ABSTRACT
In mammalian testes, “A-single” spermatogonia function as stem cells that sustain sperm production for fertilizing eggs. Yet, it is not understood how cellular niches regulate the developmental fate of A-single spermatogonia. Here, immunolabeling studies in rat testes define a novel population of ERBB3+ germ cells as approximately 5% of total SNAP91+ A-single spermatogonia along a spermatogenic wave. As a function of time, ERBB3+ A-single spermatogonia are detected during a 1- to 2-day period each 12.9-day sperm cycle, representing 35%–40% of SNAP91+ A-single spermatogonia in stages VIII–IX of the seminiferous epithelium. Local concentrations of ERBB3+ A-single spermatogonia are maintained under the mean density measured for neighboring SNAP91+ A-single spermatogonia, potentially indicative of niche saturation. ERBB3+ spermatogonia also synchronize their cell cycles with epithelium stages VIII–IX, where they form physical associations with preleptotene spermatocytes transiting the blood-testis barrier and Sertoli cells undergoing sperm release. Thus, A-single spermatogonia heterogeneity within this short-lived and reoccurring microenvironment invokes novel theories on how cellular niches integrate with testicular physiology to orchestrate sperm development in mammals.
Keywords: A-single spermatogonia, epithelial cycle, ERBB3, germline stem cell, HER3, seminiferous, SNAP91, spermatogenesis, spermatogonia, spermatogonial stem cells, stem cell niche, stem cells, testis
Subtypes of rat A-single spermatogonia that inhabit seminiferous epithelium at stages VIII–IX unveil novel theories on how germline stem cell niches work in mammals.
INTRODUCTION
It has been known for more than a century that spermatozoa develop from germline stem cells in mammalian testis through the process of spermatogenesis (for review, see [1]). Still, it remains a mystery how complex microenvironments within mammalian testes instruct stem cells to initiate spermatogenesis. Mammalian germline stem cells unique to testes are generally termed spermatogonial stem cells [2–4]. Spermatogonial stem cells maintain spermatogenesis within tubular structures comprising the seminiferous epithelium, which undergoes a precisely timed cycle of events during spermatozoan maturation [5]. As a result, stem cell-derived spermatozoa are systematically released from testes each seminiferous epithelial cycle by a process known as spermiation [5]. Cycle time is set within a species due to germline-intrinsic factors [6] and ranges from 8.6 days in mice to 12.9 days in rats, 16 days in humans, and 17 days in hamsters [7].
A widely accepted theory in mammals is that type “A-single” spermatogonia harbor the spermatogonial stem cell population [8–10]. Hence, A-single spermatogonia have long been postulated to proliferate as stem cells and to generate early spermatozoan progenitors termed “A-paired” spermatogonia [8–10]. Type A-paired spermatogonia are derived from dividing A-single spermatogonia that do not complete cytokinesis. This yields two germline cells connected to each other by a cytoplasmic bridge. Amplifying divisions beyond the A-paired stage also do not complete cytokinesis and continue to generate longer syncytial chains containing 4, 8, and 16 cells, termed “A-aligned” spermatogonia, which are progressing through early steps in spermatogenesis [8–10]. Thus, cytoplasmic bridges of A-aligned spermatogonia are considered a distinguishing trait of early spermatozoan progenitors.
Developmentally, most A-paired and A-aligned spermatogonia appear distinct from A-single spermatogonia, because A-paired and A-aligned spermatogonia are more susceptible to being transformed into type A1 spermatogonia during stages VII–VIII of the seminiferous epithelial cycle [8–11]. Type A1 spermatogonia then give rise to a series of differentiating spermatogonial types (i.e., types A1 → A2 → A3 → A4 → Int → B → meiosis) that divide at specific stages of the epithelial cycle before initiating meiosis as spermatocytes [12]. Spermatocytes complete meiosis to generate male haploid gametes, which morph into flagellated spermatozoa that fertilize oocytes for sexual reproduction [5].
This developmental hierarchy, which positions A-single spermatogonia as mammalian germline stem cells, is genetically confirmed in flies where “isolated” germ cells in the single-cell state function as stem cells that generate early differentiating sperm progenitors (gonialblasts) or egg progenitors (cystoblasts) that develop into syncytial chains as they progress through meiosis and form gametes [13]. However, the dogma that mammalian A-single spermatogonia purely represent stem cells has recently been challenged, because pulse-labeling experiments in genetically engineered mice predict spermatogonial stem cells to reside within a novel subpopulation of NEUROG3−, GFRα1+, NANOS2+ A-single spermatogonia [11, 14–16]. This has provided insight regarding the concept of “actual” and “potential” spermatogonial stem cell populations in mammals, and it predicts that a fraction consisting of approximately 60%–96% Ngn3-EGFP-negative, NANOS2+, GFRα1+ A-single spermatogonia is more representative of an “actual” stem cell population [11, 14–16]. Moreover, it was concluded that A-single spermatogonia may not be the sole source of male germline stem cells in adult mammals [11]. This was based on NEUROG3+ A-aligned spermatogonia that were filmed fragmenting to generate shorter syncytia, which included production of new A-single spermatogonia [11]. Time-lapse imaging in Drosophila testes has further shown fragmentation of spermatogonial syncytia containing 4–16 cells in the vicinity of germline stem cell niches after experimentally inducing severe germ cell loss [17]. Interestingly, syncytial fragmentation under these conditions yielded predominantly paired spermatogonia that reoccupied vacant germline stem cell niches [17].
Still, defining the associated cellular components that comprise a germline stem cell niche within mammalian gonads continues to evade scientists [4, 18]. This inability to pinpoint how spermatogonial stem cell fate is regulated at an anatomical level in mammals prohibits genetic analyses to more precisely elucidate how spermatogenesis is maintained and initiated in vivo. Given the cyclical nature of the seminiferous epithelium [5], extrinsic factors critical for maintenance of stem spermatogonia [19], and dependence of spermatogonial stem cell numbers on Sertoli cell numbers [20], it is reasonable to hypothesize that highly structured niches do regulate sperm stem cell fate in mammals. Moreover, in mammals, genetic or chemical depletion of endogenous germline stem cells is required for donor spermatogonia to effectively colonize recipient testes and maintain spermatogenesis [4]. This concept is clearly supported by discoveries in Drosophila where early differentiating progenitors “re-fill” vacant niches and become germline stem cells lacking syncytia [21, 22]. Thus, based on modeling in both invertebrates and vertebrates, germline stem cell niches in mammals would theoretically function to regulate the fate of A-single spermatogonia.
Here, we identify a factor related to the neuregulin receptor, ERBB3, that is transiently detected during a 1- to 2-day period each 12.9-day rat spermatogenic cycle in a rare subset of SNAP91+, ZBTB16+, SALL4+ A-single spermatogonia. Along a rat spermatogenic wave, the ERBB3+ and ERBB3− A-single spermatogonia colocalize specifically to epithelial segments of stage VIII–IX seminiferous tubules undergoing sperm release. Therein, ERBB3+ spermatogonia form direct contacts with Sertoli cells and transitioning preleptotene spermatocytes, thus mapping this novel spermatogonial type to definable microanatomy at the basement membrane of the rat seminiferous epithelium. Accordingly, selective induction of early spermatozoan progenitors from one A-single spermatogonial pool within this ephemeral environment presents a model where remaining A-single spermatogonia act as stem cells to support subsequent rounds of spermatogenesis.
MATERIALS AND METHODS
Animal Protocols
Protocols for use of wild-type (Harlan Co.) and tgGCS-EGFP [23] Sprague-Dawley rats in the present study were approved by the Institutional Animal Care and Use Committee at the University of Texas Southwestern (UTSW) Medical Center in Dallas, as certified by the Association for Assessment and Accreditation of Laboratory Animal Care International.
Analysis of A-Single Spermatogonial Subtypes
Immunofluorescence-based data on numbers of spermatogonia were collected in testis sections and seminiferous tubule whole mounts (0.5- to 2.5-cm pieces) after labeling with antibodies to spermatogonial markers, as detailed below under Immunofluorescence Labeling in Testis Sections and Immunofluorescence Labeling in Seminiferous Tubule Whole Mounts. Seminiferous tubule pieces used for analysis of segments containing ERBB3+ spermatogonia were imaged in whole mounts using a Nikon SMZ1500 fluorescence stereomicroscope equipped with ACT-1 imaging software (Nikon Instruments, Inc.). The length of each tubule piece was measured using the NeuronJ plug-in for ImageJ software (version 1.47; http://rsb.info.nih.gov/ij/) [24], which computationally traced the contour of each tubule piece to provide pixel length for normalization to total field width in pixels/mm [25]. Measurements of tubule lengths recorded using ImageJ were verified by printing tubule images on paper and tracing them individually using a Scale Master Pro-XE advanced digital plan measure with PC interface 6335 (Calculated Industries) calibrated to an internally placed scale bar.
