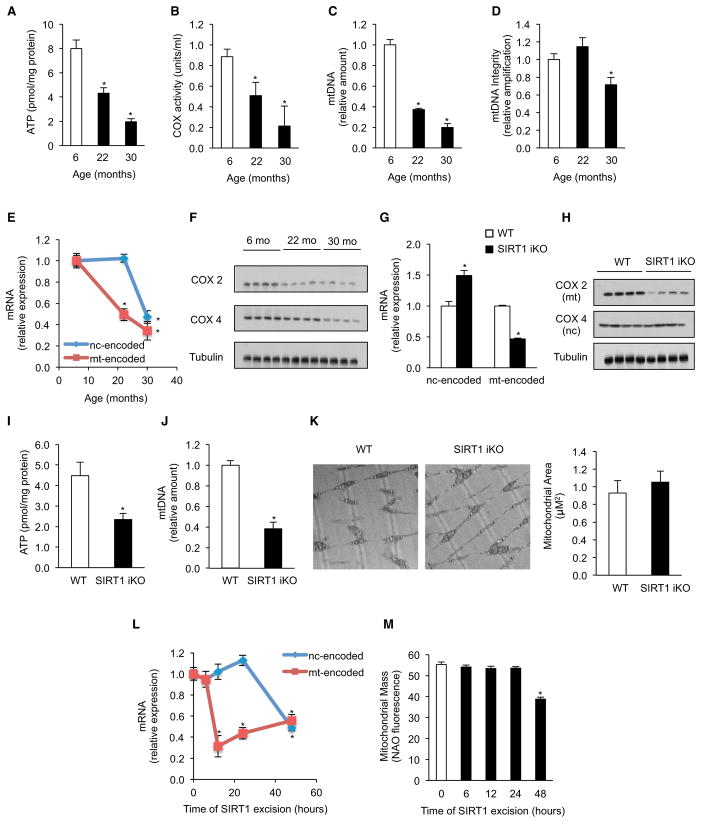
Aging is associated with disruption of mitochondrial homeostasis, but the underlying mechanisms are unclear. As in previous reports (Lanza and Nair, 2010), we observed a progressive, age-dependent decline in OXPHOS efficiency with age in skeletal muscle (Figures 1A and 1B). By 22 months of age, ATP content and complex IV (COX) activity were decreased, even more so by 30 months of age. Although mtDNA content declined at both ages, the integrity of mtDNA was only lower in the 30 month olds (Figures 1C and 1D). Together with previous reports (Lapointe and Hekimi, 2010), this suggested an aging mechanism that disrupts OXPHOS prior to the accumulation of significant mtDNA damage.
Figure 1. Aging and Loss of SIRT1 Leads to a Specific Decline in Mitochondrial-Encoded Genes and Impairment in Mitochondrial Homeostasis in Skeletal Muscle.
(A) ATP content of 6-, 22-, and 30-month-old mice (n = 5, *p < 0.05 versus 6-month-old mice).
(B) Cytochrome c oxidase (COX) activity (n = 5, *p < 0.05 versus 6-month-old animals).
(C and D) Mitochondrial DNA content (C) and DNA integrity (D) (n = 5, *p < 0.05 versus 6-month-old animals).
(E) Expression of nuclear- and mitochondrially encoded genes (n = 5, *p < 0.05 versus 6-month-old animals).
(F) Immunoblot for COX2 and COX4 in 6-, 22-, and 30-month-old mice.
(G) Expression of nuclear- (NDUFS8, NDUFAS, SDHb, SDHd, Uqcrc1, Uqcrc2, COX5b, Cox6a1, ATP5a1, and ATPc1) and mitochondrially encoded genes (ND1, ND2, ND3, ND4, ND4l, ND5, ND6, Cytb, COX1, COX2, COX3, ATP6, and ATP8) in WT and SIRT1 iKO mice (n = 5, *p < 0.05 versus WT).
(H and I) (H) Immunoblot for COX2 and COX4 and (I) ATP content in WT and SIRT1 iKO mice (n = 5, *p < 0.05 versus WT).
(J) Mitochondrial DNA content of WT and SIRT1 iKO mice (n = 5, *p < 0.05 versus WT).
(K) Electron microscopy of gastrocnemius from WT and SIRT1 iKO mice and mitochondrial area (n = 4).
(L) Expression of nuclear- and mitochondrially encoded genes in SIRT1 flox/flox Cre-ERT2 primary myoblasts treated with vehicle (0 hr) or tamoxifen (OHT) to induce SIRT1 excision for 6, 12, 24, and 48 hr (n = 4, *p < 0.05 versus vehicle).
(M) Mitochondrial mass by NAO fluorescence in SIRT1 flox/flox Cre-ERT2 primary myoblasts treated with vehicle (0 hr) or OHT to induce SIRT1 excision for 6, 12, 24, and 48 hr (n = 4, *p < 0.05 versus vehicle).
Nuclear- and mitochondrially encoded genes were ND1, Cytb, COX1, ATP6 and NDUFS8, SDHb, Uqcrc1, COX5b, ATP5a1, respectively. Tissue samples are gastrocnemius unless otherwise stated. Values are expressed as mean ± SEM. See also Figure S1.
A clue came from observations that the activity of OXPHOS complexes I, III, and IV decline with age, but complex II, the only complex composed exclusively of nuclear-encoded subunits, does not (Kwong and Sohal, 2000). Thus, we tested whether OXPHOS decline might be due to the specific loss of mitochondrially encoded transcripts. Mitochondrially encoded OXPHOS mRNAs (ND1, Cytb, COX1, ATP6) were all significantly lower at 22 months relative to 6 month olds, whereas those encoded by the nuclear genome (NDUFS8, SDHb, Uqcrc1, COX5, ATP5a) remained unchanged; but by 30 months of age, both the nuclear- and the mitochondrially encoded mRNAs were lower (Figures 1E). Protein levels of the mitochondrially encoded COX2 gene were decreased at 22 months, but COX4, a nuclear-encoded subunit, was only slightly lower. By 30 months, both proteins were reduced relative to young mice (Figure 1F). The mitochondrial unfolded protein response (mtUPR) has been recently linked to longevity (Durieux et al., 2011; Houtkooper et al., 2013; Mouchiroud et al., 2013); however, under these conditions there was no evidence of a mtUPR at 22 months of age (Figure S1A available online).
Knockout of SIRT1 Mimics Aging by Decreasing Mitochondrial, but Not Nuclear-Encoded, OXPHOS Components
We wondered whether the specific decline in mitochondrially encoded OXPHOS components in aged mice might be due, in part, to a loss of SIRT1 activity. To test this, we utilized an adult-inducible SIRT1 knockout mouse (SIRT1-iKO) (Price et al., 2012), which circumvents the developmental abnormalities of germline SIRT1 knockouts. SIRT1 was deleted at 2–4 months of age, and skeletal muscle was analyzed 2–6 months later. As expected, the mRNA levels of all 13 mitochondrially encoded OXPHOS genes and the two rRNAs were reduced in the SIRT1 iKO mice compared to wild-type controls (Figures 1G and S1B). Strikingly, there was no decrease in the expression of any of the nuclear-encoded components under fed conditions (Figure 1G). Again, protein levels of mitochondrially encoded COX2 were significantly decreased, whereas the nuclear-encoded COX4 was unaltered (Figure 1H), coincident with a decline in complex IV (COX), but not complex II (SDH), activity (Figures S1D and S1E). Similar to old mice, cellular ATP levels and mtDNA content were reduced (Figures 1I and 1J), with no apparent induction of mtUPR (Figure S1C).
Given that SIRT1 maintains mitochondrial mass by increasing PGC-1α activity, we were surprised to see that, under these basal conditions (i.e., the fed state), there was no effect of SIRT1 deletion on mitochondrial mass (Figure 1K). To understand why, we cultured SIRT1 iKO primary myoblasts and induced Cremediated deletion of the SIRT1 catalytic core ex vivo. After 12 hr, only the mitochondrially encoded OXPHOS mRNAs decreased (Figure 1L). Again, mtDNA content and mitochondrial membrane potential declined, with no change in mitochondrial mass (Figures 1M, S2A, and S2B). By 48 hr, mRNA from both the nuclear- and mitochondrially encoded genes had decreased, with a loss of mitochondrial mass and a further decrease in membrane potential (Figures 1L, 1M, S2A, and S2B). These data suggested that loss of SIRT1 results in a biphasic disruption of mitochondrial homeostasis.
