Summary
In embryonic stem (ES) cells and in early mouse embryos, the transcription factor Oct4 is an essential regulator of pluripotency. Oct4 transcriptional targets have been described in ES cell lines; however, the molecular mechanisms by which Oct4 regulates establishment of pluripotency in the epiblast (EPI) have not been fully elucidated. Here we show that neither maternal nor zygotic Oct4 are required for formation of EPI cells in the blastocyst. Rather, Oct4 is first required for development of the primitive endoderm (PE), an extraembryonic lineage. EPI cells promote PE fate in neighboring cells by secreting Fgf4, and Oct4 is required for expression of Fgf4, but we show that Oct4 promotes PE development cell-autonomously, downstream of Fgf4 and Mapk. Finally, we show that Oct4 is required for expression of multiple EPI and PE genes, as well as multiple metabolic pathways essential for the continued growth of the preimplantation embryo.
Introduction
Since ES cells are derived from the blastocyst, understanding how cell fates are established during development provides key lessons for ES cell biology. During formation of the mouse blastocyst, cell fates are selected by regulated cell fate decisions. First, the inner cell mass (ICM) is segregated from the differentiating trophectoderm (TE, future placenta) around E3.0. Subsequently, the ICM is subdivided into the pluripotent epiblast (EPI) and the primitive endoderm (PE, future yolk sac) around E3.75. Recent work has revealed that EPI cells help induce formation of PE cells by secreting Fgf4, which then induces expression of PE genes via Mapk (Chazaud et al., 2006; Guo et al., 2010; Kang et al., 2012; Nichols et al., 2009; Yamanaka et al., 2010). Thus pluripotency genes, such as Nanog, induce PE differentiation non cell-autonomously (Frankenberg et al., 2011; Messerschmidt and Kemler, 2010). Simultaneously, Nanog also represses expression of the PE gene Gata6 cell-autonomously within EPI cells (Frankenberg et al., 2011). Together, these mechanisms produce a ‘salt and pepper’ distribution of EPI and PE cells within the ICM at E3.75. Prior to this time point (E3.5), additional PE genes (Sox17 and Pdgfra) are expressed in a subset of ICM cells, and by E3.75, Gata6 is coexpressed with Sox17, Pdgfra and Gata4 in the PE (Artus et al., 2011; Niakan et al., 2010; Plusa et al., 2008). By the time of implantation, EPI and PE cells will have sorted into distinct groups of cells, and Sox7 is then expressed in PE cells by E4.0 (Artus et al., 2011).
In both the embryo and in ES cells, Oct4 is widely appreciated as an essential pluripotency factor. ES cells cannot be derived from Oct4 null embryos, owing to conversion of ICM to TE fate (Nichols et al., 1998; Niwa et al., 2000). However, not all ICM cells acquire TE gene expression in Oct4 null embryos (Ralston et al., 2010), suggesting that Oct4 may promote pluripotency in vivo by a mechanism distinct from repression of TE. Alternatively, maternal Oct4 could partially compensate for the loss of zygotic Oct4 during cell fate specification in the blastocyst (Foygel et al., 2008). Ultimately, the mechanisms by which Oct4 regulates cell fate specification during blastocyst formation are unclear, as are the Oct4-dependent genes.
Here we show that neither maternal nor zygotic Oct4 is required for expression of Nanog or Sox2, nor for formation of the blastocyst. Rather, zygotic Oct4 is required for PE cell fate. Surprisingly, the mechanism by which Oct4 promotes PE fate differs from the mechanism by which Nanog promotes PE fate. While Nanog induces PE fate non cell-autonomously, upstream of Fgf4 (Frankenberg et al., 2011; Messerschmidt and Kemler, 2010), we show that Oct4 promotes PE gene expression cell-autonomously, and is required for Fgf4/Mapk to activate expression of PE genes. Finally, by transcriptome analysis, we identify pluripotency genes whose expression is dependent on Oct4 in the blastocyst, including Esrrb, Klf2, and Zscan10. In addition, we present evidence that the developmental arrest of Oct4 embryos is associated with a failure to transcriptionally activate multiple energetic metabolism pathways, rather than apoptosis.
Results
Oct4 is required to maintain expression of Gata6 and PE cell number
Our prior work indicated that Oct4 is required to repress the TE genes Cdx2 and Gata3 in a subset of ICM cells (Ralston et al., 2010), but it was not clear whether acquisition of TE fate disrupted EPI or PE fate or both. We therefore examined EPI and PE cell fate specification (defined on the basis of Nanog and Gata6 expression) in litters collected from Oct4 zygotic null heterozygous intercrosses around the time that Nanog and Gata6 adopt a mutually exclusive expression pattern in EPI and PE cells (E3.75), and then sort into morphologically discrete groups (E4.0, E4.25). Non-mutant embryos possessed expected average cell numbers (Fig. S1A), indicating that our staging scheme can be directly compared with published analyses of preimplantation development. At E3.75, the expression of Gata6 and Nanog appeared similar between Oct4 null embryos and non-mutant littermates (Fig. 1A). However, starting at E4.0, a decline in Gata6 expression level was apparent in Oct4 null embryos. In non-mutant embryos at E4.0, Gata6-positive and Nanog-positive cells were localized to prospective PE and EPI regions, but in Oct4 null embryos, Gata6 was weakly detectable in only a few cells (Fig. 1B). In non-mutant embryos at E4.25, Gata6-positive and Nanog-positive cells had further sorted into distinct regions within the ICM. In Oct4 null embryos, which had collapsed, Gata6 was no longer detectable, and Nanog-positive cells had not sorted into a discrete EPI layer, but were intermingled with unlabeled cells (Fig. 1C). We conclude that zygotic Oct4 is not required for expression of Nanog in EPI cells, consistent with prior observations (Chambers et al., 2003), and that Oct4 is required to maintain expression of Gata6 in PE cells after E3.75.
Figure 1. Zygotic Oct4 is required for maintenance of Gata6 expression.
A–C) Gata6 and Nanog immunostaining in Oct4 null and non-mutant embryos at indicated stages (white arrowhead = unlabeled cells, dotted line = ICM, bar = 20 μm). D) Average numbers of inside and outside cells in Oct4 null and non-mutant embryos. E) Average proportions of inside and outside cells in Oct4 null and non-mutant embryos examined in panels D–I. F–H) Average numbers of ICM cells labeled by Nanog, Gata6, or neither. I) Average proportions of inside cells labeled with Nanog, Gata6, or neither. J) Average numbers of TUNEL-positive inside and outside cells in Oct4 null and non-mutant embryos. For panels D–K, n = number of embryos examined, error bars = standard deviation (s.d.), p = p-value. See also Figure S1.
