SUMMARY
Ferroptosis is a form of nonapoptotic cell death for which key regulators remain unknown. We sought a common mediator for the lethality of 12 ferroptosisinducing small molecules. We used targeted metabolomic profiling to discover that depletion of glutathione causes inactivation of glutathione peroxidases (GPXs) in response to one class of compounds and a chemoproteomics strategy to discover that GPX4 is directly inhibited by a second class of compounds. GPX4 overexpression and knockdown modulated the lethality of 12 ferroptosis inducers, but not of 11 compounds with other lethal mechanisms. In addition, two representative ferroptosis inducers prevented tumor growth in xenograft mouse tumor models. Sensitivity profiling in 177 cancer cell lines revealed that diffuse large B cell lymphomas and renal cell carcinomas are particularly susceptible to GPX4-regulated ferroptosis. Thus, GPX4 is an essential regulator of ferroptotic cancer cell death.
INTRODUCTION
Cells can undergo regulated forms of cell death in a variety of contexts (Galluzzi et al., 2012), including during development (Penaloza et al., 2006). Activation of alternative regulated cell death mechanisms may be beneficial for treating diseases such as cancer, in which apoptotic cell death mechanisms are suppressed due to genetic alterations. Indeed, activation of alternative cell death pathways may overcome the drug resistance associated with existing chemotherapeutic agents, providing new drug targets.
Regulators of apoptosis have been targeted with small molecules to induce cell death in cancer cells (Cotter, 2009). Recently, regulated, nonapoptotic cell death processes have been discovered, including necroptosis (Degterev et al., 2005) and ferroptosis (Dixon et al., 2012).
Ferroptosis is a mode of cell death involving the production of iron-dependent reactive oxygen species (ROS). In engineered human fibroblast cell lines, the small molecule erastin was found to induce preferential lethality in cells overexpressing oncogenic HRAS (Dolma et al., 2003). Erastin-induced ferroptotic cell death was distinct from apoptosis, necrosis, and autophagy, based on morphological, biochemical, and genetic criteria. Ferroptosis involves metabolic dysfunction that results in the production of both cytosolic and lipid ROS, independent of mitochondria but dependent on NADPH oxidases in some cell contexts (Dixon et al., 2012).
We have reported the identification of additional small molecules, named RSL3 (Yang and Stockwell, 2008a), ML162, and DPI10 (Weïwer et al., 2012), that display oncogenic-RAS-synthetic-lethality (the RSL phenotype) in engineered fibro-blast-derived tumorigenic cell lines. Here, we sought to test whether these and other compounds also induce ferroptosis, and whether they could be used to elucidate a central regulator of ferroptosis, which controls cell death by all FIN (ferroptosis inducing) compounds.
We focused initially on erastin and RSL3. Erastin reprograms cancer cell metabolism by modulating VDAC2/VDAC3 (Yagoda et al., 2007) and system xc– (Dixon et al., 2012) to trigger ferroptosis, whereas RSL3-induced ferroptosis is not dependent on these factors (Yang and Stockwell, 2008a) but results in a similar downstream cell death phenotype. Thus, we viewed erastin and RSL3 as ideal probes to elucidate conserved downstream regulators of ferroptosis. We used metabolomic profiling to evaluate comprehensively changes in metabolism occurring upon erastin treatment, and chemoproteomics to identify candidate target proteins for RSL3, which led to the discovery of a common pathway regulating cell death in response to all known compounds that induce ferroptosis.
RESULTS
Erastin Depletes Glutathione to Trigger Selective Ferroptosis
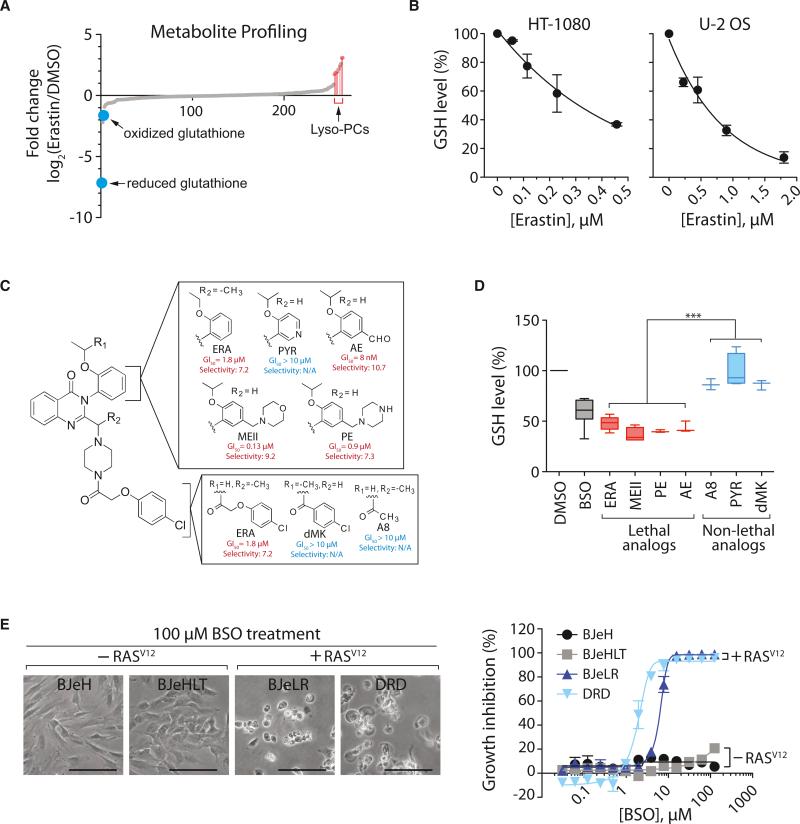
To investigate the global changes in metabolism induced by erastin, we treated HT-1080 fibrosarcoma cells with DMSO or erastin, and extracted polar and lipid metabolites. The metabolite extract was subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis to determine the quantity 149 polar and 115 lipid metabolites (Table S1 available online; Figure 1A). Both reduced glutathione (GSH) and oxidized glutathione (GSSG) were depleted significantly upon erastin treatment, whereas the level of lysophosphatidyl cholines (lysoPCs) was increased; the increase in lysoPCs may reflect the generation of lipid ROS during erastin-induced ferroptosis because PCs have been found to be converted to lysoPCs (deacylated PCs) upon lipid oxidation (Parthasarathy et al., 1985).
Figure 1. Ferroptosis Involves Generation of Lyso-PC and Depletion of Glutathione.
(A) Changes in metabolites upon erastin treatment.
(B) Dose-dependent depletion of GSH by erastin in HT-1080 cells and U-2 OS cells.
(C) Structure and activity of erastin (ERA) analogs. Potency (GI50; concentration required for 50% growth inhibition) and selectivity (ratio of GI50 in HRAS wild-type cells divided by GI50 in HRAS mutant cells) of each analog are shown. PYR, pyridine erastin; AE, aldehyde erastin; MEII, morpholine erastin II; PE, piperazine erastin.
(D) GSH depletion by erastin analogs. HT-1080 cells were incubated with 10 μM erastin analogs for 5 hr or 100 μM BSO for 12 hr. BSO was used as a positive control for GSH depletion. Data were normalized to the DMSO sample. Box-and-whisker plots (n = 3–8) are as follows: midline represents median, box is the 25th– 75th percentiles, and whiskers are minimum and maximum. ***p < 0.001.
(E) BSO induces selective lethality in BJ-derived tumorigenic cells expressing oncogenic HRAS. Scale bars, 60 μm.
In (B) and (E), data are presented as mean ± SD (n = 3). See also Figure S1 and Table S1.
The significant depletion of GSH/GSSG was consistent with the fact that erastin induces the formation of ROS, causing an oxidative cell death. GSH/GSSG constitutes a major cellular anti-oxidant system and provides reducing equivalents to eliminate oxidative species. We treated three cell lines with erastin, determined GSH levels using Ellman's reagent, and confirmed the dose-dependent, GSH-depleting effect of erastin (Figures 1B and S1A). We found that GSH depletion by erastin is necessary for erastin's lethality because supplementing the culture medium with GSH or N-acetylcysteine (NAC), a biosynthetic precursor to GSH, prevented erastin-induced cell death (Figure S1B).
We sought to test further whether the glutathione-depleting activity of erastin was essential for lethality. We established a synthetic route to create six erastin analogs (see Data S1 for synthesis) and tested these analogs for selective lethality in BJ-derived engineered cell lines (Figure 1C). Three compounds (MEII, PE, and AE) retained selective lethality, whereas three compounds (A8, PYR, and dMK) were not lethal (Figure 1C). Lethal analogs of erastin depleted cellular GSH more effectively than nonlethal analogs of erastin (Figure 1D), which further suggested that the GSH-depleting activity of erastin is necessary for erastin lethality.
We reasoned that, if GSH depletion was contributing to erastin's lethality, then GSH depletion by other reagents might partially mimic erastin's selective lethality in the BJ-cell line system, which consists of isogenic cell lines (two with and two without oncogenic HRAS), through which ferroptosis-inducing compounds such as erastin were discovered. When the four BJ-derived cells were treated with BSO, an oncogenic HRAS-selective lethal phenotype was observed under conditions of low cell density (Figure 1E), suggesting that GSH depletion is sufficient for induction of ferroptosis in BJ cells with oncogenic HRAS overexpression, although additional factors may enhance the potency and efficacy of the lethality caused by GSH depletion under more general circumstances. These results indicated that erastin depletes GSH through preventing cystine uptake via inhibition of system xc– (Dixon et al., 2012).
