ABSTRACT
Assisted reproductive technologies (ART) have enabled millions of couples with compromised fertility to conceive children. Nevertheless, there is a growing concern regarding the safety of these procedures due to an increased incidence of imprinting disorders, premature birth, and low birth weight in ART-conceived offspring. An integral aspect of ART is the oxygen concentration used during in vitro development of mammalian embryos, which is typically either atmospheric (∼20%) or reduced (5%). Both oxygen tension levels have been widely used, but 5% oxygen improves preimplantation development in several mammalian species, including that of humans. To determine whether a high oxygen tension increases the frequency of epigenetic abnormalities in mouse embryos subjected to ART, we measured DNA methylation and expression of several imprinted genes in both embryonic and placental tissues from concepti generated by in vitro fertilization (IVF) and exposed to 5% or 20% oxygen during culture. We found that placentae from IVF embryos exhibit an increased frequency of abnormal methylation and expression profiles of several imprinted genes, compared to embryonic tissues. Moreover, IVF-derived placentae exhibit a variety of epigenetic profiles at the assayed imprinted genes, suggesting that these epigenetic defects arise by a stochastic process. Although culturing embryos in both of the oxygen concentrations resulted in a significant increase of epigenetic defects in placental tissues compared to naturally conceived controls, we did not detect significant differences between embryos cultured in 5% and those cultured in 20% oxygen. Thus, further optimization of ART should be considered to minimize the occurrence of epigenetic errors in the placenta.
Keywords: assisted reproductive technologies, DNA methylation, epigenetic reprogramming, gene regulation, genomic imprinting, in vitro fertilization
Culturing mouse embryos with low and high oxygen tension promotes abnormal DNA methylation and expression profiles at imprinted genes in placental tissues; the variety of outcomes observed in different placentae suggests that these epigenetic defects arise randomly.
INTRODUCTION
Infertility is a significant problem in developed countries and has been addressed in part through the use of assisted reproductive technologies (ART). Children born through various forms of ART account for 1%–3% of total births in developed countries, and these figures are likely to rise in the coming years [1]. Although most ART-conceived children appear healthy, there is growing concern about the safety of ART. For example, ART-conceived children exhibit higher incidence rates of preterm birth, low birth weight, and rare imprinting disorders than their naturally conceived counterparts [2]. It is thought that the combined effects of infertility and advanced maternal age in patients undergoing fertility treatments are the predominant cause of these abnormal phenotypes [3]. Studies involving animal models, however, in which subfertility and advanced age are not confounding factors, demonstrate that specific components of ART procedures can influence epigenetic profiles in the early embryo [4]. Because epigenetic defects may play a role in the induction of the abnormal phenotypes observed in ART-conceived children, further investigation into the effects of fertility treatments on epigenetic gene regulation is warranted.
Following fertilization, embryos undergo extensive reprogramming that involves erasure of existing epigenetic marks, followed by establishment and subsequent maintenance of new epigenetic modifications [5]. Genomic imprinting refers to an epigenetic phenomenon whereby certain genes are expressed in a parent-of-origin-specific manner, and this process is critical for normal embryonic development [6]. The functional asymmetry of imprinted genes is regulated by allele-specific epigenetic marks, such as DNA methylation, that are established during gametogenesis in each parental germ line and transmitted to the zygote upon fertilization [6]. Importantly, differential methylation in imprinted genes must be maintained throughout embryogenesis despite the genome-wide epigenetic reprogramming that occurs during this developmental window. Given that imprinted genes play important roles in fetal growth and placental function, any mistake in epigenetic programming at these loci could adversely influence embryonic development. Indeed, aberrant expression of imprinted genes is associated with disease phenotypes and developmental defects in both humans and animal models [7]. Therefore, preserving monoallelic expression of imprinted genes is crucial for normal development of the organism.
The epigenome of the early embryo is sensitive to environmental perturbations, for example, maternal diet or exposure to endocrine disruptors can influence DNA methylation patterns in a gestating fetus [8, 9]. In addition, certain aspects of ART, such as endocrine stimulation and embryo culture, have the potential to disrupt epigenetic reprogramming during development [10–12]. These procedures coincide chronologically with key developmental stages when genome-wide epigenetic reprogramming occurs, and it is believed that the molecular machinery involved in this reprogramming process is sensitive to external influences [6]. Indeed, ART-conceived children have an increased incidence of rare epigenetic disorders, such as Beckwith-Wiedemann syndrome and Angelman syndrome [13], and a disproportionate number of these children exhibit loss of DNA methylation at genetic elements called imprinting control regions (ICRs) that are critical for regulating the monoallelic expression of imprinted loci. In addition, the increased risk of premature birth and low birth weight in ART-conceived children may also be attributed to abnormal epigenetic regulation at imprinted loci [2]. Together, these abnormal phenotypes suggest that ART procedures induce aberrant epigenetic profiles during preimplantation development. In support of this hypothesis, studies using a mouse model have demonstrated that embryo culture, an integral component of ART, disrupts normal epigenetic programming in both embryonic and extraembryonic cells [10, 14]. In addition, previous work from our laboratory showed that the midgestation placenta exhibits heightened sensitivity to in vitro culture compared to that in embryonic tissue [14, 15], which could impair fetal growth and development.
Mammalian embryos are cultured in either a physiological (5%) or an atmospheric (20%) oxygen concentration. Although both oxygen concentrations are widely used in fertility clinics, embryos cultured in 5% O2 exhibit improved embryonic development compared to those cultured in 20% O2 [16, 17]. However, the molecular mechanisms responsible for this biological phenomenon are unknown. One possible but untested explanation for the improvement in embryogenesis is that use of 5% O2 during culture provides a more permissive environment for normal epigenetic reprogramming following fertilization. We previously demonstrated that embryo culture and transfer after in vivo fertilization is associated with abnormal methylation and expression profiles for a subset of embryos [10, 15]. In the current study, we mimicked the clinical application of in vitro fertilization (IVF) in a mouse model by introducing IVF followed by in vitro culture to the blastocyst stage and nonsurgical embryo transfer. To investigate the effects of different oxygen tensions on epigenetic regulation, a comprehensive analysis was undertaken by measuring DNA methylation at multiple ICRs as well as allele-specific expression of several imprinted genes in embryonic and placental tissues from IVF-derived mouse embryos that were exposed to either 5% or 20% O2 during in vitro culture. In addition, we performed these analyses with 2 sets of mouse crosses to compare the sensitivity of different substrains to environmental perturbations associated with ART procedures. Our results show that placentae from ART-derived embryos exhibit an enhanced sensitivity to ex vivo manipulations and that the induction of epigenetic defects in these tissues arise through a stochastic process. Although culturing embryos in 20% O2 adversely influenced embryonic development in both of the mouse strains, this impairment was not associated with a statistically significant increase in epigenetic abnormalities compared to embryos cultured in 5% O2. Taken together, the results from this study suggest that the use of in vitro culture at 5% or 20% O2 during ART can increase the occurrence of random epigenetic errors in placental tissues.
MATERIALS AND METHODS
Animals
Two different mouse crosses were used to analyze epigenetic profiles in embryonic and placental tissues. B6 (CAST7) mice possess a chromosome 7 from Mus musculus subsp. castaneus (Cast) on a C57BL/6J background. B6 (CAST7) females were crossed with B6SJLF1 males (B6; Jackson Laboratory), and the ensuing F1 progeny carried several previously characterized strain-specific polymorphisms that were used to distinguish the parental alleles of imprinted genes on chromosome 7 [18]. In addition, CF1 females (Harlan Laboratories) were crossed with B6SJLF1 males, and DNA methylation analyses were performed with the resulting embryos in parallel with F1 hybrids. CF1 females also served as surrogate mothers (described below). Naturally conceived controls were generated by mating B6SJLF1 males with either B6 (CAST7) or CF1 females. The day on which a vaginal plug was observed was denoted as embryonic day (E) 0.5, and the females were euthanized 10 days later to collect E10.5 naturally conceived controls. All animal work was conducted with the approval of the Institutional Animal Care and Use Committee at the University of Pennsylvania.