Oct4 null embryos were previously reported to arrest around E4.5 (Nichols et al., 1998). Consistent with this, we noted that the average numbers of inside and outside cells were similar between Oct4 null and non-mutant embryos at E3.75 and E4.0, but both were significantly reduced in Oct4 null embryos by E4.25 (p<0.001, Fig. 1D), indicating defects in both ICM and TE development. At E4.25, the inside cell population was disproportionately smaller in Oct4 null than non-mutant embryos (p<0.001, Fig. 1E). We therefore asked whether this was due to a loss of either EPI or PE cells over the course of preimplantation development. By counting Nanog-positive, Gata6-positive, and unlabeled (Gata6-negative/Nanog-negative) cells in Oct4 null and non-mutant littermates, we determined that both the average number and the proportion of Gata6-positive cells progressively decreased in Oct4 null embryos over the course of preimplantation development (Fig. 1G, I). Comparing Oct4 null embryos at E3.75 and E4.25, the decrease in the average number of Gata6-positive cells was significant (p = 0.002), consistent with either a loss of PE cells or their conversion to EPI cell fate. However, we detected an increase in both the average number and the proportion of unlabeled cells in Oct4 null embryos from E3.75–E4.25 (Fig. 1H, I), differing significantly from non-mutant embryos at all time points (Fig. 1I). By contrast, the average number of Nanog-positive cells did not differ significantly between Oct4 null and non-mutant embryos until the latest time point, when the average number of Nanog-positive cells was significantly lower in Oct4 null embryos (p = 0.009, Fig. 1F), suggesting that PE cells do not acquire EPI cell fate in Oct4 null embryos. Since the proportion of Nanog-positive cells was significantly greater in Oct4 null than in wild type ICMs (p<0.001, Fig. 1I), this suggests that PE cells cease to proliferate or die between E4.0 and E4.25. By TUNEL assay, we compared the number of apoptotic cells between Oct4 null and non-mutant littermates, and observed no significant difference in the average number of TUNEL-positive cells in Oct4 null blastocysts at E4.0 or E4.25 compared to non-mutants (Fig. 1J and Fig. S1B). These results indicate that Oct4 is required to maintain expression of Gata6 in the fully expanded blastocyst and to promote proliferation of EPI, PE, and TE cells prior to implantation.
Oct4 is required for expression of multiple PE fate markers
Development of the PE lineage is characterized by the sequential activation of multiple genes. Subsequent to Gata6 expression, Sox17 and Pdgfra are detected starting at E3.5, followed by expression of Gata4, which is initiated at E3.75, and Sox7, which is initiated at E4.0 (Artus et al., 2011; Niakan et al., 2010; Plusa et al., 2008). Interestingly, neither Sox17 nor Pdgfra were detectable above background in Oct4 null embryos at E3.5 or later (Fig. 2A, B and Fig. S2). Low levels of Gata4 were detectable in Oct4 null embryos at E3.75, but Sox7 was not detected in Oct4 null embryos at E4.0 (Fig. 2C, D). These observations indicate that Oct4 promotes expression of some PE genes (Sox17 and Pdgfra) as early as E3.5, in addition to its role promoting other PE genes (Gata6, Gata4, and Sox7) by E3.75–E4.0. Thus Oct4 is required for initial development of PE cell fate, and not simply its maintenance.
Figure 2. Expression of PE genes is disrupted in Oct4 null embryos and is not due to ectopic expression of Cdx2.
A–D) Pdgfra, Sox17, Gata4, and Sox7 immunostaining in Oct4 null and non-mutant embryos at indicated stages. E) Nanog and Cdx2 immunostaining in Oct4 null embryo and non-mutant embryos at E4.0. (Dotted line = ICM, arrow = Cdx2-negative PE cell. In panels A–E white bar = 20 μm. F) Average number of Oct4 null ICM cells expressing Cdx2 or Nanog at E4.0 (n = 10 embryos). Cdx2 was detected in only about half of PE (Nanog-negative) cells, indicating that ectopic Cdx2 cannot account for the loss of PE gene expression in all PE cells in Oct4 null embryos. By comparison, Cdx2 is never detected in non-mutant ICM cells at E4.0 (Strumpf et al., 2005; Ralston et al., 2008; Ralston et al., 2010). See also Figure S2.
We next asked whether PE development is disrupted in Oct4 null embryos because the ICM differentiates into trophoblast (Nichols et al., 1998). We previously showed that Oct4 represses expression of Cdx2 in the ICM (Ralston et al., 2010). If TE fate disrupts PE fate specification, then we expected to detect elevated Cdx2 in all PE cells of Oct4 null embryos at E4.0. To identify presumptive PE cells in the Oct4 null embryos, we used Nanog to label the EPI cells, leaving the presumptive PE cells unlabeled. If acquisition of Cdx2 disrupts PE gene expression, then we would expect to observe an inverse correlation between Nanog and Cdx2 expression in Oct4 null embryos. However, we did not observe an inverse correlation between Nanog and Cdx2 expression in Oct4 null embryos. Rather, Cdx2 was ectopically expressed in both EPI (Nanog-positive) and PE (Nanog-negative) populations with equal frequency (Fig. 2E, F; p=0.35, Fischer’s test) in Oct4 null embryos. Moreover, Cdx2 was only detected in about half of Oct4 null PE cells (Fig. 2F). Therefore, acquisition of Cdx2 is not correlated with loss of PE gene expression in Oct4 null embryos. We conclude that Oct4 promotes expression of PE genes through a Cdx2-independent mechanism.
Maternal Oct4 is dispensable for development
We next asked whether the initial expression of Gata6 and Nanog depends on maternal Oct4, since Oct4 is present in oocytes (Rosner et al., 1990) and siRNA knockdown of maternal (m) and zygotic (z) Oct4 leads to developmental arrest prior to the blastocyst stage (Foygel et al., 2008). We used the female germline-expressed Zp3-Cre (de Vries et al., 2000) to delete a conditional allele of Oct4 during oogenesis (Fig. 3A). By qPCR, we verified that Oct4 transcript levels were reduced to undetectable levels in oocytes from Oct4 germline-deleted females (Fig. 3B). When Oct4 germline-deleted females were mated to wild type males, live pups lacking m Oct4 were born (24/24 pups genotyped), and litters from these crosses were not significantly smaller than control crosses (10.3+/−1.5 and 7.0+/−0.8 pups/litter, respectively). Since only half of the Oct4 m null pups had inherited the Zp3-Cre transgene (13/24 pups), Oct4 deletion must have occurred during oogenesis. We conclude that m Oct4 is not required for fetal development.
Figure 3. Repression of Nanog and expression of Gata6 is independent of maternal Oct4.
A) Breeding strategy for generating m and mz null Oct4 offspring (fl = floxed; del = deleted). B) qPCR analysis of average Oct4 transcript levels in wild type and Oct4 maternal null oocytes (n = 3 pools of 10 oocytes per genotype). C) Average numbers of inside and outside cells in blastocysts of indicated genotypes. D) Gata6 and Nanog immunostaining in Oct4 m null and Oct4 mz null embryos (bar = 20 μm. E) Average numbers of Gata6 or Nanog-positive cells in blastocysts of indicated genotypes (in panels B, C, and E error bars = s.d.). See also Figure S3.