Spermatogonial counts in whole mounts were normalized per average spermatogenic wave length in adult Sprague-Dawley rats (2.6 cm/wave) [26]. Spermatogonial counts in adolescent (age, 30–31 days) rats were also normalized to 2.6 cm/unit length; however, it should be noted that spermatogenic waves have yet to develop to adult length in adolescent rats [27]. Images from fluorescence labeling were acquired using an IX70 fluorescence microscope (Olympus, Inc.) equipped with Simple-PCI software (Compix, Inc., Imaging Systems). Areas of imaged ERBB3+ spermatogonia were also analyzed in ImageJ by enhancing contrast within its linear range and use of the “wand tool” to select and measure the total pixels per selected image composing each cell. Real area was obtained by the product of cell area in pixels and total real area covered by each pixel magnified 200-fold.
Frequency of ERBB3+ A-Single Spermatogonia/Epithelial Cycle
To estimate relative numbers of ERBB3+ A-single spermatogonia within the total population of SNAP91+ A-single spermatogonia during stages VIII–IX of spermatogenesis, the time of ERBB3+ A-single spermatogonia detection/cycle of the rat seminiferous epithelium was estimated to be between 1 and 2 days. This was based on 1) specific localization to stage VIII–IX tubular segments and 2) duration of stage VIII plus stage IX of rat spermatogenesis (i.e., 30 h + 7 h) as an approximate period of ERBB3 detection in A-single spermatogonia [5, 7]. Given this, 37-h period of ERBB3 detection/310-h cycle = 11.9% total cycle time × ∼340 SNAP91+ A-single cells scored/wave in 120-day-old rats = ∼40 SNAP91+ A-single cells/segment containing ERBB3+ spermatogonia; then, ∼15 ERBB3+ A-single spermatogonia/wave/42 SNAP91+ A-single cells/segment = 35.7% of the A-single spermatogonia population.
The frequency of ERBB3+ A-single spermatogonia within the total SNAP91+ A-single population during stages VIII–IX was also estimated by capturing images along segments of seminiferous tubule whole mounts where greater than 90% of contiguous microscopic fields (0.15 mm2) contained at least one ERBB3+ A-single spermatogonium. This was considered a more accurate measurement of local A-signal spermatogonia concentrations during stages VIII–IX, because several tubule pieces contained proximal regions during stage VII–VIII transitions that harbored one or more small clusters of ERBB3+ A-single spermatogonia separated by two to four microscopic field-lengths (∼0.9–1.8 mm) from regions ahead of where most contiguous fields contained ERBB3+ A-single spermatogonia. Images were acquired using an LCPlanFl 20×/0.40 Ph1 Japan (Cap-Pl I C.5) objective (Olympus) on the IX70 fluorescence microscope system described above. This was done by scanning along lengths of seminiferous tubule fragments using the 20× objective and capturing overlying fluorescence images, first in red (ERBB3+ cells) and then in green (SNAP91+ cells).
Staging ERBB3+ Spermatogonia
Spermatogenic stages were scored in paraformaldehyde-fixed, frozen histological sections (section thickness, 8 μm) prepared from testes of wild-type rats after staining by the periodic acid-fuchsin sulfurous acid technique [28] but modified for frozen sections (John M. Shelton and James A. Richardson, UTSW Molecular Pathology Core Laboratory) [29]. Spermatid development was used as the primary criterion to identify cellular associations defining each of the 14 spermatogenic stages in Sprague-Dawley rats [5, 7, 30]. Adjacent, unstained sections were colabeled with antibodies to ERBB3 and ZBTB16 (also called PLZF), together with Hoechst 33342 dye, as described below under Immunofluorescence Labeling in Testis Sections.
Immunofluorescence Labeling in Testis Sections
Testes were isolated from wild-type (Harlan Co.) and tgGCS-EGFP [23] Sprague-Dawley rats and fixed for approximately 18 h at 4°C in 0.1 M sodium phosphate buffer (pH 7.2) containing 4% paraformaldehyde. Fixed testes were equilibrated through a 10%, 18%, and 25% sucrose (w/v, dissolved in 1× PBS [catalog no. 14040-182; Invitrogen, Inc.]) gradient by sequential overnight incubations (∼24 h) at 4°C in 15 ml of each respective sucrose solution. Once equilibrated to 25% sucrose, testes were embedded in tissue freezing medium (catalog no. 72592; Electron Microscopy Sciences, Inc.) and frozen using a cryobath (catalog no. 45972; Shandon Lipshaw). Frozen testes were used to prepare a parallel series of cryosections (section thickness, 8 μm). Frozen sections were stored at −40°C until use in antibody-labeling assays or stained by the periodic acid-fuchsin sulfurous acid technique described above. Before antibody labeling, sections were equilibrated in air to approximately 22–24°C for 15 min, hydrated in PBS (catalog no. D8537; Sigma) at 22–24°C for 10 min, heat-treated at 80°C for 8 min in 10 mM sodium citrate (pH 6.0), and then incubated for 1 h at 22–24°C in blocking buffer (Roche Blocking Reagent [1% v/v] diluted in 0.1 M sodium phosphate buffer [pH 7.2] containing Triton X-100 [0.1% v/v]). Sections were then treated for 18–24 h at 22–24°C with respective antibodies diluted in blocking buffer at the following concentrations: anti-ERBB3, 0.25 μg/ml (catalog no. sc-285; Santa Cruz Inc.); anti-SNAP91, 1:1500 dilution from manufacture (catalog no. GTX11329, clone ID AP180-1; GeneTex); anti-CD9, 2 μg/ml (catalog no. 551808, clone RPM.7; BD Biosciences); anti-PLZF, 0.2 μg/ml (catalog no. OP128, clone 2A9; Calbiochem); anti-SALL4, 0.2 μg/ml (catalog no. H0005716-M03, clone 6E3; Abnova), anti-pS6 protein Ser235/236 epitope, 0.4 μg/ml (catalog no. 2011 or catalog no. 4857, clone 91b2; Cell Signaling Technology); anti-pS6 protein Ser240/244 epitope, 0.4 μg/ml (catalog no. XP5364, clone D68F8, or catalog no. XP4278, clone 61H9; Cell Signaling Technology). After treatment with primary antibodies, sections were washed three times for 10 min/wash in 50 ml of PBS and then incubated for 40 min at 22–24°C with respective Alexa Fluor 594 (Invitrogen, Inc.) or Alexa Fluor 488 (Invitrogen, Inc.) secondary antibodies diluted to 4 μg/ml in PBS containing 5 μg/ml of Hoechst 33342 dye (catalog no. H3570; Molecular Probes). After treatment with secondary antibodies, sections were washed three times for 10 min/wash in 50 ml of PBS. After the third wash in PBS, sections were cover-slipped for viewing using Fluorogel mounting medium (catalog no. 17985-10; Electron Microscopy Sciences). Images were acquired using the IX70 fluorescence microscope as described above.
5-Ethynyl-2′-Deoxyuridine Incorporation
Wild-type Sprague-Dawley rats (n = 3/group) were administered 5 mg/kg of 5-ethynyl-2′-deoxyuridine (EDU) dissolved in PBS by one or two i.p. injections (120-day-old rats: one injection, 0 h; 31-day-old rats: two injections, 0 and 6.5 h). Testes were harvested from 31-day-old rats 24 h following their initial injection with EDU and from 120-day-old rats 44 h following their single injection with EDU. Seminiferous tubules were processed according to the manufacturer's instructions for labeling cells with EDU (Click-IT EdU Alexa Fluor 488 Imaging Kit, catalog no. C10337; Invitrogen), followed by immunofluorescence with antibodies to testis cell markers, as described below under Immunofluorescence Labeling in Seminiferous Tubule Whole Mounts.
