Abstract
Mitochondria are a common energy source for organs and organisms; their diverse functions are specialized according to the unique phenotypes of their hosting environment. Perturbation of mitochondrial homeostasis accompanies significant pathological phenotypes. However, the connections between mitochondrial proteome properties and function remain to be experimentally established on a systematic level. This uncertainty impedes the contextualization and translation of proteomic data to the molecular derivations of mitochondrial diseases. We present a collection of mitochondrial features and functions from four model systems, including two cardiac mitochondrial proteomes from distinct genomes (human and mouse), two unique organ mitochondrial proteomes from identical genetic codons (mouse heart and mouse liver), as well as a relevant metazoan out-group (drosophila). The data, composed of mitochondrial protein abundance and their biochemical activities, capture the core functionalities of these mitochondria. This investigation allowed us to redefine the core mitochondrial proteome from organs and organisms, as well as the relevant contributions from genetic information and hosting milieu. Our study has identified significant enrichment of disease-associated genes and their products. Furthermore, correlational analyses suggest that mitochondrial proteome design is primarily driven by cellular environment. Taken together, these results connect proteome feature with mitochondrial function, providing a prospective resource for mitochondrial pathophysiology and developing novel therapeutic targets in medicine.
Keywords: mitochondrial proteome, mitochondrial function, heart diseases, intergenomic, intragenomic, proteomic comparisons
INTRODUCTION
Mitochondria are complex and intricately calibrated systems designed for the inception and perpetuation of life in a majority of eukaryotic organisms.1-4 They are explicitly engineered to transect many aspects of cellular biology such as bioenergetics, metabolism, calcium signaling, reactive oxygen species (ROS) generation, and apoptosis. Mitochondrial dysfunctions have emerged as the underlying causes of various complex diseases including cancer, diabetes, obesity, neurodegenerative diseases, aging, and multiple forms of cardiomyopathy.2,5-16 For instance, during heart failure, energy production decreases amid abnormal mitochondrial metabolic activity; however, on a molecular level, the mitochondrial proteome selectively alters its expression profile, including the redistribution of respiratory chain subunit abundances, as well as decreases in fatty acid oxidation proteins.7,9,11,17-20 Therefore, advancing our knowledge of cellular functions in health and disease requires a thorough understanding of the mitochondrial proteome and its definitive relationship to biological function.
Previous studies have reported protein compositions of mitochondria in yeast21,22 as well as in mouse,23-26 human,27,28 rat,1,29-31 rabbit,32 and drosophila33,34 tissues. However, it is evident that there are inconsistencies and discrepancies among some of the data sets. The direct link between mitochondrial proteome biology and pathological phenotypic observations remains ambiguous, which has impeded diagnostic interpretations of large-scale proteomics data. This lack of clarity is partially due to fragmented efforts in assessing mitochondrial functionalities and proteome heterogeneity in various systems. Questions of how closely proteome parameters such as diversity and abundance conform to, or predict, biological functions still remain unanswered. It is clear that an unbiased characterization of the functional proteome of mitochondria would undoubtedly further our insight into complex mitochondrial functions and mitochondrial-associated diseases. In this regard, experimental proteomics data offer an indispensable insight into organelle biology in parallel to the transcription-oriented efforts, alleviating potential issues arising from algorithms predicting mitochondrial-targeted genes and their lack of tissue specificity.
In this study, we propose a strategy that combines functional assays and protein expression measurements through label-free quantification analyses; using NSAF values35-37 to characterize multiple aspects of mitochondrial pathways has been the only feasible and economical approach for analyzing large-scale proteomic data sets from multiple model systems. We utilized this strategy in mitochondria isolated from human hearts and compared them with data from mouse hearts because of its prevalence as a proxy for human hearts; mouse livers, used as a comparison with organ mitochondria; and drosophila, serving as both a nonmammalian metazoan out-group and an experimental model found to correlate with some human diseases.38-40
Our investigation reveals intergenomic and intragenomic correlations for proteome diversity, proteome abundance, and mitochondrial function. Through correlational analyses, we delineate that the molecular mechanisms of the mitochondrial proteome are primarily dictated by its cellular environment. Furthermore, the mitochondrial proteome is predominantly composed of a small number of vastly abundant proteins involved in the principal processes of cellular survival, including oxidative phosphorylation and metabolism. With our most comprehensive mitochondrial protein repertoire to date, we established a core mitochondrial proteome of 419 conserved proteins across four model systems. Additionally, we determined that fundamental mitochondrial functions are defined by the evolutionary conservation of a core mitochondrial proteome across different species, whereas organ-derived proteins, whether by novel genetic information or cell-specific translocation, reflect the diversification in the heterogeneity of mitochondrial populations. Collectively, this study provides a useful resource and network for future studies on mitochondrial biology. The definitive characteristics of mitochondrial proteomes afford great opportunities in developing targeted treatment for human disease.