We next examined whether z Oct4 compensates for loss of m Oct4 by generating Oct4 mz null embryos (Fig. 3A). At E3.75, Oct4 mz null embryos had formed a blastocyst structure, and PCR genotyping confirmed that Oct4 mz null blastocysts were detected within litters at the expected frequency (16 m null: 12 mz null, Fig. 3C). We observed no significant difference in the average numbers of inside and outside cells in Oct4 z null, Oct4 m null, and wild type blastocysts (Fig. 3C). Moreover, Gata6 and Nanog were both still detectable in Oct4 mz null embryos (Fig. 3D), and the numbers of Gata6 and Nanog-expressing cells did not differ significantly between Oct4 mz null and z null embryos (Fig. 3E). We conclude that both blastocyst formation and the initial regulation of Gata6 and Nanog expression are Oct4-independent. In addition, we determined that expression of Sox2, another EPI marker (Aksoy et al., 2013; Guo et al., 2010), was unaffected in Oct4 mz null embryos (Fig. S3). Thus neither m nor z Oct4 are required for formation of the blastocyst, nor for the initial segregation of EPI and PE cell types.
Oct4 is not required for Mapk-dependent regulation of Nanog
Our results showed that neither m nor z Oct4 are required for the salt and pepper regulation of Nanog and Gata6 in the ICM. The salt and pepper regulation of Nanog and Gata6 is known to depend on Fgf4/Mapk signaling, which represses Nanog and promotes Gata6 in PE cells (Nichols et al., 2009; Yamanaka et al., 2010). Since this salt and peppering can occur in the absence of Oct4, we hypothesized that Fgf/Mapk signaling is active in Oct4 null embryos. To test this hypothesis, we treated Oct4 null embryos and non-mutant embryos with inhibitors of Fgfr/Mapk from E2.75–E4.0 (Yamanaka et al., 2010), with the expectation that this treatment should elevate Nanog and repress Gata6 expression if Fgfr/Mapk signaling is intact in Oct4 null embryos. In non-mutant embryos treated with Fgfr/Mapk inhibitors, Nanog was ectopically expressed and Gata6 repressed throughout the ICM as expected (Fig. 4A, B). In Oct4 null embryos treated with Fgfr/Mapk inhibitors, Nanog was also ectopically expressed throughout the ICM (Fig. 4A, B). Thus Fgfr/Mapk signaling regulates Nanog expression in PE cells in an Oct4-independent manner.
Figure 4. Oct4 is required for Fgf/Mapk-dependent expression of PE genes.
A) Nanog and Gata6 immunostaining in Oct4 null embryos cultured from E2.75 to E4.25 in control medium or medium containing Fgfr/Mapk inhibitors. B) Average proportions of inside cells expressing Nanog or Gata6 in Oct4 null and non-mutant embryos following culture in control medium or Fgfr/Mapk inhibitors. D–F) Gata6, Sox17, and Nanog immunostaining in Oct4 null embryos and wild type embryos cultured E2.75–E4.5 in Fgf4/Hep. G–I) Average proportions of ICM cells expressing Gata6, Sox17, or Nanog in Oct4 null and wild type embryos cultured from E2.75 to E4.5 in Fgf4/Hep or control medium. J) Average proportions of Oct4 z null or mz null ICM cells in which Sox17 or Nanog is weakly detected after culturing in Fgf4/Hep or control medium E2.75–E4.5. Error bars = s.d. See also Figure S4.
Surprisingly, Fgfr/Mapk inhibitors did not repress Gata6 expression in Oct4 null embryos as it did in non-mutant embryos. In fact, both the intensity of Gata6 immunostaining and the average number of Gata6-positive cells appeared elevated in Oct4 null embryos treated with Fgfr/Mapk inhibitors (Fig. 4A, C). This was unexpected, because ectopic Nanog should have repressed Gata6 (Frankenberg et al., 2011; Yamanaka et al., 2010) in the PE cells of inhibitor-treated Oct4 null embryos. Our results therefore suggest that Oct4 participates in Nanog-mediated repression of Gata6. These results are consistent with the proposal that Fgfr/Mapk signaling promotes Gata6 in PE cells by relieving Nanog-mediated repression of Gata6 (Frankenberg et al., 2011). Moreover, we observed Gata6 expression in Oct4 null embryos whether the inhibitor treatment was started at E2.5 or E2.75 (Fig. S4A, B), consistent with the finding that early Gata6 expression is Fgf4-independent (Kang et al., 2012). By contrast, other PE genes (Sox17 and Sox7) were not ectopically expressed in inhibitor-treated Oct4 null embryos (Fig. S4C, D). Rather, Sox17 and Sox7 were repressed in both Oct4 null and non-mutant embryos following treatment with Fgfr/Mapk inhibitors. Together, these data suggest that in Oct4 is not required for Fgfr/Mapk-dependent repression of Nanog, but Oct4 is required for Nanog-dependent repression of Gata6 in the ICM.
Oct4 is required for Fgf4-induced expression of multiple PE genes
Our results suggested that Fgfr/Mapk signaling actively repressed Nanog in PE cells of Oct4 null embryos, in spite of the fact that Fgf4 mRNA levels are lower in Oct4 null embryos (Nichols et al., 1998). However, it was unclear whether the reduced level of Fgf4 mRNA in Oct4 embryos could explain the loss of PE genes, such as Sox17, Sox7, and eventually Gata6. We next tested whether PE gene expression could be rescued in Oct4 null embryos by treatment with exogenous Fgf4. Expression of PE genes is induced in wild type, Fgf4 null and Nanog null embryos cultured in exogenous Fgf4 and Heparin (Fgf4/Hep) (Frankenberg et al., 2011; Kang et al., 2012). In non-mutant embryos, Fgf4/Hep treatment from E2.75 to E4.5 led to ectopic expression of Gata6 and Sox17 throughout the ICM (Fig. 4D, E, G, H), as expected. However, in Oct4 null embryos, Fgf4/Hep treatment failed to expand the expression domains of Gata6 or Sox17 (Fig. 4D, E, G, H). In Oct4 null embryos, Fgf4/Hep treatment induced Sox17 at low levels in 1–2 ICM cells of some embryos (Fig. 4E, H), but this level of PE gene expression was not adequate to facilitate outgrowth of PE cells in cultured Oct4 null blastocyst (Fig. S4E). Notably, Fgf4/Hep treatment was able to induce similar low levels of Sox17 in Oct4 mz null and Oct4 z null embryos (Fig. 4J). Thus moderate levels of Sox17 expression can be induced by high levels of Fgf4/Hep in the complete absence of Oct4, but Oct4 is required for robust activation of PE genes in response to exogenous Fgf4. These results are consistent with Oct4 acting downstream of Fgf4, but indicate that exogenous Fgf4 can induce modest PE gene expression in parallel to the Oct4-dependent pathway that normally activates expression of PE genes.
