ABSTRACT
Continual sperm production relies on germ cells undergoing spermatogenesis asynchronously. As a result, the testis always contains a mixed population of germ cells at different stages of their differentiation process. The heterogeneous nature of the testis makes profiling gene expression within Sertoli cells or specific populations of germ cells impossible when a wild-type testis is assessed. We recently reported a unique method for synchronizing spermatogenesis without affecting fertility by manipulating RA levels within the neonatal testis. Using this protocol, combined with the RiboTag transgenic mouse line, we have mapped the Sertoli and germ cell translatome during the initial synchronized wave of spermatogenesis. Using microarray analysis, we identified 392 and 194 germ cell and Sertoli cells transcripts, respectively, that dynamically change during spermatogonial differentiation, division, and the onset of meiosis. Functional annotation clustering revealed that transcripts enriched in germ cells were mostly associated with meiosis (21 transcripts), chromatin organization (12 transcripts), and cell cycle (3 transcripts). In addition, glycoproteins (65 transcripts), cell adhesion (15 transcripts), and cell junction (13 transcripts) transcripts were overrepresented in the Sertoli cell-enriched list. These datasets represent the first transcriptional analysis of spermatogonial differentiation, division, and meiotic onset. These data suggest that several of the genes encoding meiotic proteins are expressed and are actively being translated well before germ cells enter meiosis. In addition, this study provides novel candidate genes, Asf1b and Esyt3, that may be involved in the regulation of spermatogonial chromatin reorganization, germ-Sertoli cell interactions, and/or blood-testis barrier formation.
Keywords: gene expression, germ cells, retinoic acid, retinoids, Sertoli cells, spermatogonia, testis, translational profiling
WIN 18,446/RA treatment of neonatal mice was used to synchronize the initial wave of spermatogenesis and identify novel messages expressed within either germ or Sertoli cells as spermatogonia enter meiosis.
INTRODUCTION
Spermatogenesis is the tightly regulated process during which the somatic Sertoli cells of the seminiferous epithelium support the differentiation of spermatogonial stem cells into mature spermatozoa. For decades, the first wave or onset of spermatogenesis was considered by some investigators to be synchronous [1, 2]. However, recent reports have shown that the first wave of spermatogenesis is in fact asynchronous and occurs faster than subsequent waves [3–6]. In mice, the first wave is initiated a few days after birth, when gonocytes transition into differentiating A2 spermatogonia that divide and subsequently give rise to preleptotene spermatocytes by 8 days postpartum (dpp) [3, 7].
Several studies have mapped gene expression within the developing postnatal testis [8–13]. Most recently, Laiho et al. [9] used deep sequencing analysis to investigate gene expression and splicing variants in the whole testis at multiple time points during the first wave of murine spermatogenesis. They identified 2494 differentially expressed genes associated mostly with meiosis, piwi-interacting RNA metabolism, nucleus condensation, and flagella formation [9]. In addition, a microarray study using whole embryonic gonads and juvenile testes was used to identify potential genes involved in meiosis [13] and a mouse testis developmental time-course microarray database was developed to characterize the expression profile of the testis as whole across the first wave of spermatogenesis [8]. While these expression studies have enhanced our understanding of testis development, they relied on gene expression data collected from the whole testis, and, as a result, provide no information regarding the cell-specific gene expression events occurring during the first wave of spermatogenesis. Given the complexity of the juvenile testis, molecular signals from any individual cell type are difficult to discern when the entire testis is assessed. While testis cell-specific gene expression profiles have been generated for the neonatal testis [14], these experiments relied on examining cells following lengthy isolation experiments and may not accurately reflect their expression profiles in vivo.
The continuation of spermatogenesis requires the active metabolite of vitamin A, retinoic acid (RA). RA is required for germ cell differentiation (reviewed in [15]) as animals that are vitamin A deficient develop a block during spermatogonial differentiation and are subsequently infertile [8, 16]. Recently, we developed a technique using the bis-(dichloroacetyl)-diamine compound WIN 18,446, which inhibits RA production [17], followed by an injection of RA to synchronize spermatogonial differentiation. By using the WIN 18,446/RA synchronization method, spermatogenesis begins, but there is no production of differentiating cells resulting in an absence of the first wave of differentiating cells. This treatment creates a unique environment in which the germ cells are functionally normal, but specific germ cell populations are greatly enriched during the first wave of spermatogenesis [18]. Therefore, using this treatment, we are able to observe Sertoli/germ cell interactions in a synchronized environment.
Over the past decade, significant advancements have been made to determine gene expression within the testis [8, 9, 11, 12]. However, determining in vivo cell-type-specific gene expression has been difficult due to lack of proper genetic tools. The majority of studies that investigated cell-type specific gene expression in the testis have utilized cell isolation techniques [12, 19, 20]. In order to examine gene expression in vivo specifically in germ cells and in Sertoli cells, we took advantage of the recently constructed RiboTag mouse [21, 22] that allows for the isolation of polyribosomes from specific cell types using a Cre/Lox system. This procedure allows the identification of transcripts being actively translated in a cell-specific manner. Combining this unique system with the WIN 18,446/RA synchronization technique, we have, for the first time, analyzed in vivo gene expression changes in a synchronized environment from neonatal Sertoli and germ cells during spermatogonial differentiation, division, and the onset of meiosis. Specifically, we have isolated 392 and 194 actively translated transcripts that are differentially expressed in germ and Sertoli cells, respectively. We have identified 49 and 98 germ cell and Sertoli cell transcripts that respond to RA and identified novel candidate genes such as Asf1b and Esyt3 that may be involved in the regulation of spermatogonial chromatin reorganization and blood-testis barrier formation. These data provide the first in-depth analysis of germ and Sertoli cell gene expression during spermatogonial differentiation, division, and the onset of meiosis within a synchronized neonatal testis.
MATERIALS AND METHODS
Animals and Tissues
All the animal experiments were approved by Washington State University Animal Care and Use Committees and were conducted in accordance with the guiding principles for the care and use of research animals of the National Institutes of Health. The mouse colony was maintained in a temperature- and humidity-controlled environment with food and water provided ad libitum. Germ cell-specific (RiboTag/Stra8-cre or RiboTag/Ngn3-cre) and Sertoli cell-specific (RiboTag/ Amh-cre) RiboTag mice were obtained by crossing hemizygous RiboTag female mice [21] with Stra8-cre [23], Ngn3-cre [5], or Amh-cre [24] male mice. The RiboTag mouse line was provided by Dr. Stanley McKnight and Dr. Paul Amieux [21], and the cre mouse lines, Stra8-cre (008208), Ngn3-cre (006333), and Amh-cre (007915), were purchased from Jackson Laboratories. The animals were euthanized by CO2 asphyxiation followed by decapitation (0–10 dpp) or cervical dissociation (10 dpp adult), and their testes were dissected. Samples for immunoprecipitation (IP) followed by RNA preparation were snap frozen immediately after collection, and the IP experiment was performed on the same day as tissue collection. Testis samples for morphological and immunohistochemical analyses were fixed in Bouin fixative between 2 and 8 h, depending on the sample size, and then dehydrated through a graded ethanol series before being embedded in paraffin wax. Testis sections (4 μm) were placed on Superfrost Plus slides (Menzel-Glaser) for analysis.
WIN 18,446 and RA Treatment
Neonatal RiboTag/cre-positive mice were treated with WIN 18,446 and RA to synchronize spermatogenesis essentially as described by Hogarth et al. [18]. Briefly, 1 dpp mice were fed 100 μg/g body weight WIN 18,446, suspended in 1% gum tragacanth, for 9 consecutive days. At 9 dpp (Day 9 of treatment), mice were either sacrificed (0 h WIN 18,446-only treated mice) or intraperitoneally injected with 200 μg of RA in 10 μl of dimethylsulfoxide and then left to recover for either 4 h, 12 h, or 1, 2, 4, 6, or 8 days to obtain samples that spanned spermatogonial differentiation, division, and the onset of meiosis (Fig. 1).
