Abstract
Malignant peripheral nerve sheath tumors (MPNSTs) are genetically diverse, aggressive sarcomas that occur sporadically or in association with neurofibromatosis type 1 syndrome. Reduced TP53 gene expression and amplification/overexpression of the epidermal growth factor receptor (EGFR) gene occur in MPNST formation. We focused on determining the cooperativity between reduced TP53 expression and EGFR overexpression for Schwann cell transformation in vitro (immortalized human Schwann cells) and MPNST formation in vivo (transgenic mice). Human gene copy number alteration data, microarray expression data, and TMA analysis indicate that TP53 haploinsufficiency and increased EGFR expression co-occur in human MPNST samples. Concurrent modulation of EGFR and TP53 expression in HSC1λ cells significantly increased proliferation and anchorage-independent growth in vitro. Transgenic mice heterozygous for a Trp53-null allele and overexpressing EGFR in Schwann cells had a significant increase in neurofibroma and grade 3 PNST (MPNST) formation compared with single transgenic controls. Histological analysis of tumors identified a significant increase in pAkt expression in grade 3 PNSTs compared with neurofibromas. Array comparative genome hybridization analysis of grade 3 PNSTs identified recurrent focal regions of chromosomal gains with significant enrichment in genes involved in extracellular signal–regulated kinase 5 signaling. Collectively, altered p53 expression cooperates with overexpression of EGFR in Schwann cells to enhance in vitro oncogenic properties and tumorigenesis and progression in vivo.
Malignant peripheral nerve sheath tumors (MPNSTs) are aggressive, malignant tumors of Schwann cell origin that compose approximately 10% of diagnosed soft tissue sarcomas. MPNSTs arise spontaneously or in association with the inherited tumor predisposition syndrome neurofibromatosis type 1 (NF1). Sporadic MPNSTs have a 0.001% incidence in the general population, whereas individuals with NF1 (1 in 3500 people) have an 8% to 13% lifetime risk of developing MPNSTs.1–3 MPNSTs are the most common malignancy in adults with NF1 and the leading cause of NF1-related mortality. Because of the invasive nature and high incidence of metastasis of MPNSTs, surgical resection, radiotherapy, and chemotherapeutic treatments have proved to be ineffective for long-term treatment, resulting in 5-year survival rates of approximately 40%.1,4–6 The severity and lack of adequate treatments for MPNSTs emphasize the need for improved understanding of the genetic basis of these tumors.
Currently, few genetic drivers are implicated in benign neurofibroma formation and further progression into MPNSTs. The most commonly altered known gene is NF1, which encodes the neurofibromin 1 protein, an RAS-GTPase–activating protein that causes NF1 syndrome when inherited in a mutated form.7–10 Mutations in the NF1 gene are also observed in approximately 40% of sporadic MPNSTs.11 Deletion or mutation of the NF1 gene in cells causes increased and aberrant signaling through progrowth and proproliferation signaling pathways [RAS/mitogen-activated protein kinase (MAPK)/extracellular signal–regulated kinase (ERK) and phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR)] in human neurofibromas and MPNST-derived cell lines.12–14 However, NF1 gene loss alone likely is not sufficient for MPNST formation on the basis of results from genetically engineered mouse models (GEMMs).15 Increased expression of growth factor receptors and ligands, such as epidermal growth factor receptor (EGFR), NRG, PDGF, HGF, SCF, and TGFβ1, is also stimulated in neurofibromas and MPNSTs with NF1 mutation.16–21 In addition to NF1 mutations, few genomic aberrations have been identified in neurofibromas.22 However, genomic aberrations, such as copy number alterations (CNAs), commonly occur in MPNSTs, suggesting that progression from benign to malignant tumor formation requires many cooperating genomic alterations.22 Deletions and/or mutations of cell cycle regulators TP53, RB1, and CDKN2A and gene amplification of growth factor receptor genes ERBB2, EGFR, KIT, MET, and PDGFR are identified in human MPNSTs.23–34 However, identification of genetic drivers of MPNST formation is hindered because of the hyperdiploid or near-triploid genomes of MPNSTs.35–42
In addition to NF1 mutations, genetic alterations in TP53 and EGFR genes frequently occur in human MPNSTs. Deletions and/or point mutations of TP53 occur in approximately 75% of human MPNSTs, but rarely inactivate both alleles, suggesting haploinsufficiency is sufficient for MPNST formation.43 Moreover, a GEMM with cis-linked Nf1- and Trp53-null mutations (NPcis mice) rapidly develops sarcomas, including MPNSTs, in which not all tumors undergo loss of heterozygosity (LOH) of the wild-type (WT) Nf1 and Trp53 alleles.44,45 EGFR gene amplification and/or overexpression occur in 25% to 75% of human MPNSTs.25,46–48 Transgenic mice overexpressing human EGFR in Schwann cells and their precursors display a nerve hyperplasia phenotype with features of early-stage neurofibroma pathogenesis and rare incidence of benign neurofibroma formation, but no MPNST.49 Furthermore, inhibition of EGFR signaling in NPcis mice with a hypomorphic allele of EGFR increased survival compared with NPcis mice with intact EGFR signaling.49 Finally, inhibition of EGFR kinase activity in cell culture–based assays reduced migration of MPNST cells.50 These results suggest that aberrant EGFR expression is involved in MPNST progression, but only in the context of other mutations. For example, in human esophageal cancer, EGFR overexpression and TP53 mutations frequently co-occur, and in vitro human esophageal epithelial cells can be transformed by overexpression of WT EGFR, activation of telomerase reverse transcriptase, and reduced TP53 expression by RNA interference.51,52 Anecdotally, a human cell line derived from an NF1-associated MPNST had EGFR gene amplification and deletion of exons 5 to 8 within the TP53 gene.53
Herein, we assessed the cooperativity of WT EGFR overexpression and reduced TP53 expression in a CDK4 and telomerase reverse transcriptase immortalized human Schwann cell line (iHSC1λ) and with GEMMs. HSC1λ cells overexpressing EGFR with reduced TP53 expression have a significant increase in proliferation and anchorage-independent growth, phenotypes characteristic of oncogenic transformation. Transgenic mice heterozygous for Trp53 and overexpressing EGFR in Schwann cells have a significant increase in Schwann cell tumorigenesis compared with single transgenic controls. Schwann cell tumors in these mice histologically resemble human neurofibromas and MPNSTs. Genetic analysis of tumors and tumor-derived cell lines demonstrate frequent loss of the Trp53 WT allele and a high incidence of aneuploidy with CNA gains on chromosomes 4, 5, 8, and 15. Collectively, the data demonstrate cooperativity between EGFR overexpression and p53 haploinsufficiency for Schwann cell tumorigenesis.
Materials and Methods
Gene Expression Data Analysis
Published data from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession number GSE14038; Affymetrix GeneChip HU133 Plus 2.0) were used for gene expression pattern analysis. For gene annotation, custom CDF (custom GeneChip library file), on the basis of reference sequence target definitions (Hs133P REFSEQ version 8, University of Michigan, Ann Arbor, MI), was downloaded and used to provide accurate interpretation of GeneChip data.54
Statistical comparisons were done using R/Bioconductor packages and GeneSpring GX, version 7.3.1 (Agilent Technologies, Santa Clara, CA). Differentially expressed genes were defined as genes with expression levels at least threefold higher or lower in target groups (MPNST) compared with normal human Schwann cells after applying Benjamini and Hochberg55 false-discovery rate correction (P ≤ 0.05).
MPNST Whole-Methylome Data Analysis
Feber et al56 published unbiased whole-methylome data of normal primary human Schwann cells (NHSCs), neurofibromas, and MPNST genomes in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession number GSE21714). We adopted the Feber et al56 method for detecting differentially methylated regions (DMRs) in MPNST compared with NHSC.57 Briefly, Batman methylation scores per 100 bp were averaged for each 1000-bp window. A conservative threshold for DMR calling was used, on the basis of the 95th percentile of the difference in methylation score. DMR regions were mapped to human genome build version hg18 (National Center for Biotechnology Information-36).
The nearest CpG island shores (CpG-IS) to the transcription start sites of each gene/miRNA were scanned.56,57 We defined CpG-IS as areas up to 2 kilobases in distances from CpG islands. We considered the nearest upstream CpG-IS from the transcription start site, within 5000 bp, ranges from each transcription start sites. The genomic coordinates of miRNAs, genes, and CpG islands (National Center for Biotechnology Information 36/hg18) were extracted from corresponding tracks of the University of California, Santa Cruz, Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway, last accessed January 9, 2013). In case of intragenic miRNAs, we assumed that their expression is influenced by the nearest CpG-IS to the transcription start site of their host gene.
CNA Data Analysis
CNA data on 51 primary MPNSTs were from the published GSE33881 data set [Agilent Human Genome CGH Microarray kit (4 × 44,000), http://www.ncbi.nlm.nih.gov].58,59 A circular binary segmentation algorithm was applied to the log2 ratios of intensity values from tumor and normal to reduce local noise effects. Circular binary segmentation calculates a likelihood-ratio statistic for each array probe by permutation to locate change points.60 After the segmentation step, a CGH call algorithm was used to assign each segment an aberration label: gain, loss, or normal.61 A visualization program was written in R (http://www.r-project.org) to present overall gain/loss patterns of all 51 MPNSTs. Genomic coordinates used in plots were based on hg19/GRCh37.p8 (R/Bioconductor biomaRt package; Fred Hutchinson Cancer Research Center, Seattle, WA).62
TMA Data
Representative areas of disease were identified on H&E-stained sections for 30 dermal neurofibromas, 31 plexiform neurofibromas, and 32 MPNSTs. Blocks consisting of duplicate 1.0-mm core samples were constructed with a manual tissue arrayer (Beecher Instruments Inc., Sun Prairie, WI) and limited to 64 cores per recipient block. Unstained tissue microarray (TMA) sections (4 μm thick) were deparaffinized and rehydrated using standard methods. Slides went through antigen retrieval and were incubated in pH 6.0 buffer (Reveal Decloaking reagent; Biocare Medical Inc., Concord, CA) in a steamer for 30 minutes at 95°C to 98°C, followed by a 20-minute cool down period. Slides were rinsed in running tap water, followed by immersion in 1× Tris-buffered saline/0.1% Tween-20 (TBST; pH 7.4). Endogenous peroxidase activity was quenched by slide immersion in 3% hydrogen peroxide solution (Peroxidazed; Biocare Medical Inc.) for 10 minutes, followed by a TBST rinse. A serum-free blocking solution (Background Sniper; Biocare Medical Inc.) was placed on sections for 30 minutes. Blocking solution was removed, and slides were incubated in primary antibody diluted in 10% blocking solution/90% TBST. Anti-EGFR (1:500; Sigma Prestige, St. Louis, MO) was incubated overnight at 4°C, followed by a TBST rinse and detection with Alexa Fluor 488 (1:500; Invitrogen, Grand Island, NY) for 2 hours at room temperature. Next, slides were immersed in TBST. A second application of blocking solution (Background Sniper) was applied for 10 minutes, followed by p53 (1:2000; Dako, Carpinteria, CA), incubated for 60 minutes at room temperature, and rinsed in TBST. Alexa Fluor 555 (1:500; Invitrogen) was applied for 2 hours at room temperature. Slides were rinsed in TBST, followed by distilled water, and coverslipped using ProLong Gold Anti-fade with DAPI (Invitrogen).
Cell Cultures/Assays
Immortalized human Schwann cells (iHSC1λ) were acquired from the laboratory of Dr. Margaret Wallace. iHSC1λ cells were cultured in complete media [1× Dulbecco's modified Eagle's medium (DMEM), 10% fetal bovine serum, and 1× penicillin/streptomycin] and grown at 37°C in 5% CO2. Proliferation assays were set up in a 96-well plate format with 500 cells per well in full DMEM media containing 1 μg/mL of puromycin (Life Technologies, Carlsbad, CA). Proliferation was assessed every 24 hours for 5 days using the CellTiter 96 AQueous One Solution Cell Proliferation assay (Promega, Madison, WI) following the manufacturer's protocols. Cultured cells were plated at 500 cells per well of a 96-well plate in replicates of 10. Measurements were taken on days 0, 1, 2, 3, and 4 using the BioTek Synergy MX automated plate reader (Bio-Tek, Winooski, VT). Experiments were performed in triplicate. Soft agar anchorage-independent colony formation assays were performed as previously described.63 After 2 weeks of growth, cells were fixed in 10% formalin containing 0.005% crystal violet for 1 hour at room temperature. Formalin was removed, and colonies were imaged on a Leica S8 AP0 microscope (Leica Microsystems Inc., Buffalo Grove, IL). Twelve images per cell line were taken, and automated colony counts were done using ImageJ software version 1.48 (NIH, Bethesda, MD).