Erastin Inactivates GPX Enzymes through GSH Depletion
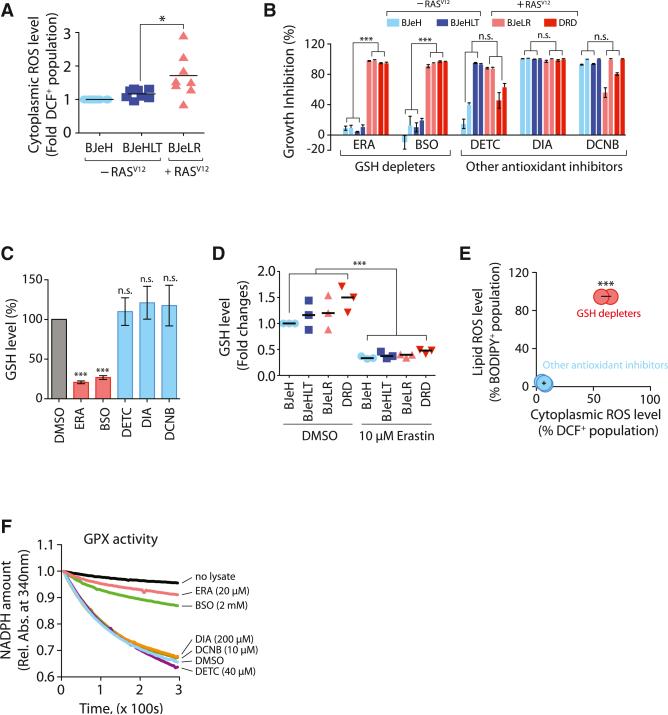
Given that we found GSH depletion to be critical for erastin-induced ferroptosis, we investigated how GSH depletion by erastin was able to induce selective lethality in the engineered BJ cell lines. It has been hypothesized that most cancer cells, including RAS-transformed fibroblasts (Irani et al., 1997), are under high levels of oxidative stress (Szatrowski and Nathan, 1991), which therefore needs to be counteracted by increasing the ROS-scavenging capacity, to prevent oxidative damage (Hussain et al., 2003). In this model, targeting ROS-scavenging systems through multiple points, including GSH depletion, would cause an imbalance in this equilibrium, leading to oxidative cell death (Trachootham et al., 2006). In order to test whether this simple hypothesis could explain erastin's selective lethality, we examined basal ROS levels in the BJ-derived engineered cell lines using H2DCF, a cytosolic ROS sensor, and confirmed that BJeLR cells have modestly elevated ROS levels compared to BJeH (BJ-TERT) and BJeHLT (BJ-TERT/LT/ST) cells (Figure 2A). We then treated the four BJ-derived cell lines with an SOD inhibitor (DETC), a thiol-reactive reagent (DIA), a thioredoxin reductase inhibitor (DCNB), or a catalase inhibitor (ATZ) (Figures 2B and S2). Three of these antioxidant inhibitors (DETC, DIA, and DCNB) killed BJ-derived cells, but they neither depleted GSH nor displayed selective lethality, which was in contrast to the two GSH-depleting reagents, erastin and BSO (Figures 2B, 2C, and S2). The results indicate that it is not possible to induce selective ferroptosis in the BJ-derived cell lines by simply targeting the antioxidant network. Instead, unique biochemical and metabolic changes downstream of GSH depletion were likely to be responsible for the selective induction of ferroptosis.
Figure 2. GSH Depletion Inactivates GPX Enzymes to Induce Ferroptosis.
(A) Basal ROS levels among BJ-derived cell lines were compared (n = 8).
(B) The growth inhibition effect of antioxidant-targeting compounds was determined in the four BJ-derived cells (n = 3). The bar graph indicates growth inhibition at two different concentrations (2× GI50 and 4× GI50 for each compound in BJeLR cells).
(C) Other antioxidant inhibitors do not deplete GSH during cell death (n = 3).
(D) Erastin depletes cellular GSH equally in the four BJ-derived cell lines.
(E) GSH-depleting reagents elevated both cytosolic and lipid ROS level, whereas other antioxidant inhibitors did not (n = 3).
(F) GSH-depleting reagents (ERA and BSO) inhibited GPX activity. Rel. Abs., relative absorbance.
n.s., not significant; *p < 0.05; ***p < 0.001. Error bars in (B), (C), and (E) represent mean ± SD. See also Figure S2.
The degree of GSH depletion upon erastin treatment in the four BJ cell lines was also examined (Figure 2D). We found that these four BJ-derived cell lines contained varying amounts of basal GSH in the absence of any treatment, as reported previously by Kang and Enger (1992), but were depleted of GSH to a similar extent upon erastin treatment. The concentration of erastin used in this experiment (10 μM) was lethal to BJeLR and DRD cells (expressing HRASV12) but was not lethal to BJeH and BJeHLT cells (with wild-type RAS), even upon prolonged incubation (Figure 2B). Therefore, the selective lethality among these cell lines was not caused by differential depletion of GSH or by differences in the basal level of GSH.
We then treated BJeLR cells with either GSH-depleting reagents (erastin or BSO) or other antioxidant-targeting re-agents (DETC, DIA, and DCNB), and stained cells with either BODIPY-C11, a membrane-targeted lipid ROS sensor, or H2DCF, a cytoplasmic ROS sensor, to detect changes in ROS accumulation upon compound treatment (Figure 2E). GSH-depleting reagents strongly increased BODIPY-C11 and H2DCF signals, whereas other antioxidant inhibitors did not increase the fluorescence signals from either ROS sensor, indicating that both cytosolic and lipid ROS levels were increased selectively by GSH depletion in the oncogenic HRAS-containing cell lines (Figure 2E).
We considered that one consequence of GSH depletion could be inactivation of glutathione-dependent peroxidases (GPXs). GPXs catalyze the reduction of hydrogen peroxide and organic hydroperoxides to water or the corresponding alcohols, using GSH as an essential cofactor (Brigelius-Flohé and Maiorino, 2013).
The total activity of GPXs in BJeLR cells was examined using tert-butylhydroperoxide (tBuOOH) as a substrate, by monitoring the rate of NADPH oxidation, which is coupled to the tBuOOH-reducing activity of GPXs in cell lysates. When BJeLR cell lysates treated with vehicle only (0.08% DMSO) were added to the GPX activity assay, we observed a decrease in the amount of NADPH, indicating that tBuOOH was reduced by GPXs in the cell lysate (Figure 2F). Treatment of BJeLR cells with antioxidant inhibitors (DETC, DIA, and DCNB) did not affect GPX activity because the rate of NADPH oxidation was similar to the vehicle-only-treated sample.
When lysates from BJeLR cells that had been treated with GSH-depleting reagents (erastin or BSO) were analyzed, NADPH oxidation was prevented, indicating that GPXs were inactivated upon GSH depletion (Figure 2F). Taken together, these data indicate that erastin and BSO inactivate cellular GPXs, leading to the generation of cytoplasmic and lipid ROS. Other antioxidant inhibitors did not deplete GSH, an essential cofactor for GPX enzyme activity, and therefore, did not inhibit GPX activity and did not cause accumulation of peroxides.
RSL3 Binds to and Inactivates GPX4
Having determined that erastin acts by depleting GSH and inhibiting GPXs, we examined the mechanism of action of RSL3, another ferroptosis inducer (Yang and Stockwell, 2008a). Cell death induced by erastin and RSL3 shared common ferroptotic features, such as iron, MEK, and ROS dependence; however, RSL3 was not dependent on VDAC2/VDAC3 or system xc– implying that a different initiating mechanism could converge on a similar form of ferroptotic cell death. We thus used RSL3 as a probe to illuminate the shared downstream mechanism involved in executing ferroptosis.