FIG. 1.
WIN 18,446/RA treatment of neonatal mice during a synchronized first wave of spermatogenesis. Neonatal mice were fed vehicle (gum tragacanth) or WIN 18,446 for nine consecutive days and either sacrificed or injected with vehicle (dimethylsulfoxide) or RA. STRA8 localization in cross-sections of vehicle control (top) and WIN 18,446/RA (bottom) treated animals. Testes were collected prior to injection (0 h) or 1, 2, 4, 6, and 8 days postinjection. Green arrows denote STRA8-negative spermatogonia, red arrows denote STRA8-positive spermatogonia, and black arrows denote STRA8-positive preleptotene spermatocytes. Bars = 100 μm.
Immunohistochemistry
Immunohistochemistry using the anti-hemagglutinin (anti-HA) antibody (MMS-101R; Covance) was performed as described by Hogarth and Griswold [25]. Antigen retrieval was performed in 0.01 M citrate (pH 6; >90°C maintained for 5 min). Anti-HA antibody was applied at 500 ng/ml and incubated overnight at room temperature in 5% normal goat serum/0.1% bovine serum albumin/PBS. Control sections were incubated without primary antibody. Binding of primary antibodies was detected using 2 μg/ml biotinylated goat anti-mouse antibody (B7151; Sigma). Antibody binding was detected as a brown precipitate following development with DAB-Plus substrate kit (002020; Invitrogen). The sections were mounted under glass coverslips in DPX mountant (VWR International). Germ and somatic cell types were identified in Bouin fixed tissue on the basis of their nuclear morphology and position within the developing testis [26]. In the postnatal testis, sections from at least three RiboTag/cre-positive animals were analyzed for epitope-Tag localization.
Immunoprecipitation Assay
Testes from RiboTag/Stra8-cre or RiboTag/Ngn3-cre were pooled at the early time points in order to obtain sufficient RNA yields. Six testes from RiboTag/Stra8-cre or RiboTag/Ngn3-cre mice were pooled for the 0 h to 2 days post-RA injection time points whereas only one testis was required for the 4–8 days time points. One testis was used for all RiboTag/Amh-cre time points. Testes were homogenized at 3% weight/volume in 50 mM Tris, 100 mM KCl, 12 mM MgCl2, and 1% NP-40 buffer, and IP was performed as previously described [21] with a few modifications. Briefly, 50 μl of cleared homogenate was frozen at −20°C and saved for the total whole testis lysate RNA. Five microliters of anti-HA antibody was added to the remaining cleared homogenate and incubated at 4°C for 4 h. Four hundred microliters of washed beads (10004D; Life Technologies) were added to the antibody-homogenate solution and incubated overnight at 4°C. After incubation, the beads were washed in high salt buffer three times for 10 min, resuspended in RLT lysis buffer with beta-mercaptoethanol that was supplied within the Qiagen RNeasy Mini Kit (74104; Qiagen), and RNA was extracted.
RNA Extraction
RNA from the total whole testis lysate (50 μl) and the IP was obtained using the RNeasy mini kit (Qiagen) according to manufacturer's instructions. RNA was quantified using a NanoDrop 1000 Spectrophotometer (Thermo Scientific), and its quality was assessed using an Agilent 2100 Bioanalyzer with the RNA 6000 Nano kit (Agilent Technologies).
Microarray Analysis
One hundred nanograms of either total or IP and extracted RNA was amplified and labeled using NuGen Ovation labeling kit and hybridized to Affymetrix GeneChip Mouse Gene 1.0 ST Arrays. Array output was normalized using the Robust Multiarray Average algorithm, and data analysis was conducted using GeneSpring (Version 12.5 GX; Agilent Technologies). Duplicate samples were used in every time point across the synchronized spermatogonial differentiation until meiotic onset, and the mean values of the two replicates were used in subsequent analyses. Genes were considered to be expressed if they 1) had a raw score of greater than 200 in at least one time point, 2) were determined to be significantly different versus control (WIN 18,446-only, 0 h) treatment by ANOVA (P < 0.05), and 3) showed a 1.5-fold change for IP versus total RNA analysis. The expression cutoff of 200 was based on the expression of ovary-specific genes (Zp1, Zp3, and Gdf9). Functional annotation clustering was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources 6.7, freely available at http://david.abcc.ncifcrf.gov/. Transcripts that passed a moderated t-test [27] and a 2-fold change when comparing total RNA from time 0 h and 4 h post-RA injection were considered to be RA-regulated transcripts. Additionally, to determine if the transcripts associated differently with ribosomes during the first wave of spermatogenesis, the ratio of IP/total RNA was calculated and then the standard deviation was determined between each time point for each transcript. The Affymetrix Raw.CEL files have been deposited with the National Center for Biotechnology Information Gene Expression Omnibus, accession number GSE54408.
RESULTS
WIN 18,446/RA Treatment Synchronized Neonatal Spermatogenesis
The first wave of spermatogenesis is very complex because once RA triggers spermatogonial differentiation, it continues to occur in several sections of the seminiferous tubule at various times [3, 4]. Recently, we developed a procedure using the bis-(dichloroacetyl)-diamine compound WIN 18,446, which inhibits retinaldehyde dehydrogenase activity and RA production [17], followed by an injection of RA to synchronize spermatogonial differentiation [18]. WIN 18,446 treatment held the spermatogonia in an undifferentiated state, and once RA is administered, it triggers the A aligned (Aal) spermatogonia to differentiate (Fig. 1) [18]. Histological analysis of our synchronized samples indicated that a slightly more differentiated spermatogonial population, culminating in the appearance of preleptotene spermatocytes at 6 days post-RA injection, appeared at each collection point (Fig. 1). Undifferentiated germ cells were enriched in WIN 18,446-only treated animals (Fig. 1) whereas differentiating A spermatogonia were the most advanced cell types at 4 h through 1 day post-RA injection (Fig. 1). Intermediate and B spermatogonia were enriched at 2 and 4 days post-RA injection, respectively (Fig. 1). Preleptotene and leptotene spermatocytes accumulated by 6 and 8 days post-RA injection, respectively (Fig. 1). In addition to preleptotene spermatocytes, a renewed population of differentiating and undifferentiated spermatogonia is also enriched at 6 days post-RA injection. Because the A-A1 transition is occurring at 1 day post-RA injection, the expression difference between the germ cell population at 1 day and 6 day post-RA injection may be attributed to the appearance of the preleptotene population. This treatment replicated a rapid first wave of spermatogenesis indicated by appearance of preleptotene spermatocytes at 6 days post-RA injection (Fig. 1, black arrows). This result was consistent with observations from Drumond et al. [3].