Cell Culture Constructs
EGFR cDNA (Addgene, Cambridge, MA) and the shTP53 (OpenBiosystems, Pittsburg, PA) were cloned into the Gateway Vector System (Life Technologies) and subcloned into a piggyBac (PB) transposon vector, as previously described.64 The PB control vector contains the Luciferase and Gfp reporter genes. Cells were transfected with 2 μg of EGFR/shTP53, EGFR, shTP53, or Luciferase PB transposon (Supplemental Figure S1A) and 500 ng of PB7 transposase plasmid using the NEON transfection system, following the manufacturers' protocols (Life Technologies). Successfully transfected cells were enriched with 1 μg/mL puromycin. Transcription activator-like effector nucleases (TALENs) were generated against the human TP53 locus using a previously established protocol.65 Briefly, the first coding exon of the TP53 gene was targeted with TALENs to introduce mutations near the translational start (Supplemental Figure S1D). TALEN left (direction refers to the orientation that the TALENs bind relative to the site to be cut), 5′-GGAGGAGCCGCAGTCA-3′; TALEN left RVD sequence, NNNNNINNNNNINNHDHDNNHDNINNNGHDNI; TALEN right, 5′-CCCCCTCTGAGTCAGG-3′; and TALEN right, HDHDNGNNNIHDNGHDNINNNINNNNNNNNNN. TALEN plasmids were transfected into HSC1λ cells with neon transfection (Life Technologies), followed by subsequent single-cell cloning and analysis by a PCR-based assay (CEL-I) and sequencing to identify TALEN-induced mutations. The following CEL-I primers were used: sense, 5′-TGGGTTGTGGTGAAACATTG-3′; antisense, 5′-TCCCACAGGTCTCTGCTAGG-3′.
Generation of Transgenic Animals
Generation of transgenic mice carrying the 3′-cyclic nucleotide 3′-phosphodiesterase gene (Cnp) promoter driving the human EGFR cDNA (Cnp-EGFR) has been previously described.49 Transgenic mouse harboring a conditional Trp53 allele possessing an R270H point mutation in exon 8 has been previously described.66 For our experiments, this was never bred to a Cre-expressing mouse. Therefore, the dominant negative allele was never expressed but used as a marker for the null allele, essentially making every cell in the mouse heterozygous for Trp53. We refer to this allele as Trp53+/− to indicate it is not expressed because of the presence of a floxed stop cassette in intron 1.66 Single transgenic mice were crossed to obtain double transgenic experimental mice possessing one allele of each transgene. All animal work was conducted under an institutionally approved animal welfare protocol.
PCR Genotyping
Genotypes of transgenic mice were determined using a PCR-based approach: First, genomic DNA was isolated from tail clippings using standard proteinase K treatment, phenol-chloroform extraction, and ethanol precipitation. Genomic DNA was resuspended in sterile Tris-EDTA buffer [10 mmol/L Tris-HCl (pH 7.5) and 1 mmol/L EDTA (pH 8)] and quantified using a Nanodrop spectrophotometer (Thermo Scientific, Waltham, MA). PCR genotyping was performed using 100 ng of diluted genomic DNA as template in a 25-μL PCR volume. PCR primers used for Cnp-EGFR were as follows: forward, 5′-TGACATCTCCTCCTCCCTTC-3′; and reverse, 5′-TGCCCAACTGCGTGAGC-3′ (amplicon, 380 bp). Trp53R270H floxed alleles were as follows: WT, 5′-TTACACATCCAGCCTCTGTGG-3′ (forward) and 5′- CTTGGAGACATAGCCACACTG-3′ (reverse); and flox, 5′-AGCTAGCCACCATGGCTTGAGTAAGTCTGCA-3′ (forward) (WT amplicon, 170 bp; and floxed allele amplicon, 270 bp). PCR conditions for ReddyMix (Thermo Scientific, Waltham, MA) were used according to the manufacturer's instructions with an initial denaturing step of 95°C for 2 minutes; 30 to 35 cycles of denaturing at 95°C for 25 seconds, annealing at 55°C for 35 seconds, and extension at 72°C for 65 seconds; followed by a final extension at 72°C for 10 minutes. PCR products were separated on a 1.5% agarose gel and genotype determined by the absence or presence of expected amplicons.
Tumor Analysis
Mice were monitored three times a week for changes in mobility (paralysis), frank tumor development >1 cm, or moribundity. If mice met one of the criteria, they were culled and tumors were carefully removed from the sacrificed animal under a dissecting microscope (Leica Microsystems Inc.), washed, and placed in cold PBS. These separated tumors were split into samples for DNA, RNA, and protein extraction. Tissue samples for RNA were stored at −80°C in RNAlater (Sigma-Aldrich, St. Louis, MO) to prevent RNase contamination and degradation. DNA extraction was done as previously described in the PCR genotyping section. Extraction of RNA was done using the TRIzol reagent (Life Technologies) using protocols described by the manufacturer. Protein extraction was performed using standard isolation techniques. Histological sections were only taken for larger tumors (>2 mm in diameter). Formalin-fixed, paraffin-embedded sections from various tissues were divided into sections (5 μm thick) using a standard microtome (Leica Microsystems Inc.), mounted, and heat fixed onto glass slides. Tissue section slides were either stained with H&E using standard protocols or used for immunohistochemistry (IHC) as described in the next section.
IHC Data
Tissues were fixed in 10% buffered formalin and embedded in paraffin blocks. Sections were cut (5 μm thick) and rehydrated through a series of graded ethanols. Slides underwent antigen retrieval by boiling for 30 minutes in antigen unmasking solution (Vector Laboratories Inc., Burlington, CA). Endogenous peroxidases were quenched with 3% hydrogen peroxide solution for 10 minutes. For antibody staining, a M.O.M. kit (Vector Laboratories Inc.) was used for blocking and antibody incubations. Primary antibodies used were as follows: mouse anti–Ki-67 (1:100; Leica Microsystems Inc.), rabbit anti-S100β (1:100; Santa Cruz Biotechnology, Dallas, TX), rabbit anti-EGFR (1:100; Cell Signaling Technology, Danvers, MA), rabbit anti–phosphorylated (phospho)-EGFR (1:400; Cell Signaling Technology), mouse anti-p53 (1:100; Cell Signaling Technology), mouse anti-p21 (1:100; BD Pharmingen, San Jose, CA), rabbit anti–phospho-Erk (1:100; Cell Signaling Technology), and rabbit anti–phospho-Akt (1:100; Cell Signaling Technology). After a series of washes, slides were incubated with the corresponding anti-mouse biotinylated or anti-rabbit biotinylated secondary antibody (1:250; Vector Laboratories Inc.). Slides were washed, incubated with the vectastain avidin-biotin complex kit (Vector Laboratories Inc.) for 30 minutes at room temperature, washed again, and stained using peroxidase substrate kit diaminobenzidine (Vector Laboratories Inc.). Finally, slides were counterstained with hematoxylin, dehydrated, cleared with citrosolv, and mounted with Permount (Fisher Scientific, Waltham, MA).
Pathological Analysis of Nerve-Associated Tumors
H&E-stained tissue sections were evaluated for degree of cellularity, nuclear atypia, necrosis, hemorrhage, and myxoid background, on the basis of a scale of none, low, medium, or high for each phenotype. Slides immunostained for the proliferative marker Ki-67 were evaluated and graded on the scale of none, low, medium, or high with a subgrading of 0 to 3. Neurofibromas tend to have none to low-grade 1 Ki-67 staining, whereas grade 3 PNSTs had more proliferating cells and a medium- to high-grade 2 to 3 Ki-67 staining index. In addition, mitotic figures were counted on the basis of the H&E stain for one tissue section per tumor. Also, tumors were assessed for the presence of mast cells by toluidine blue staining and scored on the basis of presence or absence of mast cells. S100β IHC was evaluated for negative, focal, or diffuse staining pattern.
Generation of Cell Lines
Large nerve-associated tumors were carefully dissected using aseptic techniques. The tumor was sliced into small pieces, then placed into 1× DMEM containing 2 μg/mL collagenase A. This solution was incubated at 37°C in 5% CO2 for 3 hours to allow dissociation of cells from the bulk tumor. Before plating, the solution was triturated and placed through a 33-μm filter. Cells were plated on 10-cm dishes containing complete media (1× DMEM, 10% fetal bovine serum, and 1× penicillin/streptomycin) and grown at 37°C in 5% CO2. These cell strains were transduced with a lentivirus containing an enhanced green fluorescent protein (eGFP) and luciferase reporter transgene. Successfully transduced cells, as assessed by eGFP expression, were cell sorted using FACSAria (BD Biosciences, San Jose, CA). The top 5% eGFP+ cells and eGFP− cells were collected into 96-well plates. eGFP− clones that grew from the cell sort were assessed by immunofluorescence with rabbit anti-S100β (1:100; Santa Cruz Biotechnology), rabbit anti-human EGFR (1:100; Cell Signaling Technology), and rabbit anti-Olig2 (1:100; Abcam, Cambridge, MA) to demonstrate Schwann cell origin and expression of human EGFR.
Trp53 LOH Analysis
The analysis was performed as previously described.67 Briefly, genomic DNA isolated from tumor cell lines underwent PCR amplification for exon 8 of Trp53. Purified PCR products underwent a restriction enzyme digest with Msl1. The Trp53R270H allele possesses an Msl1 recognition sequence in exon 8, not observed in the WT allele. When separated on a 2% agarose gel, three products may be present after restriction endonuclease digest: a higher-molecular-weight band (WT allele) and two lower-molecular-weight bands (the digested Trp53R270H allele).
Cytogenetic Analysis
Spectral karyotyping analysis (SKY) was performed by the University of Minnesota (Minneapolis) Cytogenetics core. SKY was performed on early-passage cell lines (three to seven passages) derived from Cnp-EGFR;Trp53+/− mouse tumors. Cells were treated for 3.25 hours with colcemid, then harvested according to standard cytogenetic protocols. Eleven metaphases were analyzed by G-banding. An additional eight metaphase cells were examined by multicolor fluorescence in situ hybridization with spectral karyotyping. The G-banding and fluorescence in situ hybridization results were integrated for final karyotype interpretation. For array comparative genomic hybridization (aCGH), mouse tumor DNA was restriction digested and labeled with fluorochrome cyanine-5 using random primers and exo-Klenow fragment DNA polymerase. Control tail DNA from the same mouse was labeled concurrently in cyanine-3. The sample and control DNAs were combined, and aCGH was performed with a microarray constructed by Agilent Technologies, Inc., that contains approximately 170,000 distinct biological oligonucleotides spaced at an average interval of 10.9 kb. The ratio of sample/control DNA for each oligo was calculated using Feature Extraction software version 10.5 (Agilent Technologies, Inc.). The abnormal threshold was applied using Genomics Workbench version 7.0 (Agilent Technologies, Inc.). A combination of several statistical algorithms was applied. A minimum of three oligos that have a minimum absolute ratio value of 0.1 [on the basis of a log(2) ratio] is required for reporting of a copy number loss or gain.
Allografts
Cultured cells were trypsinized, resuspended at 1 × 106 in 1× PBS, and injected s.c. into severe combined immunodeficiency (SCID)/BIEGE mice. After 1 month of growth or when tumors reached 10% of body weight, tumors were harvested, and wet weights were taken and then fixed in 10% buffered formalin for histological analysis.
Results
EGFR and TP53 Expression Alterations Co-Occur in Human MPNSTs
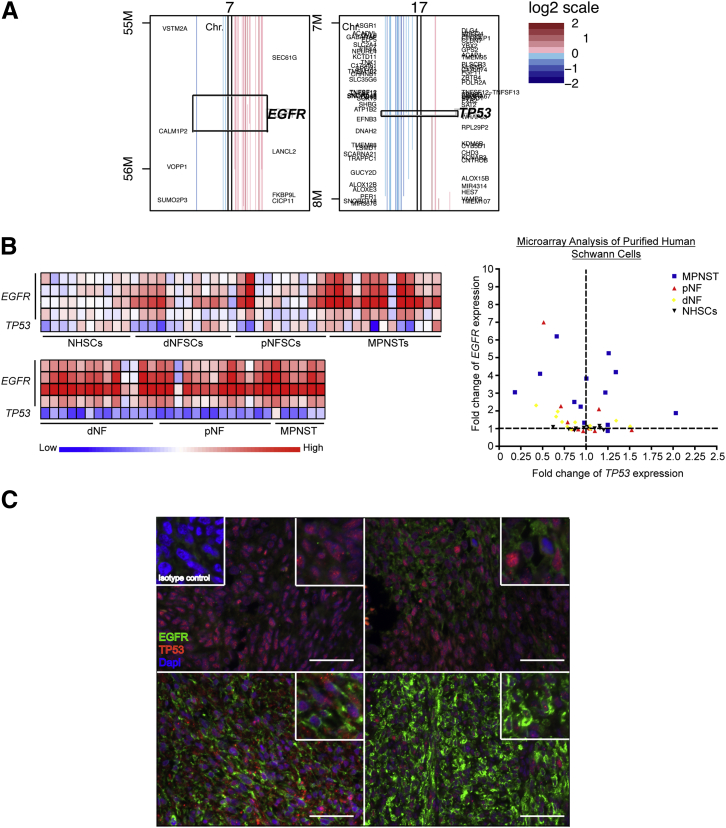
To determine whether there are co-occurring changes of EGFR and TP53 genomic CNAs and gene expression patterns in human MPNSTs, we used previously generated human aCGH, human microarray expression, methylome data, and a TMA (Figure 1, A–C). Data sets are generated from independent analysis of nonoverlapping, unique human patient samples. By using aCGH data (GSE33881), we identified EGFR gene CNA gains in approximately 37% of samples (19 of 51 patients) and TP53 CNA losses in approximately 29% of samples (15 of 51 patients) (Figure 1A).59 EGFR CNA gains tend to occur in the context of near complete amplification of the entire chromosome 7, whereas TP53 CNA losses tend to coincide with loss of chromosome arm 17p. Of 51 patients, 5 had co-occurring EGFR gene CNA gains and TP53 gene CNA losses [odds ratio with 95% CI of 39.92298, Fisher's exact test (FET) P = 3.53924 × 10−6]. To determine whether the CNAs observed reflect alterations in gene expression, we analyzed methylome, microarray expression, and TMA data.