Nuclear NAD+ Levels Regulate Mitochondrially Encoded Genes
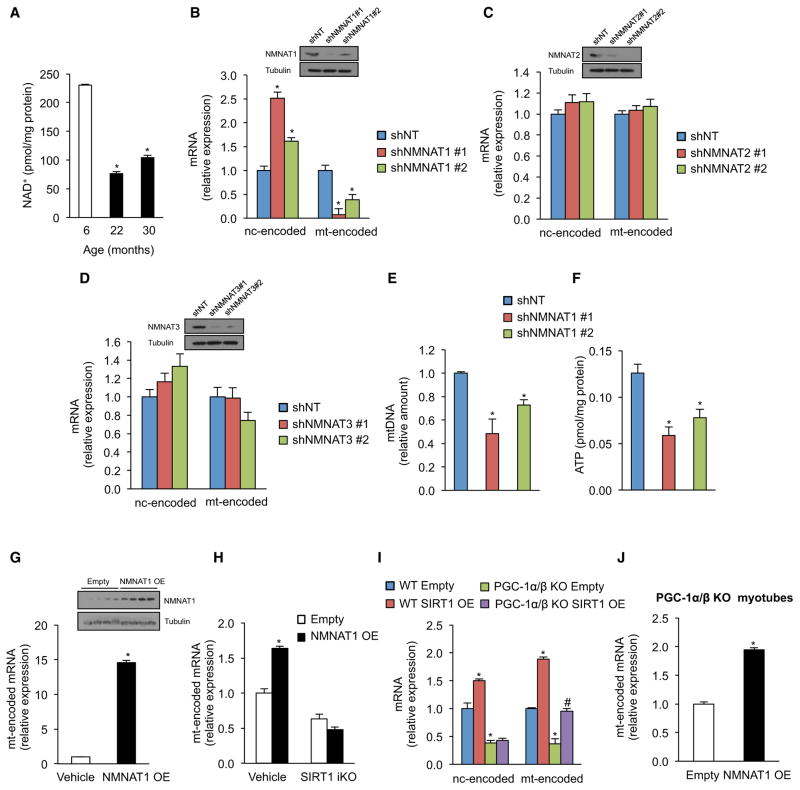
Because there was no decline in SIRT1 protein with age (Figure S2E), we hypothesized that SIRT1 activity might be compromised in old mice due to a paucity of NAD+. Recent studies show that NAD+ levels are regulated independently in different cell compartments and that overall NAD+ levels decline during aging (Braidy et al., 2011; Massudi et al., 2012; Yang et al., 2007). However, it is not clear in which cellular compartment(s) is NAD+ relevant to aging (Cantó and Auwerx, 2011). Consistent with other reports (Braidy et al., 2011; Massudi et al., 2012), there was less total NAD+ in the skeletal muscle of elderly mice (Figure 2A). To determine which compartment might be responsible, we manipulated NAD+ levels in the different compartments by independently knocking down isoforms of nicotinamide mononucleotide adenylyltransferase, which regulate NAD+ levels in the nucleus (NMNAT1), golgi/cytoplasm (NMNAT2), and mitochondria (NMNAT3) (Berger et al., 2005). Knockdown of NMNAT2 or NMNAT3 had no effect on OXPHOS genes, whereas knockdown of NMNAT1 resulted in a specific reduction in the expression of mitochondrially encoded OXPHOS, mtDNA content, and ATP levels (Figures 2B–2F).
Figure 2. Nuclear NAD+ Levels Regulate Mitochondrial-Encoded Genes and Mitochondrial Homeostasis through SIRT1, Independently of PGC-1α/β.
(A) NAD+ levels in gastrocnemius of 6-, 22-, and 30-month-old mice (n = 5, *p < 0.05 versus 6-month-old mice).
(B–D) Expression of nuclear- and mitochondrially encoded genes in primary myoblasts transduced with NMNAT1 (B), NMNAT2 (C), NMNAT3 (D), or nontargeting shRNA (n = 4, *p < 0.05 versus shNT).
(E and F) Mitochondrial DNA content and (H) ATP content (I) in primary myoblasts transduced with NMNAT1 or nontargeting shRNA (n = 4, *p < 0.05 versus shNT).
(G) Expression of mitochondrially encoded genes in tibialis of 10- to 12-month-old mice overexpressing NMNAT1 compared to the contralateral tibialis muscle treated with vehicle (n = 4, *p < 0.05 versus vehicle).
(H) Expression of mitochondrially encoded genes in SIRT1 flox/flox Cre-ERT2 primary myoblasts treated with vehicle or OHT to induce SIRT1 excision infected with adenovirus overexpressing NMNAT1 or empty vector (n = 4, *p < 0.05 versus vehicle empty vector).
(I and J) Expression of nuclear- and mitochondrially encoded genes in WT and PGC-1α/β knockout myotubes treated with adenovirus overexpressing SIRT1 (I) or NMNAT1 (J) (n = 4, *p < 0.05 versus WT empty; #p < 0.05 versus PGC-1α/β KO empty).
Nuclear- and mitochondrially encoded genes were ND1, Cytb, COX1, ATP6 and NDUFS8, SDHb, Uqcrc1, COX5b, ATP5a1, respectively. Tissue samples are gastrocnemius muscle unless otherwise stated. Values are expressed as mean ± SEM. See also Figure S2.
These results indicated that increasing the production of NAD+ within the nuclear pool might stimulate mitochondria. Overexpression of NMNAT1 in skeletal muscle of 10- to 12-month-old mice dramatically increased the expression of mitochondrially encoded OXPHOS genes (Figure 2G). Overexpression of NMNAT1 in primary myoblasts produced a similar effect that was SIRT1 dependent (Figure 2H). Together, these data indicated that mitochondria are regulated by nuclear NAD+ and that the impairment in OXPHOS function during aging may be precipitated by depletion of the nuclear NAD pool.
SIRT1 Can Regulate Mitochondria through a PGC-1α/β-Independent Pathway
A central dogma in the sirtuin field is that SIRT1 promotes mitochondrial function in response to fasting and CR by deacetylating PGC-1α (Gerhart-Hines et al., 2007; Rodgers et al., 2005). Consistent with this, SIRT1 iKO animals failed to upregulate both nuclear- and mitochondrially encoded OXPHOS genes in response to fasting (Figure S2C). However, our findings in fed animals (see Figure 1) indicated that SIRT1 can regulate mitochondrial genes independently of PGC-1α. To test this, we examined primary myotubes from PGC-1α/β knockout (KO) mice (Zechner et al., 2010) and from PGC-1α muscle-specific null mice (Handschin et al., 2007), and we saw no defect in the ability of SIRT1 and NMNAT1 to induce mitochondrially encoded OXPHOS genes (Figures 2I and 2J). Thus, SIRT1 can induce OXPHOS genes in the absence of PGC-1α/β (Figure S2D).