RiboTag Activation in Sertoli and Germ Cells
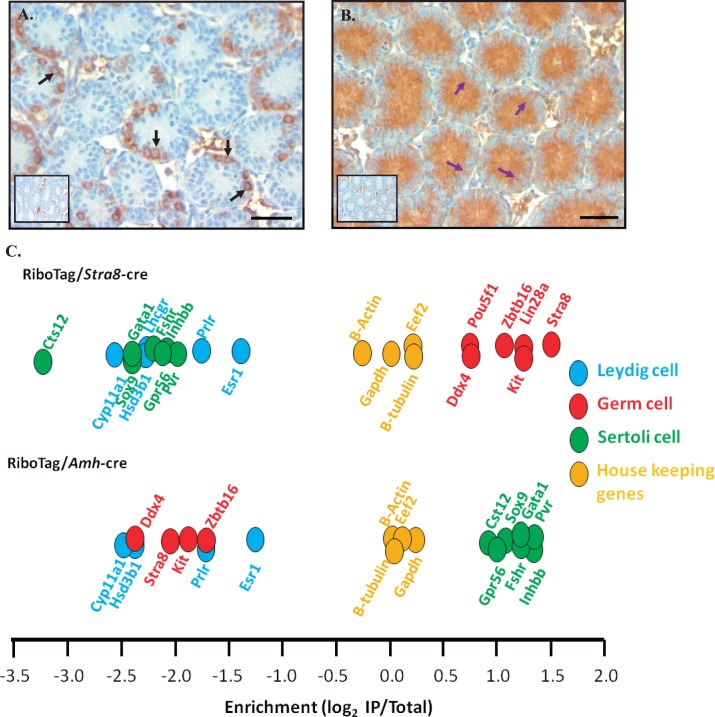
Previous publications have shown that breeding the RiboTag mouse line with mice carrying a cell-specific cre recombinase transgene expresses a HA tag on the ribosomal component RPL22 in a cell-specific manner [21, 22]. This mouse line was created by Sanz et al. [21] in which the targeted allele consists of a duplicated HA-tagged exon 4 that is preceded by the loxP flanked wild-type exon 4. Using the Cre/Lox system, the targeting vector expresses the wild-type RPL22 protein in the absence of cre recombination, and after recombination expresses the tagged RPL22 protein [21]. The RiboTag mouse line bred to cell-specific cre recombinase mice have been used to isolate forebrain, midbrain, Leydig, and Sertoli cell-specific mRNAs from adult mouse tissues in vivo [21, 22]. This study utilized RiboTag/cre-positive mice to insert a tag on ribosome that were actively translating transcripts specifically within germ or Sertoli cells during spermatogonial differentiation, division, and meiotic onset. Cre recombinase under the direction of the Stra8 promoter has been shown to excise differentiating A, intermediate, type B, and a subset of undifferentiated A spermatogonia [23, 28]. In addition, cre recombinase was expressed in Sertoli cells from Embryonic Day 15 onward using the Amh-cre mouse line [29]. To ensure that tagged RLP22 was being integrated into polyribosomes in a cell-specific manner, immunohistochemistry was performed on testes isolated from RiboTag/Stra8-cre- or Amh-cre-positive mice using a commercially available antibody against the HA tag. The tag was detected only within the cytoplasm of germ (Fig. 2A, black arrows) or Sertoli (Fig. 2B, purple arrows) cells and not within any other cell types of the testis. To further validate that cell-specific transcripts were enriched following IP and RNA extraction, an enrichment value was calculated by comparing the expression of well characterized testis cell-specific transcripts within mRNA that was associated with HA-tagged polyribosomes (IP mRNA) to the total mRNA samples following microarray analysis. The enrichment values for transcripts isolated from RiboTag/Stra8-cre-positive and RiboTag/Amh-cre-positive mice are illustrated in Figure 2. Known germ cells transcripts, including Stra8, Lin28a, and Kit, were found to be significantly enriched in the RiboTag/Stra8-cre-positive IP mRNA samples, whereas Sertoli and Leydig cell markers were not enriched (Fig. 2C). Additionally, by utilizing the RiboTag/Amh-cre mouse line, Sertoli cell transcripts were enriched and germ and Leydig cell transcripts were decreased within the IP mRNA (Fig. 2C). These results are consistent with previously published data [21, 22] and indicate that testis cell-specific transcripts were isolated using the RiboTag/cre-positive system.
FIG. 2.
Activation of the RiboTag in germ or Sertoli cells of the testis. RiboTag mice were bred with either a germ cell-specific cre line (Stra8-cre; A) or a Sertoli cell-specific cre line (Amh-cre; B). RiboTag activation was verified via immunohistochemistry using an anti-HA antibody in synchronized neonatal testis sections. Black arrows represent immunopositive spermatogonia, and purple arrows represent immunopositive Sertoli cell cytoplasm. No primary antibody negative control is represented within the inset. Bars = 50 μm. C) Microarray analysis results confirm the enrichment for well-known germ, Sertoli, Leydig, or housekeeping genes in the IP from RiboTag/Stra8-cre (top) or RiboTag/Amh-cre (bottom) mouse lines.
Identification of Actively Translated Germ and Sertoli Cell Transcripts During Spermatogonial Differentiation, Division, and the Onset of Meiosis
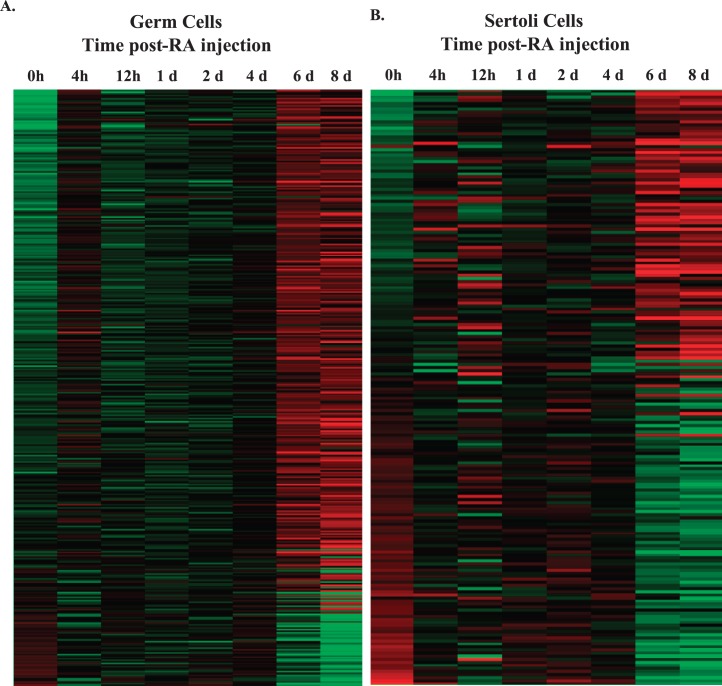
In order to accurately investigate changes in germ or Sertoli cell gene expression across spermatogonial differentiation, division, and the onset of meiosis, microarray analysis was performed on total and IP mRNA samples isolated from RiboTag/Stra8-cre or RiboTag/Amh-cre, respectively, synchronized neonatal mouse testes. We generated a list of 2345 germ cell and 1149 Sertoli cell actively translated transcripts that were enriched by 1.5-fold in the IP fraction compared to total mRNA sample at one or more time points during spermatogonial differentiation, division, and meiotic onset (Supplemental Tables S1 and S2; Supplemental Data are available online at www.biolreprod.org). Using an ANOVA test, the expression of all eight time points was compared to the WIN 18,446-only (0 h) treated sample. We found 392 and 194 transcripts that significantly changed in expression in germ and Sertoli cells, respectively, as germ cells differentiated and Sertoli cells matured during neonatal spermatogenesis (Fig. 3, A and B). Interestingly, the majority of germ cell transcripts (331) increased in expression (Fig. 3A), whereas a larger number of Sertoli cell transcripts, 101, decreased in expression across spermatogonial differentiation, division, and meiotic onset (Fig. 3B). We believe these observations reflect germ cells differentiating to form preleptotene spermatocytes and the shifting of Sertoli cells from a mitotically active state toward terminal differentiation during spermatogenesis [30, 31].
FIG. 3.
Heat map of germ cell- or Sertoli cell-enriched transcripts during the first wave of spermatogenesis. Relative expression levels of 392 germ cell-enriched (A) and 192 Sertoli cell-enriched (B) transcripts that significantly change during a synchronized first wave of spermatogenesis. Significance was determined by ANOVA (P < 0.05) and a greater than 2-fold change compared to WIN 18,446-only (0 h) treated mice. Expression was normalized to the mean intensity of the probe set, and these probes were ordered by expression (green = low; red = high). Data were visualized in GeneSpring GX Version 12.5; h = hour, d = days.