Immunofluorescence Labeling in Seminiferous Tubule Whole Mounts
Seminiferous tubules were dissected from rat testes in 10-cm dishes containing Dulbecco modified Eagle medium:Ham F12 medium (1:1; DHF12 medium, catalog no. D8437; Sigma) at approximately 22–24°C. Individual tubules were teased apart from testes by blunt dissection, rinsed in fresh DHF12 medium, and fixed for 1 h at 4°C in 20 ml of 0.1 M sodium phosphate buffer (pH 7.2) containing 4% paraformaldehyde. Optimal colabeling with antibodies to SNAP91 and ERBB3 was obtained by suspending the paraformaldehyde-fixed tubules in ice-cold methanol and then immediately incubating them for additional 20 min at −20°C (applied to obtain spermatogonial frequency data from rats aged 104–126 days). Following fixation, tubules were washed three times in 20 ml of PBS for approximately 10 min/wash at 22–24°C and then stored at 4°C for up to 2 wk before use. For indirect labeling with antibodies, tubule pieces (length, 0.5–2.5 cm) were aliquoted into microfuge tubes containing 800 μl of 10 mM sodium citrate (pH 6.0) and incubated at 80°C for 6–8 min. After heat treatment, tubules were immediately washed by dipping in PBS at 22–24°C and then incubated for 1 h at 22–24°C in blocking buffer before treating with respective primary and secondary antibodies as described above in Immunofluorescence Labeling in Testis Sections. Colabeling with antibodies raised in the same species (i.e., cytoplasmic SNAP91 or CD9 with nuclear PLZF or SALL4 mouse immunoglobulin Gs) required sequential overnight incubations. Labeled tubules were sealed between microscope slides (25 × 75 × 1 mm; catalog no. 48311-703; Super Frost Plus White Micro Slides; VWR International) using Fluorogel mounting medium, and images were acquired as described above.
Transcript Profiles in Spermatogenic Cell Cultures
Snap91 and Sall4 profiles were extracted from National Center for Biotechnology Information Gene Expression Omnibus (GEO) DataSets, series GSE830, which profiled changes in transcript abundance in flow-sorted GCS-EGFP rat spermatogonia that maintained or lost stem cell function over time in culture [31]. The MSC-1 Sertoli cell line [32] was a gift from Michael D. Griswold (Washingtion State University, Pullman, WA); the SNL fibroblast line was a gift from Allan Bradley (Welcome Trust Sanger Institute, Hixton CB10, United Kingdom).
RESULTS
Identification of ERBB3+ A-Single Spermatogonia in Rats
Because neuregulin-1 stimulated spermatogonial development in culture [33], immunolabeling experiments were initiated to study neuregulin receptors in testis sections prepared from adult transgenic rats that express a germ cell-specific reporter (tgGCS-EGFP) [23]. Labeling for the neuregulin receptor, ERBB3, localized to plasma membrane and cytoplasm of individual EGFP+ spermatogenic cells at the basal lamina of seminiferous tubules (Fig. 1A, arrow). ERBB3-labeling also localized to a distinct population of interstitial somatic cells (negative for tgGCS-EGFP) (Fig. 1, A and B, asterisks). In wild-type rat testis sections, the ERBB3+ spermatogenic cells exhibited labeling for the nuclear transcription factor, PLZF (ZBTB16) (Fig. 1C). PLZF is a molecular marker for type A spermatogonia in testes [34, 35]. In contrast, interstitially localized ERBB3+ somatic cells did not exhibit PLZF labeling (Fig. 1D). Due to cytoplasmic extensions, the ERBB3+ spermatogonia were observed between 30 and 60 μm in length (Fig. 1, A and C).
FIG. 1.
Identification of ERBB3+ A-single spermatogonia in rats. A) Testis sections from an approximately 120-day-old tgGCS-EGFP transgenic rat exhibiting EGFP in spermatogenic cells (green). Note the immunolabeling for ERBB3 (red) overlaying an EGFP+ spermatogonium (arrow), interstitially localized EGFP− somatic cells (asterisk), and Hoechst 33342-labeled nuclei (blue). Bar = 25 μm. B) Additional examples of ERBB3 labeling (red) in interstitially localized somatic cells (asterisk) of the same rat testis shown in A. Bar = 25 μm. C) ERBB3+ spermatogonium (arrow) in a seminiferous tubule cross section from a 9-mo-old wild-type rat. Left) ERBB3 labeling (red cytoplasmic) overlaying PLZF labeling (green nuclear). Right) Same section shown at left overlaying Hoechst 33342-labeled nuclei (blue). Bar = 20 μm. D) PLZF+ spermatogonium (green nuclear; arrow) and ERBB3+ interstitial somatic cells (red cytoplasm; asterisk) in a seminiferous tubule cross section from an adult rat. Bar = 40 μm. E) PLZF+, ERBB3+ A-single spermatogonium (arrow) and PLZF+, ERBB3− A-single-like spermatogonium (*) in seminiferous tubule whole mount from an adult rat. Note the PLZF+, ERBB3− chains of type A-aligned spermatogonia. Bar = 20 μm. F) Rare ERBB3+ “A-paired-like” spermatogonia at the periphery of a rat seminiferous tubule displaying PLZF+ nuclei. Arrow points to structure resembling a cytoplasmic bridge. Bar = 20 μm.
When analyzed in whole mounts of seminiferous tubules (Supplemental Fig. S1A; all Supplemental Data are available online at www.biolreprod.org), the ERBB3+, PLZF+ type A spermatogonia were highly restricted to individual cells lacking syncytial formation, indicating they represented A-single spermatogonia (Fig. 1E and Supplemental Fig. S1B). However, a smaller fraction of ERBB3+, PLZF+ spermatogonia (<5%) were syncytial pairs representing A-paired spermatogonia (Fig. 1F and Supplemental Fig. S1C). ERBB3+ A-single spermatogonia also exhibited labeling for the transcription factor, SALL4 (Fig. 2A), which like testicular PLZF is selectively detected in type A spermatogonia [2]. ERBB3+ A-single spermatogonia were clearly distinct from longer chains of PLZF+ (Fig. 1E) and SALL4+ (Fig. 2A) aligned spermatogonia. Thus, immunolabeling for ERBB3 was highly localized to A-single spermatogonia in adult rats.
FIG. 2.
ERBB3 and SNAP91 labeling in SALL4+ rat spermatogonia. A) Colabeling for ERBB3 (red) and SALL4 (green) in a seminiferous tubule from a 120-day-old rat. Note the ERBB3-labeling localized to cytoplasm of A-single and paired-like spermatogonia and nuclear localization of SALL4. Bar = 40 μm. B) SALL4 (top) and SNAP91 (bottom) mRNA abundance rapidly declined in type A spermatogonia after culture for 20 days on SNL mouse fibroblasts in serum-containing medium (left). Sp, freshly isolated differentiating spermatogenic cells; USg, freshly isolated undifferentiated type A spermatogonia; MSC+USg, undifferentiated type A spermatogonia after culture for 20 days on the MSC-1 cell line in serum-containing medium (right). Note that tgGCS-EGFP rat germ cells were used to enable fluorescence-activated cell sorting purification from SNL and MSC-1 cell lines before mRNA isolation [31]. Data from National Center for Biotechnology Information GEO DataSets, series GSE830. C) Colabeling for SNAP91 (red) and SALL4 (green) in a seminiferous tubule from a 120-day-old rat. Note the endosome-like vesicles in cytoplasm marked by SNAP91 and nuclear SALL4 in A-single and A-paired spermatogonia. Bar = 20 μm. D) Numbers of PLZF+ spermatogonia per 2.6-cm length of seminiferous tubules from 33-day-old rats colabeled with antibodies to ERBB3 or SNAP91 (mean ± SEM, n = 3 rats).
Molecular Marker Profile for ERBB3+ A-Single Spermatogonia
Initial studies in adult rats (age, 90–175 days) indicated ERBB3+ spermatogonia were quite rare compared to reports on the relative abundance of rat A-single spermatogonia (Supplemental Fig. S1C) [9]. Therefore, additional molecular markers were identified from microarray data available in silico [31] and used to evaluate populations of A-single spermatogonia in the rat. SNAP91 provided an interesting candidate stem cell marker, because transcripts encoding SNAP91 copurify in fractions of testis cells enriched with spermatogonial stem cells [31, 36]. Similar to Sall4, the relative abundance of Snap91 dropped rapidly in spermatogonia cultured under conditions that promoted differentiation (GEO DataSets, series GSE830) (Fig. 2B). However, SNAP91 was detected in cytoplasmic granules of SALL4+ (Fig. 2C) and PLZF+ (Fig. 2D) A-single and A-paired spermatogonia. Detection of SNAP91 clearly waned in aligned spermatogonia beyond the 2-cell step in development (Fig. 2D). Consistent with estimates obtained in adult rats (Supplemental Fig. S1C), the SNAP91+, PLZF+ A-single spermatogonia were substantially more abundant (10-fold) than ERBB3+, PLZF+ A-single spermatogonia in 33-day-old rats (Fig. 2D).