Finally, we examined expression of Nanog in Fgf4/Hep-treated Oct4 null embryos. In non-mutant embryos, Fgf4/Hep treatment abolished expression of Nanog (Fig. 4F,I), as expected. However, in Oct4 null embryos, the proportion of Nanog-expressing cells was unchanged by Fgf4/Hep treatment. These observations indicate that Oct4 is required for exogenous Fgf4 to repress Nanog in EPI cells, which could explain the failure of exogenous Fgf4 to induce Gata6 expression in Oct4 null EPI cells. However, in Fgf4-treated Oct4 null PE cells, which do not express Nanog, Oct4 is required for expression of Gata6 and Sox17.
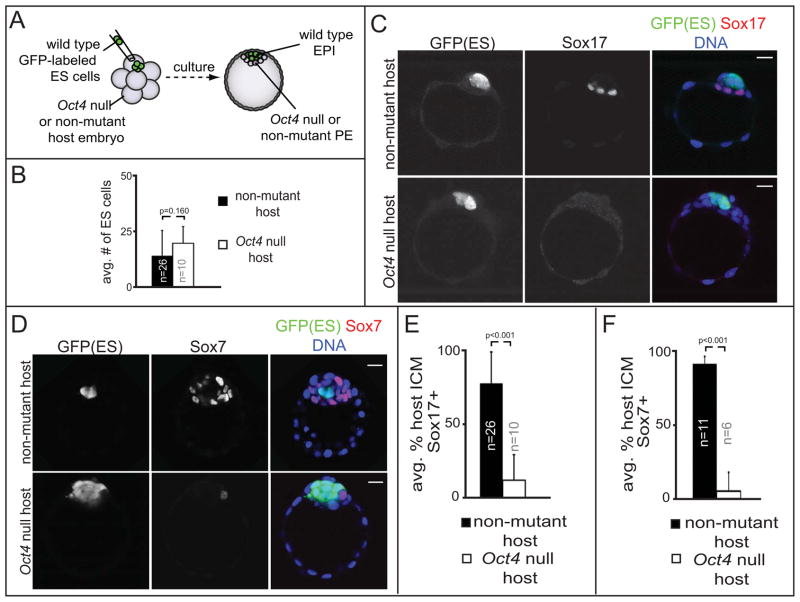
Oct4 is required cell-autonomously for PE gene expression
Our observations that Oct4 is required for ICM cells to respond to exogenous Fgf4, along with the observation that Oct4 is expressed in both PE and EPI cells (Palmieri et al., 1994), suggested that Oct4 acts cell-autonomously in PE cells. To test whether Oct4 is required cell-autonomously in PE cells, we generated chimeric embryos by aggregating fluorescently-labeled, wild type ES cells with Oct4 null or non-mutant host embryos at the precompacted 8-cell stage (E2.5), which leads to almost entirely ES cell-derived fetus (Poueymirou et al., 2007), and examined PE rescue in chimeric embryos at E4.25 (Fig. 5A). We predicted that if Oct4 were required cell-autonomously in PE cells, then wild type ES cells would fail to rescue PE gene expression in Oct4 null chimeras. We first established that the contribution of ES cells to Oct4 null and non-mutant host embryos was equivalent (Fig. 5B). Next, we determined that Sox17 and Sox7 were each strongly induced by ES cells in non-mutant host embryos (Fig. 5C–F). However, ES cells failed to strongly induce either Sox17 or Sox7 in Oct4 null chimeras (Fig. 5C–F). In some Oct4 null chimeras, we detected 1–2 cells expressing Sox17 or Sox7 (6/10 and 1/6 chimeras, respectively) (Fig. 5D–F). Importantly, the low level of Sox17 or Sox7 expression was not correlated with the number of ES cells present in the chimera (Fig. S5A), nor was the low level of Sox17 expression dependent on maternal Oct4, since Sox17 was also induced inefficiently in Oct4 mz null embryos complemented with wild type ES cells (Fig. 5SB). The low levels of PE gene expression in Oct4 null chimeras are consistent with the notion that Fgf4 can induce modest PE gene expression in parallel to the Oct4-dependent pathway. However, full activation of PE gene expression requires Oct4 cell-autonomously, since wild type ES cells failed to rescue the Oct4 null PE phenotype.
Figure 5. Oct4 cell-autonomously promotes expression of PE genes.
A) Schematic of method used to generate chimeric embryos. B) Comparable contribution of ES cells to Oct4 null or non-mutant embryo hosts. C) Sox17 and GFP in control and Oct4 null chimeras. D) Sox7 and GFP in control an Oct4 null chimeras. Note presence of rare Sox7-positive cell in Oct4 null chimeras. F, G) Average proportions of ICM cells in which Sox17 or Sox7 are detected in chimeras generated from Oct4 null or non-mutant host embryos. Error bars = s.d. See also Figure S5.
Oct4 regulates expression of multiple PE and EPI genes
To gain more comprehensive insight into the role of Oct4 in blastocyst formation, we compared the transcriptomes of Oct4 null and wild type embryos by RNA-sequencing individual whole embryos at time points before, during, and after EPI/PE specification (E2.5, E3.75, and E4.5). We first evaluated differences in global gene expression patterns between genotypes at each stage by multidimensional scaling, which showed separation between genotypes for all individuals at each embryonic stage, however, embryos showed tighter clustering within genotypes at E4.5 (Fig. 6A), the stage at which Oct4 null embryos begin to arrest (Nichols et al., 1998). Accordingly, we were able to resolve greater numbers of statistically significant Oct4-dependent genes prior to E4.5 (Fig. 6B and Table S1). Notably, Oct4 was significantly lower in Oct4 null embryos at all stages, and were the levels of Sox17, Pdgfra, and Gata4, consistent with our immunofluorescence analyses, and other PE genes such as Sparc, Lama1, Lamb1, and Lamc1 (Miner et al., 2004; Niakan et al., 2010). We also observed significantly lower levels of the pluripotency genes Esrrb, Klf2, and Zscan10 (Ivanova et al., 2006; Jiang et al., 2008; Martello et al., 2012; Wang et al., 2007; Zhang et al., 2006) in Oct4 null embryos. By contrast, expression levels of TE genes Cdx2, Krt8, and Gata3 were not significantly upregulated in Oct4 null embryos at any stage, although Eomes was significantly upregulated in Oct4 null embryos at E2.5. These observations suggest that our statistical threshold restricted the lists of Oct4-dependent genes to genes exhibiting strong and reproducible expression differences between whole Oct4 null and wild type embryos. In addition, since whole embryos were sequenced, we were unable to discriminate gene expression changes in a lineage-specific manner. Nevertheless, we were able to resolve differences in expression of known Oct4-dependent genes. For example, we observed significantly lower levels in Oct4 null embryos of Fgf4 at E3.75 and Spp1 at E3.75 and E4.5, consistent with prior reports (Botquin et al., 1998; Nichols et al., 1998). Unexpectedly, Gata6 was significantly upregulated at E2.5 and Nanog was significantly upregulated at E3.75 (Fig. 6B), although no difference in Gata6 or Nanog protein levels had been apparent by immunofluorescence in Oct4 null embryos during preimplantation. Curiously, Gata6 levels did not significantly differ in Oct4 null blastocysts, even though differences of Gata6 protein had been apparent. It is interesting to consider the possibility that Gata6 could be post-transcriptionally or post-translationally regulated in the blastocyst.