Functional annotation clustering using DAVID was then performed, revealing that the 392 actively translated germ cell-enriched transcripts were significantly associated (P < 0.01) with meiosis (34 transcripts), sexual reproduction (30 transcripts), DNA binding (69 transcripts), spermatogenesis (11 transcripts), chromosome segregation (10 transcripts), and DNA packaging and chromatin organization (12 transcripts) (Table 1). The 194 Sertoli cell transcripts were associated with biological terms and processes such as glycoprotein (65 transcripts), cell adhesion (15 transcripts), membrane (74 transcripts), cell-substrate adhesion (5 transcripts), cell junction (13 transcripts), and adherens junction (4 transcripts) (Table 2). All of the biological terms and processes linked with Sertoli cells are known to be involved in forming the blood-testis barrier [30, 32]. Seven Sertoli cell transcripts displayed a similar expression pattern to known blood-testis barrier mRNAs, such as N-cadherin [33], with an increase in expression between 4 and 12 h, a decrease at 4 days, and then an increase again at 6 days post-RA injections (Fig. 4). These results imply that these novel transcripts may play a role in forming the blood-testis barrier.
TABLE 1.
Identification of biological processes associated with germ cell-enriched transcripts.
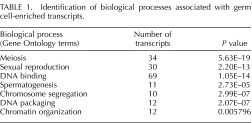
TABLE 2.
Identification of biological processes associated with Sertoli cell-enriched transcripts.
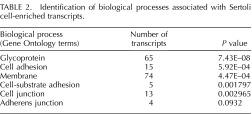
FIG. 4.
Identification of candidate blood-testis barrier transcripts within Sertoli cells. Graphs depicting the microarray expression pattern of Sertoli cell-enriched transcripts that are associated processes involved in blood-testis barrier formation. Raw expression values are represented on the y-axis, and time post-RA injection is on the x-axis.
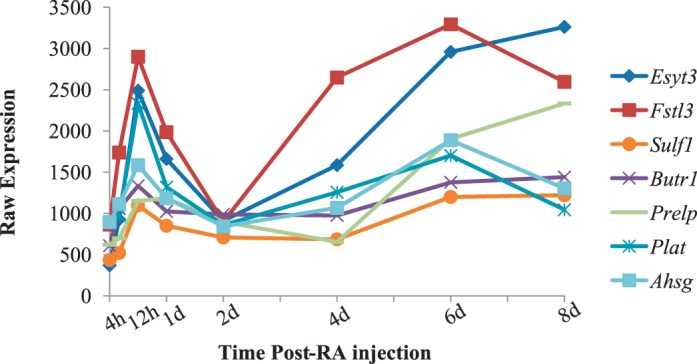
Spermatogonia undergo dramatic chromatin reorganization as they differentiate. To further investigate the transcripts associated with DNA packaging and chromatin reorganization identified by the DAVID analysis, their expression was plotted across the first wave of spermatogenesis. Six of those transcripts were highly expressed (greater than 500 in at least one time point) and demonstrated changes in expression as spermatogenesis progressed (Fig. 5A). We also found three histone variants in the testis that were actively transcribed and exhibited significant changes in expression as the germ cells undergo differentiation (Fig. 5B), which support results from Sun and Qi [34]. Products from these transcripts may play a role in remodeling chromatin within spermatogonia as they differentiate to form meiotic spermatocytes.
FIG. 5.
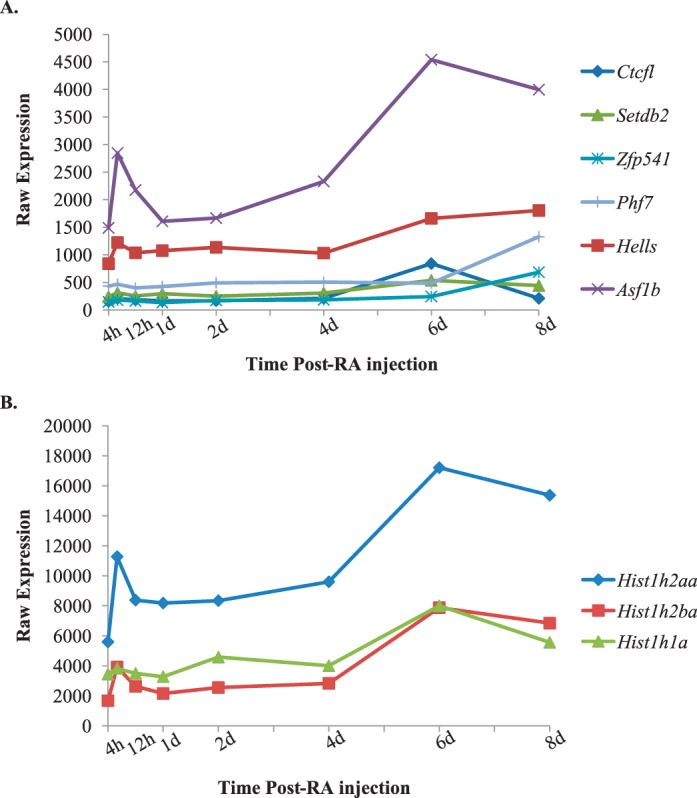
Identification of candidate chromatin modifying transcripts within germ cells. Graphs depicting the microarray expression pattern of germ cell-enriched transcripts that are associated with DNA packaging and chromatin organization (A) and histone modifications (B) across the first wave of synchronized spermatogenesis. Raw expression values are represented on the y-axis, and time post-RA injection is given on the x-axis.
Germ and Sertoli Cell RA-Responsive Transcripts
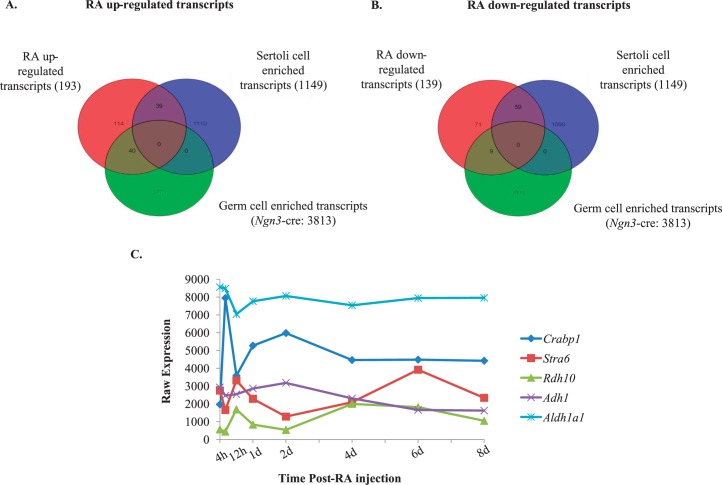
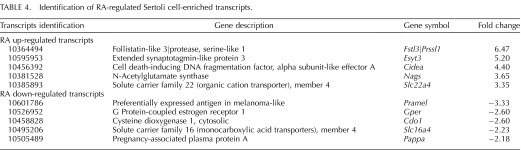
RA is necessary for spermatogonial differentiation, yet whether it acts within Sertoli cells, germ cells, or both to coordinate this process is still to be fully understood as are the downstream consequences of RA signaling to control this differentiation step. Yoshida et al. [5] demonstrated that cre recombinase under the direction of the Ngn3 promoter was expressed in A singles (As), A pair (Apr), and Aal spermatogonia. Therefore, we utilized this mouse, bred to the RiboTag mouse line, to excise the wild-type RPL22 protein and express the HA-tagged RPL22 protein [21] within germ cells prior to the A to A1 spermatogonia transition, allowing for the investigation of the undifferentiated spermatogonial response to RA in vivo. IP mRNA was compared to total mRNA to determine which germ cell transcripts were enriched in WIN 18,446-only and WIN 18,446/RA-treated testes left to recover for 4 h. These comparisons revealed 3813 transcripts that were enriched in germ cells when using the Ngn3-cre mouse line (Fig. 6A). To ascertain which transcripts were regulated by RA, total RNA from the RiboTag/Ngn3-cre WIN 18,446-only treated testes were compared to total RNA from testes collected from WIN 18,446/RA-treated animals that were left to recover for 4 h. Using a moderated t-test and requiring a greater than 1.5-fold change, we found 193 and 139 transcripts that were up- and down-regulated, respectively, with RA treatment (Supplemental Table S3). To ascertain which of these were germ or Sertoli cell transcripts, Venn diagrams were used to determine all the possible relations between up-and down regulated RA-responsive and germ cell- and Sertoli cell-enriched mRNAs (Fig. 6, A and B). We discovered 40 germ cell- and 39 Sertoli cell-enriched genes that were up-regulated with RA (Fig. 6A). Furthermore, we found 9 germ cell- and 59 Sertoli cell-enriched transcripts that were down-regulated with RA (Fig. 6B). The top five up- and down-regulated RA-responsive germ cell- or Sertoli cell-enriched transcripts are given in Tables 3 and 4, respectively. Interestingly, these lists include Gadd45b and 6330545A04Rik (Rsg1) that have not previously been shown to be RA-responsive in germ cells [14] although Gadd45b has been shown to be RA-responsive in other tissues [35, 36]. Rsg1 has been shown to be important for cytoplasmic localization and membrane trafficking in Xenopus multiciliated cells [37]; however, the role of Rsg1 in mammalian cells is unknown.