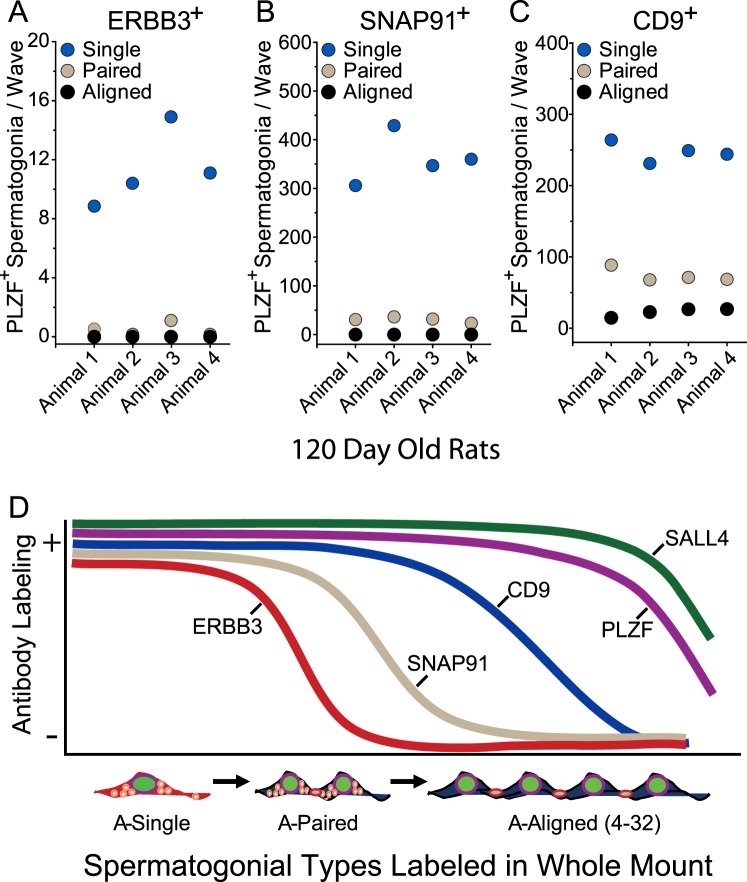
Antibodies to CD9 were previously used to isolate enriched fractions of rat spermatogonial stem cells [37]. Here, CD9 was selectively detected in the cytoplasm and at the plasma membrane of A-single and A-paired spermatogonia of 120-day-old rats, but also in aligned spermatogonia at the 4- to 8-cell stages of development (Supplemental Fig. S2A). In most tubular segments from 120-day-old rats, labeling intensity of CD9 sharply declined within longer chains of greater than four PLZF+ spermatogonia but became detectable again in later steps of differentiating spermatogonia/early spermatocytes, as initially reported in rodents [37]. In testes of 31-day-old rats, a similar antibody labeling profile was observed for CD9 in A-single and A-paired spermatogonia. However, in contrast to adult rats, vivid CD9 labeling was observed in syncytial chains of type A spermatogonia consisting of 4 to more than 32 cells in 31-day-old rats (Supplemental Fig. S2B). Thus, like SNAP91, CD9 provided an additional cytoplasmic marker to study properties of rat A-single spermatogonia.
Analysis of PLZF+ spermatogonia in 120-day-old rats by colabeling with antibodies to ERBB3 (Fig. 3A), SNAP91 (Fig. 3B), and CD9 (Fig. 3C) provided estimates that numbers of ERBB3+ A-single spermatogonia scored per wave were approximately 3.2% of that scored for SNAP91+ A-single spermatogonia and approximately 4.8% of that scored for CD9+ A-single spermatogonia (Supplemental Table S1). Likewise, numbers of ERBB3+ A-paired spermatogonia scored per wave were approximately 1.1% of that scored for SNAP91+ A-paired spermatogonia and approximately 0.6% of that scored for CD9+ A-paired spermatogonia. ERBB3+ A-paired spermatogonia represented 3.4% of total ERBB3+ clonal units per wave, and ERBB3 was not detected in syncytia containing more than two cells. Thus, the antibody to ERBB3 selectively marked rare A-single spermatogonia in rats based on labeling profiles generated with spermatogonial marker proteins (Fig. 3D).
FIG. 3.
ERBB3+ A-single spermatogonia are rare. A–C) Relative numbers of PLZF+, ERBB3+ spermatogonia (A); PLZF+, SNAP91+ spermatogonia (B); and PLZF+, CD9+ spermatogonia (C) per wave of spermatogenesis scored in four separate 120-day-old rats (also see statistical summary in Supplemental Table S1). D) Antibody-labeling profiles for marker proteins in A-single, A-paired, and A-aligned spermatogonia observed by fluorescence microscopy in seminiferous tubule whole mounts prepared from 120-day-old rats. +, positive for antibody labeling; −, negative for antibody labeling (arbitrary scale).
Identification of SNAP91+ A-Single Spermatogonial Subtypes in Rat Testes
Next, the relative abundance of ERBB3+ spermatogonia within the SNAP91+ spermatogonial population was directly investigated in 31- and 120-day-old rats. Colabeling seminiferous tubules for SNAP91 and ERBB3 demonstrated distinct populations of SNAP91+, ERBB3+ and SNAP91+, ERBB3− A-single spermatogonia in rats of each age (Fig. 4A, Supplemental Fig. S3, and Supplemental Table S2). All ERBB3+ spermatogonia scored in both 31- and 120-day-old rats were SNAP91+. In 120-day-old rats, ERBB3+ A-single spermatogonia represented approximately 4.5% of SNAP91+ A-single spermatogonia, and ERBB3+ A-paired spermatogonia represented approximately 4.3% of SNAP91+ A-paired spermatogonia (Fig. 4B and Supplemental Table S2). SNAP91+, ERBB3+ spermatogonia were not detected in syncytia containing more than two cells. Thus, colabeling experiments using cytoplasmic markers defined ERBB3+ A-single spermatogonia as a subtype of SNAP91+ A-single spermatogonia in rats.
FIG. 4.

ERBB3+ spermatogonia are a subtype of A-single spermatogonia. A, Left) A-single spermatogonia observed after colabeling for SNAP91 and ERBB3 in whole-mount seminiferous tubules from 120-day-old rats. Bar = 20 μm. A, Right) Higher-magnification image of an ERBB3+, SNAP9+ rat A-single spermatogonium. Note the intricate ERBB3+ lamellipodia and SNAP91+ endosome-like vesicles in cytoplasm. B) Numbers of ERBB3+ or ERBB3−, SNAP91+ A-single (As), A-paired (Apr), and A-aligned (Aal) spermatogonia per 2.6-cm length of seminiferous tubules from 31-day-old (mean ± SEM, n = 3) and 120-day-old (mean ± SEM, n = 4) rats (also see statistical summary in Supplemental Table S2).
ERBB3+ A-Single Spermatogonia Do Not Incorporate EDU Efficiently
To determine the percentage of ERBB3+ spermatogonia in S phase, seminiferous tubule whole mounts were prepared from 120-day-old rats 44 h after a single injection of the thymidine analog, EDU. Interestingly, no ERBB3+ spermatogonia were observed with incorporated tracer after scoring 137 ± 22 ERBB3+ clonal units/rat (mean ±SEM, n = 3 rats) (Fig. 5A). Within the same tubular segments, chains of ERBB3−, EDU+ aligned-like spermatogonia clearly incorporated tracer (Fig. 5A). Similarly, in tubule whole mounts prepared from 31-day-old rats 24 h after administering the first of two EDU injections (6.5 h apart), only 0.66% ± 0.62% of ERBB3+ A-single spermatogonia and no ERBB3+ A-paired spermatogonia were scored EDU+ (498 total clonal units scored, with 493 A-single and 5 A-paired) (Fig. 5B). In seminiferous tubules from the same rats, 14.3% ± 4.0%, 12.2% ± 5.6%, and 5.7% ± 3.8% of respective CD9+ A-single, paired, and aligned spermatogonia were scored EDU+ (580 total clonal units scored, with 160 A-single, 165 A-paired, and 255 A-aligned) (Fig. 5, B and C). Thus, in 310-day-old rats, greater than 20-fold more CD9+ A-single spermatogonia incorporated EDU during a 24-h treatment period than did ERBB3+ A-single spermatogonia (P < 0.009).
FIG. 5.
Percentage of CD9+ and ERBB3+ spermatogonia in S phase. A) ERBB3 labeling in cytoplasm of A-single spermatogonia (red) and EDU labeling in nuclei (green) of various testis cell types in seminiferous tubule of a 120-day-old rat. Labeling was conducted 44 h after a single i.p. injection of EDU (5 mg/kg). Arrow points to a chain of type A-aligned-like spermatogonia. Bar = 40 μm. B) Mean frequencies of EDU uptake by CD9+ and ERBB3+ A-single (As), A-paired (Apr), and A-aligned-like (Aal) spermatogonia along the lengths of seminiferous tubule whole mounts from 31-day-old rats. Labeling was conducted 24 h after the first of two total i.p. injections of EDU (5 mg/kg; injection 1, 0 h; injection 2, 6.5 h). Bars represent the mean ±SEM (n = 3 rats; *P < 0.009, %EDU+, ERBB3+ vs. %EDU+, CD9+ A-single populations, Student t-test). C) Colabeling for EDU (green, nuclei) and CD9 (red, cytoplasm) in A-single (As) and A-aligned-like (Aal) spermatogonia from rats described in B. White arrows, EDU+ spermatogonia; yellow arrows, EDU− spermatogonia. Bar = 20 μm.