SIRT1 Regulates Mitochondrially Encoded Genes through HIF-1α
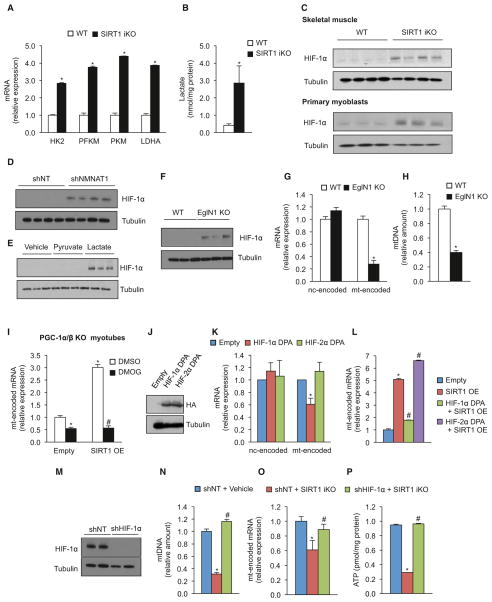
Next, we sought to understand how SIRT1 regulates mitochondria independently of PGC-1α/β. Analysis of SIRT1 iKO animals indicated that genes involved in glycolysis were upregulated, with increased lactate levels (Figures 3A and 3B) and a switch from slow-twitch oxidative fibers (MyHCIIa) to fast-twitch glycolytic fibers (MyHCIIb) (Figure S1F). These metabolic changes were reminiscent of Warburg remodeling of metabolism in cancer cells, which is known to be mediated, in part, by the stabilization of the transcription factor HIF-1α (Majmundar et al., 2010). The levels of HIF-1α and the expression of HIF-1α target genes were considerably higher in the SIRT1 iKO (Figures 3C and S3A). Despite being cultured under normoxic conditions, primary myoblasts deleted for SIRT1 also had increased HIF-1α protein levels and activity of a HIF-1α reporter (Figures 3C and S3B). Reducing NAD+ levels, either by knocking down NMNAT1 or by treating cells with lactate (which decreases the NAD+/NADH ratio), also caused HIF-1α protein stabilization (Figures 3D, 3E, and S3C).
Figure 3. Loss of SIRT1 Induces a Pseudohypoxic State that Disrupts Mitochondrial-Encoded Genes and Mitochondrial Homeostasis.
(A and B) HK2, PFKM, PKM, and LDHA mRNA (A) and lactate levels (B) of WT and SIRT1 iKO mice (n = 5, *p < 0.05 versus WT).
(C) Immunoblot for HIF-1α and tubulin in WT and SIRT1 iKO mice and in SIRT1 flox/flox Cre-ERT2 primary myoblasts treated with vehicle or OHT to induce SIRT1 excision for 24 hr (SIRT1 iKO).
(D) Immunoblot for HIF-1α and tubulin in primary myoblasts transduced with NMNAT1 or nontargeting shRNA.
(E) Immunoblot of HIF-1α and tubulin in primary myoblasts treated with pyruvate, lactate, or vehicle for 24 hr.
(F) Immunoblot for HIF-1α and tubulin in WT and EglN1 KO mice.
(G) Expression of nuclear- and mitochondrially encoded genes of WT and EglN1 KO mice (n = 5, *p < 0.05 versus WT).
(H) Mitochondrial DNA content of WT and EglN1 KO mice (n = 5, *p < 0.05 versus WT).
(I) Expression of mitochondrially encoded genes in PGC-1α/β KO myotubes treated with adenovirus overexpressing SIRT1 treated with DMSO or the HIF-stabilizing compound DMOG (n = 4, *p < 0.05 versus empty DMSO; #p < 0.05 versus SIRT1 OE DMSO).
(J) Immunoblot for HA tag and tubulin in control and C2C12 cells overexpressing either HIF-1α or HIF-2α with the proline residues mutated (HIF-1α DPA and HIF-2α DPA).
(K) Expression of nuclear- versus mitochondrially encoded genes in HIF-1α DPA or HIF-2α DPA C2C12 cells (n = 6, *p < 0.05 versus empty vector).
(L) Expression of mitochondrially encoded genes in HIF-1α DPA or HIF-2α DPA C2C12 cells with adenovirus overexpressing SIRT1 (n = 4, *p < 0.05 versus empty vector; #p < 0.05 versus SIRT1 OE).
(M) Immunoblot for HIF-1α and tubulin in SIRT1 flox/flox Cre-ERT2 primary myoblasts transduced with HIF-1α or nontargeting shRNA and treated with DMOG.
(N) Mitochondrial DNA in SIRT1 flox/flox Cre-ERT2 primary myoblasts transduced with HIF-1α or nontargeting shRNA, treated with vehicle or OHT to induce SIRT1 excision (SIRT1 iKO) (n = 4, *p < 0.05 versus shNT vehicle; #p < 0.05 versus shNT SIRT1 iKO).
(O) Expression of mitochondrially encoded genes in SIRT1 flox/flox Cre-ERT2 primary myoblasts transduced with HIF-1α or nontargeting shRNA and treated with OHT to induce SIRT1 excision (SIRT1 iKO) (n = 4, *p < 0.05 versus shNT vehicle; #p < 0.05 versus shNT SIRT1 iKO).
(P) ATP content in SIRT1 flox/flox Cre-ERT2 primary myoblasts transduced with HIF-1α or nontargeting shRNA and treated with vehicle or OHT to induce SIRT1 excision (n = 5, *p < 0.05 versus shNT vehicle; #p < 0.05 versus shNT SIRT1 iKO).
Nuclear- and mitochondrially encoded genes were ND1, Cytb, COX1, ATP6 and NDUFS8, SDHb, Uqcrc1, COX5b, ATP5a1, respectively. Tissue samples are gastrocnemius unless otherwise stated. Values are expressed as mean ± SEM. See also Figure S3.
HIF-1α has been studied extensively in cancer and during hypoxia; however, its role in normal physiology remains largely unknown. To better understand this, HIF-1α was stabilized ectopically in vivo by deleting the EglN1 gene encoding HIF prolyl hydroxylase 2 (PHD2) (Minamishima et al., 2008). Upon EglN1 deletion and HIF-1α stabilization in muscle, there was a specific decline in mtDNA content and decreased levels of mitochondrially encoded, but not nuclear-encoded, OXPHOS mRNA, paralleling the effects of SIRT1 deletion and normal aging (Figures 3F–3H). Pharmacological stabilization of HIF-1α in PGC-1α/β knockout myotubes reduced expression of mitochondrially encoded genes (Figures 3I and S3D), whereas treating PGC-1α/β KO cells with pyruvate (to increase NAD+ levels) up-regulated mitochondrially encoded genes, an effect that was prevented by stabilization of HIF-1α (Figure S3E). Stabilization of HIF-1α in primary cells and transgenic mice blocked the ability of SIRT1 to upregulate mitochondrially encoded genes and increase ATP levels, with a specific loss of mitochondrially encoded mRNAs (Figures 3I–3L and S3F–SFI). Overexpression of a stabilized mutant version of the related factor HIF-2α did not have the same effect (Figures 3J–3L and S3I), demonstrating that the inhibition of OXPHOS and mitochondrially encoded genes is HIF-1α specific. In primary myoblasts lacking HIF-1α, deletion of SIRT1 had no effect on mtDNA content, mitochondrially encoded gene expression, or ATP levels (Figures 3M–3P). Together, our results show that HIF-1α, but not HIF-2α, regulates mitochondria in response to SIRT1 activity, which is under the control of nuclear NAD+ levels.
SIRT1 Stabilizes HIF-1α via VHL
HIF-1α can be stabilized by ROS originating from complex III of the ETC as part of retrograde response (Bell et al., 2007). Six hours after inducing SIRT1 deletion in primary myoblasts, HIF-1α levels increased (Figure 5F), and by 12 hr, mitochondrial homeostasis was impaired (Figures 1L, S2A, and S2B). Yet, ROS levels did not increase until the 24 hr time point (Figure S4A). Myoblasts depleted of mitochondrial DNA (rho0), which are unable to produce ROS and signal to the nucleus (Chandel and Schumacker, 1999), were similar to the parental control cells (Figure S4B), indicating that ROS and retrograde signaling are not the cause of HIF-1α stabilization.
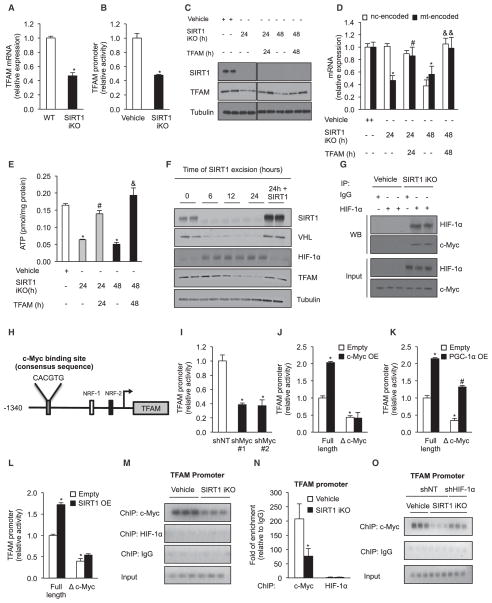
Figure 5. SIRT1 Regulates Mitochondrial Homeostasis by Modulation of the TFAM Promoter through HIF-1α/c-Myc.