FIG. 6.
Retinoic acid (RA)-regulated germ and Sertoli cell transcripts. Venn diagram for all RA up-regulated (A) or down-regulated (B) germ (RiboTag/Ngn3-cre) or Sertoli (RiboTag/Amh-cre) cell transcripts. The Sertoli cell-enriched transcripts were determined by comparing the IP mRNA to the total mRNA in all the time points assayed, whereas germ cell-enriched transcripts were identified by comparing WIN 18,446-only and WIN 18,446/RA-treated testes left to recover for 4 h. C) Graph depicting the microarray expression pattern of the top five retinoid metabolism transcripts that significantly change in expression during the first wave of synchronized spermatogenesis. Significance was determined by an ANOVA test (P < 0.05) compared to WIN 18,446-only (0 h) treatment. Raw expression values are represented on the y-axis, and time post-RA injection is given on the x-axis.
TABLE 3.
Identification of RA-regulated germ cell-enriched transcripts.
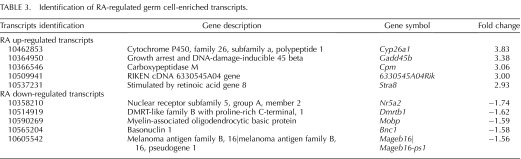
TABLE 4.
Identification of RA-regulated Sertoli cell-enriched transcripts.
Identification of Germ or Sertoli Cell Transcripts That Increase Their Association with Ribosomes During Spermatogonial Differentiation, Division, and the Onset of Meiosis
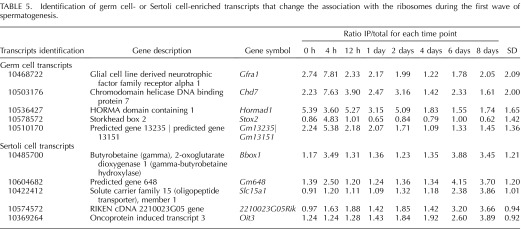
A recent report by Chappell et al. [38] revealed that germ cell and housekeeping transcripts display significant changes in translational efficiency during the onset of spermatogenesis. The RiboTag mouse line provides an alternative tool for investigating translational efficiency because RPL22 has been shown to be mostly incorporated within polyribosomes, those associated with higher levels of mRNA translation, with little incorporation into the lighter fraction of ribosomes [21], those less often associated with translation. Therefore, by comparing the IP RNA/total RNA ratio at each time point for the germ cell- or Sertoli cell-enriched transcripts and calculating the standard deviation between the ratios at all eight time points, we were able to investigate changes in mRNA association with the ribosome during a synchronized spermatogonial differentiation, division, and meiotic onset. The top five germ and Sertoli cell transcripts that have a variable IP RNA/total RNA ratio (Supplemental Table S4), signaling that their association with ribosomes changes most significantly during spermatogonial differentiation, division, and meiotic onset, are listed in Table 5. It is interesting to note that four of the five germ cell transcripts all displayed their highest ratio of IP to total RNA at 4 h post-RA injection. In addition, approximately 9.5% of the germ cell-enriched transcripts that are significantly changing during spermatogonial differentiation have the highest ratio at this time point (Supplemental Table S4). These data indicate that RA may alter the loading of germ cell mRNA onto polyribosomes. These results suggest that both germ and Sertoli cell mRNA transcripts significantly change their association with ribosomes during spermatogonial differentiation, division, and the onset of meiosis, which is consistent with previously published data [38].
TABLE 5.
Identification of germ cell- or Sertoli cell-enriched transcripts that change the association with the ribosomes during the first wave of spermatogenesis.
Both Germ and Sertoli Cells Contain Components of the Retinoid Metabolism Pathway
The active metabolite of vitamin A (i.e., RA) is a key regulator of the transition of undifferentiated A spermatogonia to differentiating A1 spermatogonia and, subsequently, the production of spermatozoa [16, 39–41]. While there is some published information regarding which retinoid metabolism and signaling proteins function within the testis (reviewed in [42]), the cell-specific localization of some of these proteins in the neonatal testis is unknown. Therefore, using the total RNA samples and a list of known retinoid metabolism genes, we determined the most dynamically changing genes encoding retinoid metabolism proteins during spermatogonial differentiation, division, and meiotic onset (Fig. 6C). Furthermore, we wanted to determine which metabolism genes are enriched in neonatal germ or Sertoli cells during synchronized spermatogenesis. A Venn diagram was used to compare a list of known retinoid metabolism genes to a list of germ cell- or Sertoli cell-enriched transcripts (Fig. 7). The retinoid metabolism transcripts enriched in germ cells were Rxrg, Rarb, Rarg, Cyp26a1, Rbp4, and Crabp1 while the retinoid metabolism transcripts enriched in Sertoli cells were Rxrb, Adh4, Lpl, Stra6, and Aldh1a2. These data demonstrate that in neonatal animals both germ and Sertoli cells contain the components required for retinoid production, signaling, and degradation.
FIG. 7.
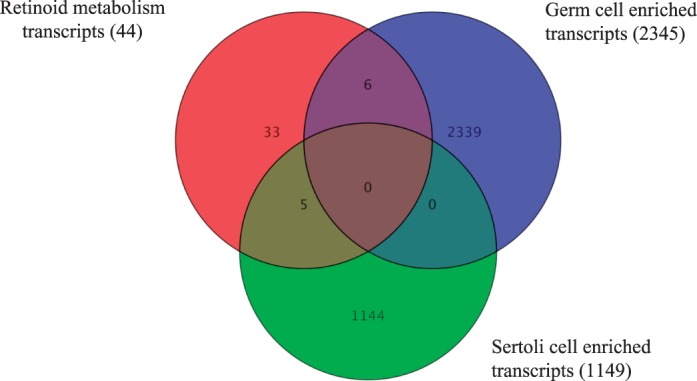
Retinoid metabolism transcripts within germ and Sertoli cell. A Venn diagram depicting all the known retinoid metabolism transcripts (red circle) that are enriched in germ cells (RiboTag/Stra8-cre; blue circle) or Sertoli cells (RiboTag/Amh-cre; green circle).