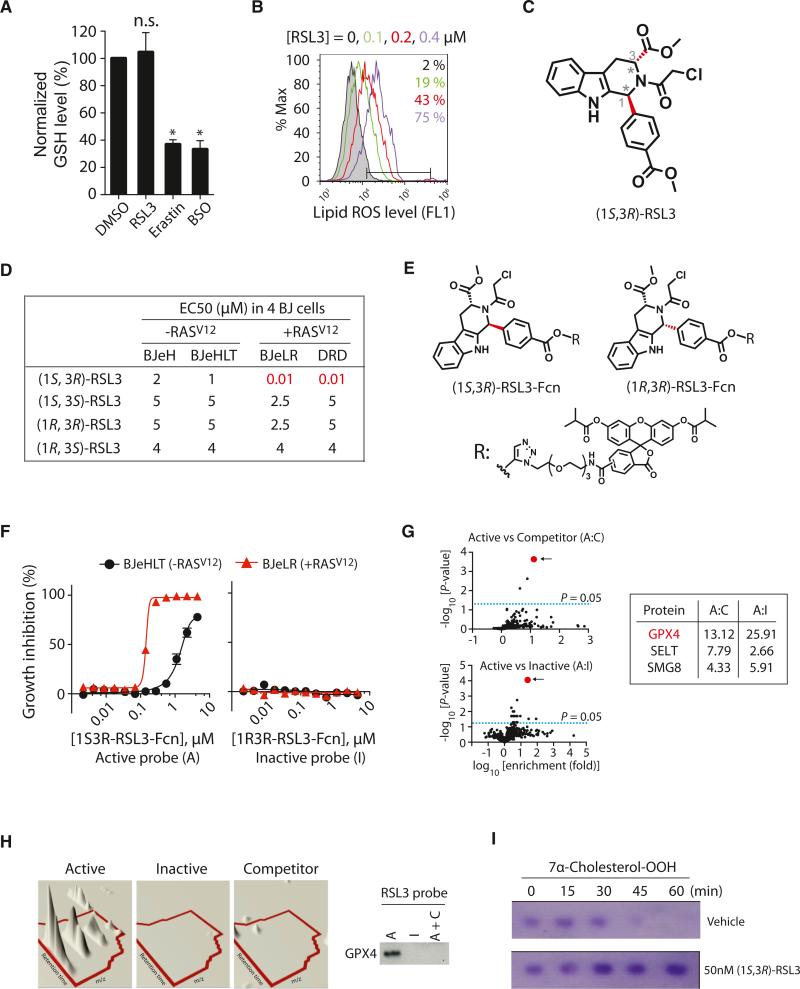
When we examined cellular GSH levels during RSL3-induced cell death, we found that GSH remained unaffected by a lethal RSL3 concentration (2 μM) in BJeLR cells, which was in contrast to erastin's effect (Figure 3A). However, BODIPY-C11 staining revealed the generation of lipid ROS in RSL3-treated BJeLR cells, which indicated that lipid oxidation is common to both erastin-induced and RSL3-induced ferroptotic cell death (Figure 3B).
Figure 3. RSL3 Binds to and Inhibits GPX4.
(A) RSL3 does not deplete GSH. The level of GSH was determined after treating with 2 μM RSL3, 10 μM erastin, or 1 mM BSO (n = 3; *p < 0.05). Error bars indicate ± SD.
(B) RSL3 treatment increased lipid ROS level, as erastin did.
(C) The structure of (1S, 3R)-RSL3 is shown.
(D) Only the (1S, 3R) diastereomer displayed selective lethality in HRASV12-expressing cells in the four BJ-derived cell lines.
(E) Structure of RSL3 affinity probes used in the chemoproteomics experiments is shown. Fcn, fluorescein affinity tag.
(F) The active affinity probe with the (1S, 3R) stereochemistry exhibited selective lethality against cells with HRASV12, whereas an affinity probe with the (1R, 3R) stereochemistry was not lethal.
(G) Affinity-based chemoproteomics identified GPX4 (red dot) as the most likely binding protein for (1S, 3R)-RSL3. Fold enrichment values of peptides at the indicated condition and their FDR-adjusted p values were represented as volcano plots. The top three candidates are shown.
(H) Confirmation of GPX4 binding to active (1S, 3R)-RSL3 affinity probe. Left panel is a 3D visualization of isotopic clusters of peptide ILAFPCNQFGK from GPX4 as rendered by TransOmics software. Right panel: cell lysates prepared from BJeLR cells treated with active probe (A), inactive probe (I), or active probe in the presence of competitor (A+C) that were affinity purified by α-fluorescein antibodies. Then, the purified protein samples were probed for GPX4 by western blot using GPX4-specific antibody.
(I) (1S, 3R)-RSL3 inhibits enzyme activity of GPX4.
See also Figure S3 and Table S2.
In order to understand the mechanistic basis of RSL3's enigmatic induction of lipid ROS in the absence of GSH depletion, we used affinity-based chemoproteomics to identify candidate target proteins for RSL3. We discovered that the chloroacetamide moiety of RSL3 was essential for its activity and that replacement with other electrophiles resulted in a loss of potency (Table S2; see Data S1 for synthesis). These data suggested that RSL3 targets an enzyme with a nucleophilic active site, such as serine, threonine, cysteine, or selenocysteine. Despite the potential for promiscuous reactivity of the chloroacetamide, we discovered that only the (1S, 3R)-RSL3 diastereomer of RSL3 exhibited selective lethality in the BJ cell system, whereas the three other diastereomers of RSL3 lacked selectivity and were more than 100-fold less potent (Figures 3C and 3D; Table S2; Data S1 for synthesis). These results suggested that (1S, 3R)-RSL3 covalently binds to one or more proteins to induce the potent and selective lethality observed in BJ cells expressing HRASV12.
Affinity reagents were synthesized by attaching an isobutyrylprotected fluorescein tag via a polyethylene glycol (PEG) linker to the phenyl substituent at the 1 position in the tetrahydro-β-carboline ring system of RSL3, which was found to tolerate structural modifications. Despite a loss in potency upon incorporation of the affinity tag, 10-fold oncogenic HRAS selectivity was retained for an affinity analog with the active (1S, 3R) stereo-chemistry, whereas an affinity analog with the (1R, 3R) stereo-chemistry showed no activity (Figures 3E and 3F; see Data S1 for synthesis).
Samples for proteomic analysis were prepared by treating intact BJeLR cells with (1S, 3R)-RSL3-fluorescein (active probe treatment), (1R, 3R)-RSL3-fluorescein (inactive probe treatment), or (1S, 3R)-RSL3-fluorescein pretreated with free (1S, 3R)-RSL3 (competitor treatment). Treated cells were lysed, and fluorescein-tagged proteins were affinity purified using Sepharose beads coupled to an anti-fluorescein antibody. Eluted proteins were identified with a Synapt G2 HDMS mass spectrometer (Waters) using a quantitative label-free shotgun proteomic strategy with data-independent scanning (MSE) and ion mobility spectrometry (see Extended Experimental Procedures).
Proteins enriched in “active probe” versus “inactive probe” and “active probe” versus “competitor” treatments were determined (Figure 3G). Three independent affinity preparations of each of these treatments were further subsampled in three LCMS/MS analyses (Table S2). Candidates were chosen on the basis of (1) exhibiting enrichment in both active:inactive and active:competitor comparisons with p < 0.01 (with false discovery rate correction; see Extended Experimental Procedures) and (2) identification and quantitation by n > 2 tryptic peptides. This analysis ranked GPX4 (glutathione peroxidase 4, PhGPx) as the top candidate protein target for (1S, 3R)-RSL3 (Figure 3G; Table S2).
It was intriguing that GPX4 was identified as the most highly ranked candidate in our unbiased chemoproteomic approach to find target proteins for RSL3 because erastin inhibited cellular GPX enzymes through GSH depletion (Figures 1 and 2). We confirmed that GPX4 was purified specifically by the affinity analog (1S, 3R)-RSL3-fluorescein, using fresh samples prepared from BJeLR cells treated with the active probe, the inactive probe, or the competitor as before, analyzed for GPX4 abundance by western blot (Figures 3H and S3A).
In order to examine whether RSL3 binding to GPX4 inactivates the peroxidase activity of GPX4, we treated a clone of the COH BR1 breast cancer cell line overexpressing GPX4 (L7G4), with (1S, 3R)-RSL3 or vehicle only, prepared cell lysates, and incubated them with 7α-cholesterol hydroperoxide (7α-cholesterol-OOH) to determine the rate of reduction to the corresponding alcohol (7α-cholesterol-OH). 7α-cholesterol-OOH is a specific substrate for GPX4; no other GPX enzyme can catalyze the reduction of 7α-cholesterol-OOH (Kriska and Girotti, 2005). In vehicle-treated samples, 7α-cholesterol-OOH decreased over time in a GSH-dependent manner, due to reduction by GPX4 in the lysate. Upon treatment with (1S, 3R)-RSL3, however, no reduction of 7α-cholesterol-OOH was observed, indicating that GPX4 was inhibited by (1S, 3R)-RSL3 (Figure 3I; see Figure S3B for quantitation data). Treatment with the inactive diastereomer (1R, 3R)-RSL3 did not inhibit GPX4 activity (Figure S3C).
We used another proteomics data analysis tool, Elucidator (Rosetta Biosoftware), and found that only GPX4 was a significantly enriched protein common to both TransOmics and Elucidator analysis algorithms (Figure S3D). Evaluation of other candidates using specific siRNAs and shRNAs failed to show either modulation of (1S, 3R)-RSL3 sensitivity or cell killing (Figures S3E–S3I), which excludes the functional role of these other binding proteins on ferroptosis induced by (1S, 3R)-RSL3.