IVF and Embryo Transfer
Mature spermatozoa were collected from the caudae epididymides of B6SJLF1 males and allowed to capacitate for 90 min in TYH (modified Krebs-Ringer bicarbonate) medium [19] at 37°C in a humidified atmosphere of 5% CO2 in atmospheric oxygen under washed mineral oil. Metaphase II eggs were recovered from CF1 or B6 (CAST7) females that had been superovulated by consecutive injections with 5 IU of equine chorionic gonadotropin and 5 IU of human chorionic gonadotropin (hCG) administered 48 h apart. Approximately 14 h after injection of hCG, cumulus-egg complexes were collected from the oviducts, washed in Whitten/HEPES buffer [20], and then transferred into a fresh drop of TYH medium under mineral oil. The eggs were then inseminated with capacitated spermatozoa for 3 h at 37°C in a reduced oxygen environment. After fertilization, zygotes were washed in Whitten/HEPES buffer to remove cumulus cells and sperm and then transferred to potassium simplex optimized medium (KSOM) containing amino acids [21] for embryo culture. Embryos were cultured for 3.5 days at 37°C in either a reduced oxygen environment (5% CO2, 5% O2, 90% N2) or a high-oxygen environment (5% CO2 in air [∼20% O2]) to obtain morula/blastocyst-stage embryos that were subsequently transferred to 2.5-day postcoitum or 3.5-day postcoitum pseudopregnant CF1 females. Embryo transfers were performed using the Non-Surgical Embryo Transfer Device (NSET; Paratechs), and a maximum of 15 morula/blastocyst-stage embryos were transferred into each surrogate mother. The day of embryo transfer was denoted as E3.5, and all CF1 recipient females were euthanized 8 days after transfer to obtain E10.5 embryos that were morphologically similar to E10.5 naturally conceived controls.
Isolation of DNA and RNA from Embryonic and Placental Tissues
Naturally conceived and IVF-derived embryos were collected at E10.5, and somites were counted for each embryo. Any embryo that had fewer than 30 somites was excluded from the experiment to facilitate an accurate comparison to the naturally conceived controls. After collection of the embryo, the placenta was mechanically separated from the maternal decidua and the embryonic yolk sac. Both the whole embryo and placenta were snap-frozen in separate tubes, using liquid nitrogen and stored at −80°C until further use. DNA and RNA were isolated simultaneously from each tissue by using phenol-chloroform extraction and TRIzol (Invitrogen), respectively. Briefly, the whole embryo or placenta was homogenized in 0.5 ml of lysis buffer (50 mM Tris, pH 8.0, 100 mM EDTA, 0.5% SDS) by using a 1-ml syringe and a 22-guage needle to obtain a homogenous cell lysate. One half of the lysate was used to isolate genomic DNA, and the other half was used to isolate total RNA. For DNA isolation, the lysate was diluted two-fold with lysis buffer, and proteinase K was added to digest the remaining tissue. After incubating the samples for 90 min, we performed phenol-chloroform extraction to isolate genomic DNA. For RNA isolation, the other half of the lysate was added to TRIzol to make the overall volume 1 ml, and total RNA was extracted using the manufacturer's instructions.
Bisulfite Pyrosequencing and LUminometric Methylation Assay
DNA methylation was measured at the ICRs of multiple imprinted genes by using bisulfite pyrosequencing. Bisulfite mutagenesis was performed with 1 μg of isolated genomic DNA from embryonic and placental tissues, using a bisulfite kit (Epitect; Qiagen). Bisulfite pyrosequencing was carried out as described previously [9, 22]; primer sequences for the pyrosequencing assays can be found in Supplemental Table S1 (all supplemental data are available online at www.biolreprod.org). The LUminometric methylation assay (LUMA) was used to assess methylation profiles at repetitive elements throughout the genome as previously described [23]. The assay uses the restriction cut sites HpaII and MspI on 1 μg of genomic DNA, followed by polymerase extension on the overhangs by using pyrosequencing technology to calculate global methylation levels in embryonic and placental tissues.
Allele-Specific Bisulfite Sequencing
Allele-specific bisulfite sequencing was performed using nested PCR for the H19/Igf2 and Peg3 ICRs as previously described [24–26]. Briefly, the same bisulfite-treated DNA was used for PCR amplification using nested primers, and the amplified products were cloned into a vector using a PCR cloning kit (Strataclone; Agilent), transformed into chemically competent Escherichia coli cells, and plated on LB plates with ampicillin (0.1 mg/ml). Recombinant plasmids were isolated and sequenced at the University of Pennsylvania DNA sequencing facility. Maternal and paternal alleles were distinguished by using multiple polymorphisms between the B6 and Cast alleles in the F1 hybrids as previously described [24, 26].
Allele-Specific Expression of Imprinted Genes
Total RNA isolated from embryonic and placental tissues (described above) was subjected to DNase treatment (Promega), and first-strand synthesis was performed using Superscript III reverse transcriptase (Invitrogen) and random hexamers. To ensure that cDNA samples were devoid of genomic DNA contamination, samples without reverse transcriptase were processed in parallel. Allele-specific expression levels of 5 imprinted genes (H19, Igf2, Peg3, Kcnq1ot1, and Cdkn1c) located on chromosome 7 were measured using RT-PCR, followed by allele-specific restriction enzyme digests [15, 27]. The digested PCR products were run on 12% polyacrylamide gels, and gel images were captured using Gel Logic 212 Pro imaging system (Carestream). Each band was quantified with Molecular Imaging software version 5.02.30 (Carestream), and the value of the repressed allele was divided by the sum of all bands to calculate the percent of expression from the repressed allele. Allele-specific expression of Snrpn was measured using the LightCycler real-time PCR system (Roche) as described previously [15, 18]. Samples that exhibited at least 10% expression from the repressed allele were considered biallelic.
Statistical Analyses
Embryo development data were analyzed using the chi-squared test. Bisulfite pyrosequencing, LUMA, and allele-specific expression data were analyzed using the variance ratio test to calculate an F statistic and determine whether there were any significant differences among naturally conceived controls and IVF tissues cultured at 5% O2 (IVF-5%), naturally conceived controls and IVF-20% O2 tissues, and IVF-5% O2 and IVF-20% O2 tissues. Because the same data set was subjected to multiple comparisons, the Bonferroni correction was used to control for the family specific error rate. All statistical analyses were performed using QI Macros SPC software for Excel version 2013.07 (Microsoft).
RESULTS
Embryo Development in 5% O2 versus 20% O2 During In Vitro Culture
Previous studies involving animal models and humans have shown that culturing mammalian embryos in atmospheric oxygen (∼20% O2) compromises preimplantation development compared to those that are cultured in a reduced oxygen environment (5% O2) [17, 28–33]. However, the studies involving mice have typically used natural fertilization to assess the effects of oxygen concentration during culture. Our goal was to recapitulate human IVF, including the use of 2 different oxygen tensions, in a mouse model and determine the consequences for embryonic development and the epigenetic regulation of imprinting. Accordingly, after in vitro fertilization, F1 hybrid embryos were transferred to KSOM and cultured at 37°C in either 5% or 20% O2 for 24 h. Embryos that had progressed to the 2-cell stage were counted and transferred into a different drop of KSOM. After 3 more days of in vitro culture, the number of embryos that developed to the blastocyst stage was determined, and those embryos were used for nonsurgical embryo transfer later that day.