Actively Translated Cell Cycle Transcripts Have Cyclic Expression During the First Wave of Spermatogenesis in Germ and Sertoli Cells
During the first wave of spermatogenesis, germ and Sertoli cells undergo multiple rounds of cellular division and differentiation to form haploid spermatozoa and mature Sertoli cells. Throughout this time, the expression of cell cycle proteins must be carefully regulated in both somatic and germ cells to ensure proper division. For that reason, we decided to investigate cell cycle transcripts during synchronized spermatogonial differentiation, division, and meiotic onset in both germ and Sertoli cells. A list of 92 genes known to play a role in controlling the cell cycle was compared to the actively translated germ or Sertoli cell transcripts that dynamically change. We discovered that there were significantly more cell cycle transcripts actively translated in germ cells (32) compared to Sertoli cells (3) (Supplemental Table S5). One of the key regulatory protein families that control cell cycle progression is the cyclins. These proteins are components of a complex that ensure DNA is properly duplicated, repaired, and segregated into new daughter cells (reviewed in [43]). We mined our synchronized neonatal microarray data to look specifically at cyclin transcripts in germ cells during spermatogonial differentiation (Fig. 8A). Within germ cells, there were six cyclins actively translated during spermatogonial differentiation, division, and meiotic onset, and five of these transcripts demonstrated cyclic changes in expression. The sixth cyclin, Ccnb3, only increased in expression in the 6 and 8 days postinjection time points, when preleptotene spermatocytes are present. These results imply that these six cyclins may play a role in regulating cell cycle progression in germ cells during neonatal spermatogenesis. Within Sertoli cells, two of the three cell cycle transcripts were highly expressed, displayed changes in expression across spermatogonial differentiation, division, and meiotic onset, and belonged to the TGF-β signaling pathway (Fig. 8B). These results indicate that that these two transcripts, Tgfb1 and Smad3, are actively translated within Sertoli cells and possibly play a role in regulating Sertoli cell proliferation during neonatal spermatogenesis.
FIG. 8.
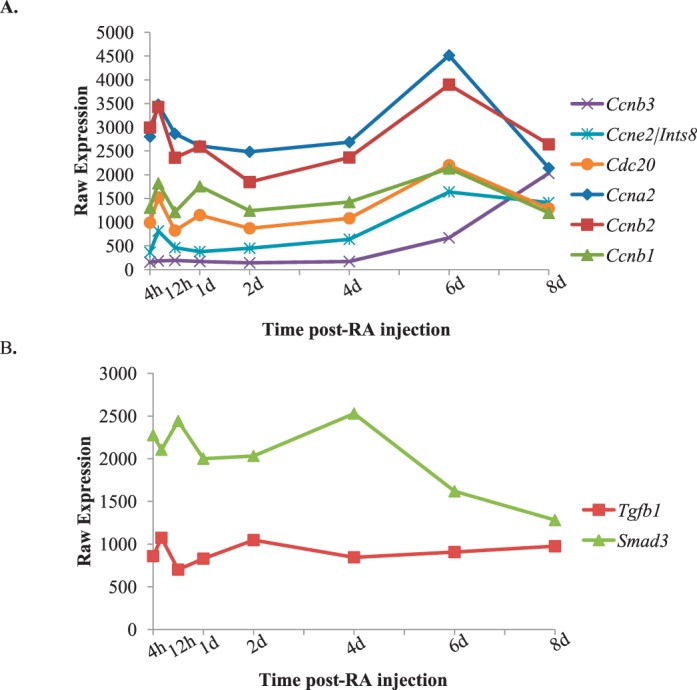
Actively translated cell cycle transcripts during the first wave of spermatogenesis. Graphs depicting the microarray expression patterns of actively translated germ cell-enriched cyclins (A) and actively translated Sertoli cell cycle transcripts, Tgfb1 and Smad3, (B) across the first wave of synchronized spermatogenesis. Raw expression values are represented on the y-axis, and time post-RA injection is given on the x-axis.
Known Meiotic Transcripts Are Expressed Prior to the Onset of Meiosis
Meiosis is essential for generation of spermatozoa and involves recombination and segregation of homologous chromosomes. One of the fundamental unanswered questions regarding spermatogenesis is at what point the initiation of meiosis begins. Does it initiate at the A to A1 transition when spermatogonia are committed to differentiation or does it begin at the first appearance of the first meiotic cell, preleptotene spermatocytes? To investigate this question, the expression of transcripts known to code for proteins associated with key meiotic processes such as synaptonemal complex formation, cohesin, or DNA double-stranded break repair were plotted across spermatogonial differentiation, division, and the onset of meiosis (Fig. 9). Of the 17 meiotic genes investigated, nine transcripts were associated with polyribosomes and were expressed before the first appearance of meiotic germ cells, which in WIN 18,446/RA-treated animals occurs at 6 days post-RA injection with the enrichment of preleptotene spermatocytes. These results suggest that meiosis may initiate when germ cells commit to differentiation prior to the start of meiosis. In addition, it is also possible that the meiotic genes are expressed within undifferentiated cells and that RA stimulation does not alter the expression of the mRNAs dramatically until the initiation of meiosis with the first appearance of preleptotene spermatocytes.
FIG. 9.
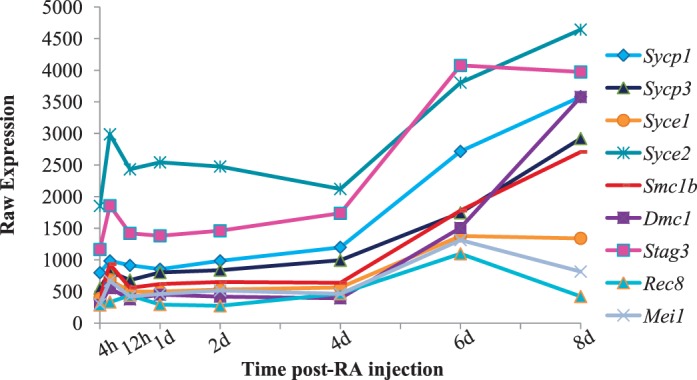
Actively translated meiotic transcripts during the first wave of spermatogenesis. Graph depicting the microarray expression pattern of known meiotic transcripts that are actively translated within germ cell across the first wave of synchronized spermatogenesis. Raw expression values are represented on the y-axis, and time post-RA injection is given on the x-axis.
DISCUSSION
This study represents the first in-depth analysis of the expression of actively translated genes in either germ or Sertoli cells during a synchronized neonatal wave of spermatogenesis. Over the past decade, there have been several investigations performed to determine gene expression within the postnatal testis [8, 9, 12]. However, these studies utilized starting material consisting of multiple cell types or testis cell populations isolated via lengthy tissue dissociation techniques. The combination of our unique WIN 18,446/RA synchronization protocol [18] and the RiboTag mouse line [21, 22] has enabled us, for the first time, to examine in vivo gene expression patterns as each differentiating germ cell initially appears during spermatogonial differentiation, division, and the onset of meiosis (Fig. 1). Using the WIN 18,446/RA synchronization method, we can recapitulate the shortened timing that is a hallmark characteristic of the first wave of spermatogenesis. Clearly, the traditional first wave consisting of gonocytes differentiating to A2 spermatogonia is blocked as there are no differentiating germ cells in the presence of WIN 18,446. However the initial wave of differential spermatogonia development that we observed likely results from releasing the block in the transition from undifferentiated to differentiated spermatogonia in a synchronized manner.
Functional annotation clustering of actively translated germ cell transcripts revealed functions such as DNA binding, DNA packaging, and chromosome organization are enriched during spermatogonial differentiation, division, and the onset of meiosis (Fig. 5A). Of these, the most interesting and novel appears to be ASF1 antisilencing function 1 homolog B (Saccharomyces cerevisiae) (Asf1b). This transcript was the most highly expressed and dynamically changing DNA packaging and chromosome organization mRNA through the first wave and has an expression profile similar to that of a RA-responsive transcript, Stra8. The expression profile for Stra8 includes peaks in expression 12 h and 6 days post-RA injection in testes of WIN 18, 446/RA-treated mice. ASF1B, also known as CIA-II, is expressed in spermatogonia and spermatocytes [44] and is a H3-H4 histone chaperone, indicating its involvement in histone changes during replication, transcription, and DNA repair (reviewed in [45]). ASF1b has no known RA response element; however, it does contain an E2F-responsive sequence that is necessary for E2F1 activation of Asf1b transcription [46]. E2F1 binding has been shown to be induced by RA [47]; therefore, it is possible that RA may play a role in driving the expression of Asf1b and subsequently regulating histone structure and function. In addition, this transcript may play an essential role in reorganizing the chromatin of spermatogonia, preparing them to enter meiosis. However, more studies need to be performed to determine whether RA directly regulates the expression of these transcripts and if their products are necessary for the progression of spermatogenesis.