RNAi-Mediated GPX4 Knockdown Induces Ferroptosis
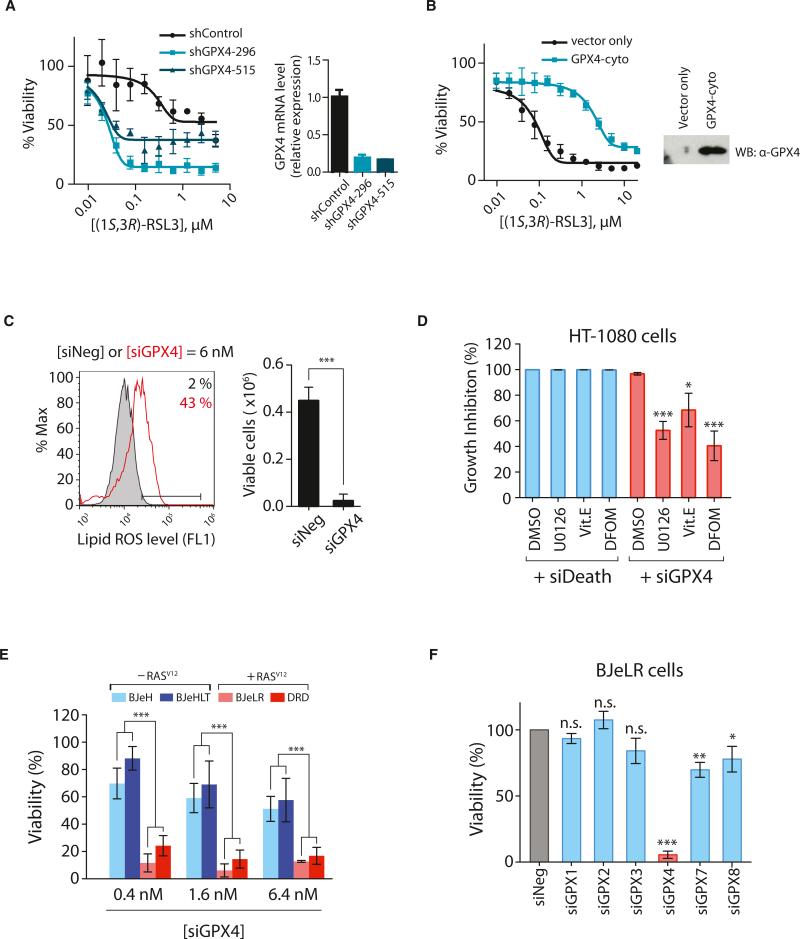
We hypothesized that, if RSL3 were to inhibit a protein essential for cancer cell viability, then reducing the levels of this protein would sensitize cells to RSL3. Silencing of GPX4 mRNA using shRNAs that cause partial knockdown of GPX4 strongly sensitized cells to (1S, 3R)-RSL3 (Figure 4A). Conversely, when we overexpressed GPX4, we found that it caused strong resistance to (1S, 3R)-RSL3 lethality (Figure 4B), as expected for a relevant target protein.
Figure 4. RSL3 Targets GPX4 to Induce Ferroptosis.
(A) Knockdown of GPX4 using shRNAs rendered HT-1080 cells hypersensitive to (1S, 3R)-RSL3 lethality.
(B) Overexpression of GPX4 rendered HT-1080 cells resistant to (1S, 3R)-RSL3 lethality.
(C) HT-1080 cells transfected with a pool of siRNAs targeting GPX4 showed increased lipid ROS level as assessed by BODIPY-C11 staining. siNeg has no homology to any known mammalian genes and was used as a negative control.
(D) Known inhibitors of ferroptosis, 10 μM U0126, 100 μM Vit. E, or 100 μM DFOM, were able to suppress siGPX4-induced cell death, whereas they could not suppress cell death induced by siDeath.
(E) Knockdown of GPX4 displayed selective lethality in the four BJ-derived isogenic cell lines.
(F) Other GPX isoforms are not relevant to ferroptotic cell death.
The values in (D)–(F) were normalized to control samples transfected with siNeg. Bar graphs in (C)–(F) are mean ± SD (n = 3). *p < 0.05; **p < 0.01; ***p < 0.001. Data in (A) and (B) are presented as mean ± SD (n = 3). See also Figure S4.
We were able to achieve a more effective knockdown of GPX4 using a pool of siRNAs targeting GPX4 (20-fold decrease in the GPX4 mRNA; Figure S4A) compared to a single clone of shRNA (5-fold decrease in the GPX4 mRNA; Figure 4A). With this more effective knockdown, HT-1080 cells underwent cell death with accompanying lipid ROS generation (Figure 4C). Cell death induced by siGPX4 was rescued by the same suppressors of RSL3—an iron chelator (DFOM), a MEK inhibitor (U0126), and an antioxidant (vitamin E [Vit. E])—which suggested that GPX4 knockdown induced ferroptotic cell death (Figure 4D) similar to RSL3. None of these ferroptosis inhibitors suppressed cell death induced by siDeath, a control siRNA pool targeting multiple essential genes, highlighting the ferroptosis-specific action of these inhibitors (Figure 4D). Furthermore, siGPX4 induced selective cell death in BJeLR and DRD cells (with HRASV12), but not BJeH and BJeHLT cells (wild-type HRAS), which recapitulated the selective lethality of erastin and RSL3 (Figures 4E and S4B). These data suggested that GPX4 is the primary target of RSL3, mediating its ability to induce ferroptosis specifically in the oncogenic HRAS-containing BJ-derived fibroblasts.
There are eight isoforms of GPXs in humans with different tissue expression and substrate specificities. In BJeLR cells, six GPX isoforms are expressed (GPX1, GPX2, GPX3, GPX4, GPX7, and GPX8) as determined by RT-qPCR (Figure S4C). Knockdown of each isoform affected cell viability to varying levels; however, GPX4 knockdown was the most lethal to BJeLR cells, which highlights the prominent role of GPX4 inhibition in inducing cell death, as compared to other GPX enzymes (Figures 4F and S4D).
Taken together, these data suggest that GPX4 is a central regulator of ferroptosis induced by erastin and RSL3. Cell death was enhanced in the BJ-derived cell lines expressing HRASV12 due to the increased basal ROS (Figure 2A) and enhanced lipid peroxidation after GPX4 inhibition, which caused selective lethality in this engineered isogenic cell line model.
GPX4 Regulates Ferroptosis Induced by 12 Divergent Compounds
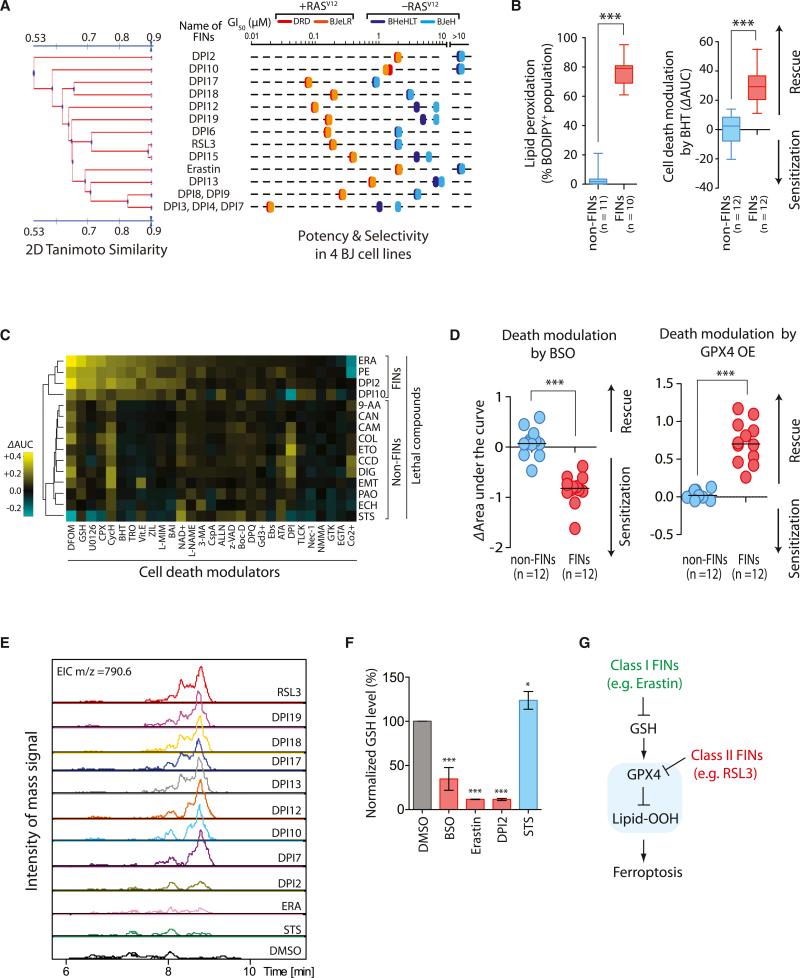
In a larger screening campaign to find additional FIN compounds, 14 candidate compounds were discovered out of more than a million tested (Figure 5A; see Figure S5 for structures) (Weïwer et al., 2012; Yang et al., 2012). These 14 compounds displayed selective lethality in HRASV12-expressing cells in the four BJ-derived cell lines (Figure 5A; Table S3). We defined ten structurally diverse FIN groups, not including erastin or RSL3 (Figure 5A), to use in subsequent experiments.
Figure 5. Ferroptosis Occurs through a GPX4-Regulated Pathway.
(A) Discovery of additional FINs based on selective lethality in the four BJ cell lines.
(B) FINs (red) are distinct from non-FINs (blue) in accompanying lipid ROS generation during the cell death process and in death suppression by an antioxidant, BHT.
(C) Modulatory profiling (Wolpaw et al., 2011) with erastin, PE, DPI2, DPI10, and other lethal molecules confirmed that PE, DPI2, and DPI10 induced a similar form of cell death as erastin in HT-1080 cells. ΔAUC with a positive sign indicates suppression of cell death, whereas a negative sign indicates sensitization by cell death modulators upon lethal compound treatment.
(D) Inhibition of GPX4 by BSO sensitized cells to death induced by 12 FIN compounds, whereas activation of GPX4 by cDNA overexpression rescued cells from the lethality of FIN compounds.
(E) Eight structurally diverse FIN compounds inhibited GPX4, whereas two FIN compounds, DPI2 and erastin, and the negative control staurosporine (STS), a non-FIN compound, did not show direct GPX4 inhibition in this LC-MS-based assay.