In total, 253 embryos from the 5% O2 group and 298 embryos from the 20% O2 group were transferred into recipient females (Table 1). At E10.5, we collected 30 5% O2 embryos from 14 different litters and found that most of the litters had 1 or 2 pups. Similarly, we collected 16 20% O2 embryos from 8 different litters and observed that these litters had 1 or 2 pups as well. These observations are in contrast to the naturally conceived controls, from which we collected 16 embryos from 3 different litters, and each litter had either 5 or 6 pups. It should be noted that all embryos included in this analysis were morphologically normal. Notably, F1 hybrid embryos that were cultured in 20% O2 exhibited reduced development to the blastocyst stage compared to those that were cultured at 5% O2 (Table 1). In addition, the embryos cultured at 20% O2 resulted in fewer embryos at E10.5 than the 5% O2 group (Table 1). This finding is similar to that of a previous report which showed that culturing mouse embryos in 20% O2 compromises the developmental potential of the resulting blastocysts after embryo transfer [34].
TABLE 1.
Effect of oxygen concentration on embryonic development.
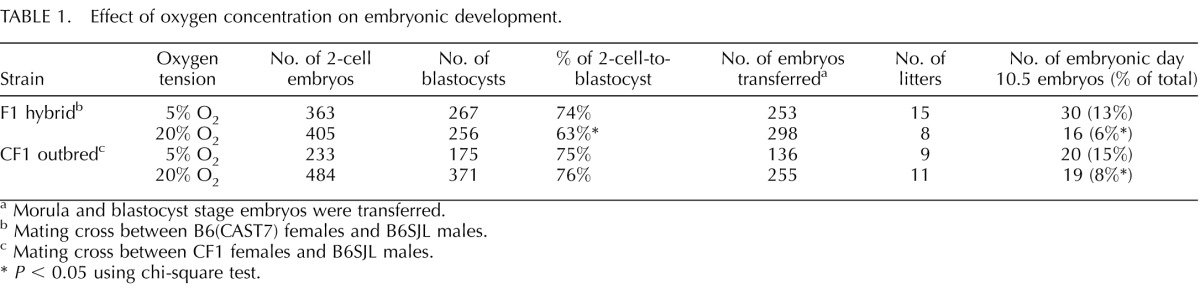
Morula and blastocyst stage embryos were transferred.
Mating cross between B6(CAST7) females and B6SJL males.
Mating cross between CF1 females and B6SJL males.
P < 0.05 using chi-square test.
DNA Methylation Analysis of Multiple ICRs in Embryonic and Placental Tissues
To determine whether the embryos isolated at E10.5 that appeared morphologically normal exhibited any epigenetic abnormalities, we analyzed DNA methylation profiles by using bisulfite pyrosequencing assays for 2 paternally methylated ICRs (H19/Igf2 and Dlk1/Gtl2) and 4 maternally methylated ICRs (Snrpn, Peg3, Kcnq1ot1, and Peg1). Methylation at these regions is established in either the male or female germ line during gametogenesis and then stably maintained throughout embryogenesis and postnatal development [5]. Importantly, abnormal methylation at some of the human orthologous ICRs has been observed in ART-conceived children [1].
We assessed DNA methylation at 6 ICRs using bisulfite pyrosequencing assays in both embryonic and placental tissues from naturally conceived and IVF-derived embryos that were exposed to either 5% or 20% O2 during in vitro culture. Each pyrosequencing assay measured methylation levels at 5 or 6 individual CpG sites located within the ICR (Supplemental Table S1), and the average levels for all of the CpG sites in an individual ICR were reported. The parental allele-specific methylation at ICRs results in an overall methylation profile of approximately 50% in both the embryo and placenta. Because both human and animal studies have shown that only a subset of IVF-derived offspring exhibit epigenetic abnormalities [4], we used the variance to compare significance between naturally conceived and IVF-derived samples. In total, we analyzed 16 naturally conceived embryos, 18 IVF-derived embryos cultured in 5% oxygen (IVF-5% O2), and 16 IVF-derived embryos cultured in 20% oxygen (IVF-20% O2). Twelve of the IVF-5% O2 samples that we collected were excluded because IVF-20% O2 samples were not generated in parallel.
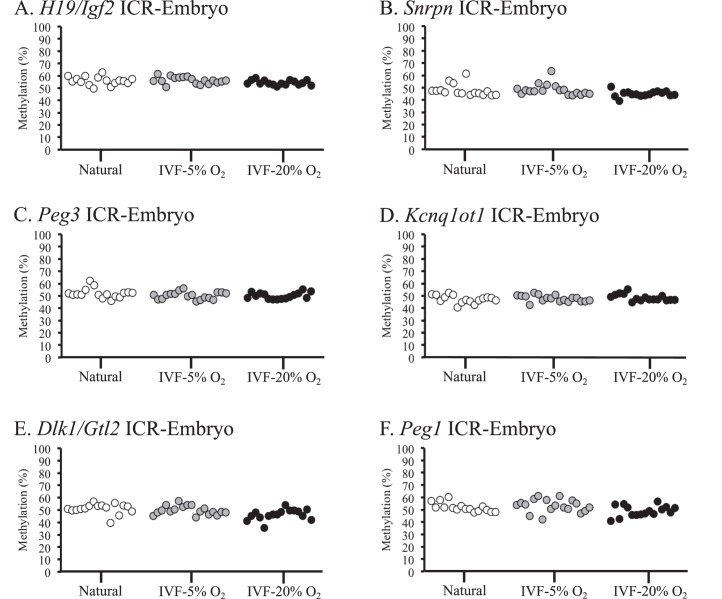
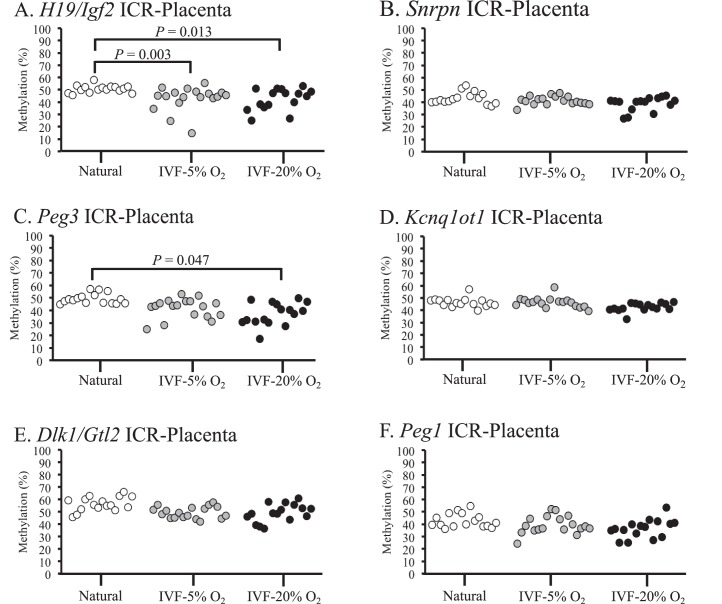
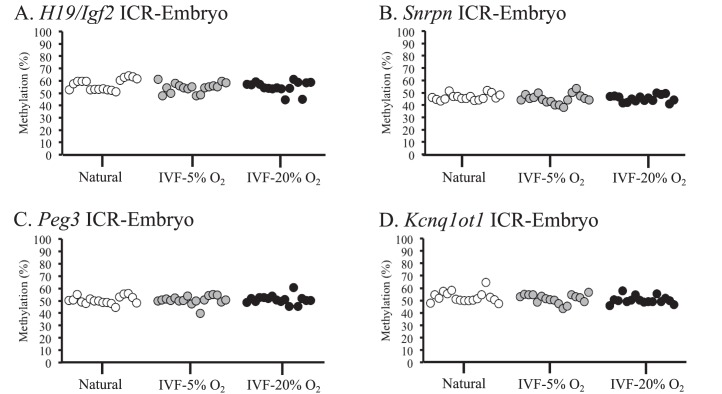
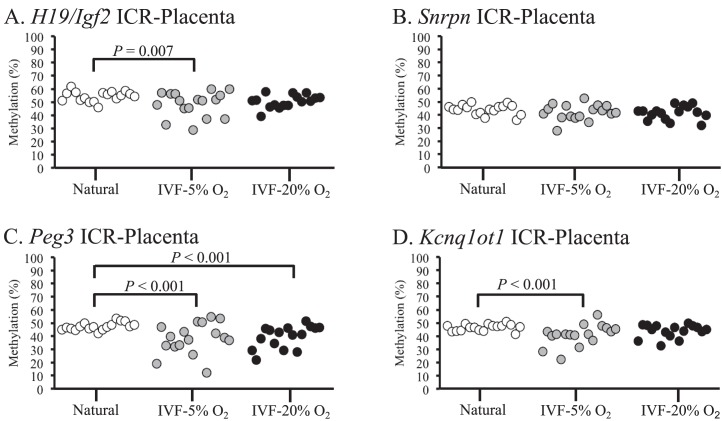
Embryonic tissues from naturally conceived and IVF-derived embryos exhibited normal methylation profiles for all 6 ICRs, and no statistical differences in variance were detected (Fig. 1, A–F). Because many of the IVF-derived samples that were analyzed exhibited variability in DNA methylation levels within different litters (i.e., there were samples with normal and reduced methylation profiles within 1 IVF litter), single embryos and placentae were used as statistical units. DNA methylation was assessed in the placentae from the same embryos because epigenetic regulation in placental tissues can be influenced by environmental factors during in utero development [9, 35]. Most placentae from IVF-derived concepti exhibited methylation profiles similar to those of naturally conceived controls (Fig. 2). However, some placentae from IVF-5% O2 and IVF-20% O2 embryos had reduced methylation compared to those of controls, and a few of these tissues showed reduced methylation at multiple ICRs (Supplemental Table S2). Although several placentae had markedly reduced methylation in the IVF-derived groups, there were no statistically significant differences detected at the Snrpn, Kcnq1ot1, Peg1, and Dlk1/Gtl2 ICRs (Fig. 2, B, D, E, and F). Placental tissues from the 5% and 20% O2 group, however, were significantly more variable at the H19 ICR compared to those of controls (Fig. 2A). In addition, placentae from IVF-20% O2 embryos were statistically different from those of controls at the Peg3 ICR, whereas no significant differences were detected between the 5% O2 group and controls (Fig. 2C). Thus, the methylation profiles at ICRs in the placenta are more susceptible to perturbation by ART-associated manipulations than embryonic tissues, and the H19 ICR appears to be particularly sensitive to external factors.
