Medulloblastoma is the most common paediatric malignant tumour. To identify altered genetic events in a comprehensive manner, we recently performed exome sequencing of a series of medulloblastomas [1]. This study identified mutations in genes involved in chromatin modification in 20% of patients examined, including the myeloid/lymphoid or mixed lineage leukemia (MLL) family genes MLL2 and MLL3, which were not previously known to be associated with medulloblastoma [1]. The majority of those alterations were nonsense or frameshift mutations, indicating that MLL2 and MLL3 are new medulloblastoma tumour suppressor genes [1]. Subsequent exome sequencing studies further validated MLL2 pathway mutations as medulloblastoma driver events [2-4]. In this report, we present detailed histopathological characteristics of three cases with MLL2/3 gene mutations.
The male patient discussed in case #1 initially presented as a 5-year-old with a profound frontal headache associated with nausea and vomiting, following receipt of an immunization booster. Five days later the headache returned, and he was noted to have a gait imbalance; a magnetic resonance imaging scan showed a fourth ventricular mass (Figure 1A). Histopathological analysis revealed a medulloblastoma. Therapy consisted of craniospinal irradiation with a posterior fossa boost and chemotherapy consisting of a bone marrow transplant protocol of vincristine, amifostine, cisplatin, and cyclophosphamide. He is now 5 years post therapy without evidence of disease.
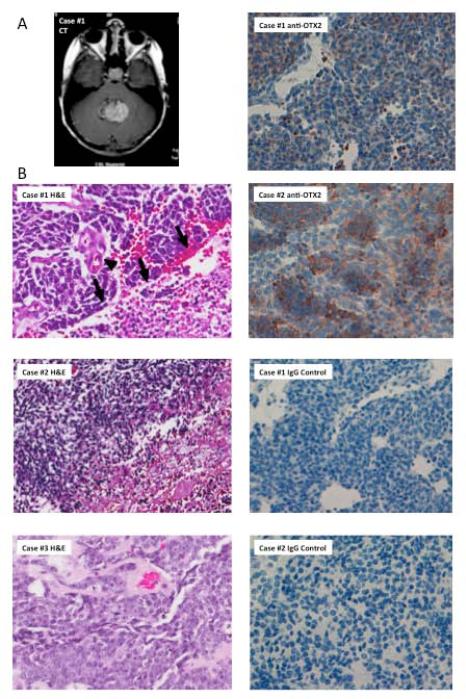
Figure.
A) CT demonstrates a hyperdense 3x4 centimeter midline posterior fossa mass that appears to fill the fourth ventricle, containing punctate calcifications and cystic components. B) Haematoxylin and eosin staining of histological sections from Case #1, Case #2, and Case #3. Note that Case #1 demonstrates hyperchromatic, molded nuclei associated with vascular proliferation (arrowhead) and geographic necrosis (arrows) indicative of a severely anaplastic medulloblastoma; geographic necrosis also suggests a lack of differentiation. C) Immunohistochemical assay shows positive OTX2 expression in Case #1 (Score:40% 2+) and Case #2 (Score: 50% 2+).
Case #2 is of a male patient who presented as an 11-year-old who began to experience decreased appetite and headaches that awoke him, associated with nausea and vomiting. A computed tomography scan showed marked hydrocephalus with a 4 cm mass in the posterior fossa. Histopathological analysis identified a medulloblastoma. Post-operatively, he underwent cranio-spinal radiation therapy and chemotherapy with vincristine, cisplatin, and cyclophosphamide supplemented with hyperalimentation via gastric tube placement. Now at six years post-diagnosis, he is doing well at recent follow-up.
Case #3 is a female patient who presented as a 7-year-old with a three-week history of headache associated with morning nausea and vomiting, dizziness and recent onset of double vision. Radiographic studies revealed an enhancing mass lesion in the fourth ventricle. Axial and sagittal gadolinium-enhanced images demonstrated diffuse leptomeningeal spread of disease. Histopathological analysis disclosed a medulloblastoma. Cytological examination of her post-operative cerebrospinal fluid revealed malignant cytology. The patient began craniospinal X-ray therapy. Three months following initial diagnosis, she died of disease. Postmortem examination of the brain and spinal cord revealed extensive spread along the subarachnoid space of the cerebellum, forebrain, brain stem, and spinal cord.