EXPERIMENTAL PROCEDURES
Experimental procedures on human tissues were approved by the UCLA Human Subjects Protection Committee (HSPC) and the UCLA Institutional Review Boards (IRBs). The experimental procedures on animals were performed in accordance with the Animal Research Committee guidelines at UCLA and the Guide for the Care and Use of Laboratory Animals, published by the National Institutes of Health. An overview of our experimental workflow is shown in Figure S1 in the Supporting Information. A comprehensive Methods Section is available in the Supporting Information.
Tissue Sample Collection
Tissue samples were procured from each model system. Mouse heart and liver tissue samples were obtained from thirty 8–10 week old ICR strain mice. Human heart samples from the free anterior left ventricular wall were collected from individuals (average age = 49 ± 8 years, n = 5, 4 males and 1 female) previously treated with a left ventricular assist device (LVAD). These individuals exhibited normal left ventricular end diastolic dimension (LVEDD) after LVAD treatment. This improvement is featured in Figure S2 in the Supporting Information. Additionally, approximately 1000 adult wild-type Drosophila melanogaster (Oregon R strain) were immobilized by chilling prior to mitochondria extraction. More information regarding methods can be found in S2 in the Supporting Information.
Isolation and Purification of Functional, Viable Mitochondria from Human Heart, Mouse Heart, Mouse Liver, and Drosophila
Mitochondria were isolated from freshly collected mouse hearts, mouse livers, human hearts, and Drosophila melanogaster by differential centrifugation as described.26,41,42 The freshly isolated mitochondria were subjected to a series of functional and structural validations.26
Assessment of Mitochondrial Function
The activities of the mitochondrial electron transport chain (ETC) complexes I (C–I) and V (C–V) were assessed in vitro by spectrophotometric measurements.43-46 Pharmacological inhibitors were employed to determine the inhibitor-insensitive background of each complex. Pyruvate dehydrogenase (PDH) activity, proteolytic activity, and glutathione reductase activity assays were performed according to the manufacturer’s instructions. Mitochondrial O2 consumption and the susceptibility of mitochondria to calcium-induced injury were determined as described.26,41 A detailed explanation of the assay methods is available in the Supporting Information.
Quantitative Proteomic Profiling of Mitochondrial Proteomes
SDS-PAGE, LC–MS/MS, and spectral analyses were performed as described.26,41 Details regarding sample separation, chromatography, instrumentation settings, database searching, and protein identification criteria are detailed in the Supporting Information S4. Mitochondrial protein abundances were assessed according to normalized spectral abundance factors (NSAF);35-37,41 this was then compared across all biological samples. The spectral counts for peptides shared among multiple proteins were divided proportionally according to the total spectral count of each protein’s unique peptides, with proteins possessing a greater amount of unique spectral counts acquiring a larger portion of the shared peptide’s spectral count. Figure S3 in the Supporting Information summarizes the mass spectrometry (MS) experiments.
Bioinformatics and Statistical Analyses
The molecular properties of the mitochondrial proteome, including molecular weight (MW), isoelectric point (pI), transmembrane domains, and mitochondrial target sequences, were analyzed using the UniProt Knowledgebase26,41,47 and TargetP.48 Biological information for individual proteins was extrapolated from gene ontology annotations (biological processes and molecular functions). Protein orthologs among human, mouse, and drosophila were identified via BioMart.49 In addition, the involvement of mitochondrial proteins in diseases was determined by searching through the Online Mendelian Inheritance in Man (OMIM) and peer-reviewed publications on PubMed.50 Correlational analyses were calculated using Spearman’s coefficient. Finally, the IntAct database (IntAct database release 164b)51 was used to determine the known mitochondrial protein–protein interactions among C–I, C–V, redox, and their associated partner proteins. Cytoscape 3.0 Network Data Integration, Analysis, and Visualization software52 was subsequently employed to depict these interactions. Swelling assay data and spectra analyses results were reported as mean ± SEM. Differences among the experimental groups were analyzed using one-way ANOVAs with posthoc contrasts utilizing the Student’s t test.53 The Mann–Whitney U test was used to determine the significance of protein abundance distribution differences. Values of p < 0.05 were recognized as significant.