FIG. 1.
DNA methylation profiles in embryonic tissues from naturally conceived and IVF-derived F1 hybrid embryos are shown. Bisulfite pyrosequencing was used to measure DNA methylation in embryonic tissues at the H19/Igf2 ICR (A), Snrpn ICR (B), Peg3 ICR (C), Kcnq1ot1 ICR (D), Dlk1/Gtl2 ICR (E), and Peg1 ICR (F). Each dot represents the average methylation from all of the CpG sites analyzed in an individual sample. Statistical significance was analyzed using variance of average methylation values from individual samples.
FIG. 2.
DNA methylation profiles in placental tissues from naturally conceived and IVF-derived F1 hybrid embryos are shown. Bisulfite pyrosequencing was used to measure DNA methylation in placental tissues at the H19/Igf2 ICR (A), Snrpn ICR (B), Peg3 ICR (C), Kcnq1ot1 ICR (D), Dlk1/Gtl2 ICR (E), and Peg1 ICR (F). Each dot represents the average methylation from all of the CpG sites analyzed in an individual sample. Brackets indicate statistical differences between 2 groups, using variance. P values are given where statistical differences were observed (P < 0.05).
To confirm that the reduced methylation profiles observed in IVF-derived placentae were bona fide epigenetic errors and not the result of PCR bias, allele-specific bisulfite sequencing was performed with a subset of the placental samples that exhibited both normal and abnormal methylation at the H19/Igf2 and Peg3 ICRs in the pyrosequencing analysis (Fig. 2, A and C; and see Supplemental Table S2). For the H19/Igf2 ICR, we analyzed 3 naturally conceived placentae, 3 IVF-5% O2 placentae, and 3 IVF-20% O2 placentae. All of the naturally conceived placentae exhibited typical methylation profiles at the H19/Igf2 ICR, with the paternal allele fully methylated and the maternal allele unmethylated (Supplemental Fig. S1A). Importantly, the IVF-5% O2 and IVF-20% O2 placentae that exhibited a reduction in methylation according to the pyrosequencing analysis also had reduced methylation on the paternal allele when allele-specific bisulfite sequencing was performed (Supplemental Fig. S1A). We performed the same analysis for the Peg3 ICR using different samples and successfully confirmed our pyrosequencing results for this region as well (Supplemental Fig. S1B). Thus, these results demonstrate that the methylation abnormalities observed by pyrosequencing were due to a loss of methylation on the normally repressed allele.
Global Methylation Levels in Embryonic and Placental Tissues
To ascertain whether the observed abnormalities at the H19/Igf2 and Peg3 ICRs were the result of a global failure in methylation maintenance, we assessed genomic methylation in the same embryonic and placental tissues. To measure global methylation levels, we used LUMA, which uses the MspI and HpaII restriction cut sites, followed by polymerase extension and pyrosequencing to assess DNA methylation at repetitive elements throughout the genome. We detected a high level of methylation in embryonic tissues (∼80% methylation) and a lower level of methylation in placental tissues (∼60% methylation), similar to previous reports [36]. Notably, no significant differences were observed in either embryonic or placental tissues from naturally conceived and IVF-derived embryos (Fig. 3). In addition, the placentae that exhibited reduced methylation at multiple ICRs showed methylation levels similar to those of naturally conceived controls (Fig. 3; Supplemental Table S2), which suggests that the previously observed epigenetic errors are specific to certain loci in the genome and are not due to an impairment of the methylation machinery during the early stages of development.
FIG. 3.
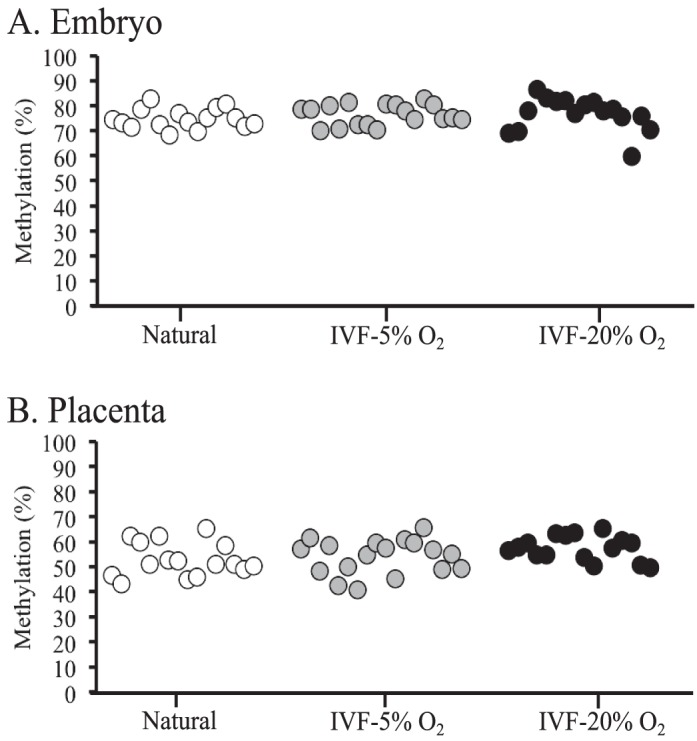
Global methylation levels for embryonic and placental tissues from F1 hybrid embryos are shown. DNA methylation was measured at repetitive elements throughout the genome, using LUMA in embryonic (A) and placental (B) tissues from naturally conceived and IVF-derived embryos. Each dot represents the average methylation of an individual sample from 2 replicates.