(A) TFAM mRNA analyzed by qPCR in gastrocnemius of WT and SIRT1 iKO animals. Values were normalized to WT mice (n = 5, *p < 0.05 versus WT).
(B) TFAM promoter activity in SIRT1 flox/flox Cre-ERT2 primary myoblasts treated with vehicle or OHT to induce SIRT1 excision for 24 hr (SIRT1 iKO). Relative luciferase values were normalized to vehicle cells (n = 6, *p < 0.05 versus vehicle).
(C) Immunoblot for SIRT1, TFAM, and tubulin in SIRT1 flox/flox Cre-ERT2 primary myoblasts treated with vehicle or OHT to induce SIRT1 excision (SIRT1 iKO) for 24 or 48 hr, after which cells were infected with control or TFAM adenovirus.
(D) Expression of nuclear- versus mitochondrially encoded genes in SIRT1 flox/flox Cre-ERT2 primary myoblasts treated with OHT to induce SIRT1 excision (SIRT1 iKO) for 24 or 48 hr, after which cells were infected with TFAM adenovirus. Values were normalized to vehicle cells (n = 4, *p < 0.05 versus vehicle; #p < 0.05 versus SIRT1 iKO 24 hr; &p < 0.05 versus SIRT1 iKO 48 hr).
(E) ATP content in SIRT1 flox/flox Cre-ERT2 primary myoblasts treated with vehicle or OHT for 24 or 48 hr as in (D) (n = 4, *p < 0.05 versus vehicle; #p < 0.05 versus SIRT1 iKO 24 hr; &p < 0.05 versus SIRT1 iKO 48 hr).
(F) Immunoblot of SIRT1, VHL, HIF-1α, TFAM, and tubulin in SIRT1 flox/flox Cre-ERT2 primary myoblasts treated with vehicle or OHT to induce SIRT1 excision and in cells treated with OHT for 24 hr, after which SIRT1 was added back by adenoviral infection.
(G) Interaction of HIF-1α and c-Myc determined by immunoprecipitation of HIF-1α in SIRT1 flox/flox Cre-ERT2 primary myoblasts treated with OHT to excise SIRT1.
(H) The c-Myc-binding site on the TFAM promoter.
(I) TFAM promoter activity in primary myoblasts transduced with c-Myc or nontargeting shRNA (n = 4, *p < 0.05 versus shNT).
(J–L) TFAM promoter full-length activity or with mutation of c-Myc-binding site (Δ c-Myc) in primary myoblasts overexpressing c-Myc (J), PGC-1α (K), SIRT1 (L), or empty vector (n = 4, *p < 0.05 versus empty; #p < 0.05 versus Δ c-Myc empty).
(M and N) Chromatin immunoprecipitation (ChIP) (M) and respective quantification by qPCR (N) of c-Myc and HIF-1α to the TFAM promoter in SIRT1 flox/flox Cre-ERT2 primary myoblasts treated with vehicle or OHT to induce SIRT1 excision (n = 3, *p < 0.05 versus vehicle).
(O) ChIP of c-Myc to the TFAM promoter in SIRT1 flox/flox Cre-ERT2 primary myoblasts transduced with HIF-1α or nontargeting shRNA treated with vehicle or OHT to induce SIRT1 excision for 24 hr (SIRT1 iKO). Mitochondrially encoded genes were ND1, Cytb, COX1, ATP6.
Values are expressed as mean ± SEM. See also Figure S5.
HIF-1α stability has been previously reported to be regulated by acetylation of lysine 709 (Geng et al., 2011). To test whether SIRT1-mediated deacetylation was the mechanism, we mutated K709 to glutamine (an acetylation mimetic) or to arginine (nonacetylated mimetic), with K674 serving as a negative control (Lim et al., 2010). Neither of the K709 substitutions stabilized HIF-1α, nor were they affected by SIRT1 deletion (Figure S4C), indicating that SIRT1 does not regulate HIF-1α protein stability by deacetylating K709.
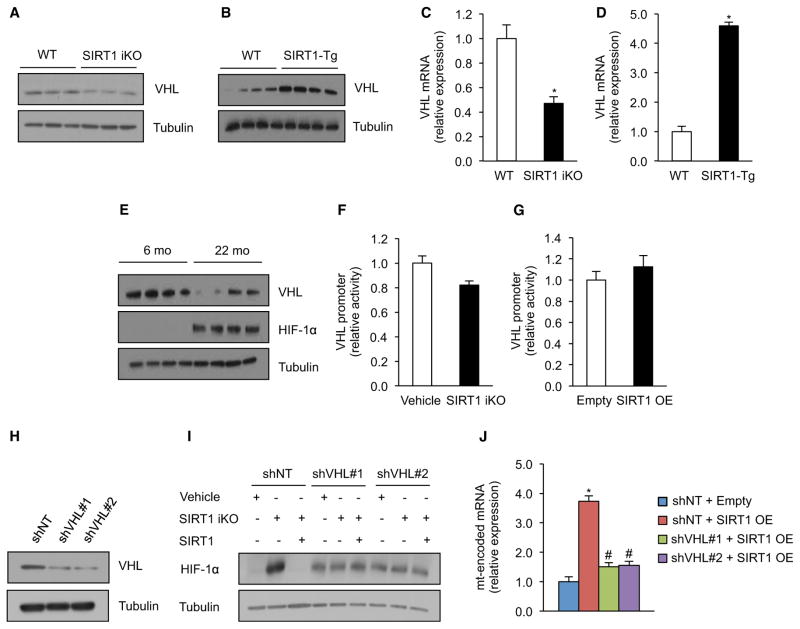
HIFα proteins are regulated by a proteasomal degradation mechanism mediated by the Von Hippel-Lindau (VHL) E3 ubiquitin ligase that recognizes hydroxylated proline residues on HIFα (Kaelin, 2008). Knockout of SIRT1 did not affect HIF-1α hydroxylation (Figure S4D), but in the SIRT1 iKO mouse and transgenic overexpressor the levels of SIRT1 correlated with VHL levels (Figures 4A–4D). VHL promoter activity was not altered by SIRT1 deletion, suggesting posttranscriptional regulation (Figures 4F and 4G). HIF-2α was also stabilized by SIRT1, though HIF-2α target genes were not upregulated (Figures S4E and S4F). The re-establishment of SIRT1 eliminated HIF-1α protein and restored levels of mitochondrial OXPHOS mRNA in SIRT1 iKO myoblasts, but these effects were lost when VHL was knocked down (Figures 4H–4J; also see Figure 5F). Thus, SIRT1 is constantly required to maintain mitochondrial homeostasis by inducing VHL and by ensuring that HIF-1α is degraded efficiently.
Figure 4. SIRT1 Regulates HIF-1α Stabilization in Skeletal Muscle through Regulation of VHL Expression.
(A and B) Immunoblot for VHL and tubulin in gastrocnemius of WT and SIRT1 iKO mice (A) and WT and SIRT1-Tg mice (B).
(C and D) VHL mRNA in WT and SIRT1 iKO mice (C) and WT and SIRT1-Tg mice (D). Values normalized to WT mice (n = 5, *p < 0.05 versus WT).
(E) Immunoblot for VHL, HIF-1α, and tubulin in gastrocnemius of 6- and 22-month-old mice.
(F) VHL promoter activity in SIRT1 flox/flox Cre-ERT2 primary myoblasts treated with vehicle or OHT for 24 hr to induce SIRT1 excision (SIRT1 iKO). Luciferase values normalized to vehicle cells (n = 5).
(G) VHL promoter activity in primary myoblasts with adenovirus expressing SIRT1 or empty vector. Luciferase values normalized to empty vector cells (n = 5).
(H) Immunoblot for VHL and tubulin in SIRT1 flox/flox Cre-ERT2 primary myoblasts transduced with VHL or nontargeting shRNA.