RESULTS
Heterogeneic Programming of Mitochondrial Function across Organs and Organisms
We compiled a panel of biochemical assays to evaluate several functional parameters of intact and viable mitochondria isolated from the four model systems. To assess mitochondrial bioenergetics and other biological functions, we determined the reaction rate of the respiratory chain complexes, the respiratory control index (RCI), PDH activity, proteolytic capacity, redox regulation, and susceptibility to calcium-induced stress (Figure 1).
Figure 1.
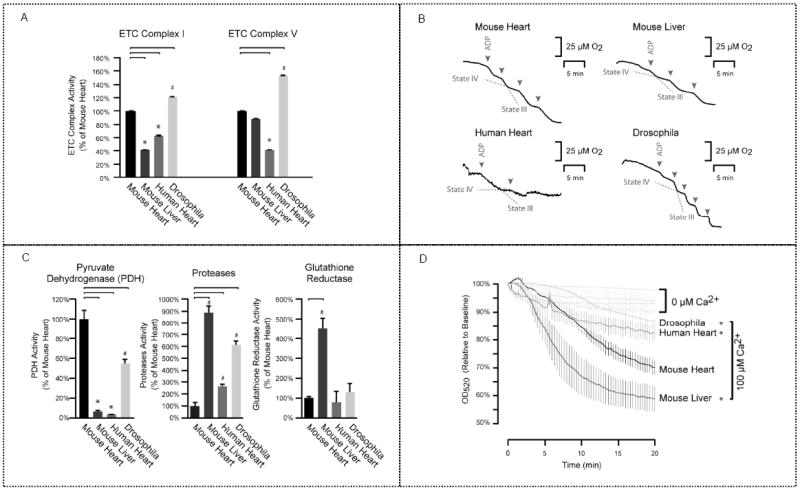
Mitochondrial functional characterization. (a) Electron production rate per microgram of mitochondrial proteins was analyzed to determine ETC complex activity and subsequently normalized to mouse cardiac mitochondria. Mouse liver mitochondria showed lower C–I activity and drosophila demonstrated higher C–I activity compared with the mouse heart. In addition, drosophila exhibited the highest C–V activity overall, with the lowest C–V activity in human heart mitochondria. These differences in respiratory flux and ATP generation can be attributed to cellular environment and genetic background. * or # represents p < 0.05 versus mouse heart; n = 4 per group. (b) RCI tracers are presented; they were measured as a ratio of the oxygen consumption rate by an initial O2 concentration of 220 mmol·L−1. Drosophila mitochondria were the most tightly coupled (RCI: 12.0), followed by the human heart (8.9), mouse heart (7.6), and mouse liver (7.0). A higher RCI value indicates tighter coupling of oxidation and phosphorylation processes. (c) Additional key enzymatic mitochondrial functions revealed similar intra- and intergenomic variability. Mouse liver mitochondria exhibited the highest proteolytic and glutathione reductase activities, whereas the mouse heart had the highest PDH activity, which may be explained by the altered production of reducing equivalents. * or # represents p < 0.05 versus mouse heart; n = 4 per group. (d) Calcium-induced mitochondrial swelling was measured as a reduction of optical density. Mouse liver mitochondria displayed the highest susceptibility to calcium-induced swelling. * represents p < 0.05 versus mouse heart with calcium overload; n = 4 per group.
The activities of ETC C–I (NADH dehydrogenase) and C– V (F1F0 ATP synthase) were assessed. Results were normalized, by tissue weight, to mouse cardiac mitochondria for comparison (Figure 1a). Notably, mouse liver mitochondria demonstrated substantially lower C–I activity, human heart mitochondria showed the lowest C–V activity, and drosophila exhibited the highest Complex activity for C–I and C–V. These higher activities in drosophila suggest increases in both the delivery of reducing equivalents and ATP production due to the high energy turnover of insect flight muscles. In addition, the differential C–I activity levels between mouse heart and mouse liver indicate that cellular environment plays a significant role in the regulation of ETC function. Together, our data highlighted the heterogeneity of these two respiratory complexes and underscored the effect of cellular environments on functional activities.
We next examined the RCI as an indicator of coupling tightness for the respiratory circuitry. The human heart had an RCI of 8.9, whereas mouse heart and mouse liver had RCI values of 7.6 and 7.0, respectively (Figure 1b). Drosophila mitochondria exhibited the highest RCI (12.0), demonstrating a tighter coupling of oxidation and phosphorylation processes.