Allele-Specific Expression of Imprinted Genes
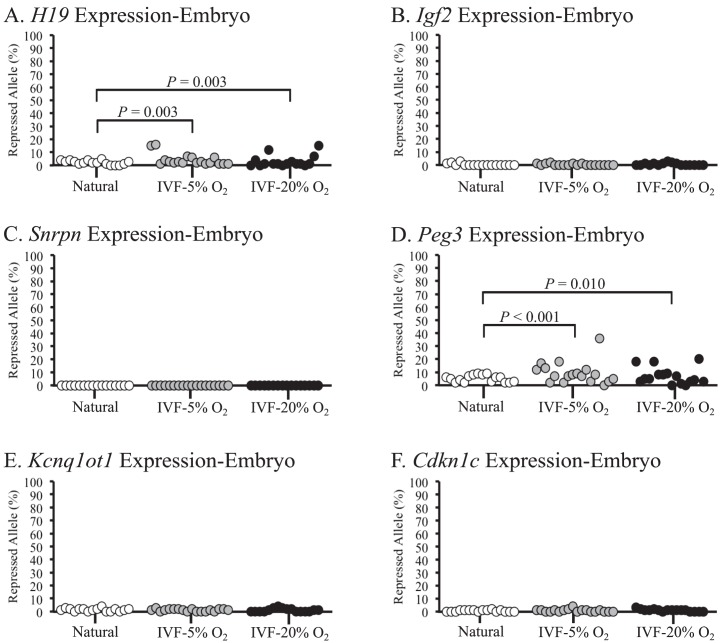
We analyzed allele-specific expression of six imprinted genes (H19, Igf2, Snrpn, Peg3, Kcnq1ot1, and Cdkn1c) in the same embryonic and placental tissues to determine whether abnormal methylation at an ICR was correlated with aberrant expression of an imprinted gene. These genes were chosen because they reside on chromosome 7 and are regulated by the previously assayed ICRs. The H19/Igf2 ICR regulates the expression of H19 and Igf2, whereas the Kcnq1ot1 ICR regulates the expression of Kcnq1ot1 and Cdkn1c. The Snrpn and Peg3 ICRs regulate the expression of Snrpn and Peg3, respectively. Four of these genes, namely Igf2, Snrpn, Peg3, and Kcnq1ot1, are expressed from the paternal allele, whereas H19 and Cdkn1c are expressed from the maternal allele. Notably, the monoallelic expression levels of Igf2, Snrpn, Kcnq1ot1, and Cdkn1c were maintained in all naturally conceived and IVF-derived embryonic tissues (Fig. 4, B, C, E, and F), whereas a significant increase in biallelic expression of H19 and Peg3 was observed in IVF-derived embryonic tissues cultured at both of the oxygen concentrations compared to those in naturally conceived controls (Fig. 4, A and D). Surprisingly, the samples that exhibited biallelic expression of H19 and Peg3 had normal methylation profiles at the corresponding ICRs (Supplemental Tables S2 and S3). These data are consistent with those from our previous studies of cultured embryos showing that allele-specific expression of H19 and Peg3 is aberrantly regulated in a subset of ART-derived embryonic tissues [14, 15]. The data further suggest that mis-expression of imprinted genes is not always correlated with abnormal methylation of the ICRs.
FIG. 4.
Allele-specific expression levels of imprinted genes in embryonic tissues from F1 hybrid embryos are shown. Allele-specific expression levels of H19 (A), Igf2 (B), Snrpn (C), Peg3 (D), Kcnq1ot1 (E), and Cdkn1c (F) were assessed in embryonic tissues from naturally conceived and IVF-derived embryos. Each circle represents the proportion of expression from the normally repressed allele in an individual sample. Samples that exhibit expression of 10% or more from the repressed allele are considered biallelic. See the legend to Figure 2 for more details.
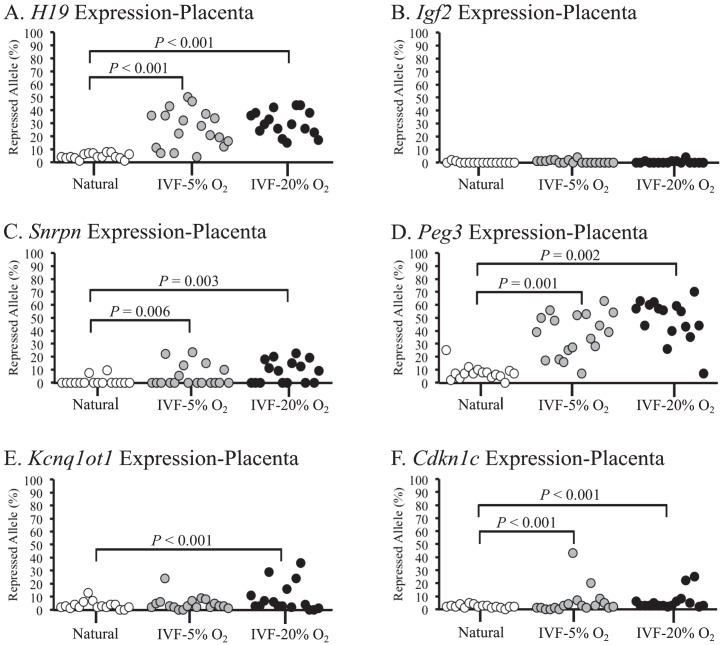
Allele-specific expression of natural and IVF-derived placental tissues revealed normal, monoallelic expression of Igf2, whereas aberrant expression profiles were observed in IVF-derived placentae for all other imprinted genes analyzed (Fig. 5). Placental tissues from embryos that were exposed to 5% and 20% O2 exhibited significant differences for the allele-specific expression levels of H19, Snrpn, Peg3, and Cdkn1c (Fig. 5, A, C, D, and F). Interestingly, Kcnq1ot1 showed significant changes in expression in placentae obtained following embryo culture in 20% O2 but not in 5% O2 compared to placentae from naturally conceived controls. In addition, much like the embryonic tissues, many of the placental tissues that exhibited biallelic expression of an imprinted gene had normal methylation at the corresponding ICR (Supplemental Tables S2 and S3). Collectively, these data show that IVF induces loss of imprinted expression in placental tissues at a much higher frequency than embryonic tissues when embryos are cultured in either 5% or 20% O2.
FIG. 5.
Allele-specific expression levels of imprinted genes in placental tissues from F1 hybrid embryos are shown. Allele-specific expression levels of H19 (A), Igf2 (B), Snrpn (C), Peg3 (D), Kcnq1ot1 (E), and Cdkn1c (F) were assessed in embryonic tissues from naturally conceived and IVF-derived embryos. Each circle represents the proportion of expression from the normally repressed allele in an individual sample. See legends to Figures 2 and 4 for more details.
Embryonic Development and DNA Methylation Analysis of Multiple ICRs Using the CF1 Outbred Mouse Strain
The F1 hybrid embryos analyzed in this study were generated using oocytes from a C57BL/6 and Mus castaneus hybrid mouse strain, which may increase their susceptibility to environmental perturbations. To determine whether our results were a peculiarity of the strain, we determined whether similar epigenetic abnormalities are detected when an outbred mouse strain was used to generate the oocytes for IVF-derived embryos. To this end, we collected oocytes from superovulated CF1 mice and produced IVF-derived embryos in parallel with the F1 hybrids by using the same capacitated sperm and culture conditions to facilitate an accurate comparison. We collected 20 E10.5 embryos from the 5% O2 group and 19 E10.5 embryos from the 20% O2 group (Table 1). The litter sizes were similar to those seen with the F1 hybrids in that most litters had 1 or 2 pups. In addition, we analyzed 14 naturally conceived controls that were collected from 3 different litters. Notably, culturing these embryos in different oxygen concentrations did not influence embryonic development from the 2-cell to blastocyst stage, but we did observe a significant reduction in the number of embryos that were collected at E10.5 when a high-oxygen concentration was used during culture (Table 1). Thus, these results suggest that the CF1 outbred strain appears to have a protective effect on preimplantation development throughout embryo culture, but postimplantation development is adversely affected by a high oxygen tension during early embryogenesis.
