Abstract
Background
Two polymorphisms, -260C/T and -651C/T, in the CD14 gene have been implicated in susceptibility to cancer. However, the results remain inconclusive. This meta-analysis aimed to investigate the association between the two polymorphisms and risk of cancer.
Methods
All eligible case-control studies published up to March 2014 were identified by searching PubMed, Web of Science, CNKI and WanFang database. Pooled odds ratio (OR) with 95% confidence interval (CI) were used to access the strength of this association in fixed- or random-effects model.
Results
17 case-control studies from fourteen articles were included. Of those, there were 17 studies (4198 cases and 4194 controls) for -260C/T polymorphism and three studies (832 cases and 1190 controls) for -651C/T polymorphism. Overall, no significant associations between the two polymorphisms of CD14 gene and cancer risk were found. When stratified by ethnicity, cancer type and source of control, similar results were observed among them. In addition, in further subgroups analysis by Helicobacter pylori (H. pylori) infection status and tumor location in gastric cancer subgroup, we found that the CD14 -260C/T polymorphism may increase the risk of gastric cancer in H. pylori-infected individuals.
Conclusions
This meta-analysis suggests that the CD14 -260C/T polymorphism may increase the risk of gastric cancer in H. pylori-infected individuals. However, large and well-designed studies are warranted to validate our findings.
Introduction
Cancer is a major public health problem worldwide and about 12.7 million cancer cases and 7.6 million cancer deaths were reported based on GLOBOCAN 2008 [1]. It is well known that cancer is a multistep process resulting from complex interactions between genetic and environmental factors [2], [3]. Despite the latter play important roles in the development of cancer. Host genetic factors are closely related to the pathophysiology of many human cancers [4]. Variants in several innate immunity genes have been identified as biologically plausible candidates for effects on cancer, such as CD14.
The CD14 gene is localized on chromosome 5q31.1, which encodes a receptor protein that binds to lipopolysaccharide (LPS), its primary ligand, and interacts with co-receptors toll-like receptor 4 (TLR4) and lymphocyte antigen 96 (LY96) [5], [6]. CD14 is expressed on the surface of monocytes, macrophages, and neutrophils as membrane CD14 (mCD14) and in the serum as soluble CD14 (sCD14) and its expression may be partially regulated at the genetic level [7], [8]. There are several polymorphism sites in the CD14 gene, and two well-studied common SNPs in the promoter region of CD14, -260C/T (rs2569190; also reported as CD14 -159) and -561C/T (rs5744455), are investigated extensively to the susceptibility of cancer [9]–[27]. However, the results remain controversial. In this study, we conduct a meta-analysis to evaluate the association between the two polymorphisms and cancer risk.
Materials and Methods
Search strategy
We searched the PubMed, Web of Science, CNKI and WanFang database before March 1, 2014, by using the key subjects “cancer”, “carcinoma”, “genetic polymorphism”, “polymorphism”, “variant” in combination with “cluster of differentiation 14”, “CD14”. Additional studies were identified by a hand search of references of original or review articles on this topic. Search results were restricted to human populations and articles were written in English or Chinese. If more than one geographic or cancer type was reported in one report, each was extracted separately. If data or data subsets were published in more than one article, only the publication with the largest sample size was included.
Inclusion criteria and exclusion criteria
Studies were included according to the following criteria: (1) studies that evaluated the association between the CD14 polymorphisms and cancer, (2) designed in case-control study, and (3) detailed genotype frequency of cases and controls were provided directly or could be calculated from the article text. Studies were excluded when they were: (1) case-only study, case reports, and review articles, (2) studies without the raw data of the -260C/T genotype of CD14, (3) repetitive publications, and (4) studies deviated from the Hardy-Weinberg equilibrium (HWE), (5) animal studies.
Data extraction
For each study, the following data were extracted independently by two investigators: the first author's name, year of publication, country of origin, ethnicity of study population, cancer type, source of control, genotype method, number of cases and controls and HWE in controls (P value). The results were compared, and disagreements were discussed among all authors and resolved with consensus.
Statistical analysis
The risk of cancer associated with the CD14 polymorphisms was estimated for each study by odds ratio (OR) and 95% confidence interval (CI). Four different ORs were calculated: dominant model (CT+TT vs. CC), recessive model (TT vs. CT+CC), heterozygote comparison (CT vs. CC), and homozygote comparison (TT vs. CC). A χ2-test-based Q statistic test was performed to assess the between-study heterogeneity [28]. When a significant Q test (P>0.1) indicated homogeneity across studies, the fixed effects model was used [29], otherwise, the random effects model was applied [30]. We also quantified the effect of heterogeneity by I 2 test (I 2<25%: no heterogeneity; I 2 = 25–50%: moderate heterogeneity; I 2 = 50–75%: large heterogeneity, I 2>75%: extreme heterogeneity) [31]. HWE among controls for each study was examined by χ2 test. We performed stratification analyses on ethnicity, tumor type and source of control. If any cancer type less than three studies was combined into “other” cancers. Additionally, we also conducted subgroup analysis by H. pylori infection status and tumor location in gastric cancer group. Analysis of sensitivity was performed to evaluate the stability of the results, namely, a single study in the meta-analysis was deleted each time to reflect the influence of the individual data set to the pooled OR. Finally, potential publication bias was investigated using Begg' funnel plot and Egger's regression test [32], [33]. P<0.05 was regarded as statistically significant.