Functional annotation clustering of the 194 Sertoli cell transcripts that significantly changed during spermatogonial differentiation, division, and meiotic onset revealed biological functions that are all important for the formation of the blood-testis barrier. While the blood-testis barrier has been well characterized, the proteins responsible for its formation and maintenance are still being elucidated. We identified five novel transcripts that have a similar expression profile to that of known blood-testis barrier transcripts (Fig. 4), but a function for their gene products in Sertoli cells has yet to be described. Of most interest in the Sertoli-enriched gene set is Esyt3 because it displayed a similar expression profile to that of RA-responsive mRNAs. Esyt3 codes for a calcium-regulated intrinsic membrane protein. Increases in intracellular calcium concentration causes changes in Sertoli cell cytoskeletal structure as well as Sertoli-Sertoli cell junction dynamics (reviewed in [48]). Therefore, it is possible that Esyt3 may play an essential role in regulating changes in Sertoli cells dynamics via controlling calcium concentrations; however, further studies need to be performed to determine its exact function within the testis. Furthermore, we found that two cell cycle transcripts, Tgfb1 and Smad3, which are associated with the TGF-β signaling pathway, are highly induced in Sertoli cells after stimulation with RA and during the initial differentiation of spermatogonia. Smad3 is known to transmit TGF-β and activin signaling, known to be necessary for multiple processes within the developing testis, including regulating testis development, germ cell numbers, and testicular architecture (reviewed in [49]). Our results support the conclusion from Itman et al. [50] that Smad3 is expressed within Sertoli cells, drives normal Sertoli cell proliferation, and is involved in regulating the timing of spermatogonial differentiation, division, and the onset of meiosis.
This study also investigated the expression of known meiotic transcripts in order to elucidate when the initiation of meiosis begins. Of the 17 known meiotic transcripts investigated, nine were found to be expressed and associated with polyribosomes in germ cells before the onset of meiosis. These data are consistent with previous reports indicating that synaptonemal complex proteins are expressed in proliferating spermatogonia [51, 52]. These data suggest that once the germ cells are committed to differentiation, at the A to A1 transition, they begin to build and load mRNAs encoding meiotic proteins onto ribosomes. In addition, it is possible that these transcripts are involved in the mitotic cell cycle because these transcripts are highly expressed in spermatogonia that are undergoing several rounds of division. Furthermore, the meiotic genes studied may be expressed in undifferentiated spermatogonia that are present at all the time points investigated, and the relatively low levels of expression may be due to the mitotic proliferation of undifferentiated spermatogonia. It is possible that RA stimulation does not alter the expression of the mRNAs dramatically until the initiation of meiosis with the first appearance of preleptotene spermatocytes. We believe these results are novel because they indicate that over 50% of the meiotic transcripts examined are present prior to the formation of meiotic spermatocytes. Nevertheless, more studies need to be performed to determine the precise point in which spermatogonia commit to undergoing meiosis.
This study analyzed, for the first time, in vivo gene expression changes within germ or Sertoli cells during neonatal spermatogenesis. Our results support the hypothesis that the WIN 18,446/RA synchronization method forces neonatal germ cells to differentiate in a uniform manner throughout the testis. This analysis also identified novel candidate transcripts that may play are role in chromatin organization and blood-testis barrier formation within the neonatal testis. Interestingly, the transcripts that were identified as possible players in chromatin organization and blood-testis barrier formation have expression patterns similar to that observed for RA-responsive transcripts. Therefore, it is possible that RA may regulate these transcripts. These data provide additional support that RA plays a role in spermatogonial differentiation, cell cycle, and blood-testis barrier formation [43, 53, 54]. This study also indicates that both germ and Sertoli cells have the components necessary to synthesize, signal, and degrade RA. Lastly, we have determined that meiotic transcripts are expressed and associate with polyribosomes prior to the onset of meiosis. These data provide the first in-depth analysis of germ and Sertoli cell gene expression during spermatogonial differentiation, division, and meiotic onset within a synchronized neonatal environment.
Supplementary Material
ACKNOWLEDGMENT
We would like to thank Drs. Paul Amieux and Stanley McKnight for gifting us the RiboTag mouse line and for technical support from the University of Washington, Seattle, WA. Special thanks go to Dr. Jon Amory at the University of Washington, Seattle, WA, for providing the WIN 18,446. Additionally, we would like to thank Derek Pouchnik in the Laboratory for Bioanalysis and Biotechnology I (LBBI) at Washington State University, Pullman, WA, for microarray hybridization and scanning.
Footnotes
Supported by HD 10808, U54 42454, and NIGMS training grant 5T32GM083864 from the NIH.
REFERENCES
- Nebel BR, Amarose AP, Hacket EM. Calendar of gametogenic development in the prepuberal male mouse. Science 1961; 134: 832–833. [DOI] [PubMed] [Google Scholar]
- McCarrey J. Cell and Molecular Biology of the Testis. New York: Oxford University Press; 1993: 58–89. [Google Scholar]
- Drumond AL, Meistrich ML, Chiarini-Garcia H. Spermatogonial morphology and kinetics during testis development in mice: a high-resolution light microscopy approach. Reproduction 2011; 142: 145–155. [DOI] [PubMed] [Google Scholar]
- Snyder EM, Small C, Griswold MD. Retinoic acid availability drives the asynchronous initiation of spermatogonial differentiation in the mouse. Biol Reprod 2010; 83: 783–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Takakura A, Ohbo K, Abe K, Wakabayashi J, Yamamoto M, Suda T, Nabeshima Y. Neurogenin3 delineates the earliest stages of spermatogenesis in the mouse testis. Dev Biol 2004; 269: 447–458. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Sukeno M, Nakagawa T, Ohbo K, Nagamatsu G, Suda T, Nabeshima Y. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development 2006; 133: 1495–1505. [DOI] [PubMed] [Google Scholar]
- de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl 2000; 21: 776–798. [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod 2004; 71: 319–330. [DOI] [PubMed] [Google Scholar]
- Laiho A, Kotaja N, Gyenesei A, Sironen A. Transcriptome profiling of the murine testis during the first wave of spermatogenesis. PLoS One 2013; 8: e61558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Lin Y, Wu J. Long non-coding RNA expression profiling of mouse testis during postnatal development. PLoS One 2013; 8: e75750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Nie R, Li Y, Friel P, Mitchell D, Hess RA, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (Stra8) in spermatogenic cells induced by retinoic acid: an in vivo study in vitamin A-sufficient postnatal murine testes. Biol Reprod 2008; 79: 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston DS, Wright WW, Dicandeloro P, Wilson E, Kopf GS, Jelinsky SA. Stage-specific gene expression is a fundamental characteristic of rat spermatogenic cells and Sertoli cells. Proc Natl Acad Sci U S A 2008; 105: 8315–8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogarth CA, Mitchell D, Evanoff R, Small C, Identification Griswold M. and expression of potential regulators of the mammalian mitotic-to-meiotic transition. Biol Reprod 2011; 84: 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li Y, Nie R, Friel P, Mitchell D, Evanoff RM, Pouchnik D, Banasik B, McCarrey JR, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (Stra8) and maturation of murine gonocytes and spermatogonia induced by retinoic acid in vitro. Biol Reprod 2008; 78: 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Koopman P. Retinoic acid, meiosis and germ cell fate in mammals. Development 2007; 134: 3401–3411. [DOI] [PubMed] [Google Scholar]