Figure 1.
Analysis of human Schwann cell tumors for co-occurring EGFR and TP53 alterations. A: CNA data from 51 human patient MPNST samples at a 1-MB resolution for EGFR and TP53. Copy number is represented by a log2 scale. B: Microarray expression analysis was performed on purified Schwann cells (top panel) from normal sciatic nerves (NHSCs; n = 10), dermal neurofibromas (dNFSCs; n = 11), plexiform neurofibromas (pNFSCs; n = 11), and MPNSTs (n = 13). In addition, bulk tumors (bottom panel) of dermal neurofibromas (dNFs; n = 13), plexiform neurofibromas (pNFs; n = 13), and MPNSTs (n = 6) were also assessed for EGFR and TP53 expression. Displayed are four probes for EGFR and a single probe for TP53. A scatterplot of the purified Schwann cells from each tumor type with the TP53 expression (x axis) plotted against EGFR expression (y axis). C: Dual immunofluorescent analysis of EGFR and TP53 expression in human MPNSTs. Depicted are four representative images of EGFR (green) and TP53 (red) expression with the nuclear DAPI marker from a human TMA. The isotype control is depicted in the top left corner. Original magnification, ×400 (C). Scale bar = 50 μm (C).
Promoter CpG-IS methylation status is predictive of gene expression: hypermethylation is associated with gene silencing, and hypomethylation is associated with gene expression. MPNST whole-methylome data (GSE21714, n = 10 MPNST samples) were analyzed for methylation of the promoter CpG-IS regions for EGFR and TP53.56 No significant alterations in promoter CpG-IS methylation of EGFR and TP53 genes in human MPNSTs were identified compared with normal human Schwann cells (data not shown).
Previously reported microarray expression data analysis on purified Schwann cells and bulk tumors for nerves, neurofibromas, and MPNSTs were used to assess EGFR and TP53 mRNA levels (GSE14038). The microarray expression data are similar to previous reports of a significant increase in EGFR mRNA levels in MPNST formation compared with neurofibromas (P = 0.0002 for purified Schwann cells, P < 0.0001 for bulk tumors, Student's t-test), whereas TP53 mRNA levels were not significantly altered in tumor progression from neurofibromas (Figure 1B).68 Of 13 MPNST cell lines, 3 had a 0.75-fold or greater reduction in TP53 expression and a threefold increase or greater in EGFR mRNA expression, whereas nearly all bulk tumor samples (including neurofibromas) possessed co-alterations in TP53 and EGFR mRNA expression (Figure 1B). This discrepancy may reflect alterations in additional cellular components, such as mast cells, macrophages, fibroblasts, axons, and endothelial cells, within the bulk tumor.
To determine whether alterations in mRNA expression reflect protein expression, dual immunofluorescent staining for EGFR and TP53 was performed on a human TMA containing dermal neurofibromas (n = 30), plexiform neurofibromas (n = 31), and MPNSTs (n = 32) (Figure 1C). All tumors stained positive with varying intensities for TP53 expression, whereas EGFR expression was observed in only 50% of MPNSTs. Figure 1C shows representative images of the 32 MPNST samples assessed. In some cases, EGFR expression, when present, was not uniform throughout the tumor but occurred in focal regions. TP53 expression was variegated throughout the tumors, with rare TP53-negative cells (Figure 1C).
To determine if alterations in EGFR and TP53 coexist in other cancer types, we analyzed The Cancer Genome Atlas (TCGA) database for several cancer types.69 EGFR and TP53 alterations (mutations, CNAs, and microarray expression) had a tendency for co-occurrence in bladder urothelial carcinoma (FET P = 0.44), acute myeloid leukemia (FET P = 0.22), lung squamous cell carcinoma (FET P = 0.113), lung adenocarcinoma (FET P = 0.015), sarcoma (FET P = 0.052), and breast cancer (FET P = 0.000003) data sets. Glioblastoma multiforme, ovarian serous cystadenocarcinoma, prostate adenocarcinoma, skin cutaneous melanoma, colon and rectum adenocarcinoma, uterine corpus endometrial carcinoma, stomach adenocarcinoma, and head and neck squamous cell carcinoma had no association or tendency for mutual exclusivity (Table 1). Collectively, the data suggest that EGFR and TP53 alterations occur in a subset of human MPNSTs and other cancers, supporting a cooperative interaction.
Table 1.
EGFR and TP53 Co-Alteration Analysis from TCGA
| Cancer type | Odds ratio | 95% CI | P value | Association |
|---|---|---|---|---|
| Bladder urothelial carcinoma | 2.6 | 0.205–32.905 | 0.44 | Tendency toward co-occurrence |
| Acute myeloid leukemia | 6.071429 | 0.518–71.178 | 0.22 | Tendency toward co-occurrence |
| Lung squamous cell carcinoma | 4.285714 | 0.547–33.555 | 0.113 | Tendency toward co-occurrence |
| Lung adenocarcinoma | 2.291667 | 1.135–4.624 | 0.015 | Tendency toward co-occurrence |
| Sarcoma | 5.085714 | 1.133–22.818 | 0.052 | Tendency toward co-occurrence |
| Breast invasive carcinoma | 11.519126 | 3.344–39.684 | 0.000003 | Tendency toward co-occurrence |
| Glioblastoma multiforme | 0.2222 | 0.123–0.401 | 0 | Toward mutual exclusivity |
| Prostate adenocarcinoma | 0 | 0-NaN | 0.475 | Toward mutual exclusivity |
| Skin cutaneous melanoma | 0.845455 | 0.0939–7.605 | 0.680 | No association |
| Uterine corpus endometrial carcinoma | 1.276923 | 0.310–5.257 | 0.494 | No association |
| Stomach adenocarcinoma | 1.811765 | 0.767–4.279 | 0.125 | No association |
| Head and neck squamous cell carcinoma | 1.616 | 0.7101–3.679 | 0.167 | No association |
| Colon and rectum adenocarcinoma | 0.550725 | 0.151–2.01 | 0.277 | No association |
| Ovarian serous cystadenocarcinoma | 0.4887 | 0.0978–2.441 | 0.314 | Toward mutual exclusivity |
| Kidney | 0 | 0-NaN | 0.978 | Toward mutual exclusivity |
NaN, not a number.
EGFR Overexpression and Reduced TP53 Expression Cooperate to Increase Proliferation and Anchorage-Independent Growth in Vitro
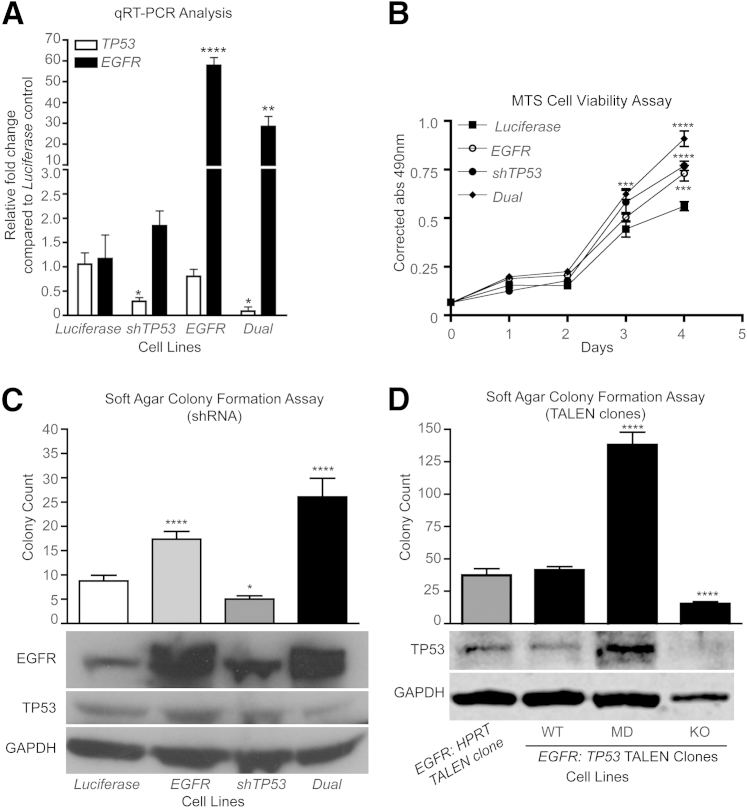
To assess the cooperativity of EGFR and TP53 expression for cellular transformation, overexpression of the full-length EGFR cDNA and shRNA-targeted knockdown of TP53 were performed in iHSC1λ cells either alone or in combination (dual) (Supplemental Figure S1A).70 A luciferase cDNA served as a control. Quantitative RT-PCR (RT-qPCR) and Western blot analysis demonstrated that EGFR overexpression and the shTP53 constructs functioned properly (Figure 2, A and C). Analysis of protein expression indicates a significant increase in EGFR (P < 0.05) and an approximately 25% reduction in TP53 expression in the dual targeted cell line (Supplemental Figure S1B). Functionally, dual EGFR overexpression and reduced TP53 expression significantly enhanced proliferation by MTS cell viability assay and anchorage-independent growth (two-tailed t-test P < 0.0001) by a soft agar colony formation assay compared with the single transgene controls (Figure 2, B and C). To validate these findings, mutations into the TP53 locus were introduced into EGFR-overexpressing iHSC1λ cells via TALENs. TALENs targeting the inert HPRT locus served as a control. Single-cell clones were selected and analyzed for mutations at the TP53 locus by sequencing. Sequencing results identified mutations that caused complete knockout (KO) of TP53 and complex mutations in which each allele possessed different mutations [called mutation detected (MD)] (Figure 2D and Supplemental Figure S1D). Western blot analysis revealed that clones with complex mutations had increased TP53 protein, but the molecular weight of the band was shifted downward, suggesting the presence of a mutated protein (Figure 2D). The KO clone displayed no protein product. Individual clones were seeded in soft agar colony formation assays to assess anchorage-independent growth capacity. Clones containing complex mutations (MD) developed significantly more colonies than the controls (Figure 2D). However, clones that had complete loss of TP53 protein formed significantly fewer colonies compared with the control. Interestingly, regardless of EGFR or TP53 status, the colony sizes were not significantly altered (Supplemental Figure S1, C and E). Collectively, these data demonstrate that increased EGFR expression and reduced/altered TP53 expression cooperate to increase oncogenic properties of immortalized human Schwann cells in vitro, but that complete loss of p53 protein may not do so. These data also suggest that additional changes are necessary to fully transform cells.
Figure 2.
Combined EGFR overexpression and reduced TP53 expression in iHSC1λ cells increase proliferation and anchorage-independent growth. A: RT-qPCR for EGFR and TP53 expression in each iHSC1λ cell line targeted with EGFR overexpression (EGFR), a TP53 shRNA (shTP53), or both (dual). A luciferase cDNA (Luciferase) served as a control. Statistical analysis performed with Student's t-test relative to Luciferase control. B: Graph depicts results from an MTS cell viability assay during the course of 4 days for each of the four cell lines. Data are a representative of three independent experiments. Statistics were performed using an unpaired Student's t-test. C: Bar graph depicts results from a soft agar colony formation assay. Experiments were performed in triplicate (n = 12 measurements for each experiment). Statistics were performed using an unpaired Student's t-test relative to a Luciferase control. Western blot analysis depicts EGFR, TP53, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression in each of the four cell lines. D: Bar graph depicts results from a soft agar colony formation assay. Experiments were performed in triplicate (n = 12 measurements for each experiment). Statistics were performed using an unpaired Student's t-test relative to an EGFR cell line targeted with HPRT TALENs (EGFR). EGFR:TP53 TALEN clones listed include clones in which the TP53 locus was not modified (WT), multiple mutations in different alleles are present (MD), and homozygous mutations that knocked out TP53 expression (KO). Statistics were performed using an unpaired Student's t-test relative to EGFR:HPRT control. Western blot analysis depicts TP53 and GAPDH expression in the EGFR-overexpressing cell lines. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.005, and ∗∗∗∗P < 0.0001.