(I) Representative immunoblot for HIF-1α in SIRT1 flox/flox Cre-ERT2 primary myoblasts transduced with VHL or nontargeting shRNA and treated with OHT for 24 hr (SIRT1 iKO), after which SIRT1 was added back by adenoviral infection.
(J) Expression of mitochondrially encoded genes in primary myoblasts transduced with VHL or nontargeting shRNA and treated with adenovirus expressing SIRT1 or empty vector (n = 5, *p < 0.05 versus shNT empty; #p < 0.05 versus shNT SIRT1 OE). Mitochondrially encoded genes were ND1, Cytb, COX1, and ATP6.
Values are expressed as mean ± SEM. See also Figure S4.
SIRT1-HIF-1α Regulates Mitochondria by Modulating c-Myc’s Ability to Activate TFAM
These results raised the question of how HIF-1α, a nuclear protein, inhibits mitochondrial OXPHOS genes. Analysis of gene expression in SIRT1 iKO mice identified the nuclear-encoded mitochondrial factor TFAM as a candidate (Figures 5A and S5A). Consistent with this, TFAM promoter activity in SIRT1 iKO myoblasts was greatly reduced (Figure 5B), the reintroduction of TFAM into SIRT1 iKO cells restored levels of mitochondrially encoded mRNAs and ATP (Figure 5C–E), and in time course studies, TFAM levels declined 6 hr after VHL and HIF-1α (Figure 5F).
Knockdown of ARNT, a HIF-1α transcriptional binding partner (Wang et al., 1995), had no appreciable effect on mitochondrially encoded OXPHOS genes and ATP levels (Figures S5B–S5D), indicating that HIF-1α acts via a different mechanism. In cancer cells, metabolic reprogramming is mediated by crosstalk between HIF-1α and c-Myc (Gordan et al., 2007), raising the possibility that c-Myc was the missing factor. In fact, c-Myc DNA-binding sites are found at mitochondrial biogenesis genes (Kim et al., 2008; Li et al., 2005). Deletion of SIRT1 in primary myoblasts increased the binding between HIF-1α and c-Myc and reduced c-Myc reporter activity (Figures 5G and S5E). Similarly, knockdown of c-Myc completely blocked the ability of SIRT1 to induce mitochondrially encoded mRNAs and mtDNA (Figures S5F–S5H). Conversely, overexpression of c-Myc in myoblasts treated with a SIRT1 inhibitor, EX-527, prevented loss of mtDNA, mitochondrially encoded mRNA, and cellular ATP levels (Figures S5I–S5L).
We tested whether c-Myc directly controls TFAM promoter in myoblasts and is modulated by SIRT1-HIF-1α. TFAM is known to be regulated by PGC-1α, which interacts with NRF 1/2 bound at positions −311 and −154 in the TFAM promoter (Figure 5H). Knockdown of c-Myc reduced TFAM promoter activity (Figure 5I), consistent with a study in cancer cells (Li et al., 2005). We identified a putative c-Myc consensus sequence, CACGTG, 1,028 bp upstream of the ATG site—the mutation of which decreased promoter activity by about half without affecting PGC-1α-mediated induction (Figures 5J and 5K). Overexpression of SIRT1 also induced the TFAM promoter reporter, and mutation of the c-Myc binding site blocks this effect (Figure 5L). Chromatin IP experiments detected an interaction between c-Myc and the TFAM promoter, which was markedly reduced when SIRT1 was deleted, but not when HIF-1α was also knocked down (Figures 5M–O). We did not detect direct HIF-1α binding to TFAM (Figures 5M and 5N), with LDHA as a positive control (Figures S5M and S5N). Together, these data provide the first direct link between HIF-1α and the regulation of mitochondrially encoded genes in skeletal muscle and identify a mechanism of PGC-1α/β-independent regulation of mitochondrial function.
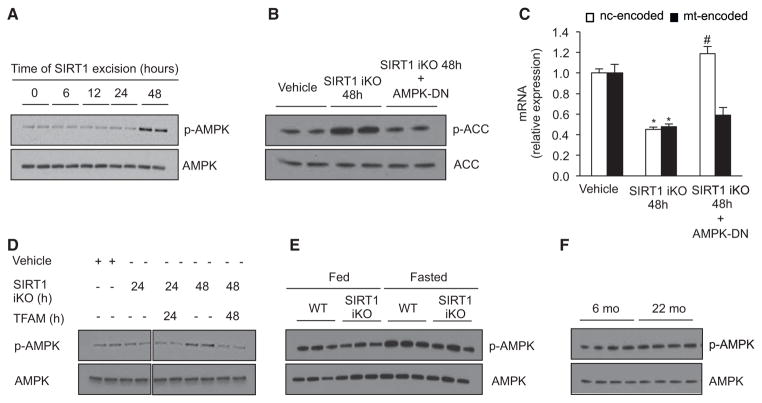
AMPK Functions as a Switch between PGC-1α-Dependent and -Independent Pathways Driven by SIRT1
Next, we determined the mechanisms that determine whether SIRT1 utilizes the PGC-1α-dependent or -independent pathways. Under conditions of low energy, AMPK-mediated phosphorylation of PGC-1α allows it to be deacetylated and activated by SIRT1 (Cantó et al., 2009; Gerhart-Hines et al., 2007; Rodgers et al., 2005), whereas under basal conditions, acetylation status is primarily regulated by the acetyltransferase GCN5 (Fernandez-Marcos and Auwerx, 2011). We speculated that the biphasic decline in OXPHOS subunits (in Figure 1L) might be due to AMPK. In time course experiments following SIRT1 deletion, AMPK activation occurred after 48 hr, well after the decline in VHL-TFAM and mitochondrial genes (Figures 1L–1M and 6A) but coincident with the decline in nuclear-encoded OXPHOS genes and mitochondrial mass (see Figures 1L–1M). An AMPK dominant-negative adenovirus (AMPK-DN) prevented the decline of nuclear OXPHOS mRNAs at 48 hr (Figures 6B and 6C), whereas forced maintenance of TFAM prevented AMPK activation (Figures 6D, 5D, and 5E). Together, these results strongly suggest that AMPK is the switch between the PGC-1α-dependent and -independent pathways. In this model, AMPK activation occurs in the absence of SIRT1 only when ATP levels fall below a threshold. Consistent with this, AMPK was unchanged under fed conditions in the SIRT1 iKO mice and 22-month-old wild-type mice but was markedly increased in fasting animals, when we observe changes in both nuclear- and mitochondrially encoded OXPHOS genes (Figure 6E and 6F).
Figure 6. AMPK Activity Regulates Switch between PGC-1α-Dependent and -Independent Mechanisms of Mitochondrial Regulation by SIRT1.
(A) Immunoblot for p-AMPK (Thr172) and AMPK in SIRT1 flox/flox Cre-ERT2 primary myoblasts treated with vehicle (0 hr) or OHT to induce SIRT1 excision.
(B) Immunoblot for p-ACC (Ser79) and ACC in SIRT1 flox/flox Cre-ERT2 primary myoblasts treated with vehicle or OHT for 48 hr (SIRT1 iKO) and infected with AMPK-DN adenovirus.
(C) Expression of nuclear- and mitochondrially encoded genes as in SIRT1 flox/flox Cre-ERT2 primary myoblasts treated with vehicle or OHT (SIRT1 iKO) and infected with empty or AMPK-DN adenovirus for the same period of time (n = 4, *p < 0.05 versus vehicle; #p < 0.05 versus SIRT1 iKO).
(D) Immunoblot for p-AMPK (Thr172) and AMPK in SIRT1 flox/flox Cre-ERT2 primary myoblasts treated with vehicle or OHT for 24 or 48 hr, after which cells were infected with control or TFAM adenovirus.
(E) Immunoblot for p-AMPK (Thr172) and AMPK in gastrocnemius of WT and SIRT1 iKO mice under fed and fasted conditions.
(F) Representative immunoblot for p-AMPK (Thr172) and AMPK in gastrocnemius of 6- and 22-month-old mice. Nuclear- and mitochondrially encoded genes were ND1, Cytb, COX1, ATP6, and NDUFS8, SDHb, Uqcrc1, COX5b, and ATP5a1, respectively.