All statistical analyses were performed using the Cochrane Collaboration RevMan 5.2 and STATA package version 12.0 (Stata Corporation, College Station, Texas).
Results
Study characteristics
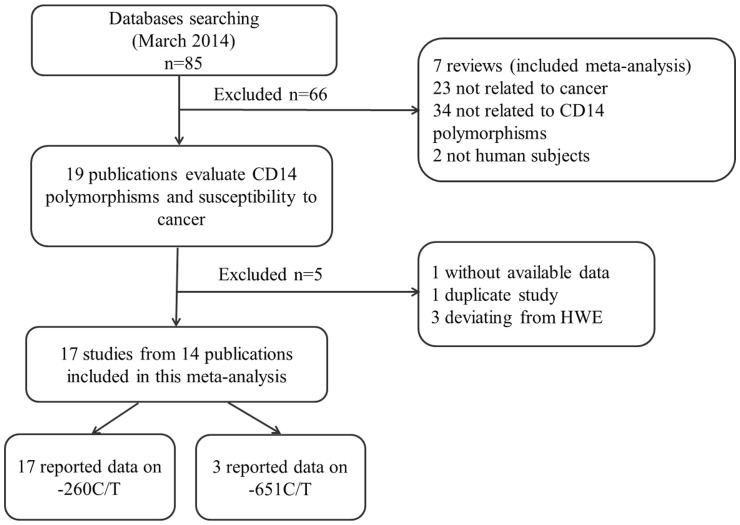
Following the searching strategy, 85 potentially relevant studies were retrieved. According to the inclusion criteria, 19 publications [9]–[27] with full-text were selected and were subjected to further examination. Because the studies [14], [19] included two tumor types respectively and the study by Hold et al [14] included two populations, we treated them separately in this meta-analysis. We excluded one study because they did not present detailed genotyping information [23]. We also excluded one study [24] because it included the overlapped data with those included in the analysis [12]. Furthermore, we removed 3 studies because their genotype distributions among the controls deviated from HWE [25]–[27]. The flow chart of study selection in summarized in Figure 1. As shown in Table 1, therefore, a total of 17 studies from 14 publications were included. Of those, there were 17 studies with 4198 cases and 4194 controls concerning -260C/T polymorphism and three studies with 832 cases and 1190 controls concerning -651C/T polymorphism. Among 17 case-control studies, ten studies were conducted in Asians and seven in Caucasians. Two cancer types were addressed: nine studies on gastric and eight on other cancers (2 on colorectal, acute lymphoblastic leukemia (ALL), lymphomas and one on esophageal, prostate, separately).
Figure 1. Flow chart showing study selection procedure.
Table 1. Characteristics of studies included in the meta-analysis.
| Study | Year | Country | Ethnicity | Cancer type | Source of controls | Genotype methods | Genotype (case/control) | HWE (P value) | |||
| Total | CC | CT | TT | ||||||||
| -260C/T | |||||||||||
| Andrie [9] | 2009 | USA | Caucasian | Childhood lymphomas | HB | PCR | 83/83 | 31/18 | 39/42 | 13/23 | 0.886 |
| Castano-Rodriguez [10] | 2013 | Malaysia, Singapore | Asian | Gastric | HB | Real time-PCR | 70/214 | 18/34 | 38/108 | 14/72 | 0.537 |
| Companioni [11] | 2013 | Mixed | Caucasian | Gastric | PB | Illumina Beadstation | 1192/352 | 307/103 | 621/173 | 264/76 | 0.833 |
| Guo [12] | 2006 | China | Asian | Colorectal | PB | PCR-RFLP | 110/160 | 35/25 | 34/77 | 41/58 | 0.947 |
| Hao [13] | 2010 | China | Asian | Gastric | HB | PCR-RFLP | 90/100 | 7/18 | 45/52 | 38/30 | 0.581 |
| Hold [14] | 2009a | Polish | Caucasian | Gastric | PB | TaqMan | 327/389 | 110/131 | 134/176 | 83/82 | 0.112 |
| Hold [14] | 2009b | USA | Caucasian | Gastric | PB | TaqMan | 306/211 | 91/52 | 147/108 | 68/51 | 0.730 |
| Hold [14] | 2009c | USA | Caucasian | Esophageal | PB | TaqMan | 158/211 | 50/52 | 74/108 | 34/51 | 0.730 |
| Landi [15] | 2006 | Spain | Caucasian | Colorectal | HB | TaqMan | 281/265 | 62/65 | 151/137 | 68/63 | 0.580 |
| Miedema [16] | 2012 | Nertherland | Caucasian | Childhood ALL | PB | PCR | 186/182 | 46/28 | 81/101 | 59/53 | 0.077 |
| Min [17] | 2012 | China | Asian | Prostate | HB | PCR-LDR | 168/208 | 73/102 | 71/80 | 24/26 | 0.105 |
| Tahara [18] | 2007 | Japan | Asian | Gastric | HB | PCR-RFLP | 149/94 | 37/14 | 80/53 | 32/27 | 0.147 |
| Wu [19] | 2006a | China | Asian | Gastric | HB | PCR | 204/210 | 52/54 | 102/102 | 50/54 | 0.679 |
| Wu [19] | 2006b | China | Asian | MALT lymphomas | HB | PCR | 70/210 | 17/54 | 29/102 | 24/54 | 0.679 |
| Yu [20] | 2011 | China | Asian | ALL | PB | PCR-RFLP | 174/539 | 29/80 | 55/259 | 90/200 | 0.796 |
| Zhang [21] | 2011 | China | Asian | Gastric | HB | PCR-RFLP | 160/296 | 85/141 | 61/135 | 14/20 | 0.102 |
| Zhao [22] | 2007 | China | Asian | Gastric | PB | PCR-RFLP | 470/470 | 33/56 | 225/227 | 212/187 | 0.305 |
| -651C/T | |||||||||||
| Miedema [16] | 2012 | Nertherland | Caucasian | ALL | PB | PCR | 188/181 | 108/96 | 66/77 | 14/8 | 0.124 |
| Yu [19] | 2011 | China | Asian | ALL | PB | PCR-RFLP | 174/539 | 99/287 | 60/213 | 15/39 | 0.952 |
| Zhao [22] | 2007 | China | Asian | Gastric | PB | PCR-RFLP | 470/470 | 257/257 | 191/183 | 22/30 | 0.735 |
HWE: Hardy-Weinberg equilibrium; PB: population-based; HB: hospital-based; PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism; PCR-LDR: polymerase chain reaction-ligase detection reaction;
ALL: acute lymphoblastic leukemia; MALT lymphomas: gastric mucosa-associated lymphoid tissue lymphoma.