Trp53 Haploinsufficiency and EGFR Overexpression Cooperate in Vivo to Significantly Increase Neurofibroma and Induce Grade 3 PNST
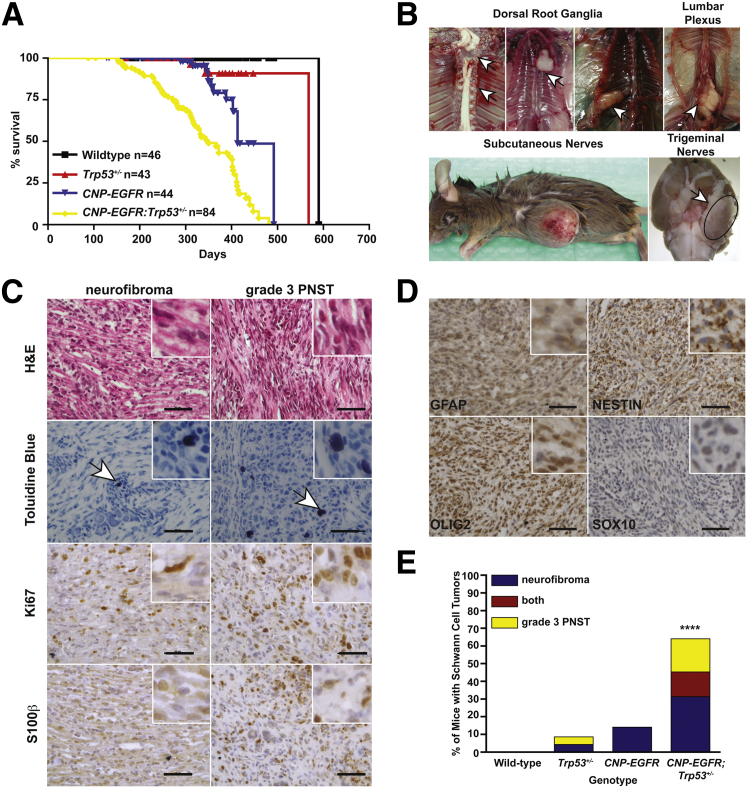
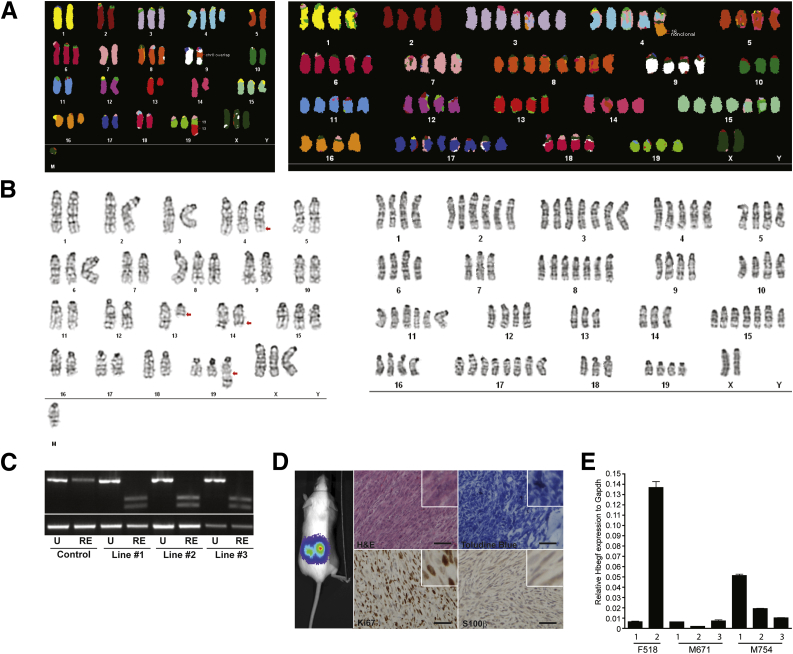
To assess cooperativity of EGFR overexpression and reduced TP53 expression in tumor formation in vivo, transgenic mice overexpressing the human EGFR gene (Cnp-EGFR) were bred to Trp53 heterozygous mice (Trp53R270H/+, from here on called Trp53+/−). Cnp-EGFR;Trp53+/− mice had a significantly reduced median survival (349 days, P < 0.0001) compared with single transgenic controls (Cnp-EGFR; 413 days) (Figure 3A). Cnp-EGFR;Trp53+/− mice developed a variety of nerve-associated tumors associated with paraspinal dorsal root ganglion (54%), trigeminal nerves (5.5%), subcutaneous nerves (1.7%), and brachial plexi (1.4%) (Figure 3B). Nerve-associated tumors were histologically evaluated for cellularity, nuclear atypia, necrosis, hemorrhage, myxoid background, and tumor border on the basis of H&E staining (Figure 3C and Supplemental Table S1). H&E staining identified features found in neurofibromas (low-medium cellularity, no-medium nuclear atypia, no necrosis, no hemorrhage, low-medium myxoid background, and no mitotic figures) and grade 3 PNSTs (MPNSTs in humans, high cellularity, high nuclear atypia, some necrosis, rare hemorrhage, and presence of mitotic figures) (Supplemental Table S1). Toluidine blue staining identified the presence of mast cells, which are often associated with neurofibroma and grade 3 PNSTs (Figure 3C). IHC for Schwann cell markers S100β, Gfap, Nestin, Olig2, and Sox10 confirmed that the nerve-associated tumors were derived from the Schwann cell lineage (Figure 3, C and D, and Supplemental Figure S2D). To determine whether mice developed true neurofibromas versus microMPNSTs, we IHC evaluated tumors for the presence of axons (neuronal nuclei), fibroblasts (vimentin), and macrophages (CD45) (Supplemental Figure S2A). This analysis demonstrated the presence of additional cell types consistent with neurofibroma formation. IHC for the proliferation marker Ki-67 indicated the presence of some proliferating cells in neurofibromas (0 to medium-grade 1) and many proliferating cells in grade 3 PNSTs (medium grade 2 to high grade 3) (Supplemental Table S1). The proliferative index of the nerve-associated tissues (normal nerve, nerve hyperplasia, neurofibromas, grade 2 PNSTs, and grade 3 PNSTs) was assessed by counting mitotic figures on the basis of H&E staining (Supplemental Figure S2B). Only grade 3 PNSTs displayed mitotic figures by H&E staining, with an average of approximately two mitotic figures per field (Student's t-test P = 0.0052). Collectively, the histological data suggest that the nerve-associated tumors represent various grades of human Schwann cell tumor formation: nerve hyperplasia, neurofibroma, grade 2 PNSTs, and grade 3 PNSTs. Neurofibromas were predominantly observed in the paraspinal dorsal root ganglion, whereas grade 3 PNSTs were observed more frequently in all other peripheral nerves. In addition, 40% (n = 11 of 27 analyzed) of grade 3 PNSTs contained rare regions of additional cellular phenotypes (epithelioid-like cells, matrix-producing cells, and wreath-like giant cells) observed in some human MPNSTs (Supplemental Figure S2C).71,72 Collectively, these data suggest that the nerve-associated tumors resemble human neurofibromas and MPNSTs.
Figure 3.
Trp53 haploinsufficiency and EGFR overexpression cooperated for grade 3 PNST formation in vivo. A: Kaplan-Meier survival plot of the four genetic cohorts analyzed: WT (n = 46), Trp53+/− (n = 43), CNP-EGFR (n = 44), and CNP-EGFR;Trp53+/− (n = 84). B: Necropsy images depict the peripheral nerves targeted for tumor development: dorsal root ganglia, lumbar plexus, subcutaneous nerves, and trigeminal nerves. Arrows indicate tumor location. C: IHC analysis of nerve-associated tumors for H&E staining, toluidine blue (mast cells), Ki-67 (proliferation marker), and S100β (Schwann cell marker) indicates the tumors have features of various grades of Schwann cell tumors. The arrows in indicate Toluidine Blue positive cells. D: IHC for Schwann cell markers Gfap, Nestin, Olig2, and Sox10. Images depicted are from a grade 3 PNST. E: Bar graph depicts breakdown of neurofibroma, grade 3 PNST, or both tumors in each mouse genetic cohort. Tumor type was determined on the basis of histological analysis performed (C and D, Supplemental Figure S2A, and Supplemental Table S1). Statistics were performed with a Fisher's exact test. ∗∗∗∗P < 0.0001. Original magnification, ×400 (C and D). Scale bar = 50 μm (C and D).
Cnp-EGFR;Trp53+/− mice had significantly increased neurofibroma formation (Cnp-EGFR FET P = 0.0003; Trp53+/− FET P < 0.0001) and grade 3 PNST formation (Cnp-EGFR FET P < 0.0001; Trp53+/− FET P < 0.0001) compared with single transgenic mice (Figure 3E). Tumor penetrance for each genotype is summarized in Supplemental Table S2. Interestingly, we observed a 15% penetrance of neurofibroma formation on the Cnp-EGFR background compared with previous reports of nerve hyperplasia with 5% neurofibroma formation.49 Discrepancies in these observations may be due, in part, to differences in mouse strain. Our mouse model was generated by backcrossing the Cnp-EGFR transgene from a C57BL/6 background to an FVB/N background (five to seven generations). Previous reports used C57BL/6-SJL mice with backcross onto C57BL/6. FVB/N mice are more susceptible to tumor formation than C57BL/6 in the context of GEMMs for tumor formation, most likely attributed to differences in genetic modifiers (Jackson Laboratories, Bar Harbor, ME).
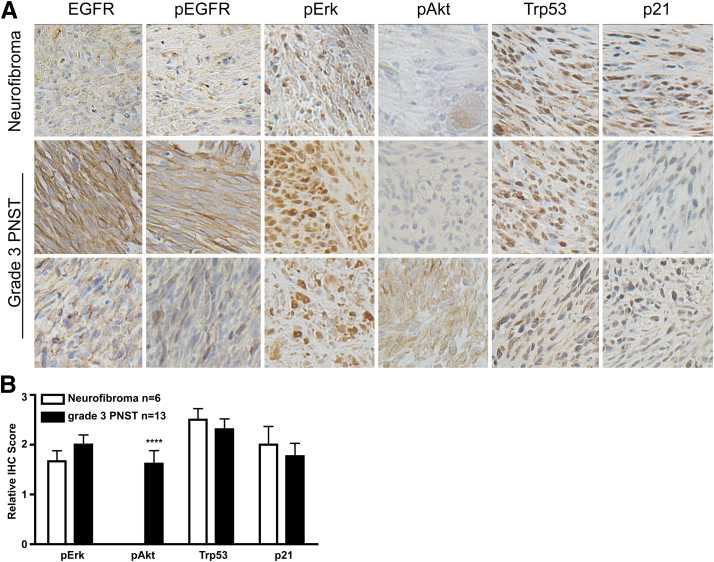
To determine whether the downstream signaling pathways of EGFR and Trp53 were altered in the Cnp-EGFR;Trp53+/− mouse tumors, histological analysis of downstream signaling pathways of EGFR (pEGFR, pErk, and pAkt) and Trp53 (p21 expression) were assessed in neurofibromas (n = 6 mice) and grade 3 PNSTs (n = 13 mice) (Figure 4A and Supplemental Figure S2D). EGFR, pEGFR, pErk, Trp53, and p21 were present in both neurofibromas and grade 3 PNSTs, with more intense EGFR, pEGFR, and pErk staining observed in grade 3 PNSTs. pAkt was absent in neurofibromas, and variable expression was observed in grade 3 PNSTs (Student's t-test P < 0.0001) (Figure 4B). These data support the previous finding that the PI3K/AKT pathway is important in progression from neurofibroma to grade 3 PNST.70,73
Figure 4.