- Griswold MD, Bishop PD, Kim KH, Ping R, Siiteri JE, Morales C. Function of vitamin A in normal and synchronized seminiferous tubules. Ann N Y Acad Sci 1989; 564: 154–172. [DOI] [PubMed] [Google Scholar]
- Amory JK, Muller CH, Shimshoni JA, Isoherranen N, Paik J, Moreb JS, Amory DW, Sr, , Evanoff R, Goldstein AS, Griswold MD. Suppression of spermatogenesis by bisdichloroacetyldiamines is mediated by inhibition of testicular retinoic acid biosynthesis. J Androl 2011; 32: 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogarth CA, Evanoff R, Mitchell D, Kent T, Small C, Amory JK, Griswold MD. Turning a spermatogenic wave into a tsunami: synchronizing murine spermatogenesis using WIN 18,446. Biol Reprod 2013; 88: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkinaki M, Lee TL, He Z, Jiang J, Golestaneh N, Hofmann MC, Chan WY, Dym M. Age affects gene expression in mouse spermatogonial stem/progenitor cells. Reproduction 2010; 139: 1011–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Oatley MJ, Avarbock MR, Tobias JW, Brinster RL. Colony stimulating factor 1 is an extrinsic stimulator of mouse spermatogonial stem cell self-renewal. Development 2009; 136: 1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A 2009; 106: 13939–13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Evanoff R, Quintana A, Evans E, Miller JA, Ko C, Amieux PS, Griswold MD, McKnight GS. RiboTag analysis of actively translated mRNAs in Sertoli and Leydig cells in vivo. PLoS One 2013; 8: e66179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 2008; 46: 738–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdcraft RW, Braun RE. Androgen receptor function is required in Sertoli cells for the terminal differentiation of haploid spermatids. Development 2004; 131: 459–467. [DOI] [PubMed] [Google Scholar]
- Hogarth CA, Griswold MD. Immunohistochemical approaches for the study of spermatogenesis. Methods Mol Biol 2013; 927: 309–320. [DOI] [PubMed] [Google Scholar]
- Russell LD, Ettlin RA, SinhaHikim AP, Clegg ED. Histological and Histopathological Evaluation of the Testis. Clearwater, FL: Cache River Press; 1990. [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004; 3: Article3. [DOI] [PubMed] [Google Scholar]
- Gallagher SJ, Kofman AE, Huszar JM, Dannenberg JH, DePinho RA, Braun RE, Payne CJ. Distinct requirements for Sin3a in perinatal male gonocytes and differentiating spermatogonia. Dev Biol 2013; 373: 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecureuil C, Fontaine I, Crepieux P, Guillou F. Sertoli and granulosa cell-specific Cre recombinase activity in transgenic mice. Genesis 2002; 33: 114–118. [DOI] [PubMed] [Google Scholar]
- Russell LD, Griswold MD. The Sertoli Cell. Clearwater, FL: Cache River Press; 1993. [Google Scholar]
- Skinner MK, Griswold MD. Sertoli Cell Biology. San Diego, CA: Elsevier Academic Press; 2005. [Google Scholar]
- Byers S, Pelletier R, Suarez-Quian C. Sertoli-Sertoli cell junctions and the seminiferous epithelium barrier. : LD Russell, Griswold MD. (eds.), The Sertoli Cell, 1st ed. Clearwater, FL: Cache River Press; 1993: 431–446. [Google Scholar]
- Kopera IA, Bilinska B, Cheng CY, Mruk DD. Sertoli-germ cell junctions in the testis: a review of recent data. Philos Trans R Soc Lond B Biol Sci 2010; 365: 1593–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Qi H. Dynamic expression of combinatorial replication-dependent histone variant genes during mouse spermatogenesis. Gene Expr Patterns 2014; 14: 30–41. [DOI] [PubMed] [Google Scholar]
- Sherman H, Gutman R, Chapnik N, Meylan J, le Coutre J, Froy O. All-trans retinoic acid modifies the expression of clock and disease marker genes. J Nutr Biochem 2012; 23: 209–217. [DOI] [PubMed] [Google Scholar]
- Yi YW, Kim D, Jung N, Hong SS, Lee HS, Bae I. Gadd45 family proteins are coactivators of nuclear hormone receptors. Biochem Biophys Res Commun 2000; 272: 193–198. [DOI] [PubMed] [Google Scholar]
- Brooks ER, Wallingford JB. The small GTPase Rsg1 is important for the cytoplasmic localization and axonemal dynamics of intraflagellar transport proteins. Cilia 2013; 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell VA, Busada JT, Keiper BD, Geyer CB. Translational activation of developmental messenger RNAs during neonatal mouse testis development. Biol Reprod 2013; 89: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Pelt AM, de Rooij DG. Synchronization of the seminiferous epithelium after vitamin A replacement in vitamin A-deficient mice. Biol Reprod 1990; 43: 363–367. [DOI] [PubMed] [Google Scholar]
- Hogarth CA, Griswold MD. The key role of vitamin A in spermatogenesis. J Clin Invest 2010; 120: 956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogarth CA, Evanoff R, Snyder E, Kent T, Mitchell D, Small C, Amory JK, Griswold MD. Suppression of Stra8 expression in the mouse gonad by WIN 18,446. Biol Reprod 2011; 84: 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogarth CA, Amory JK, Griswold MD. Inhibiting vitamin A metabolism as an approach to male contraception. Trends Endocrinol Metab 2011; 22: 136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolgemuth DJ, Manterola M, Vasileva A. Role of cyclins in controlling progression of mammalian spermatogenesis. Int J Dev Biol 2013; 57: 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara T, Horikoshi M. Transcription initiation factor IID-interactive histone chaperone CIA-II implicated in mammalian spermatogenesis. J Biol Chem 2003; 278: 35660–35667. [DOI] [PubMed] [Google Scholar]
- Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta 2004; 1677: 3–11. [DOI] [PubMed] [Google Scholar]
- Hayashi R, Goto Y, Tanaka R, Oonogi K, Hisasue M, Yoshida K. Transcriptional regulation of human chromatin assembly factor ASF1. DNA Cell Biol 2007; 26: 91–99. [DOI] [PubMed] [Google Scholar]
- Kunigal S, Ponnusamy MP, Momi N, Batra SK, Chellappan SP. Nicotine, IFN-gamma and retinoic acid mediated induction of MUC4 in pancreatic cancer requires E2F1 and STAT-1 transcription factors and utilize different signaling cascades. Mol Cancer 2012; 11: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker WH, Cheng J. FSH. and testosterone signaling in Sertoli cells. Reproduction 2005; 130: 15–28. [DOI] [PubMed] [Google Scholar]
- Itman C, Mendis S, Barakat B, Loveland KL. All in the family: TGF-beta family action in testis development. Reproduction 2006; 132: 233–246. [DOI] [PubMed] [Google Scholar]
- Itman C, Wong C, Hunyadi B, Ernst M, Jans DA, Loveland KL. Smad3 dosage determines androgen responsiveness and sets the pace of postnatal testis development. Endocrinology 2011; 152: 2076–2089. [DOI] [PubMed] [Google Scholar]
- Zheng YH, Rengaraj D, Choi JW, Park KJ, Lee SI, Han JY. Expression pattern of meiosis associated SYCP family members during germline development in chickens. Reproduction 2009; 138: 483–492. [DOI] [PubMed] [Google Scholar]
- Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT. Postmeiotic sex chromatin in the male germline of mice. Curr Biol 2006; 16: 660–667. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Saga Y. Retinoic acid signaling in Sertoli cells regulates organization of the blood-testis barrier through cyclical changes in gene expression. Development 2012; 139: 4347–4355. [DOI] [PubMed] [Google Scholar]
- Kubota H, Chiba H, Takakuwa Y, Osanai M, Tobioka H, Kohama G, Mori M, Sawada N. Retinoid X receptor alpha and retinoic acid receptor gamma mediate expression of genes encoding tight-junction proteins and barrier function in F9 cells during visceral endodermal differentiation. Exp Cell Res 2001; 263: 163–172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.