Quantitative data synthesis
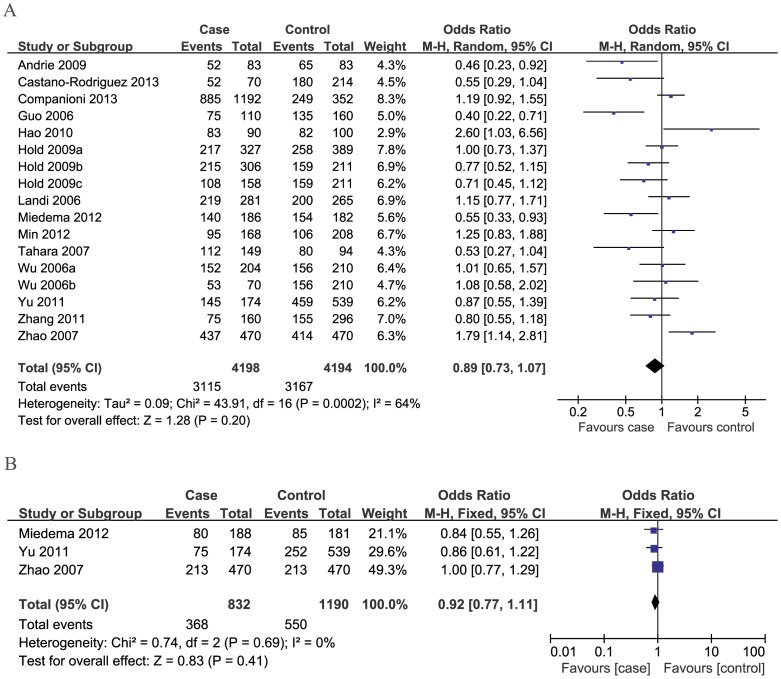
For 260C/T polymorphism, overall, no significant associations between the CD14 -260C/T polymorphism and cancer risk were found (dominant model: OR = 0.89, 95%CI: 0.73–1.07; recessive model: OR = 1.08, 95%CI: 0.93–1.25; CT vs. CC: OR = 0.85, 95%CI: 0.70–1.03; TT vs. CC: OR = 0.95, 95%CI: 0.76–1.19) (Table 2, Figure 2A).
Table 2. Summary of ORs of the CD14 polymorphisms and cancer risk.
| Variables | na | dominant model | recessive model | CT vs. CC | TT vs. CC | ||||||||
| OR(95% CI) | P b | I 2 | OR(95% CI) | P b | I 2 | OR(95% CI) | P b | I 2 | OR(95% CI) | P b | I 2 | ||
| -260C/T | |||||||||||||
| Total | 17 | 0.89(0.73,1.07) | 0.0002 | 64 | 1.08(0.93,1.25) | 0.03 | 44 | 0.85(0.70,1.03) | 0.0004 | 62 | 0.95(0.76,1.19) | 0.0005 | 61 |
| Ethnicity | |||||||||||||
| Asian | 10 | 0.92(0.68,1.25) | 0.0006 | 69 | 1.15(0.91,1.45) | 0.02 | 55 | 0.86(0.62,1.17) | 0.0008 | 68 | 1.03(0.71,1.49) | 0.0008 | 68 |
| Caucasian | 7 | 0.85(0.67,1.08) | 0.02 | 59 | 1.01(0.87,1.18) | 0.39 | 4 | 0.84(0.66,1.08) | 0.04 | 55 | 0.88(0.67,1.15) | 0.07 | 49 |
| Cancer type | |||||||||||||
| Gastric | 9 | 0.99(0.77,1.26) | 0.005 | 63 | 1.04(0.86,1.26) | 0.09 | 42 | 0.98(0.78,1.22) | 0.03 | 52 | 1.03(0.75,1.43) | 0.002 | 67 |
| Others | 8 | 0.78(0.58,1.04) | 0.009 | 63 | 1.12(0.87,1.44) | 0.05 | 51 | 0.70(0.51,0.97) | 0.005 | 65 | 0.86(0.63,1.18) | 0.04 | 53 |
| Source of control | |||||||||||||
| PB | 8 | 0.86(0.65,1.14) | 0.0005 | 73 | 1.18(1.04,1.33) | 0.15 | 34 | 0.78(0.56,1.07) | 0.0001 | 76 | 0.98(0.75,1.30) | 0.01 | 61 |
| HB | 9 | 0.91(0.70,1.19) | 0.02 | 55 | 0.97(0.74,1.27) | 0.05 | 48 | 0.93(0.78,1.12) | 0.15 | 34 | 0.91(0.61,1.36) | 0.004 | 65 |
| -651C/T | |||||||||||||
| Total | 3 | 0.92(0.77,1.11) | 0.69 | 0 | 1.02(0.70,1.49) | 0.21 | 36 | 0.92(0.76,1.11) | 0.37 | 0 | 0.98(0.67,1.44) | 0.35 | 5 |
Number of comparison,
Test for heterogeneity.