Stage-Specific Detection of ERBB3+ A-Single Spermatogonia
The low relative abundance ERBB3+ A-single spermatogonia and their distinct EDU incorporation profile prompted studies to investigate their association with other cell types within rat testes. Interestingly, in adult rats, ERBB3 was highly localized to A-single spermatogonia during stages VIII–IX of a seminiferous epithelial cycle (Fig. 6A). As a result of this stage-specific localization, ERBB3+ spermatogonia were consistently identified in associations containing type A1 spermatogonia, preleptotene spermatocytes, pachytene spermatocytes, step 8–9 round spermatids, step 19 elongated spermatids, and Sertoli cells (Supplemental Fig. S4). Potential stage-specific associations between interstitial somatic cells and ERBB3+ A-single spermatogonia were noted (Supplemental Fig. S4), but these remain to be analyzed with additional lineage markers.
FIG. 6.
ERBB3+ spermatogonia localize to stages VIII–IX of spermatogenesis. A) Distribution of ERBB3+, PLZF+ spermatogonia in stages of the seminiferous epithelial cycle scored within testis cross sections from 90-day-old rats. Bars show total ERBB3+ spermatogonia scored from three sections per rat (n = 3 rats; average counts per rat = 4.9 ± 0.97 ERBB3+ spermatogonia/section [mean ± SEM]). B) Colabeling for ERBB3 (red) and espin (green) in a testis section from a 45-day-old rat. Espin localizes to Sertoli-Sertoli junctions, forming the blood-testis barrier, which separates the adluminal and basal compartments of the epithelium. An intermediate compartment is formed during migration of transitioning preleptotene spermatocytes through the blood-testis barrier. Hoechst 33342-labeled nuclei are shown (blue). Bar = 50 μm. C) Schematic showing ERBB3+ (red) and ERBB3− (green) subtypes of SNAP91+ A-single spermatogonia relative to A-paired and A-aligned spermatogonia in stage VII–XII segments of a rat seminiferous tubule. A working model on how stem cell niches could regulate spermatogenesis is also illustrated, which is based on the stage-specific anatomical arrangement of ERBB3+ and ERBB3− A-single spermatogonial subtypes. In this scenario, heterogeneity would develop within populations of SNAP91+ A-single spermatogonia upon saturation of germline stem cell niches following the peak of stem cell divisions between rat epithelium stages X and III (stages X–III not depicted) [8–10, 52–55, 74]. Theoretically, this would render a distinct population of SNAP91+ A-single spermatogonia residing outside niches more susceptible to entering cell cycles that yield A-aligned spermatogonia [8–10]. Thus, as illustrated, it remains to be determined if the ERBB3+ spermatogonial population represents A-single spermatogonia inside or outside germline stem cell niches.
Colabeling for ERBB3 and espin highlighted adluminal and basal compartments of the seminiferous epithelium (Fig. 6B). Espin is a marker for Sertoli cell adherens junctions at the apical ectoplasmic specialization and blood-testis barrier [38]. This showed ERBB3+ spermatogonia and preleptotene spermatocytes located in the basal compartment, whereas pachytene spermatocytes and spermatids localized to the adluminal compartment (Fig. 6B). Preleptotene spermatocytes were frequently observed positioned adluminally to ERBB3+ spermatogonia within gaps of espin labeling in intermediate-like compartments (Fig. 6B). This highly restricted localization of ERBB3 on A-single spermatogonia during spermatogenic stages VIII–IX explains their low relative abundance compared to the total SNAP91+ A-single pool, and it unveils potential mechanisms for how A-single spermatogonial fate could be regulated during an epithelial cycle (Fig. 6C).
Frequency of ERBB3+ A-Single Spermatogonia During an Epithelial Cycle
Understanding the relative abundance of A-single spermatogonial subtypes in stage VIII–IX tubules could provide insight regarding their roles during spermatogenesis. Based on a 37-h period for stages VIII plus IX of spermatogenesis [5, 7] and mean numbers of A-single spermatogonia subtypes per wave (Figs. 3 and 4), ERBB3+ spermatogonia represented approximately 36% of SNAP91+ A-single spermatogonia scored within stage VIII–IX tubules of 120-day-old rats (see Materials and Methods for calculations).
In a second approach, local concentrations of ERBB3+ spermatogonia within the SNAP91+ population were scored directly within 0.15-mm2 microscopic fields of seminiferous tubules from 104- to 126-day-old rats (Fig. 7A). This area represented a field approximately 600-fold the mean area of an ERBB3+ A-single spermatogonium (247 ± 77 μm2 [mean ±SD, n = 64]). Images of 214 distinct fields were captured containing 3.4 ± 2.0 and 5.8 ± 2.8 ERBB3+ and ERBB3− A-single spermatogonia per field, respectively. In total, 9.2 ± 3.2 SNAP91+ A-single spermatogonia were scored per field. This approach estimated a mean of 38.5% ± 3.9% ERBB3+ A-single spermatogonia within their local SNAP91+ A-single populations (Supplemental Table S3).
FIG. 7.
Percentage of ERBB3+ A-single spermatogonia during stages VIII–IX. A) Representative 0.15-mm2 microscopic field captured to score local concentrations of SNAP91+ A-single spermatogonial subtypes in rat seminiferous tubule whole mounts. Shown is an ERBB3+ A-single spermatogonium (red) among ERBB3− (green), SNAP91+ A-single spermatogonia in a 126-day-old rat. Bar = 40 μm. B) Relative numbers of microscopic fields containing different percentages of ERBB3+, SNAP91+ A-single spermatogonia (n = 214 fields) captured from seminiferous tubule whole mounts prepared from 104- to 126-day-old rats (see Supplemental Table S3). C) Distribution plot illustrating relative numbers of A-single and A-paired spermatogonia imaged in each microscopic field (same 214 fields used in B). D) Plot of the same data shown in B and C but for numbers of each A-single spermatogonia population per field plotted independently after sorting their individual distributions in ascending order of numbers per microscopic field.
Accordingly, the relative abundance of ERBB3+ A-single spermatogonia in a majority of fields ranged between 20% and 60% of total SNAP91+ A-single spermatogonia per field (Fig. 7B). In their most populated regions, the relative abundance of ERBB3+ A-single spermatogonia did not exceed the mean number of SNAP91+ spermatogonia per field (Fig. 7C), and their numbers were maintained under this threshold at approximately half the mean density of ERBB3− A-single spermatogonia (Fig. 7D).
Modeling Cyclical Regulation of A-Single Spermatogonia Fate
To build on a hypothesis for how developmental fates of A-single spermatogonia can be regulated in mammals, phospho-epitope antibodies to the ribosomal S6 protein were used to map stage-specific changes in cell signaling that overlap colocalizing populations of ERBB3+ and ERBB3− A-single spermatogonia. Phosphorylation of S6 protein on serine residues 235 (pSer235) (Supplemental Fig. S5A) and 240 (pSer240) (Supplemental Fig. S5B) was detected in Sertoli cells specifically during stages VIII–XI of spermatogenesis. Robust expression of pSer235 in Sertoli cells was more tightly focused to late stage VIII and stage IX segments (Supplemental Fig. S5A). Sertoli cells expressing pSer235 formed direct contacts with ERBB3+ spermatogonia and preleptotene spermatocytes during late stage VIII and early stage IX of spermatogenesis (Fig. 8). Thus, waves of S6 protein phosphorylation in Sertoli cells clearly advanced through tubular segments containing ERBB3+ A-single spermatogonia but were initiated at times shortly after heterogeneity developed in the A-single pool (Fig. 9A).
FIG. 8.
Testis cells colocalized with ERBB3+ spermatogonia. Top, Left) Section through rat seminiferous tubules illustrating the phosphorylation profile of S6 protein on serine residue 235 (red) in Sertoli cells during stages VII, VIII, VIII–IX, and X of spermatogenesis. Note that pS6 protein in Sertoli cells is highly restricted to the stage VIII–IX tubule undergoing sperm release. Nuclei of all testis cells are labeled with Hoechst 33342 dye (blue). Bar = 200 μm. Top, Right) Higher-magnification image shows the same stage VIII–IX tubular segment illustrated at the top left. Note the ERBB3+ spermatogonium (Sgn; arrow) located on the basement membrane. Bar = 50 μm. Bottom) Higher-magnification image of the same tubular segment at the top shows distinct testis cell types associated with an ERBB3+ spermatogonium (Sgn; arrow). A1, A1 spermatogonia; T, transitioning preleptotene spermatocyte; P, pachytene spermatocyte; RS, step 8–9 spermatid; S19 ES, Step 19 elongated spermatids; Pt, peritubular myoid/endothelial cells; L, Leydig cell; SC, Sertoli cell. Bar = 25 μm.