DNA methylation profiles were assessed in embryonic and placental tissues from the CF1 embryos at the H19/Igf2, Snrpn, Peg3, and Kcnq1ot1 ICRs using bisulfite pyrosequencing. Embryonic tissues from these embryos exhibited normal methylation profiles at all of the regions analyzed (Fig. 6). However, similar to the F1 hybrid analysis results, DNA methylation was highly variable in placental tissues from these IVF-derived embryos (Fig. 7). No statistical differences were detected at the Snrpn ICR between the naturally conceived controls and IVF-derived placentae (Fig. 7B), but the IVF-5% O2 placentae were significantly more variable at the H19/Igf2 and Kcnq1ot1 ICRs than in the controls (Fig 7, A and D). In addition, both IVF-5% O2 and IVF-20% O2 placentae were significantly different from naturally conceived controls at the Peg3 ICR (Fig. 7C). Thus, results from this mating showing higher variance in methylation levels at ICRs in placentae were similar to data obtained from the F1 hybrid analysis (Figs. 2 and 7). There were differences in the DNA methylation analysis between CF1 and F1 hybrid IVF-derived embryos as well. Surprisingly, the IVF-20% O2 placentae were not significantly different at the H19/Igf2 and Kcnq1ot1 ICRs, whereas the IVF-5% O2 placentae exhibited significant differences at both of these regions (Fig. 7, A and D). Taken together, these results suggest that impaired embryonic development in 20% O2 and increased incidence of stochastic errors in placental tissues after ART are not strain-specific phenomena.
FIG. 6.
DNA methylation profiles in embryonic tissues from naturally conceived and IVF-derived CF1 outbred embryos are shown. Pyrosequencing was used to measure DNA methylation in embryonic tissues at the H19/Igf2 ICR (A), Snrpn ICR (B), Peg3 ICR (C), and Kcnq1ot1 ICR (D). Each dot represents the average methylation from all of the CpG sites analyzed in an individual sample.
FIG. 7.
DNA methylation profiles in placental tissues from naturally conceived and IVF-derived CF1 outbred embryos are shown. Pyrosequencing was used to measure DNA methylation in placental tissues at the H19/Igf2 ICR (A), Snrpn ICR (B), Peg3 ICR (C), and Kcnq1ot1 ICR (D). Each dot represents the average methylation from all of the CpG sites analyzed in an individual sample. See Figure 2 legend for more details.
DISCUSSION
In the present study, a mouse model was used to investigate the effects of oxygen concentration on genomic imprinting following IVF and embryo culture. Although numerous reports involving animals and humans have demonstrated that culturing embryos in a reduced oxygen concentration improves embryogenesis [17, 32, 33, 37], the effects of different oxygen environments in the context of IVF on epigenetic regulation in developing embryos is lacking. In contrast to previous studies, we performed IVF before subjecting developing embryos to different oxygen concentrations and comprehensively examined DNA methylation at 6 ICRs as well as allele-specific expression of 6 imprinted genes in both embryonic and placental tissues. To enhance the human relevance of our mouse model system, we simulated the ex vivo manipulations used during IVF, and this methodology was used with 2 strains of mice, an F1 hybrid strain and a CF1 outbred strain. The data presented herein demonstrate that placental tissues are susceptible to stochastic epigenetic errors at multiple imprinted genes during ART and that culturing embryos in 20% O2 does not induce a statistically significant increase of epigenetic abnormalities in embryonic or placental tissues compared to those cultured in 5% O2. Moreover, our results suggest that the hybrid vigor associated with an outbred mouse strain has a protective effect against environmental perturbations.
Oxygen levels in the uterus are dynamic and decrease at the time of implantation in several mammalian species [38]. Moreover, embryos are exposed to a decreasing oxygen gradient as they travel from the oviduct to the uterus [38], which suggests that the oxygen tension is carefully regulated in the female reproductive system to foster optimal intrauterine development. Therefore, culturing embryos in a high oxygen concentration throughout preimplantation development could impair embryonic development and promote suboptimal growth after implantation by exposing the embryo to an unnatural environment.
Here, we found that the use of 20% O2 during embryo culture can reduce the developmental potential of embryos in 2 different strains of mice. However, this impaired development was not associated with an increase in epigenetic abnormalities compared to embryos that were cultured in 5% O2. Use of 20% O2 during culture is correlated with increased generation of reactive oxygen species in the embryo compared to use of 5% O2, which may lead to oxidative stress in the embryo [37]. Importantly, oxidative stress can influence gene expression profiles, and culturing embryos in 20% O2 has been associated with a greater perturbation in global expression profiles [33]. Thus, the impaired development that we observed with 20% O2 was most likely due to altered expression profiles at nonimprinted genes as a response to oxidative stress. This observation suggests that maintenance methylation at ICRs is not adversely impaired by a high oxygen tension during embryo culture. It should be noted, however, that placental tissues from embryos cultured in 5% O2 exhibited aberrant expression and methylation at multiple imprinted genes, demonstrating that even culturing with a reduced oxygen tension induces epigenetic abnormalities in a subset of offspring.
Although we observed an increased frequency of epigenetic errors in placental tissues from IVF-derived embryos, there was a wide variety of outcomes in these tissues. Several IVF-derived placentae exhibited abnormal methylation and expression profiles at multiple imprinted genes, whereas other placentae showed epigenetic errors at only 1 or 2 imprinted loci. Because the placentae from F1 hybrid embryos have a high degree of genetic similarity, the seemingly random distribution of epigenetic defects in different placental tissues suggests that the induction of these epigenetic abnormalities occurs through a stochastic process. The different combinations of epigenetic errors at imprinted genes could result in a variety of placental phenotypes because these loci play important roles in placental function. Intriguingly, ART pregnancies are associated with a multitude of medical conditions that involve abnormal placentation such as pre-eclampsia, placenta previa, and placental abruption [3, 39], which may be explained partly by epigenetic errors at different imprinted genes. The higher incidence of premature birth and low birth weight observed in ART-conceived children may also be related to abnormal placental function resulting from faulty genomic imprinting at multiple genes. In support of this hypothesis, a small percentage of biopsy results from placentae of ART-conceived children with low birth weight exhibit abnormal methylation profiles at several imprinted genes in an apparent random fashion, that is, the set of affected genes differs from placenta to placenta (Sapienza and Coutifaris, unpublished observations). Together, our data suggest that the placenta is susceptible to the generation of random imprinting errors during ART and that different combinations of these abnormalities may induce suboptimal placental function and fetal growth.
Although biallelic expression of imprinted genes was common in IVF-derived placentae, many of these tissues exhibited normal methylation profiles at the corresponding ICR. One possible explanation for this discrepancy is that aberrant expression of these imprinted genes is caused by dysregulation of other epigenetic modifications besides DNA methylation at the ICR. However, the discordance between abnormal methylation and aberrant expression could also be due to the complexity of placental tissues. DNA methylation at ICRs can be measured in all cell types of the placenta, whereas allele-specific expression is detected only in the cells that express an imprinted gene. Given that many imprinted genes are highly expressed in specific cell types in the placenta [40], it seems plausible that abnormal methylation in this small number of cells could be masked by the abundance of normal methylation profiles in other cell types that do not express the gene. Thus, to accurately determine whether a discrepancy exists between methylation and expression profiles in the placenta, methodologies that measure DNA methylation and expression in purified cells or specific cell types should be used.