Figure 2. Meta-analysis of the association between CD14 polymorphisms and susceptibility to cancer under dominant model.
A: -260C/T; B: -651C/T.
In the subgroup analysis on ethnicity, similar results were observed in both Asian and Caucasian populations in all genetic models; when stratified by cancer type, we also failed to detect any association between the -260C/T polymorphism and gastric and other cancers (Table 2).
Stratification based on the source of controls showed significant associations between the -260C/T polymorphism and risk of cancer in the population-based subgroup under recessive model (OR = 1.18, 95%CI: 1.04–1.33). However, no significant association was found in the other three models and population-based subgroup (Table 2).
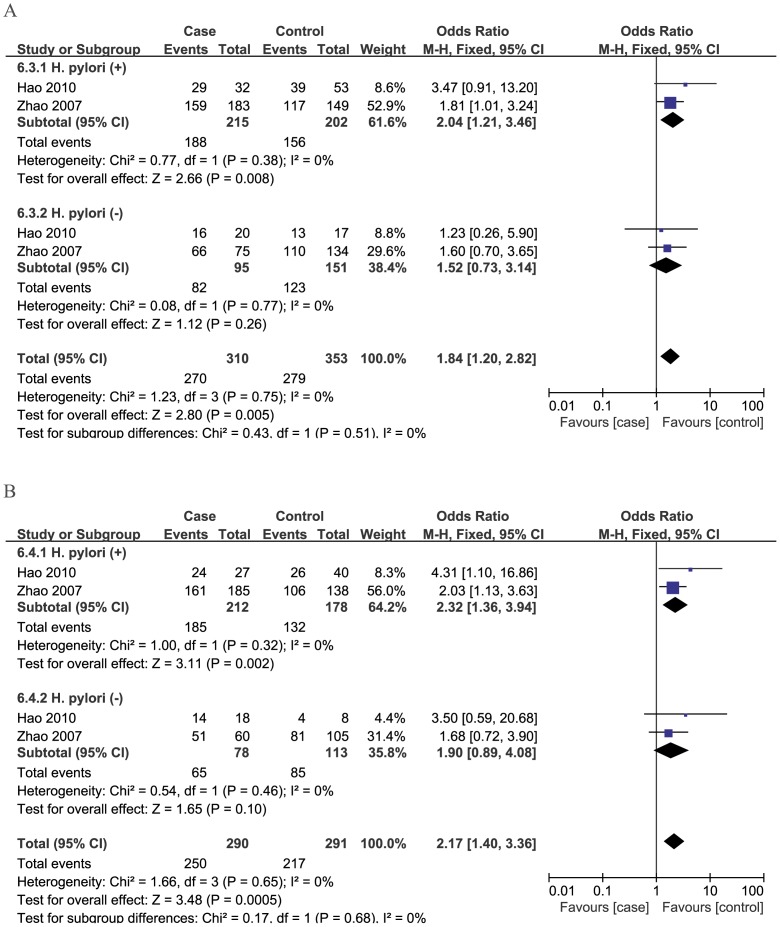
In addition, in the gastric cancer subgroup, a further stratified analysis based on H. pylori infection status and tumor location was conducted. When the analysis was stratified by H. pylori infection status, three studies [10], [13], [22] reported the available data and the pooled results showed that the -260C/T polymorphism may be a risk factor for gastric cancer in H. pylori-infected individuals (CT vs. CC: OR = 2.04, 95%CI: 1.21–3.46, TT vs. CC: OR = 2.32, 95%CI: 1.36–3.94) (Figure 3). However, in stratified analysis by tumor location, three studies [11], [14], [18] reported the available data and we found that no significant association between -260C/T polymorphism and risk of cardia and non-cardia cancers (Table 3).
Figure 3. Subgroup analysis by H. pylori infection status of odds ratios for association between CD14 -260C/T polymorphism and risk of gastric cancer.
A: CT vs CC; B: TT vs CC.