FIG. 9.
Waves of pS6 protein traverse late stage VIII seminiferous tubules. A, Top) ERBB3+ spermatogonium (Sgn) at stage VIII of spermatogenesis before sperm release. Note the pS6 protein (pSer235) at the apical tips of step 19 spermatids. Bar = 50 μm. A, Center) ERBB3+ spermatogonium during stage VIII to stage IX transition just before sperm release. Note the adjacent wave of pS6 protein in Sertoli cells of same segment. Bar = 50 μm. A, Bottom) ERBB3+ spermatogonium within pS6 protein-positive tubular segment during stage VIII–IX of spermatogenesis. Bar = 50 μm. B) Model illustrating how colocalized subtypes of A-single spermatogonia could be differentially regulated by transient fate-determining signals each seminiferous epithelial cycle.
DISCUSSION
Discovery of heterogeneity within local populations of A-single spermatogonia during a stage-specific window of the rat seminiferous epithelial cycle would invoke new theories on how cellular niches orchestrate spermatogenesis in mammals. Here, we classify distinct subtypes of SNAP91+ A-single spermatogonia that colocalize in direct proximity to each other during stages VIII–IX of the rat seminiferous epithelial cycle. Heterogeneity within the total pool of SNAP91+ A-single spermatogonia is demonstrated by differential detection of the transmembrane receptor, ERBB3. Localization of ERBB3 to the outer plasmalemma clearly outlines cytoplasmic architecture of A-single spermatogonia and distinguishes this subset of cells within a larger population of SNAP91+, PLZF+, SALL4+ A-single spermatogonia. We also found the SNAP91 protein localized to granular structures resembling endosomes [39] throughout cytoplasm of A-single and A-paired spermatogonia. To our knowledge, restricted localization of SNAP91 to rat A-single and A-paired spermatogonia is most similar to NANOS2+, GFRα1+ spermatogonia in mice [40].
In male mice, NANOS2 is essential for maintenance of spermatogonial stem cells [16] and proper meiotic entry [41, 42]. In contrast, female germ cells develop normally in NANOS2-deficient mice [42]. GFRα1 is a glycophosphatidylinositol-anchored protein that is also essential for mouse spermatogonial stem cell maintenance [43, 44]. NANOS2 expression is dependent on GFRα1, and NANOS2 partially rescues the stem cell loss phenotype in GFRα1-deficient mice [43]. Similar to NANOS2 and SNAP91 in rodents, GFRα1 is abundantly localized to A-single and A-paired spermatogonia [45]. However, GFRα1 has been commonly detected on A-aligned spermatogonia in a variety of species [16, 46–50]. Like SNAP91 and NANOS2, the ERBB3-labeling profile discovered here is highly restricted to single and paired spermatogonia, but more so to A-single spermatogonia, and, uniquely, in a stage-specific manner. Most recently, the transcription factor, ID4, was reported highly localized to PLZF+ A-single spermatogonia [51]. Stringent localization of ID4 to A-single spermatogonia, together with its requirement for spermatogonial stem cell maintenance, identified ID4 as the first fate-determining factor that specifically marks actual stem spermatogonia [51]. As a result, it will be important to determine how ID4, NANOS2, SNAP91, GFRα1, and ERBB3 are functionally linked during spermatogonial proliferation and development.
Currently, it is not known if ERBB3+ and ERBB3− A-single spermatogonia hold distinct biological roles and if they purely reflect transition states intrinsic to the A-single population during stages VIII–IX. Thus, it may well be determined that such heterogeneity in A-single spermatogonia is caused by cell-intrinsic mechanisms that are somehow buffered during other stages of an epithelial cycle. Still, because local concentrations of the ERBB3+ A-single spermatogonia appear to be maintained independent of the total A-single pool (Fig. 7), a reasonable hypothesis is that heterogeneity between neighboring SNAP91+ A-single spermatogonia during stages VIII–IX reflects differences in their developmental fates. If so, this could imply effects of germline stem cell niches during stages VIII–IX of spermatogenesis (Fig. 6C). However, A-single spermatogonia divide up to three times each epithelial cycle in rodents and, therefore, execute fate decisions upon each division to sustain a steady state of spermatogenesis [8–10]. Moreover, A-single spermatogonia undergo their cell divisions predominantly outside stages VIII–IX [8–10]. Thus, germline stem cell niches that exert effects on stem cell fates specifically during stages VIII–IX of an epithelial cycle do not seem intuitive. Yet, such models do not preclude stage-specific effects of the seminiferous epithelium on priming initial rates at which A-single spermatogonia enter cell cycles toward differentiating or self-renewing divisions (Fig. 6C).
Another related possibility is that the profile of ERBB3+ and ERBB3− A-single spermatogonia actually reflects differences in the state of their cell cycles during stages VIII–IX of spermatogenesis. However, based on [3H]thymidine incorporation assays, it is well documented that more than 95% of rat, mouse, and hamster A-single spermatogonia are not in S or M phase during late stage VIII and early stage IX of spermatogenesis [8–10, 52–56]. Comprehensive analyses in hamsters further indicate that most A-single spermatogonia are in G1 phase during stage VIII to early stage IX [54–56]. Therefore, heterogeneity between larger cohorts of A-single spermatogonia during stages VIII–IX could represent differences associated with cells in G1 phase. Interestingly, self-renewal rates of A-single spermatogonia were recently found to be dependent on the retinoblastoma tumor suppressor protein, RB [57, 58]. The RB protein is a well-established regulator of cell-cycle progression and stem cell fate [59]. Thus, the nature of A-single spermatogonia heterogeneity identified here during stages VIII–IX in rats becomes especially striking, because it overlaps a pivotal juncture in the epithelial cycle when most A-single spermatogonia appear mitotically quiescent and immediately proceeds flux of A-single spermatogonia back into more active cell cycles between stages X and III [8, 9, 54–56].
The cyclical profile of A-single spermatogonia heterogeneity identified here in rats also reveals how classically defined, stage-specific cellular associations [5, 7] could selectively emanate signals in each epithelial cycle to differentially regulate fates of stem spermatogonia. As a prerequisite for such a model, it is key that ERBB3+ and ERBB3− A-single spermatogonia persist during a restricted window of time (i.e., ∼37 h per 310-h cycle), which synchronizes their development with specific differentiation steps for distinct generations of spermatogenic cells in stage VIII–IX tubular segments. This includes 1) shedding of mature sperm from the seminiferous epithelium, 2) initiation of spermatid elongation, 3) initiation of meiotic prophase, 4) migration of spermatocytes through the blood-testis barrier, and 5) entry of type A1 spermatogonia into the cell cycle [7]. By analogy to invertebrate germline stem cell niches, it remains to be determined if or how direct associations between ERBB3+ A-single spermatogonia and other testis cell types differ from cell associations formed by ERBB3− A-single spermatogonia during epithelium stages VIII–IX. However, in a nonclassical paradigm, even if direct cellular associations formed by ERBB3+ A-single spermatogonia are not unique within the A-single pool; stage-dependent heterogeneity within the total A-single population potentially subjects these cells to differential regulation by transient, overlapping extrinsic signals (Fig. 9B).
Cellular and molecular anatomy comprising germline stem cell niches that maintain gametogenesis via specifically localized extrinsic signals are well established in Drosophila ovaries and testes [60–63]. In addition, genetic assays in mice have identified testicular factors [19, 64, 65], including Sertoli cells themselves [20, 66], required to maintain stem spermatogonia. Such paracrine effectors are postulated to interact with factors derived from testicular interstitium/vasculature to regulate spermatogonial fate [67–69]. This is because undifferentiated type A spermatogonia preferentially localize to the basement membrane of seminiferous tubules near vasculature niche-like compartments during stages VI–IX of spermatogenesis in rodents [67–69]. Accordingly, spermatogonia appear to migrate away from these vascular regions during stages VIII–IX to become more dispersed throughout the basal compartment of the epithelium as respective syncytia expand mitotically and differentiate [68]. In rats, undifferentiated spermatogonia were also reported to be significantly more proximal to interstitial regions during stages V, VII, and IX [69]. Similar to scenarios discussed above for ERBB3+ and ERBB3− A-single spermatogonia, such associations with vascular regions would occur predominantly when A-single spermatogonia are not actively dividing, but more so during transformation of A-aligned spermatogonia into type A1 spermatogonia [8–10] and reentry of A1 spermatogonia back into their cell cycle [8–10, 70].