Figure 6. Oct4 is required for expression of markers of multiple lineages and metabolic pathways.
A) MDS plots of Oct4 null (NULL) and wild type (WT) individuals generated from the top 500 genes, filtered for maximum variation between samples at each developmental stage (as indicated). B) Volcano plots for each stage highlighting key developmental genes (red) and proportion of significantly different, Oct4-dependent genes (above red dotted line, FDR<0.05). C) Venn diagrams showing degree of overlap between E3.75 Oct4-dependent genes and genes bound by Oct4 in F9 cells and F9 treated with RA for 72 hr (Aksoy et al., 2013) (similar degrees of overlap were observed between the ChIP-seq datasets and the E2.5 or E4.5 Oct4-dependent genes (not shown). D) Examples of ChIP-seq data from Aksoy et al. showing locations of Oct4 binding sites (boxed regions) near Pdgfra, Gata4, and Gata6 that exceed threshold set for the analysis shown in panel C. For reference, control (IgG ChIP) and Sox17 ChIP tracks are shown. E) Gene signature analysis showing enrichment scores for lineage-specific query signature gene sets evaluated against target gene signature distributions at each stage (* FDR<0.05). Higher enrichment scores indicate higher relative levels of lineage-specific gene expression (WT versus NULL). F) Representative GSEA plots for major and minor networks shown in panels G, H (from left to right, the first four panels are from E3.75, and the last panel from E4.5). In each panel, the asymmetric distributions of query gene sets (black vertical lines) indicate enrichment with either tail of the ranked distribution of gene expression values (shown WT to NULL, red to blue along the x-axis). The green line shows the enrichment profile, with Enrichment Score (ES) plotted on the y-axis. G, H) Enrichment map networks generated from gene set enrichment analysis results. Nodes represent individual gene sets, and node color corresponds to direction of gene expression difference between the genotypes (as indicated). Green lines indicate shared genes and line thickness is proportional to number of shared genes (TCA = tricarboxylic acid cycle; GPCR = G protein-coupled receptor). See also Figure S6 and Table S1.
We next asked what proportion of the Oct4-dependent genes might be directly transcriptionally regulated by Oct4. Because the number of EPI and PE cells is too limited to perform Oct4 ChIP-seq with blastocysts, stem cell models of pluripotent and PE-like cells have been examined. Oct4 binding sites have been identified in pluripotent F9 embryonal carcinoma (EC) cells and in F9 EC cells that have been differentiated to PE-like cells by treatment with retinoic acid (RA) (Aksoy et al., 2013). We examined intersecting sets of genes bound by Oct4 in untreated and RA-treated F9 cells (Aksoy et al., 2013), with our sets of genes that are Oct4-dependent in vivo (Table S1). This analysis indicated that around 18% of the Oct4-dependent genes identified at E3.75 are bound by Oct4 in EC cells before and/or after differentiation to PE-like cells (Fig. 6C). Notably, Oct4 binding sites were highly enriched for several important PE genes, including Pdgfra, Gata4 and Gata6 in PE-like cells, but not in the EC cells (Fig. 6D), consistent with a role for Oct4 in promoting PE gene expression cell-autonomously. Oct4 binding sites were also detected near other Oct4-dependent genes associated with PE differentiation, such as Tdgf1 and Bmp4 (Kruithof-de Julio et al., 2011; Paca et al., 2012; Plusa et al., 2008) (Fig. 6B and not shown), consistent with Oct4 regulating multiple PE pathways in parallel.
We next used the RNA-seq data to obtain a global view of how Oct4 regulates blastocyst formation. We first performed gene signature analysis to determine how EPI, PE, and TE gene signatures (Table S1) are affected by loss of Oct4. Gene signature analysis revealed that expression of genes associated with all three lineages was progressively lost from Oct4 null embryos, relative to wild type, over the course of development (Fig. 6E and Fig. S6). Curiously, PE and TE gene signatures were initially weakly associated with Oct4 null embryos at E2.5, consistent with the observed transient upregulation of Foxa2, Gata6 and Eomes in the early time point (Fig. 6B). However, the Oct4 null transcriptional profile was generally decreased at E4.5, relative to wild type (Fig. 6B), consistent with an overall developmental failure of Oct4 null embryos, rather than conversion of Oct4 null embryos to any one lineage.
We next focused on how Oct4 regulates pathways, rather than individual genes, during preimplantation development. We performed Gene Set Enrichment Analysis (GSEA) of functional pathways (KEGG, Reactome, Biocarta) to identify pathways with significantly associated gene expression profiles in Oct4 null or wild type embryos, and then visualized relationships among the significant gene sets by generating enrichment maps (Fig. 6F–H and Table S1). At E2.5, we failed to identify any strong, concordant patterns of transcriptional differences between wild type and Oct4 null embryos. However, at E3.75 and E4.25, multiple pathways were significantly disrupted in Oct4 null embryos (Fig. 6G, H). At E3.75, the strongest patterns to emerge indicated a global failure of Oct4 null embryos to upregulate oxidative phosphorylation, glycolysis, and other metabolic pathways, with concomitant aberrant upregulation of lipid metabolism and proteolytic pathways, especially those associated with promoting cell cycle progression and mRNA stability (Fig. 6F, G). This trend continued at E4.5 (Fig. 6H), which could explain the observed global decrease in transcription in Oct4 null embryos by E4.5 (Fig. 6B). In addition at E4.5, we observed an increase in proteolytic pathways associated with DNA synthesis in Oct4 null embryos (Fig. 6H), consistent with stalled cell proliferation. These results are also consistent with a failure to activate known preimplantation energetic and metabolic transcriptional programs (Leese, 2012) in the absence of Oct4, leading to an energy-starved phenotype, manifest by the misregulation of multiple basic cell biological processes and ultimately growth arrest. Consistent with this proposal, the initial increase in protein translation pathways observed at E3.75 was downregulated by E4.5. Interestingly, by E4.5, we observed a weak but significant increase in the Yap/Taz pathway, a pathway that promotes TE development (Nishioka et al., 2009). Notably, several pathway genes contained Oct4 binding sites in RA-treated F9 EC cells, including Itga3, Glud1, Eif4a2, Anapc10, Hdac2 and Tead4 (Aksoy et al., 2013), which are members of Cytoskeletal regulation, Amino acid metabolism, Protein translation, Proteasome/Cell cycle regulation, TCA/Oxidative phosphorylation, and Yap/Taz pathway clusters, respectively (Fig. 6G–H and Table S1). These observations suggest that Oct4 could regulate some pathway members cell-autonomously. Taken together, our RNA-seq analysis is consistent with a model in which Oct4 regulates expression of multiple EPI and PE genes, as well as pathways essential for the continued proliferation of the preimplantation embryo.