Table 3. Summary of ORs of the -260C/T polymorphism and gastric cancer risk by H. pylori infection status and location.
| Variables | na | dominant model | recessive model | CT vs. CC | TT vs. CC | ||||||||
| OR(95% CI) | P b | I 2 | OR(95% CI) | P b | I 2 | OR(95% CI) | P b | I 2 | OR(95% CI) | P b | I 2 | ||
| H. pylori infection | |||||||||||||
| H. pylori (+) | 3/2* | 1.51(0.58,3.92) | 0.01 | 76 | 1.28(0.95,1.73) | 0.59 | 0 | 2.04(1.21,3.46) | 0.38 | 0 | 2.32(1.36,3.94) | 0.32 | 0 |
| H. pylori (−) | 3/2* | 1.44(0.75,2.77) | 0.51 | 0 | 1.26(0.83,1.93) | 0.16 | 49 | 1.52[0. 73,3.14] | 0.77 | 0 | 1.90(0.89,4.08) | 0.46 | 0 |
| Location | |||||||||||||
| Cardia | 3 | 0.75(0.54,1.04) | 0.67 | 0 | 0.84(0.58,1.22) | 0.33 | 11 | 0.77(0.54,1.10) | 0.42 | 0 | 0.69(0.44,1.06) | 0.68 | 0 |
| Non-cardia | 3 | 0.83(0.63,1.08) | 0.33 | 9 | 0.90(0.68,1.19) | 0.25 | 28 | 0.83(0.62,1.11) | 0.56 | 0 | 0.80(0.57,1.13) | 0.15 | 48 |
Number of comparison,
Test for heterogeneity,
* 3 studies in the dominant model, 2 in the other models.
For -651C/T polymorphism, three studies were included. We found no statistical association between the -651 polymorphism and overall cancer risk in all genetic models (Table 2, Figure 2B).
Heterogeneity and sensitivity analyses
Substantial heterogeneities were observed among studies for the association between the CD14 -260C/T polymorphism and cancer risk under all genetic models (dominant model: I2 = 64%, P = 0.0002; recessive model: I2 = 44%, P = 0.003; CT vs. CC: I2 = 62%, P = 0.0004; TT vs. CC: I2 = 61%, P = 0.0005). Then, we assessed the source of heterogeneity for all genetic model comparison by ethnicity, cancer type and source of control. The heterogeneity was partly decreased in Caucasians and hospital-based populations in some models. However, there was still significant heterogeneity among Asians, gastric, population-based and other cancers. Then sensitivity analysis was performed by excluding each study individually to evaluate the stability of the results. The statistical significance of the results was not altered when any single study was omitted, confirming the stability of the results.
Publication bias
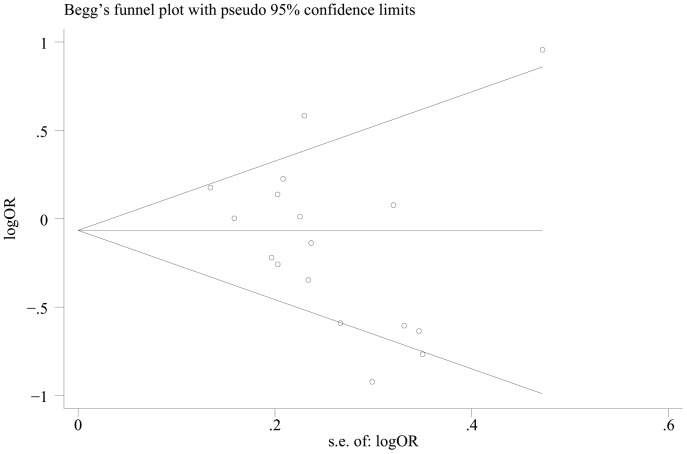
Begg's funnel plot and Egger's test were performed to assess the potential publication bias in the available literature. The shape of funnel plots did not reveal any evidence of funnel plot asymmetry (Figure 4). Egger's test also showed that there was no statistical significance for the evaluation of publication bias (dominant model: P = 0.144, CT vs. CC: P = 0.117, TT vs. CC: P = 0.141, recessive model: P = 0.123).
Figure 4. Begg's funnel plot for publication bias (dominant model).
Discussion
Genetic polymorphisms in genes whose products regulate the immune and antitumor responses in malignancies are good candidates for investigation. Many candidate genes were reported to be associated to cancer risk, such as TLRs, CD14. TLRs are pattern recognition receptors (PRR) of the innate immune system that recognise a wide variety of molecules. With respect to CD14, it is a pattern-recognition receptor that plays a central role in innate immunity and directs the adaptive immune responses [34]. As a co-receptor of TLRs, CD14 acts primarily by transferring LPS and other bacterial ligands from circulating LPS-binding protein to the TLR4/MD-2 signaling complex. Two common promoter polymorphisms have been identified in the CD14 gene at positions -260 and -651 from the AUG start codon, which correspond to -159 and -550 designated according to the transcription start site, respectively [35], [36]. With regard to -260C/T polymorphism, LeVan et al. [37] showed that the T allele has a decreased affinity for DNA/protein interactions at a GC box containing a binding site for SP1, SP2, and SP3 transcription factors and leads to an increased transcriptional activity. Consistently, Hartel et al. [38] reported that after in vitro stimulation of cord blood cultures with LPS, carriers of the -159T allele have higher levels of sCD14 compared with carriers of the -159C allele. Recently, the -260C/T polymorphism in CD14 gene has been investigated the association with many diseases, such as inflammatory bowel disease [39], alcoholic liver disease [40], tuberculosis [41], sepsis [42], coronary heart disease [43], asthma [44] and allergic rhinitis [45]. As for cancer, a previous meta-analysis conducted by Zhou et al. [46], evaluated the association between CD14 -260C/T polymorphism and risk of cancer based on 12 studies including 2498 cases and 2696 controls and reported that the CD14 -159C/T gene polymorphism is not a genetic risk factor for cancer.