To evaluate the energetic functions of mitochondria, we examined PDH, protease, and glutathione reductase activities to give a more comprehensive assessment of mitochondrial biology. We observed that mouse heart had the highest PDH activity, with significantly lower activity in mouse liver and human heart (Figure 1c). As anticipated, liver mitochondria exhibited the highest proteolytic and glutathione reductase activity. The heterogeneity of the four model systems was also confirmed by their response to calcium stress-induced injury. Liver mitochondria demonstrated a drastically higher susceptibility to calcium overload than other mitochondria (Figure 1d), likely reflecting the latter’s lack of constant calcium flux as a noncontractile organ. These results accentuate the functional contributions of molecular environment and genetic background on cellular activity.
Dynamic Mitochondrial Proteome Design across Organs and Organisms
Mitochondrial Proteome Composition
To examine the molecular basis of the observed functional heterogeneity in depth, we profiled the proteome of the four mitochondrial populations. In total, we identified 1398 unique proteins from human heart mitochondria, 1620 from mouse heart mitochondria, 1733 from mouse liver mitochondria, and 1015 from drosophila mitochondria. To our knowledge, these results represent one of the most comprehensive mitochondrial protein catalogs for each of these model systems. A complete list of identified proteins is provided in Table S1 in the Supporting Information.54 The distributions of biochemical features, including molecular weight, isoelectric point, as well as number of transmembrane domains, are shown in Figure S4 in the Supporting Information, implicating the dynamic properties of the mitochondrial proteome.
To determine how the mitochondrial proteome was partitioned among all four model systems into core proteins that preserved fundamental functions and diversified proteins that conferred specialized functions, we analyzed the number of proteins and protein orthologs shared by the four model systems. If we consider the minimal ortholog set to be common if varying numbers of homologous genes exist across organisms (Figure 2a, Table S2 in Supporting Information), 419 equivalent proteins were conserved across the model systems. Drosophila possessed the highest percentage of unique proteins (Figure 2b). Although lack of detection cannot distinguish protein absence from extremely low abundance, errors in data acquisition were minimized by comprehensive replications.
Figure 2.
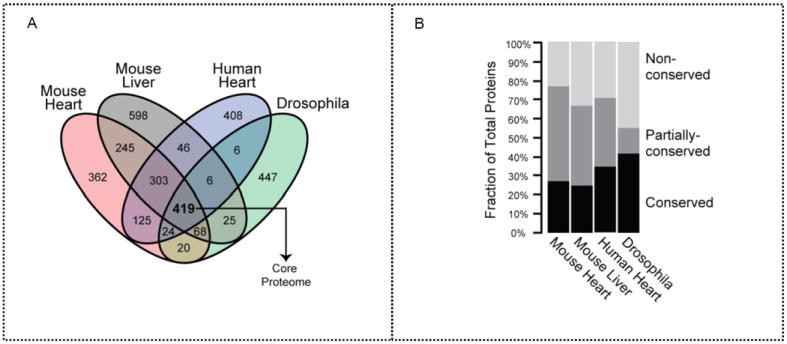
Mitochondrial proteome composition among four mitochondrial populations. (a) Protein orthologs were analyzed for each model systems. The 419 proteins identified in all four model systems represent the conserved core mitochondrial proteome. (b) Proportions of conserved and nonconserved proteins by protein number were denoted in each proteome. Drosophila demonstrated the highest protein count ratio of both conserved and nonconserved proteins.
Mitochondrial Proteome Abundance
Label-free quantification analyses performed using the NSAF values of proteins, yielded the relative abundance of each protein and allowed for cross-proteome comparison among the model systems. Although the accuracy of label-free approaches may be limited by run-to-run variability and redundant peptides among proteins,55 this technique allowed us to circumvent the challenge of synthesizing tens of thousands of stable isotope labeled peptides.56 To ensure accurate quantification, we conducted exhaustive biological and technical replicates (from 18 replicates of the mouse heart to 29 replicates of the human heart) and developed an algorithm to proportionally allocate shared peptides to their corresponding proteins based on the percentage of unique peptides per protein. Our analysis showed that the mitochondrial proteome exhibited a high dynamic range of protein expression levels, with fractional abundance spanning more than five orders of magnitude from 10−1 to 10−6 (Figure 3a), although a vast number of proteins possessed a minimum NSAF value and are in rare abundance. For example, in mouse heart, the most abundant protein accounted for ~3% of the total mitochondrial proteome, whereas the least abundant protein accounted for ~0.00003% of its total mitochondrial proteome. These examples illustrate a 105-fold difference in protein abundance. Furthermore, proteins were ranked according to their abundance (Figure 3b); the top 100 most abundant proteins in each model system represented 57–83% of the mitochondrial proteome, whereas the bottom 50% only accounted for <4% of the mitochondrial protein content (Figure 3b). Conserved proteins were highly abundant, occupying 64% of mouse heart mitochondrial protein content, 52% of mouse liver mitochondria, 64% of human heart mitochondria, and 77% of drosophila mitochondria (Figure 3c). Protein abundance distributions between human heart mitochondria and mouse heart mitochondria were more similar (Spearman’s ρ = 0.70) than those between mouse heart and mouse liver (ρ = 0.65), further supporting the notion that abundance distribution is regulated by cellular environment (Figure 3d).