(F) The two FIN compounds, DPI2 and erastin, depleted cellular GSH, which inhibits GPX4 indirectly, whereas staurosporine did not deplete GSH. Bar graph indicates mean ± SD (n = 3).
(G) Model of GPX4-regulated ferroptosis pathway. Ferroptosis inducers can be categorized into two classes based on the mode of GPX4 inhibition. *p < 0.05; ***p < 0.001. See also Figure S5 and Table S3.
BJeLR cells treated with each of the ten additional FIN compounds exhibited an increase in BODIPY-C11 fluorescence, indicating that lipid ROS were generated (Table S3 for individual flow cytometry data). We then tested 11 non-FIN compounds acting through diverse lethal mechanisms to see whether they induced lipid ROS generation using this assay (Table S3 for more information of 11 non-FIN compounds). These 11 diverse lethal compounds were confirmed to lack selective lethality in the four BJ-derived cell lines previously (Root et al., 2003), which suggested that they are not ferroptosis inducers. We found that 10 out of 11 of the non-FIN compounds did not generate lipid ROS, implying a specificity of lipid ROS generation for FIN compound-treated cells (Table S3 for individual flow cytometry data). It is likely that the reported ROS-generating activity of phenylarsine oxide was responsible for the oxidation of the BODIPY-C11 dye (Fanélus and Desrosiers, 2008).
In order to determine whether these FIN compounds genuinely induced ferroptosis, the functional requirement of lipid ROS during ferroptosis was examined by treating BJeLR cells with each lethal compound (FINs and non-FINs) in the presence of a lipophilic antioxidant, butylated hydroxytoluene (BHT). BHT strongly suppressed cell death induced by all FIN compounds (Table S3 for individual growth inhibition curves). The rescuing effect of BHT was specific to FIN compounds because BHT was not able to suppress cell death induced by 12 non-FIN compounds (Table S3 for individual growth inhibition curves). We quantified the degree of cell death suppression by calculating the normalized differences in the AUC (area under the concentration-response curve) of the compound alone and that of the compound with BHT. Combined with the BODIPY-C11 staining data, these results revealed that the FIN compounds are mechanistically distinct from the 11 non-FIN compounds (Figure 5B). An extended death mechanism analysis using the modulatory profiling approach (Wolpaw et al., 2011) with four selected FIN compounds (erastin, PE, DPI2, and DPI10) revealed that they induced a similar form of cell death that was distinct from non-FIN compounds (Figure 5C). Taken together, these data indicate that compounds that display the RSL phenotype in the four-BJ cell system are ferroptosis inducers. These data also suggest that ferroptotic cell death is not limited to erastin and RSL3 but that a number of additional small molecules can be identified that induce ferroptosis, suggesting that it may be a more generally important mechanism of lethality.
In order to determine the generality of ferroptosis regulation by GPX4, we treated HT-1080 cells with each lethal compound (FIN compound or non-FIN compound) under a GPX4-inhibited condition (using BSO treatment, which depletes glutathione and therefore inhibits all GPXs), or a GPX4-upregulated condition (by overexpressing GPX4). GPX4 inhibition using BSO enhanced ferroptotic cell death induced by all FIN compounds, whereas GPX4 overexpression suppressed ferroptosis induced by all FIN compounds (Figure 5D and Table S3). The modulation effect of BSO and GPX4 overexpression was specific to FIN compounds because their effects on cell death induced by 11 non-FIN compounds were minimal (Figure 5D).
We speculated that these additional FIN compounds acted through mechanisms similar to those induced by erastin or RSL3; we examined whether these FIN compounds inhibited GPX4 in BJeLR cells. For a side-by-side comparison of GPX4 activity in multiple samples, we used an LC-MS-based GPX4 assay in which the GPX4 enzyme activity in cell lysates was measured by their capacity to reduce exogenously added phosphatidylcholine hydroperoxide (PC-OOH), a GPX4-specific substrate. When GPX4 was inhibited, cells could not reduce PCOOH, which resulted in a signal for the [PC-OOH + H]+ ion (m/z, 790.6) in the mass chromatogram. BJeLR cells treated with any of the eight FIN compounds (DPI7, DPI10, DPI12, DPI13, DPI17, DPI18, DPI19, and RSL3) lacked GPX4 activity, being unable to reduce exogenous PC-OOH (Figure 5E). On the other hand, a control non-FIN compound, staurosporine, and two known FIN compounds, erastin and DPI2, did not inhibit GPX4 activity in the assay. The LC-MS-based assay requires addition of exogenous GSH (5 mM) in the reaction mixture; therefore, it cannot detect indirect inhibitory effects on GPX4 by GSH-depleting agents such as BSO and erastin. We then determined the cellular GSH level in BJeLR cells treated with DPI2, the one FIN compound that did not inhibit GPX4 in the LC-MS-based assay, along with BSO and erastin as positive controls, and staurosporine as a negative control (Figure 5F). DPI2 depleted 90% of cellular GSH compared to untreated BJeLR cells, indicating that it acts through a mechanism similar to erastin to induce ferroptotic cell death (Figure 5F).
These results demonstrate that GPX4-regulated ferroptosis is a common mechanism shared by multiple independent small molecule scaffolds. All FIN compounds can be categorized into two classes based on the mode of GPX4 inhibition. One class, including erastin, inhibits GPX4 through GSH depletion. The second class inhibits GPX4 without GSH depletion, such as RSL3, which inhibits GPX4 directly (Figure 5G).
Ferroptosis Inducers Inhibit Tumor Growth in a Xenograft Mouse Model
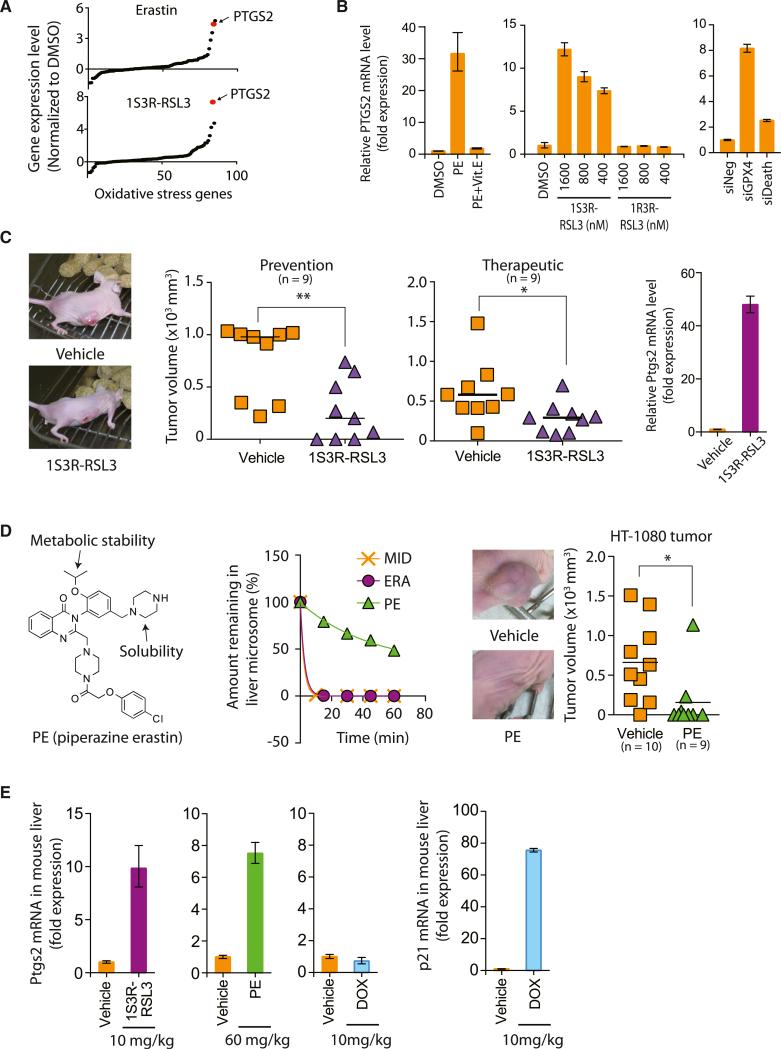
We explored whether the GPX4-regulated ferroptotic cell death pathway could be utilized to suppress the growth of tumors in a xenograft mouse model. First, we searched for a pharmacodynamic marker associated with ferroptotic cell death. A set of 83 genes that were known to be perturbed upon oxidative stress was surveyed to determine whether the expression level of any gene was affected during ferroptosis induced by erastin or RSL3 (Figure 6A; Table S4). This analysis revealed that PTGS2, a gene encoding cyclooxygenase-2 (COX-2), was the most upregulated gene in BJeLR cells upon treatment with either erastin or (1S, 3R)-RSL3 (Figure 6A). The functional relevance of PTGS2 on ferroptosis was examined using indomethacin, a PTGS-1/PTGS-2 (COX-1/COX-2) inhibitor. Ferroptotic cell death by erastin or (1S, 3R)-RSL3 was not affected by indomethacin treatment, suggesting that PTGS2 upregulation is simply a downstream marker of ferroptosis (Figure S6A).
Figure 6. Ferroptosis Suppresses Tumor Growth in a Xenograft Mouse Model.
(A) Upregulation of PTGS2 expression upon erastin and (1S, 3R)-RSL3 treatments.