In this study, we conducted a comprehensive literature search in different databases and included several additional studies, which allowed for a larger number of subjects (17 studies including 4198 cases and 4194 controls) and more precise risk estimation. Besides, we conducted a further stratified analysis based on H. pylori infection status and tumor location in gastric cancer group. In addition, we also explore the association between CD14 -651C/T polymorphism and risk of cancer based on three studies with 832 cases and 1190 controls. The pooled data demonstrated that no significant associations between the two polymorphisms of CD14 gene and cancer risk were found in overall comparison. Besides, in the subgroup analysis by ethnicity and cancer type, we also failed to detect any association between the -260C/T polymorphism and risk of Asians, Caucasians, gastric and other cancers. However, when stratified by source of control, a significant association between the -260C/T polymorphism and risk of cancer in the population-based subgroup was found under recessive model. The results seem to contradict the observations of functional studies of CD14, which had suggested that CD14 played an important role in the development of cancer. Since carcinogenesis is a multistep process involving multifactorial interplay between genetic and environmental factors that involves various genetic alterations and several biological pathways. Thus, it is unlikely that risk factors of cancer work in isolation from each other. What's more, the different linkage disequilibrium patterns usually exist in related genes and the influence of the genetic variant may be masked by other unidentified causal genes involved in carcinogenesis. In addition, only few studies on -651C/T polymorphism were included, which may also contribute to the result and it should be interpreted with caution.
As H. pylori infection is known to be the main risk factor for gastric cancer [47], we examined the potential interaction between H. pylori infection and CD14 -260C/T polymorphism in the development of gastric cancer. The pooled results showed that the -260C/T polymorphism may be a risk factor for gastric cancer in H. pylori-infected individuals. Since mCD14 is mostly expressed in monocytes/macrophages, which are accumulated in H. pylori infected mucosa [48]. That is, individual with CT/TT genotype had higher sCD14 levels compared with the carriers with C allele. The results indicate that -260C/T polymorphism might play a role in the outcome of H. pylori infection, especially the development of gastric cancer. In addition, we also explored the -260C/T polymorphism association with both anatomical localizations of gastric cancer and there was no significant association between -260C/T polymorphism and risk of cardia and non-cardia cancers. However, because only few studies were included in the above analysis, the result should be interpreted with caution, and more studies are needed.
Heterogeneity is a potential problem when interpreting the results of all meta-analysis [49]. In this meta-analysis, heterogeneity was found in overall comparison in three genetic models, when stratified by ethnicity, cancer type and source of control, the heterogeneity was partly decreased in Caucasians and hospital-based populations. However, heterogeneity still existed among Asians, population-based, gastric and other cancers. Then sensitivity analyses were conducted by successively excluding one study, the estimated pooled odd ratio changed quite little, strengthening the results from this meta-analysis. The results above suggest that the different ethnicities, cancer type and population selection might contribute to the heterogeneity observed in the meta-analysis. Besides, lifestyle, environmental background and other unknown factors may also be the source of heterogeneity. No publication bias was shown suggesting this possible true result.
In interpreting our results of the current meta-analysis, some limitations should be acknowledged. First, the controls were not uniformly defined. Some studies used a healthy population as the control group, whereas others selected patients without cancers in hospital as the reference group. Therefore, the controls may not always be truly representative in the underlying source populations, especially when the polymorphism is also expected to affect the risk of other diseases. Second, the number of published studies was not sufficiently large for a comprehensive analysis, particularly for subgroup analysis by cancer type. Thus, we may fail to explore the real association between the polymorphism and specific cancer type (such as colorectal, ALL). Third, because of the lack of original data, our results were based on single-factor estimates without adjustment for age, gender and other risk factors (e.g. smoking, drinking status), which may cause serious confounding bias.
In conclusion, this meta-analysis suggests that the CD14 -260C/T polymorphism may increase the risk of gastric cancer in H. pylori-infected individuals. However, large and well-designed studies are warranted to validate our findings. Moreover, more gene-gene and gene-environment interactions should also be considered in future analysis, which should lead to better, comprehensive understanding of the association between the CD14 polymorphisms and cancer risk.
Supporting Information
PRISMA Checklist.
(DOC)
MOOSE Checklist.
(DOC)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. Data deposition.
Funding Statement
The authors have no support or funding to report.