Activation of downstream signaling pathways in Schwann cell tumors. IHC for downstream effectors of EGFR (pEGFR, pErk, and pAkt) and Trp53 (p21) signaling. A: IHC images for either neurofibromas (n = 6) or grade 3 PNSTs (n = 13) from Cnp-EGFR;Trp53+/− mice. B: Scoring for IHC: 0, negative; 1, <25% of tumor; 2, 25% to 75% of tumor; and 3, >75% of tumor staining positive. Statistics were performed with an unpaired Student's t-test: ∗∗∗∗P < 0.0001. Original magnification, ×400 (A).
aCGH Analysis Identifies Genes Enriched in Erk5 Signaling
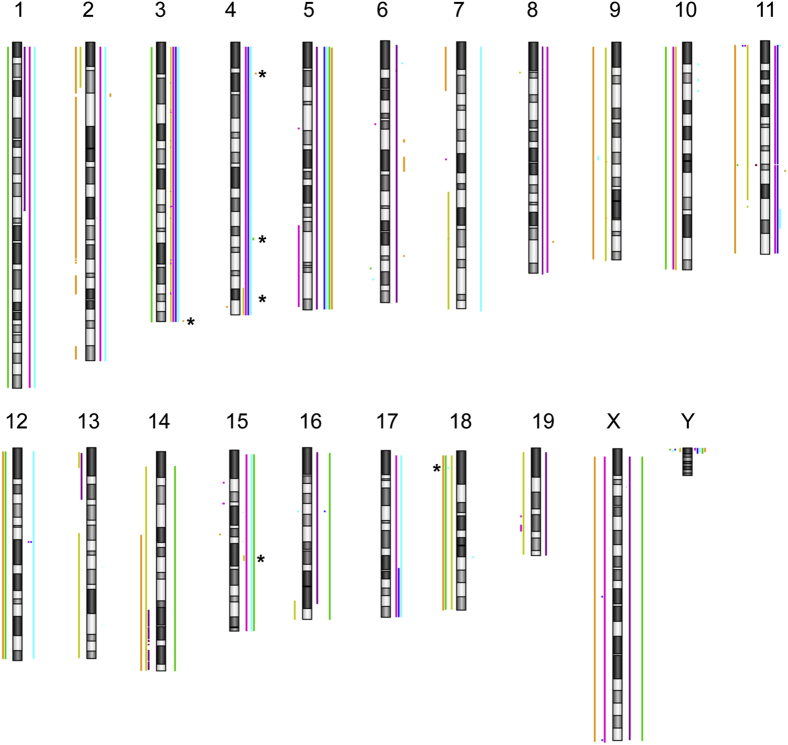
Because Cnp-EGFR;Trp53+/− mice only had a 33% penetrance of grade 3 PNST formation, additional genetic alterations must be required for the tumors to develop. LOH analysis of the Trp53 gene identified that 40% of grade 3 PNSTs lost the WT Trp53 allele (data not shown). To identify additional genetic events leading to tumor formation, we performed aCGH on seven Cnp-EGFR;Trp53+/− grade 3 PNSTs. The tumors contained a variety of chromosomal alterations, including whole chromosome gains and losses and regional gains and losses (Figure 5 and Supplemental Table S2). Whole chromosome gains occurred on chromosomes 3, 5, and 15 in four of seven mice. Three of seven grade 3 PNSTs showed whole chromosome losses on chromosomes 10 and 18. To identify candidate MPNST-driver genes, we analyzed genes found on focal chromosomal gains/losses and included tumors that also possessed whole chromosomal gains/losses that contained the focal regions of interest (Figure 5). We only assessed CNAs observed in four or more mouse tumors (Table 2). This narrowed the search down to five regions containing 2452 genes: CNAs on chromosomes 4, 15, and 18. We identified the human orthologs for the list of candidate MPNST-driver genes and assessed the CNA status on 51 human MPNST tumor samples. Comparative analysis to human CNA data identified 199 of 2452 genes with recurrent CNAs in human MPNSTs (Supplemental Table S3).
Figure 5.
aCGH analysis was performed on seven grade 3 PNSTs to identify recurrent CNAs. Depicted is an ideogram of mouse chromosomes. Individual tumors are color coded. Lines to the left of the chromosome indicate CNA losses, whereas lines to the right indicate CNA gains. The asterisks denote focal CNAs observed in at least four of seven mice that were further pursued to identify grade 3 PNST driver genes.
Table 2.
Comparative aCGH Analysis
| Chromosome No. | Region (mm9) | Size (nt) | CNAs | No. of genes | No. of human homologues | No. of human genes with recurrent CNAs |
|---|---|---|---|---|---|---|
| 3 | 87588760-95767212 | 8,178,452 | Gain | 962 | 209 | 152 |
| 4 | 87904820-90001325 | 2,096,505 | Gain | 245 | 9 | 1 |
| 4 | 128549741-137966192 | 9,416,451 | Gain | 313 | 204 | 17 |
| 4 | 140085152-155607029 | 124,578,123 | Gain | 922 | 195 | 24 |
| 15 | 60596527-63754443 | 3,157,916 | Gain | 9 | 4 | 4 |
| 18 | 9838430-10017847 | 179,417 | Loss | 1 | 1 | 1 |
| Total | 2452 | 622 | 199 |
Ingenuity Pathway Analysis (Ingenuity Systems Inc., Redwood City, CA) of the 199 genes with recurrently occurring CNAs in mouse and human MPNSTs identified eight signaling pathways significantly enriched: ERK5 signaling, ephrin receptor signaling, phospholipase C signaling, axonal guidance signaling, tec kinase signaling, ephrin A signaling, polyamine regulation in colon cancer, and P2Y purinergic receptor signaling pathway (Table 3). In addition, there was significant enrichment in genes found in normal cellular processes, such as cell cycle regulation and DNA replication, recombination, and repair (Supplemental Table S4). Moreover, 56 genes identified are implicated in cancer, including four with recurrent CNA gains (PRUNE, PTK2, NTRK1, and MYC) previously implicated in MPNST development (Supplemental Tables S3 and S4).74–77 These data suggest that EGFR overexpression and reduced Trp53 expression cooperate to generate an environment permissible for gross chromosomal alterations that enrich for protumorigenic signaling pathways for MPNST formation to occur in Schwann cells in vivo.
Table 3.
Signaling Pathway Analysis of aCGH Focal Gain Regions
| Signaling pathway | B-H corrected P value | Genes |
|---|---|---|
| ERK5 | 0.0005 | SH2D2A, MYC, RPS6KA6, MEF2D, NTRK1, RPS6KA3, CREB3L4 |
| Ephrin receptor | 0.0046 | PTK2, SHC1, GNB4, GNB3, GNB2, EFNA3, CREB3L4, EFNA4, EFNA1 |
| Phospholipase C | 0.0063 | SHC1, GNB4, PRKC1, GNB3, MEF2D, GNB2, RPS6KA3, LYN ARHGEF2, CREB3L4 |
| Axonal guidance | 0.0120 | ADAM14, EFNA3, SEMA6C, EFNA4, EFNA1, PTK2, SHC1, GNB4, PRKC1, GBN3, NTRK1, GNB2, SEMA4A |
| Tec kinase | 0.0324 | PTK2, GNB4, PRKC1, GNB3, HCK, GNB2, LYN |
| Ephrin A | 0.0380 | PTK2, EFNA3, EFNA4, EFNA1 |
| Polyamine regulation | 0.0380 | MCY, ODC1, OAZ3 |
| P2Y purinergic receptor | 0.0380 | MYC, GNB4, PRKC1, GNB3, GNB2, CREB3L4 |
B-H, Benjamini-Hochberg.
Characterization of Cnp-EGFR;Trp53+/− Grade 3 PNST-Derived Cell Lines
To identify additional mutations and to corroborate the aCGH finding, G-banding and SKY analysis were performed on single-cell clones from three early-passage (three to seven passages) grade 3 PNSTs that express Schwann cell markers (Olig2 and S100β) (data not shown). Cell clones were analyzed for cytogenetic abnormalities (G-banding and SKY), LOH of WT Trp53 allele, and allograft tumor formation. G-banding and SKY analysis from 15 and 13 cell clones, respectively, from two tumors identified variable karyotypes, including whole chromosomal gains, losses, and translocations (Figure 6, A and B). Recurrent whole chromosomal gains were observed on chromosomes 4 (27 of 28), 6 (26 of 28), 8 (27 of 28), and 15 (28 of 28). The high level of aneuploidy observed in these cell lines is similar to the incidence observed in human MPNSTs. Interestingly, 80% of the tumor-derived cell lines analyzed underwent LOH for the WT Trp53 allele within the first three passages after single-cell sorting, suggesting there is a selective pressure for the loss of the WT Trp53 allele in vivo and/or in cell culture (Figure 6C). Last, clones were injected into SCID/BIEGE mice for allograft tumor formation assays to determine whether they retain properties similar to the parental tumors (Figure 6D). Histological analysis of tumors (H&E, toluidine blue, Ki-67, and S100β) indicated that the clones produced tumors with features of grade 3 PNSTs: spindle-shaped cell morphological characteristics, mast cell infiltration, high proliferative index, and expression of a Schwann cell marker, S100β (Figure 6D). Collectively, these data suggest that EGFR overexpression and loss of Trp53 expression alone are not sufficient for MPNST formation but facilitate chromosomal aberrations that lead to MPNST formation.
Figure 6.
Characterization of grade 3 PNST-derived cell lines. A: SKY of grade 3 PNST-derived cell lines. Two clones from a single tumor are depicted. The clone on the left possesses translocations (chromosomes 9 and 19), duplications (chromosomes 3, 4, 6, 8, 15, 16, 19, and X), and deletions (chromosome 14). On the right, a clone possesses large whole chromosomal amplifications (chromosomes 1 to 19) and a translocation on chromosome 4. B: G-banding analysis of grade 3 PNST-derived cell lines. Both clones are from the same tumor in A. The clone on the left possesses a few deletions, amplifications, and translocations (arrows). The clone on the right possesses numerous whole chromosomal amplifications. C: Agarose gel electrophoresis of PCR products from a Trp53 loss-of-heterozygosity experiment. D: A band at 600 bp represents the WT Trp53 allele, whereas the double smaller bands represent the digested Trp53R270H allele. Luciferase live imaging of cell lines injected into SCID/BIEGE mice demonstrating their capacity for tumor formation. Histological analysis of tumors for H&E staining, toluidine blue staining, and IHC for Ki-67 and S100β indicates the cell lines form grade 3 PNSTs. E: Bar graph depicts RT-qPCR results for Hbegf mRNA expression relative to Gapdh on grade 3 PNST-derived cell lines from three independent tumors. Original magnification, ×400 (D). Scale bar = 50 μm (D). RE, restriction enzyme digested; U, uncut.
Discussion
We performed a comprehensive analysis of EGFR and TP53 cooperativity in human and mouse MPNST development. We identified co-occurring alterations of the EGFR and TP53 genes by CNAs, microarray expression, and dual immunofluorescent staining in human MPNSTs. Modulation of EGFR and TP53 expression in iHSCs in vitro increased proliferation and anchorage-independent growth. Transgenic mice overexpressing EGFR and haploinsufficient for Trp53 formed all grades of Schwann cell tumors. Last, cytogenetic analysis of grade 3 PNSTs from EGFR-overexpressing and TP53-haploinsufficient mice developed chromosomal aberrations that enriched for genes in the ERK5 signaling cascade.
Amplification of the EGFR gene and mutations and/or deletions of the TP53 locus are common events in human MPNST.25,46,78,79 However, there has not been a large-scale study correlating the expression of both events. Tabone-Eglinger et al47 performed a large-scale IHC analysis of EGFR expression on 52 MPNST samples (NF1 syndrome associated and sporadic) and observed that 86% of the samples overexpressed EGFR. From the 52 samples, they assessed four for co-expression of TP53 and identified half of the samples overexpressing EGFR to be associated with loss of TP53 expression.47 Comparatively, we observed TP53 expression in all MPNST samples with varying intensity of staining, whereas only 50% of samples expressed EGFR. This discrepancy with previous reports may be the result of many factors, including sample size and method. Also, immunofluorescence/IHC will not identify mutations in TP53 that may produce a mutant protein product. In addition, we analyzed DNA CNA data for the EGFR and TP53 loci in human MPNSTs and identified a subset of patients with co-occurring EGFR CNA gains and TP53 CNA losses (5 of 51 patients). Assessment of several cancer types in TCGA identified a significant co-occurrence of EGFR and TP53 aberrations (mRNA expression, CNAs, and mutations) in breast cancer and sarcomas (Table 1). Collectively, the data suggest that EGFR overexpression and reduced/impaired TP53 expression may cooperate for tumor formation in a subset of human cancers.
The reasons for this strong cooperation between EGFR overexpression and Trp53 haploinsufficiency in vivo are unclear. Some possible reasons for the cooperation include, but are not limited, to the following: Trp53 haploinsufficiency alleviates EGFR-induced senescence. Trp53 haploinsufficiency impairs DNA repair mechanisms, allowing for acquisition of rare oncogenic mutations that cooperate with EGFR overexpression. It seems possible that overexpressing EGFR activates TP53, thereby limiting cell proliferation, perhaps by up-regulating CDKN1A. Senescence is triggered by the activation of signaling pathways, such as TP53 and CDKN2A.80 TP53 loss of function and EGFR activation co-occur in other cell types, such as in human esophageal cancer, where there is a correlation between EGFR overexpression and TP53 loss of function.52,81,82 Okawa et al51 demonstrated that overexpression of EGFR, activation of telomerase reverse transcriptase, and reduction in TP53 expression were capable of transforming esophageal epithelial cells. Ohashi et al83 demonstrated that overexpression of EGFR in immortalized esophageal epithelial cells induced expression of cell cycle kinase inhibitors p15INK4B, p16INK4A, and p21. This senescent state was alleviated partially by introducing a mutant TP53, leading to increased transformation of the immortalized esophageal cells. It is possible that, in our mouse model, EGFR overexpression induces an oncogenic senescence that is partially alleviated by Trp53 haploinsufficiency, allowing for acquisition of additional mutations for grade 3 PNST formation. Because only 33% of mice developed grade 3 PNSTs, clearly additional mutations and/or genetic alterations are required for high-grade tumor formation.