Discussion
Our studies point to a novel cell-autonomous role for zygotic Oct4 in PE development. Although Oct4 is often thought of as a pluripotency factor, the idea that Oct4 has multiple developmental roles is supported by precedent in the literature. First, Oct4 is detected in both EPI and PE compartments of the blastocyst at equal levels (Guo et al., 2010; Palmieri et al., 1994). Second, Oct4 orthologues are required for definitive endoderm development in zebrafish (Onichtchouk, 2011). Third, Oct4 participates in endoderm formation in stem cell models (Aksoy et al., 2013; Gore et al., 2011; Niwa et al., 2000; Stefanovic et al., 2009; Teo et al., 2011; Zeineddine et al., 2006). Surprisingly, we did not observe a requirement for m Oct4, in contrast with the reported preimplantation knockdown phenotype (Foygel et al., 2008). Rather, our data indicate that z Oct4 promotes expression of multiple PE genes and pluripotency genes (Fig. 7A, B). Although Oct4 is required for expression of Nanog and Sox2 in ES cells (Kuroda et al., 2005; Loh et al., 2006; Tomioka et al., 2002), we found that Oct4 is not required for the initial expression of these genes in the blastocyst. Thus genetic interactions in ES cells may be maintenance, rather than initiation phases of gene regulation when compared to the blastocyst. We also showed that Oct4 simultaneously promotes expression of multiple PE genes. Knockout of Gata6, Gata4, Sox17, or Sox7 individually does not disrupt PE development prior to implantation (Artus et al., 2011; Kanai-Azuma et al., 2002; Koutsourakis et al., 1999; Kuo et al., 1997; Molkentin et al., 1997; Wat et al., 2012), consistent with genetic redundancy among PE transcription factors. Yet the Oct4 null phenotype is stronger than loss of any of these PE transcription factors. Accordingly, Sox17 is not required for initial expression of Pdgfra, Sox7, and Gata4 (Artus et al., 2011), and so loss of Sox17 alone cannot account for loss of these genes in Oct4 null embryos. Thus Oct4 regulates multiple PE genes in parallel, possibly by direct transcriptional activation.
Figure 7. Summary of Oct4 null phenotype and working model.
A) In normal embryos, Oct4 promotes survival of all three blastocyst lineages, facilitating transcription of components of multiple metabolic pathways, as well as lineage-specific, cell fate-determining transcription factors. B) In EPI cells, Oct4 induces expression of multiple genes, including signaling molecules such as Fgf4 that induce PE cell fate non cell-autonomously. In PE cells, Oct4 normally acts downstream of Mapk to promote expression of Sox17, Pdgfra, Gata4, Sox7, and eventually Gata6. In parallel, Mapk represses Nanog in an Oct4-independent manner in PE cells. Also in parallel, Oct4 represses expression of Cdx2 and other TE genes in all ICM cells.
Our data support a cell-autonomous role for Oct4 in PE development, but our model does not rule out an additional role for Oct4 in promoting PE development non cell-autonomously. Fgf4 is reduced in Oct4 null blastocysts, and we showed that treatment with exogenous Fgf4/Hep or wild type ES cells was sometimes able to induce low levels of some PE genes in a subset of cells, suggesting that Fgf4/Mapk signaling can act in parallel to Oct4 to induce some PE genes or that ICM cells are heterogeneously dependent on Oct4 for Fgf4-induced PE gene expression. Heterogeneity among PE cells is known (Takaoka et al., 2006), but it is not known whether Oct4 plays different roles within ICM populations. Ultimately, exogenous Fgf4/Hep failed to rescue most PE gene expression and outgrowth of PE cells from Oct4 null embryos, underscoring the essential cell-autonomous role for Oct4 in PE cell development.
A remaining question is, what do Oct4 null PE cells become? We find no evidence for increased apoptosis in Oct4 null embryos, and our data indicate that Oct4 null PE cells do not acquire EPI fate. We find that some, but not all, Oct4 null PE cells acquire Cdx2 expression. However, all three lineages are developmentally compromised in Oct4 null embryos, which are deficient in transcription of oxidative phosphorylation and glycolysis pathways that are especially active in TE cells (Houghton, 2006; Leese, 2012). These results point to stalled proliferation in Oct4 null embryos, and we did not observe an increase in PE differentiation genes in Oct4 null embryos by RNA-seq (Paca et al., 2012). TE proliferation can be rescued in Oct4 null ICMs by exogenous Fgf4 (Nichols et al., 1998), but it is not clear whether all ICM cells are rescued by this treatment. Future work to elucidate the developmental potential of Oct4 null ICM cells on a cell by cell level will be particularly important, given the requirement for Oct4 in repressing TE fate in ES cells (Niwa et al., 2000), and our limited understanding of the cellular origin of ES cells within the ICM.
Finally, our data suggest that Oct4 activity is regulated in a cell type-dependent manner, to cell-autonomously regulate expression of EPI or PE pathways. Several mechanisms could regulate cell type-dependent Oct4 activity. First, Oct4 activity could be regulated by phosphorylation. In support of this model, our data show that Oct4 acts downstream of Mapk, and Mapk-dependent Oct4 phosphorylation has been observed in human ES cells (Brumbaugh et al., 2012). Moreover, phosphorylation of Oct4 can alter its transcriptional activity in mouse ES cells (Saxe et al., 2009). Whether Oct4 phosphorylation contributes to lineage specification in the blastocyst is not known. Second, Oct4 nuclear export dynamics differ between presumptive EPI and PE populations (Plachta et al., 2011), and could regulate Oct4 activity in a cell type-dependent manner. However, the mechanisms regulating Oct4 import/export and the functional importance of these differences are unresolved. Finally, Oct4 transcriptional activity could be regulated by cell type-specific cofactors. For example, Oct4 could partner with Sox2 in EPI cells and with Sox17 or Sox7 in PE cells. Notably, Oct4 can bind to Sox17 in vitro, and this changes the Oct4 DNA binding pattern (Aksoy et al., 2013; Jauch et al., 2011; Ng et al., 2012). We showed here that Sox17 expression is itself dependent on Oct4, suggesting that Sox17 participates in a PE-specific feed-forward mechanism to reinforce PE cell fate in the blastocyst. Addressing these models will provide exciting new insight into an important stem cell factor and the establishment of cell lineages in the blastocyst.