Several studies have shown that certain aspects of the ART procedure can influence epigenetic profiles in a mouse model, but there is some evidence that substrains of mice exhibit different sensitivities under in vitro culture conditions [41]. Because ART is performed with genetically diverse human populations, it is important to determine whether ART-associated epigenetic defects are strain-specific. Interestingly, we found that IVF-derived CF1 concepti exhibited an increased incidence of epigenetic defects in placental tissues at multiple ICRs and that the induction of these errors occurred in a random fashion, similar to that detected in IVF-derived F1 hybrid concepti. This observation in the 2 mouse crosses suggests that the stochastic generation of epigenetic abnormalities in placental tissues during ART is not strain-specific, and therefore most likely occurs in humans as well. There were also some notable differences between F1 hybrid and CF1 IVF-derived embryos. Notably, use of 20% O2 did not impair embryonic development from the 2-cell to blastocyst stage, and the Peg3 ICR did not exhibit heightened sensitivity to high oxygen tension during culture in the CF1 placentae. These two observations may be related and imply that the outbred vigor associated with the CF1 strain confers a protective effect against environmental perturbations during the early stages of development. Although this suggests that human embryos are less sensitive to an elevated oxygen tension, it should be noted that postimplantation development of CF1 embryos was adversely affected by the use of 20% O2 during in vitro culture. Thus, while using 20% O2 may not increase the frequency of epigenetic defects at imprinted genes in human placental tissues during fertility treatments, exposure to an elevated oxygen tension throughout early embryogenesis could impair postimplantation development by influencing epigenetic regulation and expression of nonimprinted genes.
Given the increasing use of ART, it is imperative that we continue to evaluate the safety of ex vivo manipulations associated with these procedures. Several human studies have reported an increase in epigenetic errors in ART-conceived children compared to their naturally conceived counterparts, whereas other studies report no differences between the groups [42–45]. Notably, there are ethical and logistical limitations in human research that preclude the analysis of different tissues and cell types that would facilitate the detection of rare imprinting errors in ART-conceived children. The use of our mouse model minimizes confounding factors and allows us to perform allele-specific experiments on different tissues to determine the occurrence of epigenetic errors at imprinted genes in ART-derived samples. By using this strategy, we found that the use of atmospheric oxygen during embryo culture does not induce a significant increase of epigenetic defects in embryonic or placental tissues compared to embryos that were cultured in a reduced oxygen environment. However, we did observe a relatively high frequency of epigenetic abnormalities when embryos were cultured at 5% O2, which suggests that even culturing with a reduced oxygen environment is far from optimal. It is likely that other aspects of ART procedures (e.g., superovulation, light exposure, embryo transfer) also contribute to the generation of epigenetic defects in ART-conceived offspring. Indeed, a previous study from our laboratory demonstrated that performing embryo transfer with in vivo-fertilized embryos can result in loss of imprinting [15]. Interestingly, surgical embryo transfer was performed in that study, so evaluating the effect of nonsurgical embryo transfer on imprinted gene regulation will be informative because the implantation rate is lower with that methodology. The harmful effects of ART on the epigenome are most likely caused by exposing the early embryo to an unnatural environment that disrupts critical cellular processes during a developmental period when extensive epigenetic reprogramming occurs. Thus, minimizing the occurrence of epigenetic errors in placental tissues from ART-derived offspring may be a challenging endeavor that entails modifications to hormonal regimens, in vitro culture conditions, and embryo transfer protocols that are used during fertility treatments.
In conclusion, further investigation into the effects of ART procedures on epigenetic profiles in developing embryos is warranted. Many fertility clinics culture embryos to the blastocyst stage so that development can be monitored and only high-quality embryos are transferred. However, because we have observed a high frequency of epigenetic defects at imprinted genes in placental tissues, the use of short-term culture (i.e., transferring the embryo before differentiation of the trophectoderm) could decrease the incidence of epigenetic abnormalities in the placenta. Although imprinted genes are believed to exhibit heightened sensitivity to external factors because they are heavily reliant on epigenetic regulation, a recent study suggests that these genes are not more susceptible to aberrant expression than the rest of the transcriptome during the early stages of development [46]. Future studies should investigate the genome-wide effects of ART manipulations to evaluate the sensitivity of nonimprinted genes to ART procedures. In addition, it will be informative to assess profiles of epigenetic marks other than DNA methylation because histone modifications are believed to play an important role in placental tissues. Given that epigenetic abnormalities in the placenta seem to arise by a random process during ART, predicting their occurrence may be challenging. Therefore, it will be important to establish sensitive methodologies to detect the generation of epigenetic errors in human tissues. Our data suggest that epigenetic defects may occur in specific cell types in the placenta because of the discordance between abnormal methylation and expression. Evaluating whether certain cell types in the placenta are more susceptible to environmental perturbations could enhance our ability to detect ART-associated imprinting errors. Moreover, by understanding the molecular mechanisms underlying the induction of abnormal epigenetic profiles in ART-derived placental tissues, we may be able to minimize their occurrence and improve the overall health of ART-conceived children.
Supplementary Material
ACKNOWLEDGMENT
We thank Josh Plotkin for discussion and helpful comments on the statistical analysis. We also thank Erin Fischer for technical assistance.
Footnotes
Supported by grant U54-HD06817 to M.S.B., R.M.S, and C.C. and training grant T32 HD007305 to W.M. from the National Institutes of Health, and by a postdoctoral fellowship from the Lalor Foundation to E.D. Presented in part at the Specialized Cooperative Centers Program in Reproduction and Infertility research (SCCPIR) Research Meeting. Bethesda, MD, May 21, 2013.