In addition to the role TP53 has in cell cycle progression and its transcriptional activity, TP53 has a pivotal role in DNA repair mechanisms that prevent acquisition of genome mutations. Loss of Trp53 expression reduces the activity of DNA repair pathway mechanisms (base/nucleotide excision repair) that leads to the accumulation of mutations that generate genomic instability that often results in chromosomal aberrations (aneuploidy).84 This study focused on gross chromosomal alterations by cytogenetic analysis of grade 3 PNSTs and tumor-derived cell lines. The aCGH, SKY, and G-banding data demonstrated an aneuploidy phenotype in both the bulk tumors and tumor-derived cell lines, which is similar to that observed in human MPNST bulk tumors and cell lines.22 To identify potential signaling pathways that cooperate with EGFR overexpression and Trp53 haploinsufficiency for grade 3 PNST formation, we chose five focal chromosomal gains and losses observed in at least four of seven tumors analyzed by aCGH. Ingenuity Pathway Analysis of the human homologues of genes with recurrent CNAs identified eight significantly enriched signaling pathways (Table 3 and Supplemental Table S5). The top pathway, ERK5 signaling or BMK1, is an atypical MAPK signaling pathway that promotes cellular proliferation (activation of c-Jun, SAP, and MYC) and inhibits apoptosis by phosphorylating BAD and Foxo3A.85,86 Previous studies on ERK signaling in human MPNST cell lines indicate that ERK5 signaling is active and induced in the presence of EGF ligand.87 Our mouse model suggests that additional studies on ERK5/BMK1 signaling in MPNST development are warranted and may provide a potential therapeutic target for treatment of MPNSTs.
Activation of several signaling pathways known to be involved in MPNST development was also observed in our mouse model. Increased signaling through EGFR, via pAkt, was observed in Cnp-EGFR;Trp53+/− grade 3 PNSTs. These data corroborate previous reports indicating that pAkt and downstream signaling through the mTOR pathway are important in MPNST.12–14,70,73 Similar to our GEMM, NPcis tumors and tumor-derived cell lines have activated the PI3K/AKT/mTOR pathway and increased EGFR expression (Figure 6C).88 Inhibition of the PI3K/AKT/mTOR pathway in NPcis cell lines potently inhibited EGF-dependent growth.88 In addition, TP53 loss is known to induce growth factor expression, such as heparin-binding epidermal growth factor (HB-EGF).89 HB-EGF binds to EGFR and induces signaling through the MAPK and PI3K/Akt pathway. Cell lines from the NPcis mouse express HB-EGF.88 Our grade 3 PNST-derived cell lines also express HB-EGF, which may be an additional mechanism for activating the PI3K/Akt pathway in our mouse model (Figure 6E). Potentially in our model, Trp53 haploinsufficiency removes a senescent brake on Schwann cell transformation, while also causing growth factor expression to stimulate proliferation in an autocrine manner.
Recently, aCGH was performed on 11 early-passage, grade 3 PNST cell strains from the P0-GGFb3 transgenic mouse overexpressing neuregulin in the Schwann cells.74 Neuregulin is a growth factor ligand, implicated in many human cancers, including MPNSTs, that binds to and stimulates signaling through erbB receptors.17 Transgenic mice overexpressing neuregulin in Schwann cells develop both neurofibromas and frank MPNSTs. In our study, chromosomal CNAs, including whole chromosome gains of chromosomes 3, 5, 15, and 18, were identified in four of seven tumors. Interestingly, Kazmi et al74 reported whole chromosomal losses for chromosome 4, and no alterations were observed on chromosome 5. This discrepancy in CNAs may reflect differences in mouse strain background and/or activation of erbB2 and erbB3 receptor signaling pathways by neuregulin and not solely EGFR (erbB1), as in our model. Recently, Brosius et al90 demonstrated that neuregulin overexpression alone was not sufficient to generate MPNSTs on a C57BL/6J mouse strain. However, when combined with Trp53 haploinsufficiency, P0-GGFβ3 transgenic mice developed de novo MPNSTs. Interestingly, our study and Brosius et al90 identified a similar CNA gain on chromosome 15 in a region containing MYC. Moreover, MYC expression is elevated in human MPNSTs compared with neurofibromas and is directly altered by modulating components of the Wnt/β-catenin pathway.91 Collectively, the data sets support previous reports that MYC is a strong genetic driver of MPNST formation.
In summary, we determined that co-alteration of EGFR and TP53 expression in vitro and in vivo contribute to Schwann cell tumorigenesis through increased proliferation, anchorage-independent growth, PI3K/Akt pathway activation, and chromosomal alterations enriching for alterations in genes in the ERK5 signaling pathway.
Acknowledgments
We thank the following shared resources of the Masonic Cancer Center at the University of Minnesota (Minneapolis, MN): The Mouse Genetics Laboratory, Comparative Pathology, and the Tissue Procurement Facility. We thank the Research Animal Resources at the University of Minnesota, specifically Alwan Aliye for his technical support in mouse maintenance and Dr. Margaret Wallace for the gift of the human immortalized Schwann cell lines.
E.P.R., A.L.W., B.S.M., and G.M.O. performed laboratory experiments and/or analyzed the data; B.R.W. generated viral vector for single-cell cloning of tumor cells; K.C. performed bioinformatic data analysis of microarray expression data, methylome data, and CNA data; M.H.C. assessed histological and graded mouse tumors; C.L.F., A.E.R., and S.C.S. generated the TMA, performed the immunofluorescent staining, and scanned the slides; N.R. and D.A.L. supervised laboratory experiments and assisted in writing the manuscript; and E.P.R. wrote the manuscript.
Footnotes
Supported by NIH/National Institute of Neurological Disorders and Stroke grant P50 N5057531, the Margaret Harvey Schering Trust, The Zachary Neurofibromatosis Research Fund, The Jacqueline Dunlap Neurofibromatosis Research Fund, The Children's Tumor Foundation Young Investigators Award2011-01-018 (A.L.W.), and NIH/National Institute of Arthritis and Musculoskeletal and Skin Diseases Musculoskeletal Training grant AR050938 (B.S.M.).
Disclosures: D.A.L. has ownership interest (including patents) in Discovery Genomics, Inc., and NeoClone Biotechnologies International and is also a consultant/advisory board member of Discovery Genomics, Inc., and NeoClone Biotechnologies International.
Current address of E.P.R., Department of Developmental Neurobiology, St. Jude Children's Research Hospital, Memphis, Tennessee.
Supplemental Data
Modulating EGFR and TP53 expression. A: Schematics for the plasmids used to overexpress EGFR cDNA and introduce an shRNA against TP53. B: Bar graph depicts quantification of Western blot analysis in Figure 2C. The statistic used was an unpaired Student's t-test. C: Bar graph depicts size of colonies from Figure 2C. D: Sequences indicated the target sequence of the TALENs for the TP53 locus. The translational start site (ATG) is boldfaced. The TALEN binding sites are underlined. The top row depicts the WT sequence of TP53. The lower row contains dashes representing deleted nucleotides from a TP53 KO clone. E: Bar graph depicts size of colonies from Figure 2D. ∗P < 0.05.
Histological analysis of tumors. A: IHC analysis for NeuN (neurons), vimentin (fibroblasts), and CD45 (macrophages) on a neurofibroma sample. B: Graph depicts the number of mitotic events per field for each tumor type listed. C: H&E analysis of grade 3 PNSTs demonstrating a significant pleomorphism among the tumors, including variability in cell morphological characteristics and extracellular matrix content. Arrows indicate the presence of matrix in matrix-producing cells, indicating wreath-like giant cells in the last panel. D: No primary control images of tumors treated with rabbit polyclonal and mouse monoclonal antibodies. ∗∗P < 0.01, unpaired Student's t-test. Original magnification, ×400 (D).
References
- 1.Ducatman B.S., Scheithauer B.W., Piepgras D.G., Reiman H.M., Ilstrup D.M. Malignant peripheral nerve sheath tumors: a clinicopathologic study of 120 cases. Cancer. 1986;57:2006–2021. doi: 10.1002/1097-0142(19860515)57:10<2006::aid-cncr2820571022>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 2.Evans D.G., Baser M.E., McGaughran J., Sharif S., Howard E., Moran A. Malignant peripheral nerve sheath tumours in neurofibromatosis 1. J Med Genet. 2002;39:311–314. doi: 10.1136/jmg.39.5.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCaughan J.A., Holloway S.M., Davidson R., Lam W.W. Further evidence of the increased risk for malignant peripheral nerve sheath tumour from a Scottish cohort of patients with neurofibromatosis type 1. J Med Genet. 2007;44:463–466. doi: 10.1136/jmg.2006.048140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou C., Smith K.D., Liu J., Lahat G., Myers S., Wang W.L., Zhang W., McCutcheon I.E., Slopis J.M., Lazar A.J., Pollock R.E., Lev D. Clinical, pathological, and molecular variables predictive of malignant peripheral nerve sheath tumor outcome. Ann Surg. 2009;249:1014–1022. doi: 10.1097/SLA.0b013e3181a77e9a. [DOI] [PubMed] [Google Scholar]
- 5.Ferner R.E., Gutmann D.H. International consensus statement on malignant peripheral nerve sheath tumors in neurofibromatosis. Cancer Res. 2002;62:1573–1577. [PubMed] [Google Scholar]
- 6.Kolberg M., Høland M., Ågesen T.H., Brekke H.R., Liestøl K., Hall K.S., Mertens F., Picci P., Smeland S., Lothe R.A. Survival meta-analyses for >1800 malignant peripheral nerve sheath tumor patients with and without neurofibromatosis type 1. Neuro Oncol. 2013;15:135–147. doi: 10.1093/neuonc/nos287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messiaen L.M., Callens T., Mortier G., Beysen D., Vandenbroucke I., Van Roy N., Speleman F., Paepe A.D. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat. 2000;15:541–555. doi: 10.1002/1098-1004(200006)15:6<541::AID-HUMU6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 8.Leppig K.A., Kaplan P., Viskochil D., Weaver M., Ortenberg J., Stephens K. Familial neurofibromatosis 1 microdeletions: cosegregation with distinct facial phenotype and early onset of cutaneous neurofibromata. Am J Med Genet. 1997;73:197–204. doi: 10.1002/(sici)1096-8628(1997)73:2<197::aid-ajmg17>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 9.De Raedt T., Brems H., Wolkenstein P., Vidaud D., Pilotti S., Perrone F., Mautner V., Frahm S., Sciot R., Legius E. Elevated risk for MPNST in NF1 microdeletion patients. Am J Hum Genet. 2003;72:1288–1292. doi: 10.1086/374821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballester R., Marchuk D., Boguski M., Saulino A., Letcher R., Wigler M., Collins F. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 1990;63:851–859. doi: 10.1016/0092-8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- 11.Bottillo I., Ahlquist T., Brekke H., Danielsen S.A., van den Berg E., Mertens F., Lothe R.A., Dallapiccola B. Germline and somatic NF1 mutations in sporadic and NF1-associated malignant peripheral nerve sheath tumours. J Pathol. 2009;217:693–701. doi: 10.1002/path.2494. [DOI] [PubMed] [Google Scholar]
- 12.Sherman L.S., Atit R., Rosenbaum T., Cox A.D., Ratner N. Single cell Ras-GTP analysis reveals altered Ras activity in a subpopulation of neurofibroma Schwann cells but not fibroblasts. J Biol Chem. 2000;275:30740. doi: 10.1074/jbc.M001702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basu T.N., Gutmann D.H., Fletcher J.A., Glover T.W., Collins F.S., Downward J. Aberrant regulation of Ras proteins in malignant-tumor cells from type-1 neurofibromatosis patients. Nature. 1992;356:713–715. doi: 10.1038/356713a0. [DOI] [PubMed] [Google Scholar]