The term medulloblastoma describes a series of heterogeneous brain tumours originating in the cerebellum. This heterogeneity is reflected at two levels: (1) tumours are histopathologically and molecularly distinct; (2) there is a lack of tight correlation between histopathological and molecular subtypes, as tumours within each histopathological subtype are also molecularly heterogeneous. Accordingly, additional genetic alterations, and analysis of the histopathological characteristics associated with them, may provide information for improving tumour subclassification. As a first step toward that purpose, we present three medulloblastoma cases with MLL2/3 mutations. Intriguingly, all three cases demonstrate features of a moderate to severe large-cell/anaplastic subtype (Figure 1B). However, despite these similarities, clinical outcomes varied. Patient #3 had both MLL2 and MLL3 mutations and, unlike the first two patients, had a poor clinical outcome. However, Patient #3 also had MYC amplification (frequently associated with a poor prognosis [5]).
The role of MLL2/MLL3 complexes in medulloblastoma are unknown, yet genetic and biological evidence supports a tumour suppressor role [1-4, 6], and studies have identified MLL2/3 gene mutations in a variety of other cancers. MLL family genes are essential for histone modification and play roles in regulating other developmentally critical pathways [7, 8]. One of these pathways impacted by MLL2, retinoic acid signaling [9], may in turn impact orthodenticle homeobox 2 (OTX2) expression [10]. Because increased OTX2 expression was noted (Table 1, Figure 1C), it is tempting to postulate that MLL2/3 inactivation, and the subsequence changes in histone methylation, may present a mechanism for OTX2 overexpression, and thus dysregulation of OTX2-associated pathways. Additionally, it is possible that loss of MLL2/MLL3 function impairs cell differentiation and renders cells susceptible to transformation. All cases presented here demonstrated anaplastic features, geographic necrosis and characteristics of the same histopathological subclass. Molecular subclassification, completed for Cases #1 and #2, revealed Group 3 classification for both cases (classification based on Northcott et al. 2011[11]). Because of the presence of MYC amplification and the extremely poor prognosis, it is likely that the tumour in Case #3 is also a Group 3 tumor.
| Case | Age | Gender | MLL2/3 Mutation* | PTCH/CTNNB1/TP53/PTEN Status | MYC Amplification* | OTX2 Amplification* | OTX2 Expression | Histopathologic Subtype | Survival |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | Male | MLL2: missense (R4852Q) | no mutation | No | No | Positive | Severe Anaplastic | 5 years, alive |
| 2 | 11 | Male | MLL2: Frameshift | no mutation | ND | ND | Positive | Moderate Anaplastic | 6 years, alive |
| 3 | 7 | Female | MLL2: Nonsense (R4921X); MLL3: Nonsense (R2609X) | no mutation | Yes | No | ND | Large Cell | 3 months, deceased |
It is expected that improved subclassification will provide guidance for therapy and risk assessment in the clinical setting. MLL2/3 mutations add one more genetic variable for subclassification of medulloblastomas. MLL2/3 pathway mutations were found to be distributed among various histological groups in previous studies [2, 4]. Additionally, studies have found MLL2/3 mutations to be distributed among various molecular subgroups [2-4]. To clarify the subclassification issue, more detailed histopathological analysis of a large number of patients with MLL2/3 mutations will be necessary. We favour the possibility that dysregulation of the MLL2/3 pathway affects the histopathological and clinical characteristics of medulloblastoma, and we suggest an analysis of more cases is warranted.
Acknowledgements
Funding was provided by National Cancer Institute Grant R01-CA-118822-01 Supplement and the Pediatric Brain Tumor Foundation. The authors thank Lisa Ehringer, Diane Satterfield, and Ling Wang of the Preston Robert Tisch Brain Tumor Center at the Duke University Medical Center for preparing tumor samples, and Debra Fleming of the Molecular Pathology Laboratory at Duke for molecular classification of medulloblastoma samples.
Abbreviations
- MLL2
myeloid/lymphoid or mixed lineage leukemia 2
- MLL3
myeloid/lymphoid or mixed lineage leukemia 3
- 2-OTX2
orthodenticle homeobox
Footnotes
Author contributions: Gerald Grant, Herbert E Fuchs, Linda G. Leithe, Sridharan Gururangan, Darell D. Bigner, and Hai Yan provided tumors and reagents. Roger E. McLendon completed pathological analyses of samples. Giselle Lopez, Roger E.McLendon, and Yiping He contributed to the writing and editing of the manuscript.
Conflict of interest statement: The authors do not have any conflicts of interest to report.