Values are expressed as mean ± SEM.
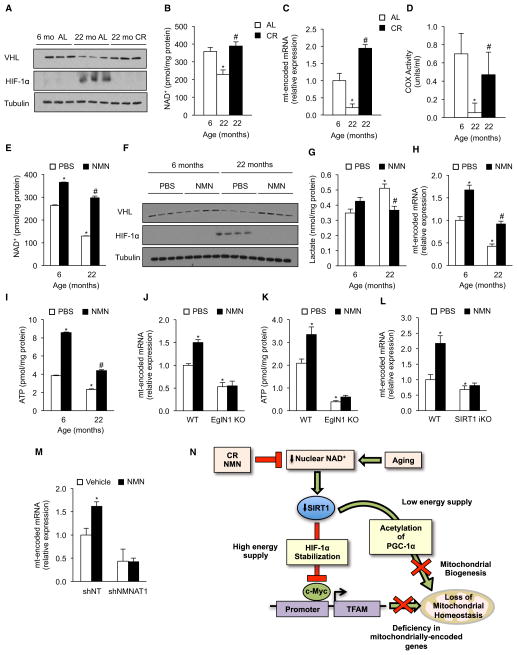
Increasing NAD+ Levels Restores Mitochondrial Homeostasis through the SIRT1-HIF-1α-c-Myc Pathway
CR is known to delay numerous diseases of aging in mammals, including cancer and type 2 diabetes. Interestingly, CR (30%–40% instituted at 6 weeks) completely prevented the decline in VHL and the increase in HIF-1α that occurs in adlibitum (AL)-fed 22-month-old mice (Figure 7A). The observed decreases in NAD+ and ATP levels, COX activity, mtDNA, and mitochondrially encoded OXPHOS components with age were also prevented by CR (Figures 7B–7D, S6A, and S6B). Unlike the accumulation of mutations in mtDNA, the pathway that we describe here should be rapidly reversible. Treatment of 22-month-old mice for 1 week with NMN, a precursor to NAD+ that increases NAD+ levels in vivo (Yoshino et al., 2011), reversed the decline in VHL and accumulation of HIF-1α (Figures 7E and 7F); reduced lactate levels; and increased ATP, COX activity, and mitochondrially encoded OXPHOS transcripts (Figures 7G–7I and S6D). In EglN1 and SIRT1 iKO mice, however, NMN failed to induce mitochondrially encoded genes or to raise ATP levels (Figures 7J–7L). Knockdown of NMNAT1 also prevented NMN from inducing mitochondrially encoded OXPHOS genes (Figure 7M), consistent with nuclear NAD+ being a key regulatory molecule. The SIRT1 iKO and the 22-month-old mice had increased levels of markers of muscle atrophy and inflammation compared to young WT mice, along with impaired insulin signaling and insulin-stimulated glucose uptake (Figures S1G–S1J and S6E–S6H). Strikingly, treatment of old mice with NMN reversed all of these biochemical aspects of aging and switched gastrocnemius muscle to a more oxidative fiber type (Figures S6E–S6H). However, we did not observe an improvement in muscle strength (data not shown), indicating that 1 week of treatment might not be sufficient to reverse whole-organism aging and that longer treatments might be required.
Figure 7. Increasing NAD+ Levels Rescues Age-Related Pseudohypoxia and OXPHOS Dysfunction through a SIRT1-HIF-1α Pathway.
(A) Immunoblot for HIF-1α and tubulin of 6- and 22-month-old AL and 22-month-old CR mice.
(B) NAD+ levels in the same cohorts as in (A) (n = 5, *p < 0.05 versus 6-month-old animals; #p < 0.05 versus 22-month-old AL).
(C) Expression of mitochondrially encoded genes of the same cohorts as in (A). Values normalized to 6-month-old mice (n = 5, *p < 0.05 versus 6-month-old mice; #p < 0.05 versus 22-month-old AL).
(D) Cytochrome c oxidase (COX) activity (n = 4, *p < 0.05 versus 6-month-old animals; #p < 0.05 versus 22-month-old AL).
(E) NAD+ levels in 6- and 22-month-old mice treated with vehicle (PBS) or NMN (n = 6, *p < 0.05 versus 6-month-old PBS; #p < 0.05 versus 22-month-old PBS).
(F) Immunoblot for VHL, HIF-1α, and tubulin of same cohorts as in (E).
(G) Lactate levels of same cohorts as in (E) (n = 6, *p < 0.05 versus 6-month-old PBS; #p < 0.05 versus 22-month-old PBS).
(H) Expression of mitochondrially encoded genes of same cohorts as in (E). Values normalized to 6-month-old PBS mice (n = 6, *p < 0.05 versus 6-month-old PBS; #p < 0.05 versus 22-month-old PBS).
(I) ATP content of same cohorts as in (E) (n = 6, *p < 0.05 versus 6-month-old PBS; #p < 0.05 versus 22-month-old PBS).
(J) Expression of mitochondrially encoded genes in WT and EglN1 KO mice treated with either vehicle (PBS) or NMN (n = 5, *p < 0.05 versus WT PBS).
(K) ATP content of same cohorts as in (J) (n = 5, *p < 0.05 versus WT PBS).
(L) Expression of mitochondrially encoded genes of WT and SIRT1 iKO mice treated with either vehicle (PBS) or NMN (n = 4, *p < 0.05 versus WT PBS).
(M) Expression of mitochondrially encoded genes in primary myoblasts transduced with NMNAT1 or nontargeting shRNA treated with either PBS or NMN (n = 4, *p < 0.05 versus shNT vehicle).
(N) Nuclear-mitochondrial communication and its decline during aging. Nuclear NAD+ levels regulate mitochondria via a PGC-1α-independent pathway that ensures the correct stoichiometry of OXPHOS subunits, but over time, a chronic pseudohypoxic response is activated, inhibiting OXPHOS. Mitochondrially encoded genes were ND1, Cytb, COX1, and ATP6.
Values are expressed as mean ± SEM. See also Figure S6.
DISCUSSION
Impairment in mitochondrial homeostasis is one of the hallmarks of aging that may underlie common age-related diseases (Lanza and Nair, 2010; Wallace, 2010). Despite its importance, there is still controversy as to why mitochondrial homeostasis is disrupted with age and whether this process can be slowed or even reversed. Here, we present evidence for a PGC-1α/β-independent pathway that ensures OXPHOS function and maintenance of mitochondrial homeostasis (Figure 7N). During aging, however, decline in nuclear energetic state or NAD+ levels reduces the activity of SIRT1 in the nucleus, causing VHL levels to decline and HIF-1α to be stabilized. This program, which likely evolved to modulate mitochondrial metabolism in response to changes in energy supply, becomes chronically activated in old mice, inducing a pseudohypoxic state that disrupts OXPHOS, a phenomenon that is consistent with antagonistic pleiotropy (Williams and Day, 2003).
One of the more surprising findings is the existence of a SIRT1-mediated pathway that regulates mitochondria independently of PGC-1α/β. The data indicate that SIRT1 can regulate these two pathways in response to the energetic state of the cell. Which one predominates depends on AMPK activity and the phosphorylation status of PGC-1α (Cantó et al., 2009).
This study shows that HIF-1α-induced metabolic reprogramming occurs in normal tissue and that it disrupts mitochondrial homeostasis. We consider the metabolic state of the old mice as pseudohypoxic because the downstream effects are similar to hypoxia but occur even when oxygen is abundant, as previously in type 2 diabetes and cancer (Ido and Williamson, 1997; Sanders, 2012; Williamson et al., 1993). An interesting implication is that reprogramming of normal tissue toward a Warburg-like state may increase ROS and establish a milieu for subsequent mutations to initiate carcinogenesis, a possibility that may help explain why cancer risk increases exponentially with age.