(B) PTGS2 expression was induced by PE, (1S, 3R)-RSL3, and siGPX4, but not by PE with Vit. E, (1R, 3R)-RSL3, and siDeath.
(C) (1S, 3R)-RSL3 inhibited tumor formation and tumor progression through induction of ferroptosis as demonstrated by upregulation of PTGS2 in the tumors. (1S, 3R)-RSL3 was administered s.c. twice a week for 2 weeks.
(D) PE showed efficacy in preventing HT-1080 tumor formation in a mouse xenograft model. The left view shows the structure of PE. The middle view is a mouse liver microsome assay demonstrating improved metabolic stability of PE over erastin. Midazolam was used as a positive control for metabolic degradation. The right view shows images representative of tumors in live mice from each treatment group. PE was delivered s.c. twice a week for 1 week and then delivered through tail vein injection once every other day for 6 days.
(E) Pharmacodynamics of PE and (1S, 3R)-RSL3 in the mouse liver tissue.
Bar graphs in (B), (C), and (E) represent mean ± SD (n = 3). In (C) and (D), the lines in the tumor volume plots indicate mean of nine data points. *p < 0.05; **p < 0.01. See also Figure S6 and Table S4.
We also developed a more effective analog of erastin that could be tested in vivo. Erastin itself has modest water solubility and is metabolically labile, precluding its use in vivo. We discovered that introduction of a piperazine moiety into the aniline ring of erastin resulted in a more water-soluble and more metabolically stable compound that was suitable for in vivo experiments; we named this compound piperazine erastin (PE). PE upregulated PTGS2 in BJeLR cells (Figure 6B). Cotreatment of Vit. E suppressed PTGS2 induction by PE, which indicates that PTGS2 upregulation is downstream of the lipid peroxidation that occurs during ferroptosis (Figure 6B). (1S, 3R)-RSL3, but not (1R, 3R)-RSL3, increased PTGS2 expression (Figure 6B). In addition, knockdown of GPX4 using siRNAs markedly increased PTGS2 mRNA abundance, whereas the effect on PTGS2 mRNA abundance of siDeath was minimal (Figure 6B). Kumagai et al. (2004) reported upregulation of PTGS2 by 4-HNE, an end product of oxidized lipids in an atherosclerosis model. Moreover, DNA microarray analysis of gene expression in skin tissue samples obtained from Gpx4-deficient mice identified Ptgs2 as a key gene upregulated as a consequence of Gpx4 loss (Sengupta et al., 2013). Taken together, these data confirm that PTGS2 upregulation is a suitable marker for the lipid peroxidation that occurs during GPX4-regulated ferroptosis.
We tested whether (1S, 3R)-RSL3 could prevent tumor growth in athymic nude mice implanted with subcutaneous (s.c.) xeno-graft tumors derived from BJeLR cells. Mice were injected with BJeLR cells s.c. and then with 100 mg/kg (1S, 3R)-RSL3 in the same site 1 day later. The (1S, 3R)-RSL3 injection was repeated twice each week for 2 weeks. Three weeks later, we observed significant prevention of tumor growth in (1S, 3R)-RSL3-treated animals (Figure 6C; p = 0.0053). Subsequently, we examined the ability of (1S, 3R)-RSL3 to shrink preexisting tumors in a therapeutic study, using s.c. xenograft tumors that had been allowed to grow for 1 week and then treated with 100 mg/kg (1S, 3R)-RSL3 twice a week for 2 weeks. Again, we observed a significant reduction in tumor volume compared with the vehicle-treated control group, with this infrequent dosing (Figure 6C; p = 0.038). The inhibition of tumor growth was likely due to the induction of ferroptosis, as determined by Ptgs2 upregulation in (1S, 3R)-RSL3-treated tumors (Figure 6C).
We then evaluated the erastin analog PE in a tumor-prevention model using nude mice into which HT-1080 cells, the human fibrosarcoma cell line, had been injected. As mentioned, PE has improved metabolic stability (Figure 6D) and water solubility (0.086 mM for erastin versus 1.4 mM for PE) compared to erastin. PE was affected similarly by cell death modulators as erastin and displayed a distinct pattern from other non-FIN lethal compounds, indicating that PE, like erastin, induces ferroptosis in HT-1080 cells (Figure 5C; Spearman correlation coefficient, 0.9291; p < 0.0001). HT-1080 cells were injected to the flank of athymic nude mice. One day later, vehicle or PE (40 mg/kg) was s.c. delivered to the nude mice with a twice-a-week schedule for 1 week. Then, vehicle or PE (30 mg/kg) was administered to the mice through tail vein once every other day for 6 days. We observed a significant delay in tumor growth in the PE-treated group compared to the vehicle-treated group (Figure 6D).
The systemic toxicity and pharmacodynamics of these two ferroptosis inducers, PE and (1S, 3R)-RSL3, were assessed by injecting the compounds through the tail vein at 60 and 10 mg/kg, respectively. At these doses, we observed upregulation of Ptgs2 in mouse liver (Figure 6E). In contrast, tail vein injection of the chemotherapeutic agent doxorubicin did not show any induction of Ptgs2 in the same tissue, confirming the specificity of Ptgs2 as a pharmacodynamic marker (Figure 6E). As a control, we detected robust upregulation of the p21 gene, a biomarker for p53 activation induced by doxorubicin-mediated DNA damage (Figure 6E). Regardless of ferroptosis activation in the liver, we observed no overt toxicity in either animal study, as assessed by the lack of acute lethality, and the lack of significant body weight reductions. Independently, NCI's DTP (developmental therapeutics program) evaluated in vivo the toxicity of intraperitoneal injection of (1S, 3R)-RSL3 and observed no toxicity up to 400 mg/kg dose, which suggested that these doses of (1S, 3R)-RSL3 were well tolerated (Figure S6).
Diffuse Large B Cell Lymphomas and Renal Cell Carcinomas Are Sensitive to Ferroptosis
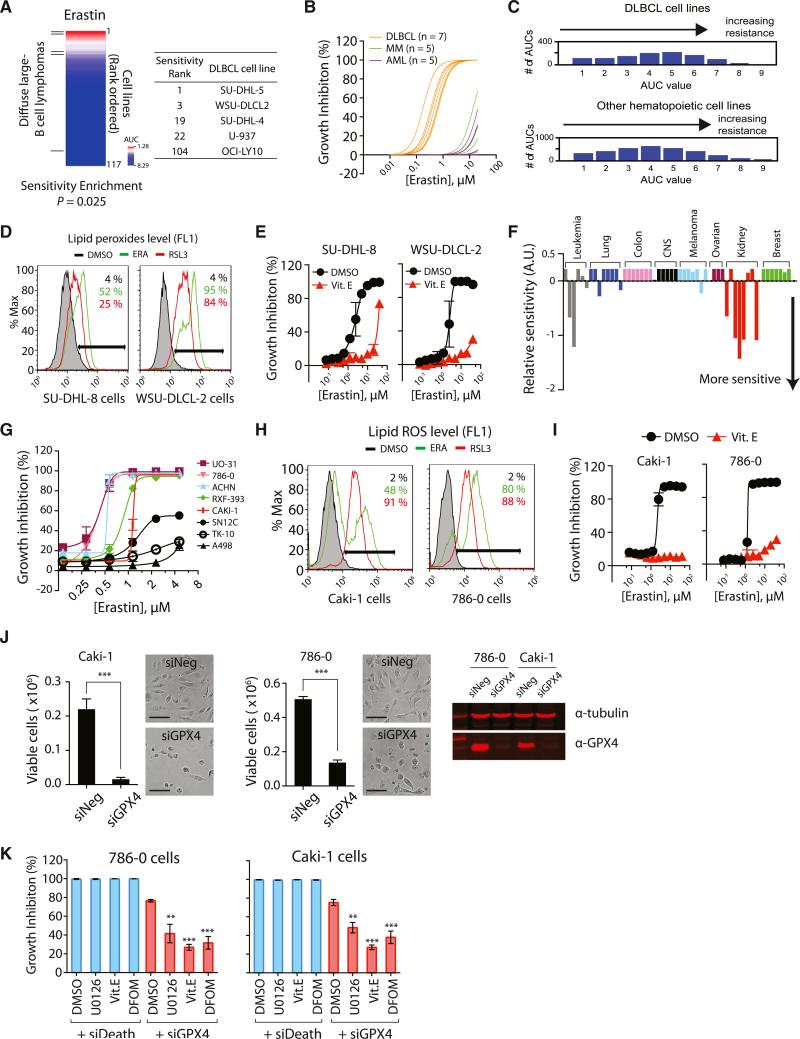
We investigated whether the mutation status of RAS genes in cancer cells is a predictor of sensitivity to erastin-induced ferroptotic cell death, by testing erastin in 117 cancer cell lines from different tissues such as hematopoietic and lymphoid tissue, large intestine, lung, ovary, and skin (Table S5). The cancer cell line panel contained 38 cancer cell lines with oncogenic-RAS mutations, which allowed us to examine the correlation between RAS mutation status and erastin potency.