References
- 1.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, Gallia GL, Jallo GI, Binder ZA, Nikolsky Y, Hartigan J, Smith DR, Gerhard DS, Fults DW, VandenBerg S, Berger MS, Marie SK, Shinjo SM, Clara C, Phillips PC, Minturn JE, Biegel JA, Judkins AR, Resnick AC, Storm PB, Curran T, He Y, Rasheed BA, Friedman HS, Keir ST, McLendon R, Northcott PA, Taylor MD, Burger PC, Riggins GJ, Karchin R, Parmigiani G, Bigner DD, Yan H, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stutz AM, Rausch T, Warnatz HJ, Ryzhova M, Bender S, Sturm D, Pleier S, Cin H, Pfaff E, Sieber L, Wittmann A, Remke M, Witt H, Hutter S, Tzaridis T, Weischenfeldt J, Raeder B, Avci M, Amstislavskiy V, Zapatka M, Weber UD, Wang Q, Lasitschka B, Bartholomae CC, Schmidt M, von Kalle C, Ast V, Lawerenz C, Eils J, Kabbe R, Benes V, van Sluis P, Koster J, Volckmann R, Shih D, Betts MJ, Russell RB, Coco S, Tonini GP, Schuller U, Hans V, Graf N, Kim YJ, Monoranu C, Roggendorf W, Unterberg A, Herold-Mende C, Milde T, Kulozik AE, von Deimling A, Witt O, Maass E, Rossler J, Ebinger M, Schuhmann MU, Fruhwald MC, Hasselblatt M, Jabado N, Rutkowski S, von Bueren AO, Williamson D, Clifford SC, McCabe MG, Collins VP, Wolf S, Wiemann S, Lehrach H, Brors B, Scheurlen W, Felsberg J, Reifenberger G, Northcott PA, Taylor MD, Meyerson M, Pomeroy SL, Yaspo ML, Korbel JO, Korshunov A, Eils R, Pfister SM, Lichter P. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, Greulich H, Lawrence MS, Lennon NJ, McKenna A, Meldrim J, Ramos AH, Ross MG, Russ C, Shefler E, Sivachenko A, Sogoloff B, Stojanov P, Tamayo P, Mesirov JP, Amani V, Teider N, Sengupta S, Francois JP, Northcott PA, Taylor MD, Yu F, Crabtree GR, Kautzman AG, Gabriel SB, Getz G, Jager N, Jones DT, Lichter P, Pfister SM, Roberts TM, Meyerson M, Pomeroy SL, Cho YJ. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, Chalhoub N, Baker SJ, Huether R, Kriwacki R, Curley N, Thiruvenkatam R, Wang J, Wu G, Rusch M, Hong X, Becksfort J, Gupta P, Ma J, Easton J, Vadodaria B, Onar-Thomas A, Lin T, Li S, Pounds S, Paugh S, Zhao D, Kawauchi D, Roussel MF, Finkelstein D, Ellison DW, Lau CC, Bouffet E, Hassall T, Gururangan S, Cohn R, Fulton RS, Fulton LL, Dooling DJ, Ochoa K, Gajjar A, Mardis ER, Wilson RK, Downing JR, Zhang J, Gilbertson RJ. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Hoff K, Hartmann W, von Bueren AO, Gerber NU, Grotzer MA, Pietsch T, Rutkowski S. Large cell/anaplastic medulloblastoma: outcome according to myc status, histopathological, and clinical risk factors. Pediatr Blood Cancer. 2010;54:369–376. doi: 10.1002/pbc.22339. [DOI] [PubMed] [Google Scholar]
- 6.Lee J, Kim DH, Lee S, Yang QH, Lee DK, Lee SK, Roeder RG, Lee JW. A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc Natl Acad Sci U S A. 2009;106:8513–8518. doi: 10.1073/pnas.0902873106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 8.Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2011;144:513–525. doi: 10.1016/j.cell.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo C, Chang CC, Wortham M, Chen LH, Kernagis DN, Qin X, Cho YW, Chi JT, Grant GA, McLendon RE, Yan H, Ge K, Papadopoulos N, Bigner DD, He Y. Global identification of MLL2-targeted loci reveals MLL2's role in diverse signaling pathways. Proc Natl Acad Sci U S A. 2012;109:17603–17608. doi: 10.1073/pnas.1208807109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di C, Liao S, Adamson DC, Parrett TJ, Broderick DK, Shi Q, Lengauer C, Cummins JM, Velculescu VE, Fults DW, McLendon RE, Bigner DD, Yan H. Identification of OTX2 as a medulloblastoma oncogene whose product can be targeted by all-trans retinoic acid. Cancer Res. 2005;65:919–924. [PubMed] [Google Scholar]
- 11.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins CE, French P, Rutka JT, Pfister S, Taylor MD. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]