Experimental Procedures
Mouse strains and genotyping
The following alleles or transgenes were maintained in an outbred (CD1) background: Pou5f1tm1Scho (Kehler et al., 2004) and Tg(Zp3-cre)93Knw (de Vries et al., 2000). Mice heterozygous for the Pou5f1 deletion allele (Oct4del+) were generated by crossing mice carrying Pou5f1tm1Scho with 129-Alpltm1(cre)Nagy (Lomelí et al., 2000). Mice were genotyped from ear punches (primer sequences provided in Supplemental Experimental Procedures). All animal research was conducted in accordance with the guidelines of the University of California Santa Cruz Institutional Animal Care and Use Committee.
Embryo collection and culture
Mice were maintained on a 12-hour light/dark cycle. Embryos were collected from timed natural matings by flushing dissected oviducts or uteri with M2 medium. Embryos were either fixed or cultured in KSOM (Millipore) at 37°C and 6% CO2. For embryos cultured in 1 μg/ml each Fgf4 recombinant human Fgf4 (R&D Systems) and Heparin (Sigma). For outgrowth assays, embryos were cultured as above until E4.5 and then transferred to individual wells of MEF-conditioned TS medium (Tanaka et al., 1998) with 1 μg/mL each Fgf4/Hep. Final concentrations of Fgfr/Mapk inhibitors were 100 nM PD173074 and 500 nM PD0325901 (Stemgent).
Immunofluorescence and confocal microscopy
Embryos were fixed, stained, imaged, and recovered for genotyping as previously described (Ralston and Rossant, 2008). Optical sections 5 μm in thickness were collected as previously described (Blij et al., 2012). Antibody information is given in the Supplemental Experimental Procedures.
Embryo and ES cell aggregation
YFP-expressing R1 ES cells (George, 2007) were cultured on MEFs in ES cell medium + 1 μM PD0325901 + 3 μM GSK3 inhibitor Chir99021 (Stemgent) (Nichols et al., 2009). Pre-compacted 4–8 cell embryos were collected from Oct4fl/del intercrosses, zonas pellucida removed with Tyrode’s Solution, and embryos aggregated with groups of 3–5 ES cells in depression wells. Aggregations were cultured in KSOM under light mineral oil at 37°C and 6% CO2. Chimeras were subsequently genotyped by PCR using primers that could distinguish wild type, floxed, and deleted Oct4 alleles (see Supplemental Experimental Procedures).
RNA isolation and cDNA preparation
Embryos were collected from natural timed matings and transferred to 50 μl Picopure extraction buffer (Acturus Biosciences). RNA isolation was carried out according to manufacturers instructions. Single blastocyst qPCR was performed as previously described (Blij et al., 2012). For each embryo examined, approximately 1kb transcripts were selectively amplified from the 3′ ends to produce four independent cDNA libraries, which were then pooled prior to sequencing (Kurimoto et al., 2007). Sequencing was performed on an Applied Biosystems SOLiD4 sequencing platform. Read depth for each embryo is provided in Table S1.
Analysis of RNA sequence data
Detailed procedures for statistical analysis of RNA-seq data, gene signature analysis, GSEA, pathway analysis, and enrichment map analysis and ChIP-seq analysis are provided in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Maternal Oct4 is dispensable for development
Oct4 promotes primitive endoderm and represses trophectoderm fates in parallel
Oct4 acts upstream of Fgf4 in epiblast and downstream of Mapk in primitive endoderm
Oct4 is required for expression of metabolic pathway genes in the blastocyst
Acknowledgments
We thank Stephanie Blij, Anthony Parenti, Daniel Carlin, Dr. Yojiro Yamanaka, and Dr. Brian J. Cox for comments on the manuscript. T.F. was supported by NIH T32 GM008646. M.A.H. was supported by CIRM TG2-01157. This study was supported by UCSC COR grants to A.R. and CIRM FA1-00617-1 major facilities grant to UCSC, and support from the Agency for Science, Technology, and Research (A*STAR) Singapore to P.R. We thank Lane Sharon, Tiffany Medina, and Eryn Wicklow for assistance with mouse breeding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aksoy I, Jauch R, Chen J, Dyla M, Divakar U, Bogu GK, Teo R, Leng NgCK, Herath W, Lili S, et al. Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm. EMBO J. 2013 doi: 10.1038/emboj.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus J, Piliszek A, Hadjantonakis AK. The primitive endoderm lineage of the mouse blastocyst: sequential transcription factor activation and regulation of differentiation by Sox17. Dev Biol. 2011;350:393–404. doi: 10.1016/j.ydbio.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blij S, Frum T, Akyol A, Fearon E, Ralston A. Maternal Cdx2 is dispensable for mouse development. Development. 2012 doi: 10.1242/dev.086025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botquin V, Hess H, Fuhrmann G, Anastassiadis C, Gross MK, Vriend G, Schöler HR. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev. 1998;12:2073–2090. doi: 10.1101/gad.12.13.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh J, Hou Z, Russell JD, Howden SE, Yu P, Ledvina AR, Coon JJ, Thomson JA. Phosphorylation regulates human OCT4. Proc Natl Acad Sci U S A. 2012;109:7162–7168. doi: 10.1073/pnas.1203874109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- de Vries WN, Binns LT, Fancher KS, Dean J, Moore R, Kemler R, Knowles BB. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis. 2000;26:110–112. [PubMed] [Google Scholar]
- Foygel K, Choi B, Jun S, Leong DE, Lee A, Wong CC, Zuo E, Eckart M, Reijo Pera RA, Wong WH, et al. A novel and critical role for Oct4 as a regulator of the maternal-embryonic transition. PLoS One. 2008;3:e4109. doi: 10.1371/journal.pone.0004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenberg S, Gerbe F, Bessonnard S, Belville C, Pouchin P, Bardot O, Chazaud C. Primitive Endoderm Differentiates via a Three-Step Mechanism Involving Nanog and RTK Signaling. Dev Cell. 2011;21:1005–1013. doi: 10.1016/j.devcel.2011.10.019. [DOI] [PubMed] [Google Scholar]
- Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong G, Wang C, Li Sun L, Clarke N, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Houghton FD. Energy metabolism of the inner cell mass and trophectoderm of the mouse blastocyst. Differentiation. 2006;74:11–18. doi: 10.1111/j.1432-0436.2006.00052.x. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Jauch R, Aksoy I, Hutchins AP, Ng CK, Tian XF, Chen J, Palasingam P, Robson P, Stanton LW, Kolatkar PR. Conversion of Sox17 into a pluripotency reprogramming factor by reengineering its association with Oct4 on DNA. Stem Cells. 2011;29:940–951. doi: 10.1002/stem.639. [DOI] [PubMed] [Google Scholar]
- Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Kanai-Azuma M, Kanai Y, Gad JM, Tajima Y, Taya C, Kurohmaru M, Sanai Y, Yonekawa H, Yazaki K, Tam PP, et al. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development. 2002;129:2367–2379. doi: 10.1242/dev.129.10.2367. [DOI] [PubMed] [Google Scholar]