Although erastin displayed synthetic lethality in the engineered cells, it did not show selective lethality in RAS-mutated cancer cell lines over RAS wild-type counterparts (Figures S7A and S7B) in this large and diverse panel of cell lines. Although RAS mutations sensitize to ferroptosis in an individual genetic context, there are other more dominant determinants of sensitivity when analyzing sensitivity across diverse contexts. Analysis of the 117-cell line erastin sensitivity data revealed that diffuse large B cell lymphomas (DLBCLs) were particularly sensitive (Figure 7A). When we divided the 117 cell lines into sensitive and resistant groups based on AUC values (sensitive if AUC < 3.5, resistant if AUC > 5.5), DLBCLs were enriched in the sensitive group (Figure 7A, p = 0.01 by chi-square test; p = 0.025 by logistical regression analysis [Basu et al., 2013]). In a larger drug-screening analysis, it was observed that suspension cell lines were generally more sensitive to growth inhibitory effects of small molecules (Basu et al., 2013). In order to examine whether the observed sensitivity enrichment in our analysis was due to the general sensitivity of suspension cell lines, we tested erastin in seven DLBCL cell lines, five acute myeloid leukemia (AML) cell lines, and five multiple myeloma (MM) cell lines. The DLBCLs displayed increased sensitivity among these suspension cell lines, which suggested that DLBCLs are particularly sensitive to ferroptotic cell death (Figure 7B; see Figure S7C for individual concentration-dependent curve).
Figure 7. DLBCLs and RCCs Are Sensitive to GPX4-Regulated Ferroptosis.
(A) Testing erastin in 117 cancer cell lines revealed DLBCLs as a cancer subtype susceptible to ferroptosis. DLBCL cell lines are marked with lines on the left. The table shows the name of DLBCL cell lines along with the sensitivity rank.
(B) DLBCLs were more sensitive to erastin than AML and MM cells.
(C) DLBCL cell lines are no more sensitive to lethal compounds than other hematopoietic cell lines. The total number of AUCs in the analysis was 3,883 (972 for DLBCL and 2,911 for other hematopoietic cell lines).
(D and E) DLBCL cells died through a mechanism characteristic of ferroptosis, as determined by lipid peroxide generation and death rescue by Vit. E.
(F) Sensitivity profile of 53 cancer cell lines in the “NCI60” cell panel against erastin. The cell lines were grouped based on their tissue origins.
(G) The eight RCC cell lines were retested with erastin to confirm their sensitivity against erastin.
(H) Erastin and RSL3 generated lipid ROS in the two RCC cell lines.
(I) Cell death was rescued by a lipophilic antioxidant, Vit. E.
(J) GPX4 depletion by siGPX4 induced cell death in RCC cell lines. The western blot (right) confirmed expression of GPX4 protein in these RCC cell lines and knockdown of GPX4 by siRNAs. Scale bars, 30 μm.
(K) Ferroptosis inhibitors suppressed cell death induced by GPX4 knockdown but could not suppress cell death induced by the control siRNAs (siDeath) that kill cells via a nonferroptotic pathway.
Data points in (E), (G), and (I)–(K) represent mean ± SD (n = 3). **p < 0.01; ***p < 0.001. See also Figure S7 and Table S5.
We further analyzed the sensitivity of DLBCLs and other hematopoietic cell lines against 203 diverse lethal compounds to see if DLBCLs are generally sensitive to lethal compounds (Figure 7C). DLBCL cell lines in fact displayed a slight resistance on average to all compounds tested compared to other hematopoietic cell lines (p < 3 3 10−4, Kolmogorov-Smirnov test). This indicates that the enhanced sensitivity of DLBCLs to erastin-induced ferroptosis is not due to a general sensitivity to all compounds.
We confirmed that erastin and RSL3 generated lipid peroxides in two DLBCL cell lines: SU-DHL-8 and WSU-DLCL-2 (Figure 7D). Moreover, erastin-induced cell death was rescued by a lipophilic antioxidant (Vit. E) in these cell lines, indicating that cells were dying through the lipid ROS characteristic of ferroptosis (Figure 7E).
The potency of erastin was also determined in a 60-cancer cell line panel (NCI60) (Shoemaker, 2006) from eight diverse tissues, which revealed increased sensitivity of renal cell carcinomas (RCCs), compared to the other tissues examined (Figures 7F and S7D). We confirmed the potency of erastin in these RCC cell lines and observed generation of lipid ROS in two representative RCC cell lines (Figures 7G and 7H). Moreover, erastin-induced death of these cell lines was suppressed by a lipophilic antioxidant (Vit. E) (Figure 7I). The two RCC cell lines expressed GPX4, as determined by western blot with a GPX4-specific antibody (Figure 7J). Knockdown of GPX4 using siRNAs decreased the level of GPX4 protein and was sufficient to kill these RCC cell lines (Figure 7J). Moreover, these cells died via a characteristic ferroptotic death upon GPX4 knockdown (Figure 7K).
DISCUSSION
Unlike other GPXs, GPX4 can catalyze the reduction of lipid peroxides in a complex cellular membrane environment (Brigelius-Flohé and Maiorino, 2013). Systemic deletion of Gpx4 in mice causes embryonic lethality, which was not observed when other Gpx genes were deleted (Ran et al., 2004), suggesting a unique role for Gpx4 in physiology. Four groups independently created conditional Gpx4 knockout mice and analyzed the cell death mechanisms after Gpx4 inhibition (Seiler et al., 2008; Sengupta et al., 2013; Ueta et al., 2012; Yoo et al., 2012). Lipid peroxidation was observed in all knockout models, highlighting the importance of Gpx4 for protecting cells from detrimental effects of lipid peroxides. Systemic deletion of Gpx4 was lethal to mice partly due to the nervous system damage caused by neuronal loss (Seiler et al., 2008; Yoo et al., 2012).
In models in which Gpx4 deletion caused lethality or cell loss, it is likely that ferroptosis had occurred. Indeed, mouse embryonic fibroblasts (MEFs) from conditional Gpx4 knockout mice died with lipid peroxide generation upon Gpx4 deletion. Supplementing Vit. E to these MEFs in culture rescued cell death (Seiler et al., 2008).
Elevation of lipid peroxidation upon GPX4 inhibition raises the question of the source of lipid peroxides in normal cellular physiology. Cellular iron may be the most important factor in lipid peroxide generation during ferroptosis. Indeed, iron chelators suppress ferroptosis (Yang and Stockwell, 2008a). Some onco-genes, including RAS and MYC, are known to alter iron metabolism by increasing iron abundance in cells and promoting transformation (Kakhlon et al., 2002;O'Donnell et al., 2006).
In summary, we have determined that Gpx4 is a central regulator of ferroptosis and that ferroptosis can be induced in mouse tumor xenografts, providing a possible therapeutic application of ferroptosis-inducing compounds. See the Extended Discussion for more information.
EXPERIMENTAL PROCEDURES
Metabolite Profiling
Two million HT-1080 cells were seeded in 10 cm culture dishes. The next day, cells were treated with 5 μg/ml erastin and incubated for 5 hr before metabolite extraction. A total of 4 ml of cold 80% methanol was added to the cell mono-layer to extract polar metabolites using a cell scraper. The cell lysate/methanol mixture was transferred to a 15 ml tube and centrifuged at 2,000 × g at 4°C for 10 min to pellet debris and proteins. The supernatant was transferred to a new tube and stored at −80°C for LC-MS/MS analysis. For lipid extract preparation, 3 ml of cold 100% isopropanol was added to the cell monolayer to scrape cells. The resulting cell lysate/isopropanol mixture was transferred to a new 15 ml tube and centrifuged at 2,000 × g at 4°C for 10 min. The cleared super-natant was transferred to a new tube and stored at −20°C for LC-MS/MS analysis.
RSL3 Target Identification
A total of 1.5 million cells were seeded into T225 flasks (Corning) 3 days before treatment in order to be confluent on the day of treatment. Three days later, 20 flasks were washed with PBS three times to remove serum proteins from the media and then treated with either 0.5 μM (1S, 3R)-RSL3-fluorescein probe (“active probe”) or 0.5 μM (1R, 3R)-RSL3-fluorescein probe (“inactive probe”) in serum-free media (DMEM) for 2 hr. For the competitor-treated samples, 20 flasks of cells were treated with 0.5 μM (1S, 3R)-RSL3 (“competitor”) for 20 min prior to treatment with 0.5 μM active probe for 2 hr. (The active and inactive probe-treated cells were treated with the equivalent amount of vehicle for the same time period of the competitor pretreatment.)
After treatment, the cells were washed once with PBS, trypsinized, and pelleted at 1,000 rpm. Following trypsinization, all the following steps were performed at 4°C. The pelleted cells were again washed in PBS twice to remove serum proteins added during the trypsinization. Cells were resuspended in nondenaturing lysis buffer for 20 min (50 mM HEPES, 40 mM NaCl, 2 mM EDTA, 0.5% Triton X-100, 1.5 mM Na3VO4, 50 mM NaF, 10 mM Na-pyrophosphate, 10 mM Na β-glycerophosphate, and Roche protease inhibitor tablet).
The resulting lysate was centrifuged at 12,000 × g for 15 min to pellet out insoluble materials, and the supernatant was removed. Protein concentration was determined using the Bradford assay (Bio-Rad). Subsequent pull-down and proteomics procedures are described in the Extended Experimental Procedures.