REFERENCES
- Eroglu A, Layman LC. Role of ART in imprinting disorders. Semin Reprod Med 2012; 30: 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skora D, Frankfurter D. Adverse perinatal events associated with ART. Semin Reprod Med 2012; 30: 84–91. [DOI] [PubMed] [Google Scholar]
- Kondapalli LA, Perales-Puchalt A. Low birth weight: is it related to assisted reproductive technology or underlying fertility? Fertil Steril 2013; 99: 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajj N, Haaf T. Epigenetic disturbances in in vitro cultured gametes and embryos: implications for human assisted reproduction. Fertil Steril 2013; 99: 632–641. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet 2005; 14: R47–R58. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS. Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev 2009; 23: 2124–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 2013; 152: 1308–1323. [DOI] [PubMed] [Google Scholar]
- McCarrey JR. The epigenome as a target for heritable environmental disruptions of cellular function Mol Cell Endocr 2012; 354: 9–15. [DOI] [PubMed] [Google Scholar]
- Susiarjo M, Sasson I, Mesaros C, Bartolomei MS. Bisphenol A exposure disrupts genomic imprinting in the mouse. Plos Genet 2013; 9: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod 2000; 62: 1526–1535. [DOI] [PubMed] [Google Scholar]
- Fortier AL, Lopes FL, Darricarrere N, Martel J, Trasler JM. Superovulation alters the expression of imprinted genes in the midgestation mouse placenta. Hum Mol Genet 2008; 17: 1653–1665. [DOI] [PubMed] [Google Scholar]
- de Waal E, Yamazaki Y, Ingale P, Bartolomei MS, Yanagimachi R, McCarrey JR. Gonadotropin stimulation contributes to an increased incidence of epimutations in ICSI-derived mice. Hum Mol Genet 2012; 21: 4460–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manipalviratn S, Decherney A, Segars J. Imprinting disorders and assisted reproductive technology. Fertil Steril 2009; 91: 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann MR, Lee SS, Doherty AS, Verona RI, Nolen LD, Schultz RM, Bartolomei MS. Selective loss of imprinting in the placenta following preimplantation development in culture. Development 2004; 131: 3727–3735. [DOI] [PubMed] [Google Scholar]
- Rivera RM, Stein P, Weaver JR, Mager J, Schultz RM, Bartolomei MS. Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum Mol Genet 2008; 17: 1–14. [DOI] [PubMed] [Google Scholar]
- Ciray HN, Aksoy T, Yaramanci K, Karayaka I, Bahceci M. In vitro culture under physiologic oxygen concentration improves blastocyst yield and quality: a prospective randomized survey on sibling oocytes. Fertil Steril 2009; 91: 1459–1461. [DOI] [PubMed] [Google Scholar]
- Waldenstrom U, Engstrom AB, Hellberg D, Nilsson S. Low oxygen compared with high oxygen atmosphere in blastocyst culture, a prospective randomized study. Fertil Steril 2009; 91: 2461–2465. [DOI] [PubMed] [Google Scholar]
- Mann MR, Chung YG, Nolen LD, Verona RI, Latham KE, Barolomei MS. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol Reprod 2003; 69: 902–914. [DOI] [PubMed] [Google Scholar]
- Toyoda Y, Yokoyama M, Hosi T. Study on the fertilization of mouse eggs in vitro. In vitro fertilization of eggs by fresh epididymal sperm. Jpn J Anim Reprod 1971; 16: 147–151. [Google Scholar]
- Whitten WK. Nutrient requirements for the culture of preimplantation embryos in vitro. Adv Bio Sci 1971; 6: 129–139. [Google Scholar]
- Ho Y, Wigglesworth K, Eppig JJ, Schultz RM. Preimplantation development of mouse embryos in KSOM: augmentation by amino acids and analysis of gene expression. Mol Reprod Dev 1995; 41: 232–238. [DOI] [PubMed] [Google Scholar]
- Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science 2012; 335: 1499–1502. [DOI] [PubMed] [Google Scholar]
- Pilsner JR, Lazarus AL, Nam DH, Letcher RJ, Sonne C, Dietz R, Basu N. Mercury-associated DNA hypomethylation in polar bear brains via the LUminometric Methylation Assay: a sensitive method to study epigenetics in wildlife. Mol Ecol 2010; 19: 307–314. [DOI] [PubMed] [Google Scholar]
- Tremblay KD, Duran KL, Bartolomei MSA. 5′2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol 1997; 17: 4322–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TL, Trasler JM, Moss SB, Yang GJ, Bartolomei MS. Acquisition of the H19 methylation imprint occurs differentially on the parental alleles during spermatogenesis. Genomics 1999; 58: 18–28. [DOI] [PubMed] [Google Scholar]
- Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MR. Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Human Mol Genet 2010; 19: 36–51. [DOI] [PubMed] [Google Scholar]
- Ideraabdullah F, Abramowitz LK, Thorvaldsen JL, Krapp C, Wen SC, Engel N, Bartolomei MS. Novel cis-regulatory function in ICR-mediated imprinted repression of H19. Dev Bio 2011; 355: 349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow GM, Foetal Quinn P. and placental growth in the mouse after pre-implantation development in vitro under oxygen concentrations of 5% and 20%. Aust J Biol Sci 1978; 32: 363–369. [DOI] [PubMed] [Google Scholar]
- Pabon JE, Findley WE, Gibbons WE. The toxic effect of short exposures to the atmospheric oxygen concentration on early mouse embryonic development. Fertil Steril 1989; 51: 896–900. [DOI] [PubMed] [Google Scholar]
- McKiernan SH, Bavister BD. Environmental variables influencing in vitro development of hamster 2-cell embryos to the blastocyst stage. Biol Reprod 1990; 43: 404–413. [DOI] [PubMed] [Google Scholar]
- Umaoka Y, Noda Y, Narimoto K, Mori T. Effects of oxygen toxicity on early development of mouse embryos. Mol Reprod Dev 1992; 31: 28–33. [DOI] [PubMed] [Google Scholar]
- Yuan YQ, Van Soom A, Coopman FO, Mintiens K, Boerjan ML, Van Zeveren A, de Kruif A, Peelman LJ. Influence of oxygen tension on apoptosis and hatching in bovine embryos cultured in vitro. Theriogenology 2003; 59: 1585–1596. [DOI] [PubMed] [Google Scholar]
- Rinaudo PF, Giritharan G, Talbi S, Dobson AT, Schultz RM. Effects of oxygen tension on gene expression in preimplantation mouse embryos. Fertil Steril 2006; 86: 1252–1265. [DOI] [PubMed] [Google Scholar]
- Karagenc L, Sertkaya Z, Ciray N, Ulug U, Bahceci M. Impact of oxygen concentration on embryonic development of mouse zygotes. Reprod Biomed Online 2004; 9: 409–417. [DOI] [PubMed] [Google Scholar]
- Mao J, Zhang X, Sieli PT, Falduto MT, Torres KE, Rosenfeld CS. Contrasting effects of different maternal diets on sexual dimorphic gene expression in the murine placenta. Proc Natl Sci U S A 2010; 107: 5557–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol 2002; 241: 171–182. [DOI] [PubMed] [Google Scholar]
- Takahashi M. Oxidative stress and redox regulation on in vitro development of mammalian embryos. J Reprod Dev 2012; 58: 1–9. [DOI] [PubMed] [Google Scholar]
- Fisher B, Bavister BD. Oxygen tension in the oviduct and uterus of rhesus monkeys, hamsters and rabbits. J Reprod Fertil 1993; 99: 673–679. [DOI] [PubMed] [Google Scholar]
- Fujii M, Matsuoka R, Bergel E, van der Poel S, Okai T. Perinatal risk in singleton pregnancies after in vitro fertilization. Fertil Steril 2010; 94: 2113–2117. [DOI] [PubMed] [Google Scholar]
- Lefebvre L. The placental imprintome and imprinted gene function in the trophoblast glycogen cell lineage. Reprod Biomed Online 2012; 25: 44–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kito S, Noguchi Y, Ohta Y. Developmental responses to two substrains of in vitro fertilized C57BL/6J mouse embryos to oxygen and amino acids. Exp Anim 2003; 52: 63–66. [DOI] [PubMed] [Google Scholar]
- Hiura H, Okae H, Miyauchi N, Sato F, Sato A, Van De Pette M, John RM, Kagami M, Nakai K, Soejima H, Ogata T, Arima T. Characterization of DNA methylation errors in patients with imprinting disorders conceived by assisted reproductive technologies. Hum Reprod 2012; 8: 2541–2548. [DOI] [PubMed] [Google Scholar]
- Turan N, Katari S, Gerson LF, Chalian R, Foster MW, Gaughan JP, Coutifaris C, Sapienza C. Inter- and intra-individual variation in allele-specific DNA methylation and gene expression in children conceived using assisted reproductive technology. PLoS Genet 2010; 6: e1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver VF, Miles HL, Cutfield WS, Hofman PL, Ludgate JL, Morison IM. Defects in imprinting and genome-wide DNA methylation are not common in the in vitro fertilization population. Fertil Steril 2012; 97: 147–153. [DOI] [PubMed] [Google Scholar]
- Zheng HY, Tang Y, Niu J, Li P. De-Sheng Ye, Chen X, Shi XY, Li L, Chen SL. Aberrant DNA methylation of imprinted loci in human spontaneous abortions after assisted reproduction techniques and natural conception. Hum Reprod 2013; 28: 265–273. [DOI] [PubMed] [Google Scholar]
- Radford EJ, Isganaitis E, Jimenez-Chillaron J, Schroeder J, Molla MAS, Didier N, Charalambous M, Mcewen KMG, Sassoon D, Patti ME, Ferguson-Smith AC. An unbiased assessment of the role of imprinted genes in an intergenerational model of developmental programming. Plos Genet 2012; 8: e1002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.