- Kang M, Piliszek A, Artus J, Hadjantonakis AK. FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development. 2012 doi: 10.1242/dev.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126:723–732. [PubMed] [Google Scholar]
- Kruithof-de Julio M, Alvarez MJ, Galli A, Chu J, Price SM, Califano A, Shen MM. Regulation of extra-embryonic endoderm stem cell differentiation by Nodal and Cripto signaling. Development. 2011;138:3885–3895. doi: 10.1242/dev.065656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Saitou M. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat Protoc. 2007;2:739–752. doi: 10.1038/nprot.2007.79. [DOI] [PubMed] [Google Scholar]
- Kuroda T, Tada M, Kubota H, Kimura H, Hatano S, Suemori H, Nakatsuji N, Tada T. Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol Cell Biol. 2005;25:2475–2485. doi: 10.1128/MCB.25.6.2475-2485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leese HJ. Metabolism of the preimplantation embryo: 40 years on. Reproduction. 2012;143:417–427. doi: 10.1530/REP-11-0484. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Lomelí H, Ramos-Mejía V, Gertsenstein M, Lobe CG, Nagy A. Targeted insertion of Cre recombinase into the TNAP gene: excision in primordial germ cells. Genesis. 2000;26:116–117. [PubMed] [Google Scholar]
- Martello G, Sugimoto T, Diamanti E, Joshi A, Hannah R, Ohtsuka S, Göttgens B, Niwa H, Smith A. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell. 2012;11:491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, Kemler R. Nanog is required for primitive endoderm formation through a non-cell autonomous mechanism. Developmental Biology. 2010 doi: 10.1016/j.ydbio.2010.04.020. [DOI] [PubMed] [Google Scholar]
- Miner JH, Li C, Mudd JL, Go G, Sutherland AE. Compositional and structural requirements for laminin and basement membranes during mouse embryo implantation and gastrulation. Development. 2004;131:2247–2256. doi: 10.1242/dev.01112. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Ng CK, Li NX, Chee S, Prabhakar S, Kolatkar PR, Jauch R. Deciphering the Sox-Oct partner code by quantitative cooperativity measurements. Nucleic Acids Res. 2012;40:4933–4941. doi: 10.1093/nar/gks153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niakan KK, Ji H, Maehr R, Vokes SA, Rodolfa KT, Sherwood RI, Yamaki M, Dimos JT, Chen AE, Melton DA, et al. Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev. 2010;24:312–326. doi: 10.1101/gad.1833510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Silva J, Roode M, Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Schöler H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith A. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Onichtchouk D. Pou5f1/Oct4 in pluripotency control: Insights from zebrafish. Genesis. 2011 doi: 10.1002/dvg.20800. [DOI] [PubMed] [Google Scholar]
- Paca A, Séguin CA, Clements M, Ryczko M, Rossant J, Rodriguez TA, Kunath T. BMP signaling induces visceral endoderm differentiation of XEN cells and parietal endoderm. Dev Biol. 2012;361:90–102. doi: 10.1016/j.ydbio.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Palmieri S, Peter W, Hess H, Schöler H. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol. 1994;166:259–267. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- Plachta N, Bollenbach T, Pease S, Fraser SE, Pantazis P. Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat Cell Biol. 2011;13:117–123. doi: 10.1038/ncb2154. [DOI] [PubMed] [Google Scholar]
- Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis A. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development. 2008;135:3081–3091. doi: 10.1242/dev.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poueymirou WT, Auerbach W, Frendewey D, Hickey JF, Escaravage JM, Esau L, Doré AT, Stevens S, Adams NC, Dominguez MG, et al. F0 generation mice fully derived from gene-targeted embryonic stem cells allowing immediate phenotypic analyses. Nat Biotechnol. 2007;25:91–99. doi: 10.1038/nbt1263. [DOI] [PubMed] [Google Scholar]
- Ralston A, Cox B, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper J, Rossant J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137:395–403. doi: 10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- Ralston A, Rossant J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev Biol. 2008;313:614–629. doi: 10.1016/j.ydbio.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Rosner MH, Vigano MA, Ozato K, Timmons PM, Poirier F, Rigby PW, Staudt LM. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature. 1990;345:686–692. doi: 10.1038/345686a0. [DOI] [PubMed] [Google Scholar]
- Saxe JP, Tomilin A, Schöler HR, Plath K, Huang J. Post-translational regulation of Oct4 transcriptional activity. PLoS One. 2009;4:e4467. doi: 10.1371/journal.pone.0004467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S, Abboud N, Désilets S, Nury D, Cowan C, Pucéat M. Interplay of Oct4 with Sox2 and Sox17: a molecular switch from stem cell pluripotency to specifying a cardiac fate. J Cell Biol. 2009;186:665–673. doi: 10.1083/jcb.200901040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka K, Yamamoto M, Shiratori H, Meno C, Rossant J, Saijoh Y, Hamada H. The mouse embryo autonomously acquires anterior-posterior polarity at implantation. Dev Cell. 2006;10:451–459. doi: 10.1016/j.devcel.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Teo AK, Arnold SJ, Trotter MW, Brown S, Ang LT, Chng Z, Robertson EJ, Dunn NR, Vallier L. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011;25:238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka M, Nishimoto M, Miyagi S, Katayanagi T, Fukui N, Niwa H, Muramatsu M, Okuda A. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 2002;30:3202–3213. doi: 10.1093/nar/gkf435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZX, Kueh JL, Teh CH, Rossbach M, Lim L, Li P, Wong KY, Lufkin T, Robson P, Stanton LW. Zfp206 is a transcription factor that controls pluripotency of embryonic stem cells. Stem Cells. 2007;25:2173–2182. doi: 10.1634/stemcells.2007-0085. [DOI] [PubMed] [Google Scholar]
- Wat MJ, Beck TF, Hernández-García A, Yu Z, Veenma D, Garcia M, Holder AM, Wat JJ, Chen Y, Mohila CA, et al. Mouse model reveals the role of SOX7 in the development of congenital diaphragmatic hernia associated with recurrent deletions of 8p23.1. Hum Mol Genet. 2012;21:4115–4125. doi: 10.1093/hmg/dds241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka Y, Lanner F, Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- Zeineddine D, Papadimou E, Chebli K, Gineste M, Liu J, Grey C, Thurig S, Behfar A, Wallace VA, Skerjanc IS, et al. Oct-3/4 dose dependently regulates specification of embryonic stem cells toward a cardiac lineage and early heart development. Dev Cell. 2006;11:535–546. doi: 10.1016/j.devcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Zhang W, Walker E, Tamplin OJ, Rossant J, Stanford WL, Hughes TR. Zfp206 regulates ES cell gene expression and differentiation. Nucleic Acids Res. 2006;34:4780–4790. doi: 10.1093/nar/gkl631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.