In Vivo Xenograft Mouse Study
Athymic nude mice (8 weeks; Charles River Laboratories) were injected with four million HT-1080 cells s.c. The next day, 400 μl of vehicle (0.625% DMSO/99.375% HBSS [pH 2]) or 40 mg/kg PE was delivered to the s.c. site where cancer cells were injected. Two days later, the s.c. injection was repeated. Three days later, 300 μl of vehicle or 30 mg/kg PE was administered to the mice through tail vein. Tail vein injection was repeated three more times, once every other day before the final tumor size was measured in both groups. The animal protocols containing all the procedures were approved by Columbia University's IACUC.
Statistical Analysis
All statistical analyses were performed by using Prism 6 (GraphPad Software).
Supplementary Material
ACKNOWLEDGMENTS
We thank the Broad Institute MLPCN team and GNF for assistance in screening compounds, Alan Epstein, Riccardo Dalla-Favera, Andrew Kung, James Doroshow, and the Columbia Genome Center for providing cell lines, Siu-hong Ho for help with flow cytometry, the Columbia NYSTEM Chemical Probe Synthesis Facility (NYSTEM contract No. C026715) for synthesis of RSL3 analogs, and Dr. John Decatur and the Columbia Chemistry NMR core facility (NSF grant CHE 0840451 and NIH grant 1S10RR025431-01A1). NCI60 cell line testing and some in vivo toxicity testing were carried out through NCI/DTP. We thank Ulshulaa Dholakia and Beverly Shelton for assistance with the animal studies and Terumo Miyazawa for sharing an authentic PC-OOH standard. We thank Scott Geromanos, James Langridge, LeRoy Martin, Martha Stapels, and Johannes PC Vissers (Waters) for their support and advice on mass spectrometry data collection and analysis, and Ryan Colligan for technical assistance with mass spectrometry data processing. The cancer cell line profiling was supported by the NCI's Cancer Target Discovery and Development Network (RC2-CA148399, awarded to S.L.S.). S.L.S. is an investigator at the Howard Hughes Medical Institute. A.W.G. was supported by NIH grant 5R01-CA70823-15. This research was supported by grants to B.R.S. from the US National Institutes of Health (5R01CA097061, 5R01GM085081, and R01CA161061), the Arnold and Mabel Beckman Foundation, and NYSTAR. B.R.S. is an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Discussion, Extended Experimental Procedures, seven figures, one data file, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.12.010.
REFERENCES
- Basu A, Bodycombe NE, Cheah JH, Price EV, Liu K, Schaefer GI, Ebright RY, Stewart ML, Ito D, Wang S, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–1161. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigelius-Flohé R, Maiorino M. Glutathione peroxidases. Biochim. Biophys. Acta. 2013;1830:3289–3303. doi: 10.1016/j.bbagen.2012.11.020. [DOI] [PubMed] [Google Scholar]
- Cotter TG. Apoptosis and cancer: the genesis of a research field. Nat. Rev. Cancer. 2009;9:501–507. doi: 10.1038/nrc2663. [DOI] [PubMed] [Google Scholar]
- Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N, Cuny GD, Mitchison TJ, Moskowitz MA, Yuan J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- Fanélus I, Desrosiers RR. Reactive oxygen species generated by thiol-modifying phenylarsine oxide stimulate the expression of protein L-isoaspartyl methyltransferase. Biochem. Biophys. Res. Commun. 2008;371:203–208. doi: 10.1016/j.bbrc.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry WS, Fulda S, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nat. Rev. Cancer. 2003;3:276–285. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- Irani K, Xia Y, Zweier JL, Sollott SJ, Der CJ, Fearon ER, Sundaresan M, Finkel T, Goldschmidt-Clermont PJ. Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science. 1997;275:1649–1652. doi: 10.1126/science.275.5306.1649. [DOI] [PubMed] [Google Scholar]
- Kakhlon O, Gruenbaum Y, Cabantchik ZI. Repression of ferritin expression modulates cell responsiveness to H-ras-induced growth. Biochem. Soc. Trans. 2002;30:777–780. doi: 10.1042/bst0300777. [DOI] [PubMed] [Google Scholar]
- Kang YJ, Enger MD. Buthionine sulfoximine-induced cytostasis does not correlate with glutathione depletion. Am. J. Physiol. 1992;262:C122–C127. doi: 10.1152/ajpcell.1992.262.1.C122. [DOI] [PubMed] [Google Scholar]
- Kriska T, Girotti AW. A thin layer chromatographic method for determining the enzymatic activity of peroxidases catalyzing the two-electron reduction of lipid hydroperoxides. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2005;827:58–64. doi: 10.1016/j.jchromb.2005.03.045. [DOI] [PubMed] [Google Scholar]
- Kumagai T, Matsukawa N, Kaneko Y, Kusumi Y, Mitsumata M, Uchida K. A lipid peroxidation-derived inflammatory mediator: identification of 4-hydroxy-2-nonenal as a potential inducer of cyclooxygenase-2 in macrophages. J. Biol. Chem. 2004;279:48389–48396. doi: 10.1074/jbc.M409935200. [DOI] [PubMed] [Google Scholar]
- Mackinnon AL, Taunton J. Target identification by diazirine photo-cross-linking and click chemistry. Curr. Protoc. Chem. Biol. 2009;1:55–73. doi: 10.1002/9780470559277.ch090167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Yu D, Zeller KI, Kim JW, Racke F, Thomas-Tikhonenko A, Dang CV. Activation of transferrin receptor 1 by c-Myc enhances cellular proliferation and tumorigenesis. Mol. Cell. Biol. 2006;26:2373–2386. doi: 10.1128/MCB.26.6.2373-2386.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy S, Steinbrecher UP, Barnett J, Witztum JL, Steinberg D. Essential role of phospholipase A2 activity in endothelial cell-induced modification of low density lipoprotein. Proc. Natl. Acad. Sci. USA. 1985;82:3000–3004. doi: 10.1073/pnas.82.9.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaloza C, Lin L, Lockshin RA, Zakeri Z. Cell death in development: shaping the embryo. Histochem. Cell Biol. 2006;126:149–158. doi: 10.1007/s00418-006-0214-1. [DOI] [PubMed] [Google Scholar]
- Ran Q, Liang H, Gu M, Qi W, Walter CA, Roberts LJ, 2nd, Herman B, Richardson A, Van Remmen H. Transgenic mice overexpressing glutathione peroxidase 4 are protected against oxidative stress-induced apoptosis. J. Biol. Chem. 2004;279:55137–55146. doi: 10.1074/jbc.M410387200. [DOI] [PubMed] [Google Scholar]
- Root DE, Flaherty SP, Kelley BP, Stockwell BR. Biological mechanism profiling using an annotated compound library. Chem. Biol. 2003;10:881–892. doi: 10.1016/j.chembiol.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Seiler A, Schneider M, Förster H, Roth S, Wirth EK, Culmsee C, Plesnila N, Kremmer E, Rådmark O, Wurst W, et al. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab. 2008;8:237–248. doi: 10.1016/j.cmet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Sengupta A, Lichti UF, Carlson BA, Cataisson C, Ryscavage AO, Mikulec C, Conrad M, Fischer SM, Hatfield DL, Yuspa SH. Targeted disruption of glutathione peroxidase 4 in mouse skin epithelial cells impairs postnatal hair follicle morphogenesis that is partially rescued through inhibition of COX-2. J. Invest. Dermatol. 2013;133:1731–1741. doi: 10.1038/jid.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- Szatrowski TP, Nathan CF. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 1991;51:794–798. [PubMed] [Google Scholar]
- Trachootham D, Zhou Y, Zhang H, Demizu Y, Chen Z, Pelicano H, Chiao PJ, Achanta G, Arlinghaus RB, Liu J, Huang P. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell. 2006;10:241–252. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Ueta T, Inoue T, Furukawa T, Tamaki Y, Nakagawa Y, Imai H, Yanagi Y. Glutathione peroxidase 4 is required for maturation of photo-receptor cells. J. Biol. Chem. 2012;287:7675–7682. doi: 10.1074/jbc.M111.335174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weïwer M, Bittker JA, Lewis TA, Shimada K, Yang WS, MacPherson L, Dandapani S, Palmer M, Stockwell BR, Schreiber SL, Munoz B. Development of small-molecule probes that selectively kill cells induced to express mutant RAS. Bioorg. Med. Chem. Lett. 2012;22:1822–1826. doi: 10.1016/j.bmcl.2011.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpaw AJ, Shimada K, Skouta R, Welsch ME, Akavia UD, Pe’er D, Shaik F, Bulinski JC, Stockwell BR. Modulatory profiling identifies mechanisms of small molecule-induced cell death. Proc. Natl. Acad. Sci. USA. 2011;108:E771–E780. doi: 10.1073/pnas.1106149108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagoda N, von Rechenberg M, Zaganjor E, Bauer AJ, Yang WS, Fridman DJ, Wolpaw AJ, Smukste I, Peltier JM, Boniface JJ, et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:864–868. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem. Biol. 2008a;15:234–245. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, Shimada K, Delva D, Patel M, Ode E, Skouta R, Stock-well BR. Identification of simple compounds with microtubule-binding activity that inhibit cancer cell growth with high potency. ACS Med. Chem. Lett. 2012;3:35–38. doi: 10.1021/ml200195s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SE, Chen L, Na R, Liu Y, Rios C, Van Remmen H, Richardson A, Ran Q. Gpx4 ablation in adult mice results in a lethal phenotype accompanied by neuronal loss in brain. Free Radic. Biol. Med. 2012;52:1820–1827. doi: 10.1016/j.freeradbiomed.2012.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.