All of the main players in the nuclear NAD+-SIRT1-HIF-1α-OXPHOS pathway are present in lower eukaryotes, indicating that the pathway evolved early in life’s history. This pathway may have evolved to coordinate nuclear-mitochondrial synchrony in response to changes in energy supplies and oxygen levels, and its decline may be a conserved cause of aging. In C. elegans, HIF-1α is known to be a key determinant of lifespan, though its precise role is still a matter of debate (Leiser and Kaeberlein, 2010). HIF-1α modulation may have differential effects on lifespan depending on the animal’s diet or whether the mtUPR is activated (Dillin et al., 2002; Durieux et al., 2011; Houtkooper et al., 2013). Though we did not detect mUPR in skeletal muscle, we do not exclude the possibility that mtUPR plays a role in other tissues or under different conditions.
Additional studies will be required to elucidate complex feedback loops that likely regulate the SIRT1-HIF-1α-Myc-TFAM pathway. For example, in cancer cells, SIRT1 directly regulates c-Myc transcriptional activity, either by deacetylation of c-Myc (Menssen et al., 2012) or by binding c-Myc and promoting its association with Max (Mao et al., 2011). Given that SIRT3 and SIRT6 also regulate HIF-1α and compromise respiration (Bell et al., 2011; Finley et al., 2011; Zhong et al., 2010), it will be interesting to test whether a decline in the activity of other sirtuins causes a similar loss of TFAM and mitochondrially encoded OXPHOS components.
How broadly applicable might these findings be? High-fat diet feeding increases levels of HIF-1α in liver (Carabelli et al., 2011) and white adipose tissue, the latter of which correlated with a decline in mitochondrial gene expression (Krishnan et al., 2012). Moreover, insulin-resistant human skeletal muscle has a signature reminiscent of hypoxia (Ptitsyn et al., 2006). In SIRT1 iKO mice, specific dysregulation of mitochondrial OXPHOS genes is also observed in the heart, demonstrating that the pathway is relevant not only to skeletal muscle (Figure S1K–S1N), but not in liver, WAT, or brain. In these tissues, other factors such as SIRT3 or SIRT6 may be responsible for regulation of HIF-1α, or the metabolic status of the tissue at the time of harvest may also be critical. Current dogma is that aging is irreversible. Our data show that 1 week of treatment with a compound that boosts NAD+ levels is sufficient to restore the mitochondrial homeostasis and key biochemical markers of muscle health in a 22-month-old mouse to levels similar to a 6-month-old mouse. Although further work is necessary, this study suggests that increasing NAD+ levels and/or small compounds that prevent HIF-1α stabilization or promote its degradation might be an effective therapy for organismal decline with age. In summary, these findings provide evidence for a new pathway that controls carbon utilization and OXPHOS independently of PGC-1α, a pathway that goes awry over time but is readily reversible, with implications for treating aging and age-related diseases.
EXPERIMENTAL PROCEDURES
Aging Cohorts, SIRT1-iKO, EglN1 KO, and SIRT1-Tg Mice and NMNAT1 Electroporation
Wild-type C57BL/6J mice were from the National Institutes of Aging, NIH. EglN1 KO, SIRT1-iKO, and SIRT1-Tg mice were described previously (Minamishima et al., 2008; Price et al., 2012). For NMN experiments, mice were given IP injections of 500 mg NMN/kg body weight per day or the equivalent volume of PBS for 7 consecutive days at 6 PM and 8 AM on day 8 and were sacrificed 4 hr after last injection. All animal care followed the guidelines and was approved by Institutional Animal Care and Use Committees (IACUCs).
Adenovirus Generation and Mutagenesis
Adenoviruses were generated as described before (Rodgers et al., 2005) and were used to infect cells for the time points described in the figures. HIF-1α and TFAM promoter mutants were generated using a commercially available kit (Stratagene) according to the manufacturer’s instructions. Details about these methodologies and the primers used for the mutagenesis can be found in the Extended Experimental Procedures.
Generation of Primary Myoblasts, Rho0 Cells, Cell Culture Treatments, Adenoviral Infections, and Gene Silencing
Primary myoblasts were isolated from PGC-1α/β KO, SIRT1 iKO, and PGC-1α null mice as described before (Price et al., 2012). Rho0 cells were generated by culturing the cells in media supplemented with 4 g/l glucose and 2 mM pyruvate, 50 ng/ml ethidium bromide (Alfa aesar), and 100 μg/ml uridine (Sigma-Aldrich) for 4 weeks to deplete mitochondrial DNA. Cells were treated with the NMN (Sigma-Aldrich) and DMOG (Cayman) as described in the figure legends, and details can be found in the Extended Experimental Procedures. Gene silencing was achieved using pLKO.1 shRNAs for the genes of interest, as described before (Gomes et al., 2012), and was used after selection with eukaryotic marker of the vector. Details about these methodologies can also been found in the Extended Experimental Procedures.
Mitochondrial Function
Cytochrome c oxidase and succinate dehydrogenase measurements were determined pollarographically as described before (Gomes et al., 2012). Cytochrome c oxidase was also measured spectrophotometrically using a commercially available kit (Sigma-Aldrich) according to the manufacturer’s instructions. ATP content was measured with a commercial kit (Roche) according to the manufacturer’s instructions. Mitochondrial membrane potential and reactive oxygen species were measured by FACS as described (Bell et al., 2011; Price et al., 2012). Electron microscopy was also determined as described (Price et al., 2012). Details about these methodologies can be found in the Extended Experimental Procedures.
Gene Expression and mtDNA Analysis
Total RNA and genomic DNA were isolated using a commercially available kit (QIAGEN) according to the manufacturer’s instructions. cDNA was generated using the iScript kit (BioRad). Gene expression and mtDNA were determined by qPCR as described (Price et al., 2012). Details and primer sequences can be found in the Extended Experimental Procedures.
Coimmunoprecipitation
Proteins from primary myoblasts were crosslinked using DSP (1 mM, Pierce), after which the cells were lysed in a low-stringency IP buffer (0.05% NP-40, 50 mM NaCl, 0.5 mM EDTA, 50 mM Tris-HCl [pH 7.4]) supplemented with protease inhibitor cocktail (Roche) and 25 U/ml endonuclease (Pierce). Endogenous HIF-1α protein was immunoprecipitated using anti-HIF-1α antibody (Cayman) coated A/G magnetic beads (Pierce), and anti-IgG was used as control. Imunoprecipitated proteins and input were run on SDS-PAGE and were revealed with anti-HIF-1α (Cayman) and anti-c-Myc (Abcam) antibodies. Details can be found in the Extended Experimental Procedures.
Chromatin Immunoprecipitation and Immunoblots
Chromatin immunoprecipitation was performed using a commercially available kit (Millipore) according to the manufacturer’s instructions and using anti-HIF1α (Cayman), anti-c-Myc (Abcam), and anti-IgG as control. Immunoblots were performed as described (Price et al., 2012), and details can be found in the Extended Experimental Procedures.
TFAM Promoter, VHL Promoter, HRE, and c-Myc Activity
TFAM promoter, VHL promoter, HRE, and c-Myc activity were determined using a luciferase-based system. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) with Renilla as the reference, and details about the constructs can be found in the Extended Experimental Procedures.
NAD+ Measurement
NAD+ from skeletal muscle was quantified with a commercially available kit (BioVision) according to the manufacturer’s instructions and as described before (Gomes et al., 2012).
Statistical Analysis
Data were analyzed by a two-tailed Student’s t test. Statistical analysis was performed using Excel software.
Supplementary Material
Acknowledgments
The Sinclair lab is supported by the NIH/NIA, the Glenn Foundation for Medical Research, the United Mitochondrial Disease Foundation, the Juvenile Diabetes Research Foundation, and a gift from the Schulak family. A.P.G. was supported by the Portuguese Foundation for Science and Technology (SFRH/BD/44674/2008) and B.P.H. by an NSERC PGS-D fellowship. N.T. is supported by an Australian Research Council Future Fellowship. We are grateful to Michael Bonkowski, Carlos Daniel de Magalhaes Filho, Meghan Rego, Nikolina Dioufa, and David Zhang for technical advice and experimental assistance; William Kaelin Jr. for kindly providing the EglN1 KO mice; Daniel Kelly, John Rumsay, and Teresa Leone for unpublished PGC-1α/β KO myoblasts and advice; Bruce Spiegelman for PGC-1α null myoblasts and advice; and Pere Puigserver and Zachary Gerhart-Hines for a SIRT1 adenovirus. D.A.S. is a consultant to Cohbar, OvaScience, HorizonScience, Segterra, MetroBiotech, and GlaxoSmithKline. Cohbar, MetroBiotech, and GlaxoSmithKline work on mitochondrially derived peptides, NAD+, and sirtuin modulation, respectively.