Along these lines, a recent study found no associations between changes in testicular interstitium/vasculature volume and enhanced donor spermatogonial stem cell colonization in mice with hypothyroidism [20]. However, a strong correlation was found between increased Sertoli cell numbers per testis and spermatogenic colonies formed per number of donor spermatogonia [20]. These findings could manifest from developmental differences between A-single spermatogonia and A-aligned spermatogonia [8–11], because associations between A-single spermatogonial subtypes and the testicular vasculature have yet to be mapped [71]. It would suggest that even more finely tuned anatomical compartments/signals exist to regulate spermatogonial fate within such vascular associated niches [72] and that fate of A-single spermatogonia is somehow primed within these regions before their most active periods of cell division. Thus, newfound molecular markers for subtypes of A-single and A-paired spermatogonia (i.e., ERBB3, ID4, SNAP91, and NANOS2) provide probes to potentially help advance these concepts.
Under steady-state conditions, it is estimated that approximately 50% of stem spermatogonia leave their niche and differentiate each epithelial cycle [73]. Accordingly, in adult rats, mice, and hamsters, numbers of A-single spermatogonia do not fluctuate much over the length of a spermatogenic wave (2-fold or less) despite dividing approximately two or three times per cycle, compared to clear amplification of total A-aligned/A1 spermatogonia numbers (>50-fold) [9, 10, 74, 75]. Here, we estimate ERBB3+ A-single spermatogonia to represent approximately 35%–40% of their local SNAP91+ A-single population within stage VIII–IX tubular segments. Thus, the relative abundance of ERBB3+ A-single spermatogonia (∼23/mm2) was found to be approximately half that of ERBB3− A-single spermatogonia (∼39/mm2) during this period of spermatogenesis (Fig. 7, C and D). By these counts, if heterogeneity within SNAP91+ A-single spermatogonia is ultimately verified as reflecting differences in their developmental fates, then about one-third to two-thirds of A-single spermatogonia would be destined to become A1 spermatogonia each epithelial cycle. In fact, these ratios are documented in rats, mice, and hamsters where following A-single spermatogonial divisions between stages X and IV, approximately twice as many clonal units of A-paired plus A-aligned spermatogonia were scored compared to A-single spermatogonia [9, 74, 76].
Supplementary Material
ACKNOWLEDGMENT
We thank John Shelton, James Richardson, Gerardo Medrano, Nephy John, and Bray Denard for their help with these experiments. We thank Drs. Diego Castrillon and Michael Buszczak for reading this manuscript and their helpful comments regarding this work.
Footnotes
Supported by The Eunice Kennedy Shriver National Institute of Child Health and Human Development NIH grants R01HD053889 and R01HD061575 and the Cecil H. & Ida Green Center for Reproductive Biology Sciences. Presented in part at the XXIst North American Testis Workshop, March 30–April 02, 2011, Montreal, Canada.
REFERENCES
- Regaud C. Some biological aspects of the radiation therapy of cancer. Am J Roentgen Radium Ther 1924; 12: 97–101. [Google Scholar]
- Phillips BT, Gassei K, Orwig KE. Spermatogonial stem cell regulation and spermatogenesis. Philos Trans R Soc Lond B Biol Sci 2010; 365: 1663–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Brinster RL. The germline stem cell niche unit in mammalian testes. Physiol Rev 2012; 92: 577–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proc Natl Acad Sci U S A 1994; 91: 11298–11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont Y. Kinetics of spermatogenesis in mammals: seminiferous epithelium cycle and spermatogonial renewal. Physiol Rev 1972; 52: 198–236. [DOI] [PubMed] [Google Scholar]
- Franca LR, Ogawa T, Avarbock MR, Brinster RL, Russell LD. Germ cell genotype controls cell cycle during spermatogenesis in the rat. Biol Reprod 1998; 59: 1371–1377. [DOI] [PubMed] [Google Scholar]
- Russell LD, Ettlin RA, Hikim APS, Clegg ED. (eds.) Histological and Histopathological Evaluation of the Testes. Clearwater, FL: Cache River Press; 1990. [Google Scholar]
- Oakberg EF. Spermatogonial stem-cell renewal in the mouse. Anat Rec 1971; 169: 515–531. [DOI] [PubMed] [Google Scholar]
- Huckins C. The spermatogonial stem cell population in adult rats. I. Their morphology, proliferation and maturation. Anat Rec 1971; 169: 533–557. [DOI] [PubMed] [Google Scholar]
- Lok D, Weenk D, De Rooij DG. Morphology, proliferation, and differentiation of undifferentiated spermatogonia in the Chinese hamster and the ram. Anat Rec 1982; 203: 83–99. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Sharma M, Nabeshima Y, Braun RE, Yoshida S. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science 2010; 328: 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monesi V. Autoradiographic study of DNA synthesis and the cell cycle in spermatogonia and spermatocytes of mouse testis using tritiated thymidine. J Cell Biol 1962; 14: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A, Fuller MT, Braun RE, Yoshida S. Germline stem cells. Cold Spring Harb Perspect Biol 2011; 3: a002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Wu X, Kaestner KH, Wang PJ. The pluripotency factor LIN28 marks undifferentiated spermatogonia in mouse. BMC Dev Biol 2009; 9: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Nabeshima Y, Yoshida S. Functional identification of the actual and potential stem cell compartments in mouse spermatogenesis. Dev Cell 2007; 12: 195–206. [DOI] [PubMed] [Google Scholar]
- Sada A, Suzuki A, Suzuki H, Saga Y. The RNA-binding protein NANOS2 is required to maintain murine spermatogonial stem cells. Science 2009; 325: 1394–1398. [DOI] [PubMed] [Google Scholar]
- Sheng XR, Matunis E. Live imaging of the Drosophila spermatogonial stem cell niche reveals novel mechanisms regulating germline stem cell output. Development 2011; 138: 3367–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 2008; 132: 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, Pichel JG, Westphal H, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 2000; 287: 1489–1493. [DOI] [PubMed] [Google Scholar]
- Oatley MJ, Racicot KE, Oatley JM. Sertoli cells dictate spermatogonial stem cell niches in the mouse testis. Biol Reprod 2011; 84: 639–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai T, Spradling A. Differentiating germ cells can revert into functional stem cells in Drosophila melanogaster ovaries. Nature 2004; 428: 564–569. [DOI] [PubMed] [Google Scholar]
- Brawley C, Matunis E. Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science 2004; 304: 1331–1334. [DOI] [PubMed] [Google Scholar]
- Cronkhite JT, Norlander C, Furth JK, Levan G, Garbers DL, Hammer RE. Male and female germline specific expression of an EGFP reporter gene in a unique strain of transgenic rats. Dev Biol 2005; 284: 171–183. [DOI] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A 2004; 58: 167–176. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH. Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry B, Clermont Y, Leblond CP. The wave of the seminiferous epithelium in the rat. Am J Anat 1961; 108: 47–78. [Google Scholar]
- Clermont Y, Perey B. Quantitative study of the cell population of the seminiferous tubules in immature rats. Am J Anat 1957; 100: 241–267. [DOI] [PubMed] [Google Scholar]
- Leblond CP, Clermont Y. Spermiogenesis of rat, mouse, hamster and guinea pig as revealed by the periodic acid-fuchsin sulfurous acid technique. Am J Anat 1952; 90: 167–215. [DOI] [PubMed] [Google Scholar]
- Chapman KM, Powell HM, Chaudhary J, Shelton JM, Richardson JA, Richardson TE, Hamra FK. Linking spermatid RNA binding protein and retrogene diversity to reproductive success. Mol Cell Proteomics 2013; 12: 3221–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblond CP, Clermont Y. Definition of the stages of the cycle of the seminiferous epithelium in the rat. Ann N Y Acad Sci 1952; 55: 548–573. [DOI] [PubMed] [Google Scholar]
- Hamra FK, Schultz N, Chapman KM, Grellhesl DM, Cronkhite JT, Hammer RE, Garbers DL. Defining the spermatogonial stem cell. Dev Biol 2004; 269: 393–410. [DOI] [PubMed] [Google Scholar]
- McGuinness MP, Linder CC, Morales CR, Heckert LL, Pikus J, Griswold MD. Relationship of a mouse Sertoli cell line (MSC-1) to normal Sertoli cells. Biol Reprod 1994; 51: 116–124. [DOI] [PubMed] [Google Scholar]