- 14.Cichowski K., Jacks T. NF1 tumor suppressor gene function: narrowing the GAP. Cell. 2001;104:593–604. doi: 10.1016/s0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- 15.Wu J., Williams J.P., Rizvi T.A., Kordich J.J., Witte D., Meijer D., Stemmer-Rachamimov A.O., Cancelas J.A., Ratner N. Plexiform and dermal neurofibromas and pigmentation are caused by Nf1 loss in desert hedgehog-expressing cells. Cancer Cell. 2008;13:105–116. doi: 10.1016/j.ccr.2007.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeClue J.E., Heffelfinger S., Benvenuto G., Ling B., Li S., Rui W., Vass W.C., Viskochil D., Ratner N. Epidermal growth factor receptor expression in neurofibromatosis type 1-related tumors and NF1 animal models. J Clin Invest. 2000;105:1233–1260. doi: 10.1172/JCI7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stonecypher M.S., Byer S.J., Grizzle W.E., Carroll S.L. Activation of the neuregulin-1/ErbB signaling pathway promotes the proliferation of neoplastic Schwann cells in human malignant peripheral nerve sheath tumors. Oncogene. 2005;24:5589–5605. doi: 10.1038/sj.onc.1208730. [DOI] [PubMed] [Google Scholar]
- 18.Kadono T., Kikuchi K., Nakagawa H., Tamaki K. Expressions of various growth factors and their receptors in tissues from neurofibroma. Dermatology. 2000;201:10–14. doi: 10.1159/000018421. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda T., Ichimura E., Shinozaki T., Sano T., Kashiwabara K., Oyama T., Nakajima T., Nakamura T. Coexpression of HGF and c-Met/HGF receptor in human bone and soft tissue tumors. Pathol Int. 1998;48:757–762. doi: 10.1111/j.1440-1827.1998.tb03834.x. [DOI] [PubMed] [Google Scholar]
- 20.Rao U.N., Sonmez-Alpan E., Michalopoulos G.K. Hepatocyte growth factor and c-MET in benign and malignant peripheral nerve sheath tumors. Hum Pathol. 1997;28:1066–1070. doi: 10.1016/s0046-8177(97)90060-5. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe T., Oda Y., Tamiya S., Masuda K., Tsuneyoshi M. Malignant peripheral nerve sheath tumour arising within neurofibroma: an immunohistochemical analysis in the comparison between benign and malignant components. J Clin Pathol. 2001;54:631. doi: 10.1136/jcp.54.8.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J., Du X. Genomic and molecular aberrations in malignant peripheral nerve sheath tumor and their roles in personalized target therapy. Surg Oncol. 2013;22:e53–e57. doi: 10.1016/j.suronc.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Birindelli S., Perrone F., Oggionni M., Lavarino C., Pasini B., Vergani B., Ranzani G.N., Pierotti M.A., Pilotti S. Rb and TP53 pathway alterations in sporadic and NF1-related malignant peripheral nerve sheath tumors. Lab Invest. 2001;81:833–844. doi: 10.1038/labinvest.3780293. [DOI] [PubMed] [Google Scholar]
- 24.Legius E., Dierick H., Wu R., Hall B.K., Marynen P., Cassiman J.J., Glover T.W. TP53 mutations are frequent in malignant NFI tumors. Genes Chromosomes Cancer. 1994;10:250–255. doi: 10.1002/gcc.2870100405. [DOI] [PubMed] [Google Scholar]
- 25.Perry A., Kunz S.N., Fuller C.E., Banerjee R., Marley E.F., Liapis H., Watson M.A., Gutmann D.H. Differential NF1, p16, and EGFR patterns by interphase cytogenetics (FISH) in malignant peripheral nerve sheath tumor (MPNST) and morphologically similar spindle cell neoplasms. J Neuropathol Exp Neurol. 2002;61:702–709. doi: 10.1093/jnen/61.8.702. [DOI] [PubMed] [Google Scholar]
- 26.Menon A., Anderson K., Riccardi V., Chung R., Whaley J., Yandell D., Farmer G., Freiman R., Lee J., Li F. Chromosome 17p deletions and p53 gene mutations associated with the formation of malignant neurofibrosarcomas in von Recklinghausen neurofibromatosis. Proc Natl Acad Sci U S A. 1990;87:5435. doi: 10.1073/pnas.87.14.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kourea H.P., Orlow I., Scheithauer B.W., Cordon-Cardo C., Woodruff J.M. Deletions of the INK4A gene occur in malignant peripheral nerve sheath tumors but not in neurofibromas. Am J Pathol. 1999;155:1855. doi: 10.1016/S0002-9440(10)65504-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen G.P., Stemmer-Rachamimov A.O., Ino Y., Moller M.B., Rosenberg A.E., Louis D.N. Malignant transformation of neurofibromas in neurofibromatosis 1 is associated with CDKN2A/p16 inactivation. Am J Pathol. 1999;155:1879. doi: 10.1016/S0002-9440(10)65507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mantripragada K.K., Spurlock G., Kluwe L., Chuzhanova N., Ferner R.E., Frayling I.M., Dumanski J.P., Guha A., Mautner V., Upadhyaya M. High-resolution DNA copy number profiling of malignant peripheral nerve sheath tumors using targeted microarray-based comparative genomic hybridization. Clin Cancer Res. 2008;14:1015. doi: 10.1158/1078-0432.CCR-07-1305. [DOI] [PubMed] [Google Scholar]
- 30.Mawrin C., Kirches E., Boltze C., Dietzmann K., Roessner A., Schneider-Stock R. Immunohistochemical and molecular analysis of p53, RB, and PTEN in malignant peripheral nerve sheath tumors. Virchows Arch. 2002;440:610–615. doi: 10.1007/s00428-001-0550-4. [DOI] [PubMed] [Google Scholar]
- 31.Holtkamp N., Okuducu A.F., Mucha J., Afanasieva A., Hartmann C., Atallah I., Estevez-Schwarz L., Mawrin C., Friedrich R.E., Mautner V.F. Mutation and expression of PDGFRA and KIT in malignant peripheral nerve sheath tumors, and its implications for imatinib sensitivity. Carcinogenesis. 2006;27:664. doi: 10.1093/carcin/bgi273. [DOI] [PubMed] [Google Scholar]
- 32.Storlazzi C., Brekke H., Mandahl N., Brosjö O., Smeland S., Lothe R., Mertens F. Identification of a novel amplicon at distal 17q containing the BIRC5/SURVIVIN gene in malignant peripheral nerve sheath tumours. J Pathol. 2006;209:492–500. doi: 10.1002/path.1998. [DOI] [PubMed] [Google Scholar]
- 33.Badache A., De Vries G.H. Neurofibrosarcoma-derived Schwann cells overexpress platelet-derived growth factor (PDGF) receptors and are induced to proliferate by PDGF BB. J Cell Physiol. 1998;177:334–342. doi: 10.1002/(SICI)1097-4652(199811)177:2<334::AID-JCP15>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 34.Badache A., Muja N., De Vries G.H. Expression of Kit in neurofibromin-deficient human Schwann cells: role in Schwann cell hyperplasia associated with type 1 neurofibromatosis. Oncogene. 1998;17:795–800. doi: 10.1038/sj.onc.1201978. [DOI] [PubMed] [Google Scholar]
- 35.Forus A., Weghuis D.O., Smeets D., Fodstad O., Myklebost O., van Kessel A.G. Comparative genomic hybridization analysis of human sarcomas, I: occurrence of genomic imbalances and identification of a novel major amplicon at 1q21-q22 in soft tissue sarcomas. Genes Chromosomes Cancer. 1995;14:8–14. doi: 10.1002/gcc.2870140103. [DOI] [PubMed] [Google Scholar]
- 36.Lothe R.A., Karhu R., Mandahl N., Mertens F., Saeter G., Heim S., Borresen-Dale A.L., Kallioniemi O.P. Gain of 17q24-qter detected by comparative genomic hybridization in malignant tumors from patients with von Recklinghausen's neurofibromatosis. Cancer Res. 1996;56:4778–4781. [PubMed] [Google Scholar]
- 37.Mechtersheimer G., Otano-Joos M., Ohl S., Benner A., Lehnert T., Willeke F., Moller P., Otto H.F., Lichter P., Joos S. Analysis of chromosomal imbalances in sporadic and NF1-associated peripheral nerve sheath tumors by comparative genomic hybridization. Genes Chromosomes Cancer. 1999;25:362–369. [PubMed] [Google Scholar]
- 38.Mertens F., Rydholm A., Bauer H.F., Limon J., Nedoszytko B., Szadowska A., Willen H., Heim S., Mitelman F., Mandahl N. Cytogenetic findings in malignant peripheral nerve sheath tumors. Int J Cancer. 1995;61:793–798. doi: 10.1002/ijc.2910610609. [DOI] [PubMed] [Google Scholar]
- 39.Plaat B.E., Molenaar W.M., Mastik M.F., Hoekstra H.J., te Meerman G.J., van den Berg E. Computer-assisted cytogenetic analysis of 51 malignant peripheral-nerve-sheath tumors: sporadic vs. neurofibromatosis-type-1-associated malignant schwannomas. Int J Cancer. 1999;83:171–178. doi: 10.1002/(sici)1097-0215(19991008)83:2<171::aid-ijc5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt H., Wurl P., Taubert H., Meye A., Bache M., Holzhausen H.J., Hinze R. Genomic imbalances of 7p and 17q in malignant peripheral nerve sheath tumors are clinically relevant. Genes Chromosomes Cancer. 1999;25:205–211. [PubMed] [Google Scholar]
- 41.Schmidt H., Taubert H., Meye A., Wurl P., Bache M., Bartel F., Holzhausen H.J., Hinze R. Gains in chromosomes 7, 8q, 15q and 17q are characteristic changes in malignant but not in benign peripheral nerve sheath tumors from patients with Recklinghausen's disease. Cancer Lett. 2000;155:181–190. doi: 10.1016/s0304-3835(00)00426-2. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt H., Taubert H., Wurl P., Bache M., Bartel F., Holzhausen H.J., Hinze R. Cytogenetic characterization of six malignant peripheral nerve sheath tumors: comparison of karyotyping and comparative genomic hybridization. Cancer Genet Cytogenet. 2001;128:14–23. doi: 10.1016/s0165-4608(01)00393-4. [DOI] [PubMed] [Google Scholar]
- 43.Lothe R.A., Smith-Sorensen B., Hektoen M., Stenwig A.E., Mandahl N., Saeter G., Mertens F. Biallelic inactivation of TP53 rarely contributes to the development of malignant peripheral nerve sheath tumors. Genes Chromosomes Cancer. 2001;30:202–206. [PubMed] [Google Scholar]
- 44.Cichowski K., Shih T.S., Schmitt E., Santiago S., Reilly K., McLaughlin M.E., Bronson R.T., Jacks T. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999;286:2172–2176. doi: 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 45.Vogel K.S., Klesse L.J., Velasco-Miguel S., Meyers K., Rushing E.J., Parada L.F. Mouse tumor model for neurofibromatosis type 1. Science. 1999;286:2176–2179. doi: 10.1126/science.286.5447.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tawbi H., Thomas D., Lucas D.R., Biermann J.S., Schuetze S.M., Hart A.L., Chugh R., Baker L.H. Epidermal growth factor receptor expression and mutational analysis in synovial sarcomas and malignant peripheral nerve sheath tumors. Oncologist. 2008;13:459–466. doi: 10.1634/theoncologist.2007-0166. [DOI] [PubMed] [Google Scholar]
- 47.Tabone-Eglinger S., Bahleda R., Cote J.F., Terrier P., Vidaud D., Cayre A., Beauchet A., Theou-Anton N., Terrier-Lacombe M.J., Lemoine A., Penault-Llorca F., Le Cesne A., Emile J.F. Frequent EGFR positivity and overexpression in high-grade areas of human MPNSTs. Sarcoma. 2008;2008:849156. doi: 10.1155/2008/849156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du X., Yang J., Ylipää A., Zhu Z. Genomic amplification and high expression of EGFR are key targetable oncogenic events in malignant peripheral nerve sheath tumor. J Hematol Oncol. 2013;6:93. doi: 10.1186/1756-8722-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Ling B.C., Wu J., Miller S.J., Monk K.R., Shamekh R., Rizvi T.A., Decourten-Myers G., Vogel K.S., DeClue J.E., Ratner N. Role for the epidermal growth factor receptor in neurofibromatosis-related peripheral nerve tumorigenesis. Cancer Cell. 2005;7:65–75. doi: 10.1016/j.ccr.2004.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su W., Sin M., Darrow A., Sherman L.S. Malignant peripheral nerve sheath tumor cell invasion is facilitated by Src and aberrant CD44 expression. Glia. 2003;42:350–358. doi: 10.1002/glia.10206. [DOI] [PubMed] [Google Scholar]
- 51.Okawa T., Michaylira C.Z., Kalabis J., Stairs D.B., Nakagawa H., Andl C.D., Johnstone C.N., Klein-Szanto A.J., El-Deiry W.S., Cukierman E., Herlyn M., Rustgi A.K. The functional interplay between EGFR overexpression, hTERT activation, and p53 mutation in esophageal epithelial cells with activation of stromal fibroblasts induces tumor development, invasion, and differentiation. Genes Dev. 2007;21:2788–2803. doi: 10.1101/gad.1544507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Esteve A., Lehman T., Jiang W., Weinstein I.B., Harris C.C., Ruol A., Peracchia A., Montesano R., Hollstein M. Correlation of p53 mutations with epidermal growth factor receptor overexpression and absence of mdm2 amplification in human esophageal carcinomas. Mol Carcinog. 1993;8:306–311. doi: 10.1002/mc.2940080414. [DOI] [PubMed] [Google Scholar]
- 53.Hakozaki M., Hojo H., Sato M., Tajino T., Yamada H., Kikuchi S., Abe M. Establishment and characterization of a novel human malignant peripheral nerve sheath tumor cell line, FMS-1, that overexpresses epidermal growth factor receptor and cyclooxygenase-2. Virchows Arch. 2009;455:517–526. doi: 10.1007/s00428-009-0848-1. [DOI] [PubMed] [Google Scholar]