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, et al. (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Bredberg A (2011) Cancer: more of polygenic disease and less of multiple mutations? A quantitative viewpoint. Cancer 117: 440–445. [DOI] [PubMed] [Google Scholar]
- 3. Pharoah PD, Dunning AM, Ponder BA, Easton DF (2004) Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer 4: 850–860. [DOI] [PubMed] [Google Scholar]
- 4. Lin BK, Clyne M, Walsh M, Gomez O, Yu W, et al. (2006) Tracking the epidemiology of human genes in the literature: the HuGE Published Literature database. Am J Epidemiol 164: 1–4. [DOI] [PubMed] [Google Scholar]
- 5. Gu W, Dong H, Jiang DP, Zhou J, Du DY, et al. (2008) Functional significance of CD14 promoter polymorphisms and their clinical relevance in a Chinese Han population. Crit Care Med 36: 2274–2280. [DOI] [PubMed] [Google Scholar]
- 6. Triantafilou M, Triantafilou K (2002) Lipopolysaccharide recognition: CD14, TLRs and the LPS-activation cluster. Trends Immunol 23: 301–304. [DOI] [PubMed] [Google Scholar]
- 7. Goyert SM, Ferrero E, Rettig WJ, Yenamandra AK, Obata F, et al. (1988) The CD14 monocyte differentiation antigen maps to a region encoding growth factors and receptors. Science 239: 497–500. [DOI] [PubMed] [Google Scholar]
- 8. Ulevitch RJ, Tobias PS (1995) Receptor-Dependent Mechanisms of Cell Stimulation by Bacterial Endotoxin. Annu Rev Immunol 13: 437–457. [DOI] [PubMed] [Google Scholar]
- 9. Andrie E, Michos A, Kalampoki V, Pourtsidis A, Moschovi M, et al. (2009) Genetic variants in immunoregulatory genes and risk for childhood lymphomas. Eur J Haematol 83: 334–342. [DOI] [PubMed] [Google Scholar]
- 10. Castaño-Rodríguez N, Kaakoush NO, Goh KL, Fock KM, Mitchell HM (2013) The role of TLR2, TLR4 and CD14 genetic polymorphisms in gastric carcinogenesis: a case-control study and meta-analysis. PLoS One 8: e60327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Companioni O, Bonet C, Muñoz X, Weiderpass E, Panico S, et al. (2014) Polymorphisms of Helicobacter pylori signaling pathway genes and gastric cancer risk in the European Prospective Investigation into Cancer-Eurgast cohort. Int J Cancer 134: 92–101. [DOI] [PubMed] [Google Scholar]
- 12. Guo Q, Zhu J, Xia B (2006) Polymorphism of CD14 gene but not the mutation of TLR4 gene is associated with colorectal cancer in Chinese patients. J Gastroenterol Hepatol 21: 92–97. [DOI] [PubMed] [Google Scholar]
- 13. Hao WY (2010) The influence of CD14-260 and-651C/T polymorphism in H.pylori infection-related gastric carcinoma. Nankai University (Article in Chinese). [Google Scholar]
- 14. Hold GL, Rabkin CS, Gammon MD, Berry SH, Smith MG, et al. (2009) CD14-159C/T and TLR9-1237T/C polymorphisms are not associated with gastric cancer risk in Caucasian populations. Eur J Cancer Prev 18: 117–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Landi S, Gemignani F, Bottari F, Gioia-Patricola L, Guino E, et al. (2006) Polymorphisms within inflammatory genes and colorectal cancer. J Negat Results Biomed 5: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miedema KG, Tissing WJ, Te Poele EM, Kamps WA, Alizadeh BZ, et al. (2012) Polymorphisms in the TLR6 gene associated with the inverse association between childhood acute lymphoblastic leukemia and atopic disease. Leukemia 26: 1203–1210. [DOI] [PubMed] [Google Scholar]
- 17. Min ZC, Mi YY, Shao N, Feng NH, Hua LX (2012) Association between CD14-260 polymorphism and the risk of the prostate cancer. J Mod Urol 17: 118–121 (Article in Chinese). [Google Scholar]
- 18. Tahara T, Arisawa T, Shibata T, Hirata I, Nakano H (2007) Association of polymorphism of TLR4 and CD14 genes with gastroduodenal diseases in Japan. Inflammopharmacology 15: 124–128. [DOI] [PubMed] [Google Scholar]
- 19. Wu MS, Cheng TY, Shun CT, Lin MT, Chen LC, et al. (2006) Functional polymorphisms of CD14 and toll-like receptor 4 in Taiwanese Chinese with Helicobacter pylori-related gastric malignancies. Hepatogastroenterology 53: 807–810. [PubMed] [Google Scholar]
- 20. Yu X, Zhang C, Sun A, Jiang L, Zheng J, et al. (2011) Genetic variations in CD14 promoter and acute lymphoblastic leukemia susceptibility in a Chinese population. DNA Cell Biol 30: 777–782. [DOI] [PubMed] [Google Scholar]
- 21. Zhang L (2011) Association of gene polymorphisms with helicobacter pylori related gastric cancer. Chongqing Medical University (Article in Chinese). [Google Scholar]
- 22. Zhao D, Sun T, Zhang X, Guo Y, Yu D, et al. (2007) Role of CD14 promoter polymorphisms in Helicobacter pylori infection-related gastric carcinoma. Clin Cancer Res 13: 2362–2368. [DOI] [PubMed] [Google Scholar]
- 23. Ture-Ozdemir F, Gazouli M, Tzivras M, Panagos C, Bovaretos N, et al. (2008) Association of polymorphisms of NOD2, TLR4 and CD14 genes with susceptibility to gastric mucosa-associated lymphoid tissue lymphoma. Anticancer Res 28: 3697–3700. [PubMed] [Google Scholar]
- 24. Chen LS, Guo QS, Xia B (2006) Polymorphism of CD14 gene promoter but not Toll-like receptor 4 gene is associated with colorectal cancer in Chinese patients. Chin J Dig 26: 735–738 (Article in Chinese). [Google Scholar]