- Hamra FK, Chapman KM, Nguyen D, Garbers DL. Identification of neuregulin as a factor required for formation of aligned spermatogonia. J Biol Chem 2007; 282: 721–730. [DOI] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet 2004; 36: 653–659. [DOI] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet 2004; 36: 647–652. [DOI] [PubMed] [Google Scholar]
- Hamra FK, Chapman KM, Nguyen DM, Williams-Stephens AA, Hammer RE, Garbers DL. Self renewal, expansion, and transfection of rat spermatogonial stem cells in culture. Proc Natl Acad Sci U S A 2005; 102: 17430–17435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Toyokuni S, Shinohara T. CD9 is a surface marker on mouse and rat male germline stem cells. Biol Reprod 2004; 70: 70–75. [DOI] [PubMed] [Google Scholar]
- Bartles JR, Wierda A, Zheng L. Identification and characterization of espin, an actin-binding protein localized to the F-actin-rich junctional plaques of Sertoli cell ectoplasmic specializations. J Cell Sci 1996; 109 (pt 6): 1229–1239. [DOI] [PubMed] [Google Scholar]
- Kohtz DS, Puszkin S. A neuronal protein (NP185) associated with clathrin-coated vesicles. Characterization of NP185 with monoclonal antibodies. J Biol Chem 1988; 263: 7418–7425. [PubMed] [Google Scholar]
- Suzuki H, Sada A, Yoshida S, Saga Y. The heterogeneity of spermatogonia is revealed by their topology and expression of marker proteins including the germ cell-specific proteins Nanos2 and Nanos3. Dev Biol 2009; 336: 222–231. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Saga Y. Nanos2 suppresses meiosis and promotes male germ cell differentiation. Genes Dev 2008; 22: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. Conserved role of nanos proteins in germ cell development. Science 2003; 301: 1239–1241. [DOI] [PubMed] [Google Scholar]
- Sada A, Hasegawa K, Pin PH, Saga Y. NANOS2 acts downstream of glial cell line-derived neurotrophic factor signaling to suppress differentiation of spermatogonial stem cells. Stem Cells 2012; 30: 280–291. [DOI] [PubMed] [Google Scholar]
- Naughton CK, Jain S, Strickland AM, Gupta A, Milbrandt J. Glial cell-line derived neurotrophic factor-mediated RET signaling regulates spermatogonial stem cell fate. Biol Reprod 2006; 74: 314–321. [DOI] [PubMed] [Google Scholar]
- Hofmann MC, Braydich-Stolle L, Dym M. Isolation of male germ-line stem cells; influence of GDNF. Dev Biol 2005; 279: 114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Ahn HW, Chu T, Bowden W, Gassei K, Orwig K, Rajkovic A. SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev Biol 2012; 361: 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassei K, Orwig KE. SALL4 expression in gonocytes and spermatogonial clones of postnatal mouse testes. PLoS ONE 2013; 8: e53976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Sukhwani M, Hansel MC, Orwig KE. Spermatogonial stem cells in higher primates: are there differences from those in rodents? Reproduction 2010; 139: 479–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Junior PH, Costa GM, Lacerda SM, Rezende-Neto JV, de Paula AM, Hofmann MC, de Franca LR. The spermatogonial stem cell niche in the collared peccary (Tayassu tajacu). Biol Reprod 2012; 86: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso M, Fuso A, Dovere L, de Rooij DG, Stefanini M, Boitani C, Vicini E. Distribution of GFRA1-expressing spermatogonia in adult mouse testis. Reproduction 2012; 143: 325–332. [DOI] [PubMed] [Google Scholar]
- Oatley MJ, Kaucher AV, Racicot KE, Oatley JM. Inhibitor of DNA binding 4 is expressed selectively by single spermatogonia in the male germline and regulates the self-renewal of spermatogonial stem cells in mice. Biol Reprod 2011; 85: 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckins C. The spermatogonial stem cell population in adult rats. II. A radioautographic analysis of their cell cycle properties. Cell Tissue Kinet 1971; 4: 313–334. [DOI] [PubMed] [Google Scholar]
- Huckins C. The spermatogonial stem cell population in adult rats. 3. Evidence for a long-cycling population. Cell Tissue Kinet 1971; 4: 335–349. [DOI] [PubMed] [Google Scholar]
- Lok D, de Rooij DG. Spermatogonial multiplication in the Chinese hamster. III. Labelling indices of undifferentiated spermatogonia throughout the cycle of the seminiferous epithelium. Cell Tissue Kinet 1983; 16: 31–40. [PubMed] [Google Scholar]
- Lok D, Jansen MT, de Rooij DG. Spermatogonial multiplication in the Chinese hamster. II. Cell cycle properties of undifferentiated spermatogonia. Cell Tissue Kinet 1983; 16: 19–29. [PubMed] [Google Scholar]
- Lok D, Jansen MT, de Rooij DG. Spermatogonial multiplication in the Chinese hamster. IV. Search for long cycling stem cells. Cell Tissue Kinet 1984; 17: 135–143. [DOI] [PubMed] [Google Scholar]
- Hu YC, de Rooij DG, Page DC. Tumor suppressor gene Rb is required for self-renewal of spermatogonial stem cells in mice. Proc Natl Acad Sci U S A 2013; 110: 12685–12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang QE, Gwost I, Oatley MJ, Oatley JM. Retinoblastoma protein (RB1) controls fate determination in stem cells and progenitors of the mouse male germline. Biol Reprod 2013; 89: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage J. The retinoblastoma tumor suppressor and stem cell biology. Genes Dev 2012; 26: 1409–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, McKearin D. Gene circuitry controlling a stem cell niche. Curr Biol 2005; 15: 179–184. [DOI] [PubMed] [Google Scholar]
- Decotto E, Spradling AC. The Drosophila ovarian and testis stem cell niches: similar somatic stem cells and signals. Dev Cell 2005; 9: 501–510. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Fuller MT. Asymmetric stem cell division and function of the niche in the Drosophila male germ line. Int J Hematol 2005; 82: 377–380. [DOI] [PubMed] [Google Scholar]
- de Cuevas M, Matunis EL. The stem cell niche: lessons from the Drosophila testis. Development 2011; 138: 2861–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goriely A, McVean GA, van Pelt AM, O'Rourke AW, Wall SA, de Rooij DG, Wilkie AO. Gain-of-function amino acid substitutions drive positive selection of FGFR2 mutations in human spermatogonia. Proc Natl Acad Sci U S A 2005; 102: 6051–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ouyang W, Grigura V, Zhou Q, Carnes K, Lim H, Zhao GQ, Arber S, Kurpios N, Murphy TL, Cheng AM, Hassell JA, et al. ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature 2005; 436: 1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu BY, Orwig KE, Avarbock MR, Brinster RL. Stem cell and niche development in the postnatal rat testis. Dev Biol 2003; 263: 253–263. [DOI] [PubMed] [Google Scholar]
- Chiarini-Garcia H, Hornick JR, Griswold MD, Russell LD. Distribution of type A spermatogonia in the mouse is not random. Biol Reprod 2001; 65: 1179–1185. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Sukeno M, Nabeshima Y. A vasculature-associated niche for undifferentiated spermatogonia in the mouse testis. Science 2007; 317: 1722–1726. [DOI] [PubMed] [Google Scholar]
- Chiarini-Garcia H, Raymer AM, Russell LD. Non-random distribution of spermatogonia in rats: evidence of niches in the seminiferous tubules. Reproduction 2003; 126: 669–680. [DOI] [PubMed] [Google Scholar]
- Huckins C. Cell cycle properties of differentiating spermatogonia in adult Sprague-Dawley rats. Cell Tissue Kinet 1971; 4: 139–154. [DOI] [PubMed] [Google Scholar]
- Shetty G, Meistrich ML. The missing niche for spermatogonial stem cells: do blood vessels point the way? Cell Stem Cell 2007; 1: 361–363. [DOI] [PubMed] [Google Scholar]
- de Rooij DG, Griswold MD. Questions about spermatogonia posed and answered since 2000. J Androl 2012; 33: 1085–1095. [DOI] [PubMed] [Google Scholar]
- de Rooij DG, van Beek ME. Computer simulation of the rodent spermatogonial stem cell niche. Biol Reprod 2013; 88: 131. [DOI] [PubMed] [Google Scholar]
- Tegelenbosch RA, de Rooij DG. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat Res 1993; 290: 193–200. [DOI] [PubMed] [Google Scholar]
- De Rooij DG, Janssen JM. Regulation of the density of spermatogonia in the seminiferous epithelium of the Chinese hamster: I. Undifferentiated spermatogonia. Anat Rec 1987; 217: 124–130. [DOI] [PubMed] [Google Scholar]
- De Rooij DG, Lok D. Regulation of the density of spermatogonia in the seminiferous epithelium of the Chinese hamster: II. Differentiating spermatogonia. Anat Rec 1987; 217: 131–136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.