Footnotes
Supplemental Information includes Extended Experimental Procedures, six figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.11.037.
References
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell EL, Klimova TA, Eisenbart J, Schumacker PT, Chandel NS. Mitochondrial reactive oxygen species trigger hypoxia-inducible factor-dependent extension of the replicative life span during hypoxia. Mol Cell Biol. 2007;27:5737–5745. doi: 10.1128/MCB.02265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Lau C, Dahlmann M, Ziegler M. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J Biol Chem. 2005;280:36334–36341. doi: 10.1074/jbc.M508660200. [DOI] [PubMed] [Google Scholar]
- Braidy N, Guillemin GJ, Mansour H, Chan-Ling T, Poljak A, Grant R. Age related changes in NAD+ metabolism oxidative stress and Sirt1 activity in wistar rats. PLoS ONE. 2011;6:e19194. doi: 10.1371/journal.pone.0019194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvári M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C, Auwerx J. NAD+ as a signaling molecule modulating metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:291–298. doi: 10.1101/sqb.2012.76.010439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli J, Burgueño AL, Rosselli MS, Gianotti TF, Lago NR, Pirola CJ, Sookoian S. High fat diet-induced liver steatosis promotes an increase in liver mitochondrial biogenesis in response to hypoxia. J Cell Mol Med. 2011;15:1329–1338. doi: 10.1111/j.1582-4934.2010.01128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS, Schumacker PT. Cells depleted of mitochondrial DNA (rho0) yield insight into physiological mechanisms. FEBS Lett. 1999;454:173–176. doi: 10.1016/s0014-5793(99)00783-8. [DOI] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Marcos PJ, Auwerx J. Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr. 2011;93:884S–90. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Harvey CT, Pittsenbarger J, Liu Q, Beer TM, Xue C, Qian DZ. HDAC4 protein regulates HIF1α protein lysine acetylation and cancer cell response to hypoxia. J Biol Chem. 2011;286:38095–38102. doi: 10.1074/jbc.M111.257055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AP, Duarte FV, Nunes P, Hubbard BP, Teodoro JS, Varela AT, Jones JG, Sinclair DA, Palmeira CM, Rolo AP. Berberine protects against high fat diet-induced dysfunction in muscle mitochondria by inducing SIRT1-dependent mitochondrial biogenesis. Biochim Biophys Acta. 2012;1822:185–195. doi: 10.1016/j.bbadis.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Choi CS, Chin S, Kim S, Kawamori D, Kurpad AJ, Neubauer N, Hu J, Mootha VK, Kim YB, et al. Abnormal glucose homeostasis in skeletal muscle-specific PGC-1alpha knockout mice reveals skeletal muscle-pancreatic beta cell crosstalk. J Clin Invest. 2007;117:3463–3474. doi: 10.1172/JCI31785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D. The biologic clock: the mitochondria? J Am Geriatr Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ido Y, Williamson JR. Hyperglycemic cytosolic reductive stress ‘pseudohypoxia’: implications for diabetic retinopathy. Invest Ophthalmol Vis Sci. 1997;38:1467–1470. [PubMed] [Google Scholar]
- Kaelin WG., Jr The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008;8:865–873. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee JH, Iyer VR. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One. 2008;3:e1798. doi: 10.1371/journal.pone.0001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan J, Danzer C, Simka T, Ukropec J, Walter KM, Kumpf S, Mirtschink P, Ukropcova B, Gasperikova D, Pedrazzini T, Krek W. Dietary obesity-associated Hif1α activation in adipocytes restricts fatty acid oxidation and energy expenditure via suppression of the Sirt2-NAD+ system. Genes Dev. 2012;26:259–270. doi: 10.1101/gad.180406.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong LK, Sohal RS. Age-related changes in activities of mitochondrial electron transport complexes in various tissues of the mouse. Arch Biochem Biophys. 2000;373:16–22. doi: 10.1006/abbi.1999.1495. [DOI] [PubMed] [Google Scholar]
- Lanza IR, Nair KS. Mitochondrial function as a determinant of life span. Pflugers Archiv. 2010;459:277–289. doi: 10.1007/s00424-009-0724-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Hekimi S. When a theory of aging ages badly. Cell Mol Life Sci. 2010;67:1–8. doi: 10.1007/s00018-009-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson NG. Somatic mitochondrial DNA mutations in mammalian aging. Annu Rev Biochem. 2010;79:683–706. doi: 10.1146/annurev-biochem-060408-093701. [DOI] [PubMed] [Google Scholar]
- Leiser SF, Kaeberlein M. The hypoxia-inducible factor HIF-1 functions as both a positive and negative modulator of aging. Biol Chem. 2010;391:1131–1137. doi: 10.1515/BC.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libert S, Guarente L. Metabolic and neuropsychiatric effects of calorie restriction and sirtuins. Annu Rev Physiol. 2013;75:669–684. doi: 10.1146/annurev-physiol-030212-183800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell. 2010;38:864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B, Zhao G, Lv X, Chen HZ, Xue Z, Yang B, Liu DP, Liang CC. Sirt1 deacetylates c-Myc and promotes c-Myc/Max association. Int J Biochem Cell Biol. 2011;43:1573–1581. doi: 10.1016/j.biocel.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Massudi H, Grant R, Braidy N, Guest J, Farnsworth B, Guillemin GJ. Age-associated changes in oxidative stress and NAD+ metabolism in human tissue. PLoS ONE. 2012;7:e42357. doi: 10.1371/journal.pone.0042357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen A, Hydbring P, Kapelle K, Vervoorts J, Diebold J, Lüscher B, Larsson LG, Hermeking H. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci USA. 2012;109:E187–E196. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamishima YA, Moslehi J, Bardeesy N, Cullen D, Bronson RT, Kaelin WG., Jr Somatic inactivation of the PHD2 prolyl hydroxylase causes polycythemia and congestive heart failure. Blood. 2008;111:3236–3244. doi: 10.1182/blood-2007-10-117812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Cantó C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, et al. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NL, Gomes AP, Ling AJ, Duarte FV, Martin-Montalvo A, North BJ, Agarwal B, Ye L, Ramadori G, Teodoro JS, et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012;15:675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptitsyn A, Hulver M, Cefalu W, York D, Smith SR. Unsupervised clustering of gene expression data points at hypoxia as possible trigger for metabolic syndrome. BMC Genomics. 2006;7:318. doi: 10.1186/1471-2164-7-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Sanders E. Pseudohypoxia, mitochondrial mutations, the Warburg effect, and cancer. Biomed Res. 2012;23:109–131. [Google Scholar]
- Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, Yamada KA, Imai S. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013;18:416–430. doi: 10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen. 2010;51:440–450. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Williams PD, Day T. Antagonistic pleiotropy, mortality source interactions, and the evolutionary theory of senescence. Evolution. 2003;57:1478–1488. doi: 10.1111/j.0014-3820.2003.tb00356.x. [DOI] [PubMed] [Google Scholar]
- Williamson JR, Chang K, Frangos M, Hasan KS, Ido Y, Kawamura T, Nyengaard JR, van den Enden M, Kilo C, Tilton RG. Hyperglycemic pseudohypoxia and diabetic complications. Diabetes. 1993;42:801–813. doi: 10.2337/diab.42.6.801. [DOI] [PubMed] [Google Scholar]
- Yang H, Baur JA, Chen A, Miller C, Adams JK, Kisielewski A, Howitz KT, Zipkin RE, Sinclair DA. Design and synthesis of compounds that extend yeast replicative lifespan. Aging Cell. 2007;6:35–43. doi: 10.1111/j.1474-9726.2006.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino J, Mills KF, Yoon MJ, Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner C, Lai L, Zechner JF, Geng T, Yan Z, Rumsey JW, Collia D, Chen Z, Wozniak DF, Leone TC, Kelly DP. Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metab. 2010;12:633–642. doi: 10.1016/j.cmet.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.