- 25. Chao YC, Chu HC, Chang WK, Huang HH, Hsieh TY (2005) CD14 promoter polymorphism in Chinese alcoholic patients with cirrhosis of liver and acute pancreatitis. World J Gastroenterol 11: 6043–6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mason TE, Ricks-Santi L, Chen W, Apprey V, Joykutty J, et al. (2010) Association of CD14 variant with prostate cancer in African American men. Prostate 70: 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zeljic K, Supic G, Jovic N, Kozomara R, Brankovic-Magic M, et al. Association of TLR2, TLR3, TLR4 and CD14 genes polymorphisms with oral cancer risk and survival. Oral Dis 2013 Jun 1 doi: 10.1111/odi.12144 [Epub ahead of print] PubMed PMID: 23796347 [DOI] [PubMed] [Google Scholar]
- 28. Lau J, Ioannidis JP, Schmid CH (1997) Quantitative synthesis in systematic reviews. Ann Intern Med 127: 820–826. [DOI] [PubMed] [Google Scholar]
- 29. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719–748. [PubMed] [Google Scholar]
- 30. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 31. Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21: 1539–1558. [DOI] [PubMed] [Google Scholar]
- 32. Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50: 1088–1101. [PubMed] [Google Scholar]
- 33. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pugin J, Heumann ID, Tomasz A, Kravchenko W, Akamatsu Y, et al. (1994) CD14 is a pattern recognition receptor. Immunity 1: 509–516. [DOI] [PubMed] [Google Scholar]
- 35. Baldini M, Lohman IC, Halonen M, Erickson RP, Holt PG, et al. (1999) A polymorphism in the 5′flanking region of the CD14 gene is associated with circulating soluble CD14 levels and with total serum immunoglobulin E. Am J Respir Cell Mol Biol 20: 976–983. [DOI] [PubMed] [Google Scholar]
- 36. Liang XH, Cheung W, Heng CK, Liu JJ, Li CW, et al. (2006) CD14 promoter polymorphisms have no functional significance and are not associated with atopic phenotypes. Pharmacogenet Genomics 16: 229–236. [DOI] [PubMed] [Google Scholar]
- 37. LeVan TD, Bloom JW, Bailey TJ, Karp CL, Halonen M, et al. (2001) A common single nucleotide polymorphism in the CD14 promoter decreases the affinity of Sp protein binding and enhances transcriptional activity. J Immunol 167: 5838–5844. [DOI] [PubMed] [Google Scholar]
- 38. Hartel C, Rupp J, Hoegemann A, Bohler A, Spiegler J, et al. (2008) 159C>T CD14 genotype–functional effects on innate immune responses in term neonates. Hum Immunol 69: 338–343. [DOI] [PubMed] [Google Scholar]
- 39. Wang Z, Hu J, Fan R, Zhou J, Zhong J (2012) Association between CD14 gene C-260T polymorphism and inflammatory bowel disease: a meta-analysis. PLoS One 7: e45144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zeng T, Zhang CL, Han XY, Zhao S, Xie KQ (2013) Association between CD14-159C>T polymorphisms and the risk for alcoholic liver disease: a meta-analysis. Eur J Gastroenterol Hepatol 25: 1183–1189. [DOI] [PubMed] [Google Scholar]
- 41. Areeshi MY, Mandal RK, Panda AK, Bisht SC, et al. (2013) CD14 -159 C>T gene polymorphism with increased risk of tuberculosis: evidence from a meta-analysis. PLoS One 8: e64747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang AQ, Yue CL, Gu W, Du J, Wang HY, et al. (2013) Association between CD14 promoter -159C/T polymorphism and the risk of sepsis and mortality: a systematic review and meta-analysis. PLoS One 8: e71237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pu H, Yin J, Wu Y, Zhang D, Wang Y, et al. (2013) The association between CD14 gene C-260T polymorphism and coronary heart disease risk: a meta-analysis. Mol Biol Rep 40: 4001–4008. [DOI] [PubMed] [Google Scholar]
- 44. Zhao L, Bracken MB (2011) Association of CD14 -260 (-159) C>T and asthma: a systematic review and meta-analysis. BMC Med Genet 12: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu Y, Wang J (2013) Association of CD14 gene -159C/T polymorphism with allergic rhinitis risk: a meta-analysis. Eur Arch Otorhinolaryngol 2013 Oct 30 [Epub ahead of print] PubMed PMID: 24170183. [DOI] [PubMed] [Google Scholar]
- 46. Zhou W, Jia L, Guo S, Hu Q, Shen Y, et al. (2013) The -159C/T polymorphism in the CD14 gene and cancer risk: a meta-analysis. Onco Targets Ther 7: 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wroblewski LE, Peek RM Jr, Wilson KT (2010) Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev 23: 713–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Karhukorpi J, Yan Y, Niemela S, Valtonen J, Koistinen P, et al. (2002) Effect of CD14 promoter polymorphism and H. pylori infection and its clinical outcomes on circulating CD14. Clin Exp Immunol 128: 326–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boccia S, De Feo E, Gallì P, Gianfagna F, Amore R, et al. (2010) A systematic review evaluating the methodological aspects of meta-analyses of genetic association studies in cancer research. Eur J Epidemiol 25: 765–775. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA Checklist.
(DOC)
MOOSE Checklist.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. Data deposition.