- 54.Dai M., Wang P., Boyd A.D., Kostov G., Athey B., Jones E.G., Bunney W.E., Myers R.M., Speed T.P., Akil H. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 56.Feber A., Wilson G.A., Zhang L., Presneau N., Idowu B., Down T.A., Rakyan V.K., Noon L.A., Lloyd A.C., Stupka E. Comparative methylome analysis of benign and malignant peripheral nerve sheath tumors. Genome Res. 2011;21:515–524. doi: 10.1101/gr.109678.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Irizarry R.A., Ladd-Acosta C., Wen B., Wu Z., Montano C., Onyango P., Cui H., Gabo K., Rongione M., Webster M. The human colon cancer methylome shows similar hypo-and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang J., Cogdell D., Yang D., Hu L., Li H., Zheng H., Du X., Pang Y., Trent J., Chen K. Deletion of the WWOX gene and frequent loss of its protein expression in human osteosarcoma. Cancer Lett. 2010;291:31–38. doi: 10.1016/j.canlet.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 59.Yang J., Ylipää A., Sun Y., Zheng H., Chen K., Nykter M., Trent J., Ratner N., Lev D.C., Zhang W. Genomic and molecular characterization of malignant peripheral nerve sheath tumor identifies the IGF1R pathway as a primary target for treatment. Clin Cancer Res. 2011;17:7563–7573. doi: 10.1158/1078-0432.CCR-11-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olshen A.B., Venkatraman E., Lucito R., Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 61.Van De Wiel M.A., Kim K.I., Vosse S.J., Van Wieringen W.N., Wilting S.M., Ylstra B. CGHcall: calling aberrations for array CGH tumor profiles. Bioinformatics. 2007;23:892–894. doi: 10.1093/bioinformatics/btm030. [DOI] [PubMed] [Google Scholar]
- 62.Sun W., Wright F.A., Tang Z., Nordgard S.H., Van Loo P., Yu T., Kristensen V.N., Perou C.M. Integrated study of copy number states and genotype calls using high-density SNP arrays. Nucleic Acids Res. 2009;37:5365–5377. doi: 10.1093/nar/gkp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Daniel A.R., Faivre E.J., Lange C.A. Phosphorylation-dependent antagonism of sumoylation derepresses progesterone receptor action in breast cancer cells. Mol Endocrinol. 2007;21:2890–2906. doi: 10.1210/me.2007-0248. [DOI] [PubMed] [Google Scholar]
- 64.Moriarity B.S., Rahrmann E.P., Keng V.W., Manlove L.S., Beckmann D.A., Wolf N.K., Khurshid T., Bell J.B., Largaespada D.A. Modular assembly of transposon integratable multigene vectors using RecWay assembly. Nucleic Acids Res. 2013;48:e92. doi: 10.1093/nar/gkt115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rahrmann E.P., Watson A.L., Keng V.W., Choi K., Moriarity B.S., Beckmann D.A., Wolf N.K., Sarver A., Collins M.H., Moertel C.L. Forward genetic screen for malignant peripheral nerve sheath tumor formation identifies new genes and pathways driving tumorigenesis. Nat Genet. 2013;45:756–766. doi: 10.1038/ng.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Vries A., Flores E.R., Miranda B., Hsieh H.M., van Oostrom C.T., Sage J., Jacks T. Targeted point mutations of p53 lead to dominant-negative inhibition of wild-type p53 function. Proc Natl Acad Sci U S A. 2002;99:2948–2953. doi: 10.1073/pnas.052713099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olive K.P., Tuveson D.A., Ruhe Z.C., Yin B., Willis N.A., Bronson R.T., Crowley D., Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 68.Hummel T.R., Jessen W.J., Miller S.J., Kluwe L., Mautner V.F., Wallace M.R., Lazaro C., Page G.P., Worley P.F., Aronow B.J., Schorry E.K., Ratner N. Gene expression analysis identifies potential biomarkers of neurofibromatosis type 1 including adrenomedullin. Clin Cancer Res. 2010;16:5048–5057. doi: 10.1158/1078-0432.CCR-10-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Collins F., Barker A. Mapping the cancer genome. Sci Am. 2007;296:50–57. [PubMed] [Google Scholar]
- 70.Keng V.W., Watson A.L., Rahrmann E.P., Li H., Tschida B.R., Moriarity B.S., Choi K., Rizvi T.A., Collins M.H., Wallace M.R. Conditional inactivation of Pten with EGFR overexpression in Schwann cells models sporadic MPNST. Sarcoma. 2012;2012:620834. doi: 10.1155/2012/620834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Magro G., Amico P., Vecchio G.M., Caltabiano R., Castaing M., Kacerovska D., Kazakov D.V., Michal M. Multinucleated floret-like giant cells in sporadic and NF1-associated neurofibromas: a clinicopathologic study of 94 cases. Virchows Arch. 2010;456:71–76. doi: 10.1007/s00428-009-0859-y. [DOI] [PubMed] [Google Scholar]
- 72.Pytel P., Taxy J.B., Krausz T. Divergent differentiation in malignant soft tissue neoplasms: the paradigm of liposarcoma and malignant peripheral nerve sheath tumor. Int J Surg Pathol. 2005;13:19–28. doi: 10.1177/106689690501300103. [DOI] [PubMed] [Google Scholar]
- 73.Keng V.W., Rahrmann E.P., Watson A.L., Tschida B.R., Moertel C.L., Jessen W.J., Rizvi T.A., Collins M.H., Ratner N., Largaespada D.A. PTEN and NF1 inactivation in Schwann cells produces a severe phenotype in the peripheral nervous system that promotes the development and malignant progression of peripheral nerve sheath tumors. Cancer Res. 2012;72:3405–3413. doi: 10.1158/0008-5472.CAN-11-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kazmi S.J., Byer S.J., Eckert J.M., Turk A.N., Huijbregts R.P., Brossier N.M., Grizzle W.E., Mikhail F.M., Roth K.A., Carroll S.L. Transgenic mice overexpressing neuregulin-1 model neurofibroma-malignant peripheral nerve sheath tumor progression and implicate specific chromosomal copy number variations in tumorigenesis. Am J Pathol. 2013;182:646–667. doi: 10.1016/j.ajpath.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Friedrich R.E., Holstein A., Middendorff R., Davidoff M.S. Vascular wall cells contribute to tumourigenesis in cutaneous neurofibromas of patients with neurofibromatosis type 1: a comparative histological, ultrastructural and immunohistochemical study. Anticancer Res. 2012;32:2139–2158. [PubMed] [Google Scholar]
- 76.Upadhyaya M., Spurlock G., Thomas L., Thomas N.S., Richards M., Mautner V., Cooper D.N., Guha A., Yan J. Microarray-based copy number analysis of neurofibromatosis type-1 (NF1)-associated malignant peripheral nerve sheath tumors reveals a role for Rho–GTPase pathway genes in NF1 tumorigenesis. Hum Mutat. 2012;33:763–776. doi: 10.1002/humu.22044. [DOI] [PubMed] [Google Scholar]
- 77.Hackstein J.H. The lethal prune/Killer-of-prune interaction of Drosophila causes a syndrome resembling human neurofibromatosis (NF1) Eur J Cell Biol. 1992;58:429–444. [PubMed] [Google Scholar]
- 78.Holtkamp N., Atallah I., Okuducu A.F., Mucha J., Hartmann C., Mautner V.F., Friedrich R.E., Mawrin C., von Deimling A. MMP-13 and p53 in the progression of malignant peripheral nerve sheath tumors. Neoplasia. 2007;9:671–677. doi: 10.1593/neo.07304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Upadhyaya M., Kluwe L., Spurlock G., Monem B., Majounie E., Mantripragada K., Ruggieri M., Chuzhanova N., Evans D.G., Ferner R., Thomas N., Guha A., Mautner V. Germline and somatic NF1 gene mutation spectrum in NF1-associated malignant peripheral nerve sheath tumors (MPNSTs) Hum Mutat. 2008;29:74–82. doi: 10.1002/humu.20601. [DOI] [PubMed] [Google Scholar]
- 80.Sharpless N.E., DePinho R.A. Cancer: crime and punishment. Nature. 2005;436:636–637. doi: 10.1038/436636a. [DOI] [PubMed] [Google Scholar]
- 81.Itakura Y., Sasano H., Shiga C., Furukawa Y., Shiga K., Mori S., Nagura H. Epidermal growth factor receptor overexpression in esophageal carcinoma: an immunohistochemical study correlated with clinicopathologic findings and DNA amplification. Cancer. 1994;74:795–804. doi: 10.1002/1097-0142(19940801)74:3<795::aid-cncr2820740303>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 82.Parenti A.R., Rugge M., Frizzera E., Ruol A., Noventa F., Ancona E., Ninfo V. p53 Overexpression in the multistep process of esophageal carcinogenesis. Am J Surg Pathol. 1995;19:1418–1422. doi: 10.1097/00000478-199512000-00008. [DOI] [PubMed] [Google Scholar]
- 83.Ohashi S., Natsuizaka M., Wong G.S., Michaylira C.Z., Grugan K.D., Stairs D.B., Kalabis J., Vega M.E., Kalman R.A., Nakagawa M., Klein-Szanto A.J., Herlyn M., Diehl J.A., Rustgi A.K., Nakagawa H. Epidermal growth factor receptor and mutant p53 expand an esophageal cellular subpopulation capable of epithelial-to-mesenchymal transition through ZEB transcription factors. Cancer Res. 2010;70:4174–4184. doi: 10.1158/0008-5472.CAN-09-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schmitt C.A., Fridman J.S., Yang M., Baranov E., Hoffman R.M., Lowe S.W. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 85.Nithianandarajah-Jones G.N., Wilm B., Goldring C.E., Müller J., Cross M.J. ERK5: structure, regulation and function. Cell Signal. 2012;24:2187–2196. doi: 10.1016/j.cellsig.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 86.Kato Y., Chao T., Hayashi M., Tapping R.I. Role of BMK1 in regulation of growth factor-induced cellular responses. Immunol Res. 2000;21:233–237. doi: 10.1385/IR:21:2-3:233. [DOI] [PubMed] [Google Scholar]
- 87.Mattingly R.R., Kraniak J.M., Dilworth J.T., Mathieu P., Bealmear B., Nowak J.E., Benjamins J.A., Tainsky M.A., Reiners J.J. The mitogen-activated protein kinase/extracellular signal-regulated kinase kinase inhibitor PD184352 (CI-1040) selectively induces apoptosis in malignant schwannoma cell lines. J Pharmacol Exp Ther. 2006;316:456–465. doi: 10.1124/jpet.105.091454. [DOI] [PubMed] [Google Scholar]
- 88.Li H., Velasco-Miguel S., Vass W.C., Parada L.F., DeClue J.E. Epidermal growth factor receptor signaling pathways are associated with tumorigenesis in the Nf1: p53 mouse tumor model. Cancer Res. 2002;62:4507–4513. [PubMed] [Google Scholar]
- 89.Fang L., Li G., Liu G., Lee S.W., Aaronson S.A. p53 Induction of heparin-binding EGF-like growth factor counteracts p53 growth suppression through activation of MAPK and PI3K/Akt signaling cascades. EMBO J. 2001;20:1931–1939. doi: 10.1093/emboj/20.8.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brosius S.N., Turk A.N., Byer S.J., Brossier N.M., Kohli L., Whitmire A., Mikhail F.M., Roth K.A., Carroll S.L. Neuregulin-1 overexpression and Trp53 haploinsufficiency cooperatively promote de novo malignant peripheral nerve sheath tumor pathogenesis. Acta Neuropathol. 2013;127:1–19. doi: 10.1007/s00401-013-1209-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Watson A.L., Rahrmann E.P., Moriarity B.S., Choi K., Conboy C.B., Greeley A.D., Halfond A.L., Anderson L.K., Wahl B.R., Keng V.W. Canonical Wnt/β-catenin signaling drives human Schwann cell transformation, progression, and tumor maintenance. Cancer Discov. 2013;3:674–689. doi: 10.1158/2159-8290.CD-13-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Modulating EGFR and TP53 expression. A: Schematics for the plasmids used to overexpress EGFR cDNA and introduce an shRNA against TP53. B: Bar graph depicts quantification of Western blot analysis in Figure 2C. The statistic used was an unpaired Student's t-test. C: Bar graph depicts size of colonies from Figure 2C. D: Sequences indicated the target sequence of the TALENs for the TP53 locus. The translational start site (ATG) is boldfaced. The TALEN binding sites are underlined. The top row depicts the WT sequence of TP53. The lower row contains dashes representing deleted nucleotides from a TP53 KO clone. E: Bar graph depicts size of colonies from Figure 2D. ∗P < 0.05.
Histological analysis of tumors. A: IHC analysis for NeuN (neurons), vimentin (fibroblasts), and CD45 (macrophages) on a neurofibroma sample. B: Graph depicts the number of mitotic events per field for each tumor type listed. C: H&E analysis of grade 3 PNSTs demonstrating a significant pleomorphism among the tumors, including variability in cell morphological characteristics and extracellular matrix content. Arrows indicate the presence of matrix in matrix-producing cells, indicating wreath-like giant cells in the last panel. D: No primary control images of tumors treated with rabbit polyclonal and mouse monoclonal antibodies. ∗∗P < 0.01, unpaired Student's t-test. Original magnification, ×400 (D).