Figure 3.
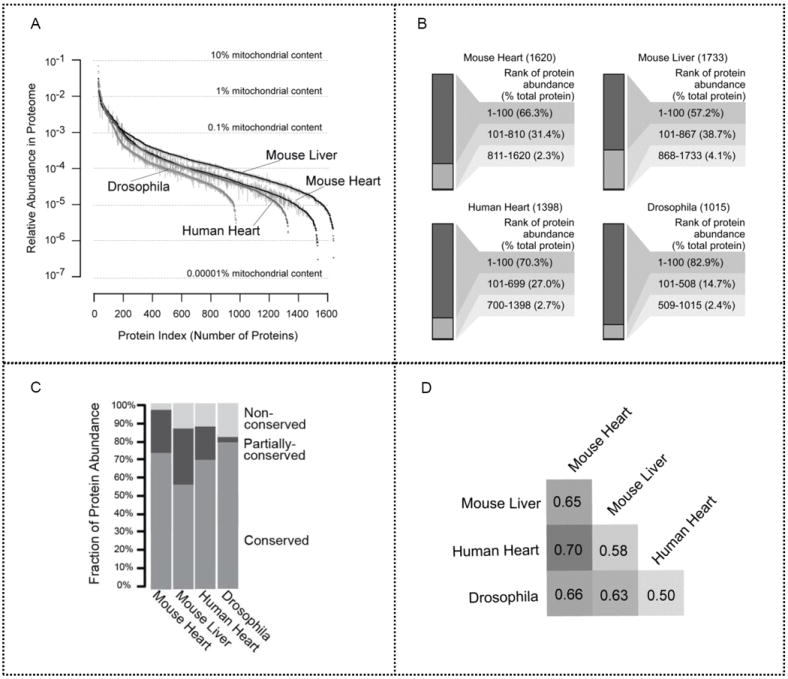
Mitochondrial proteome abundance. (a) Relative protein abundance was indexed based on NSAF values in descending order. The high dynamic range of protein expression levels for the mitochondrial proteome spans over five orders of magnitude. Error bar: SEM. (b) Proteins were ranked according to their abundance. The top 100 proteins in each model system are highly abundant and occupy a majority of the total mitochondrial protein content. The top 50% of each mitochondrial proteome accounts for over 95% of the total protein abundance, whereas the bottom 50% constitutes <4%. (c) The proportion of conserved, partially conserved, and unique proteins was illustrated in all four model systems. The conserved proteins are highly abundant, accounting for 72, 55, 68, and 78% of mitochondrial protein content in the mouse heart, the mouse liver, the human heart, and drosophila, respectively. Furthermore, nonconserved proteins in drosophila comprise the highest unique protein ratio among these organs and organisms, alluding to their specialized functionality. (d) Similarity in distribution of mitochondrial protein abundance was determined. Human heart and mouse heart mitochondrial protein abundance distribution levels are closely related, indicating that mitochondrial populations in the same organ of different species can demonstrate a stronger correlation than mitochondrial populations within different organs of the same organism. Moreover, mouse heart and mouse liver mitochondria had similar, but not identical distributions (ρ = 0.65).
Properties of Mitochondrial Proteome and Function across Organs and Organisms
Mitochondrial Proteome and Functions
Upon categorizing proteins by biological processes, we observed that highabundance clusters are particularly enriched in oxidative phosphorylation (OXPHOS), metabolism, transport, and signaling. Mouse liver possessed the highest abundance value for the metabolism cluster, reflecting its metabolic and biosynthetic specializations (Figure 4a). The mitochondrial proteome subpopulation abundances were analyzed for each functional category (Figure 4b). Among the conserved protein abundances, OXPHOS proteins as well as proteins associated with apoptosis and ETC complex assembly were more abundant in all mitochondrial populations. Proteins involved in TCA metabolic processes were also highly conserved. In contrast, proteins involved in signaling and proteolysis were less conserved among the four model systems. The mitochondrial proteome is therefore dominated by proteins involved in key fundamental metabolic processes.
Figure 4.
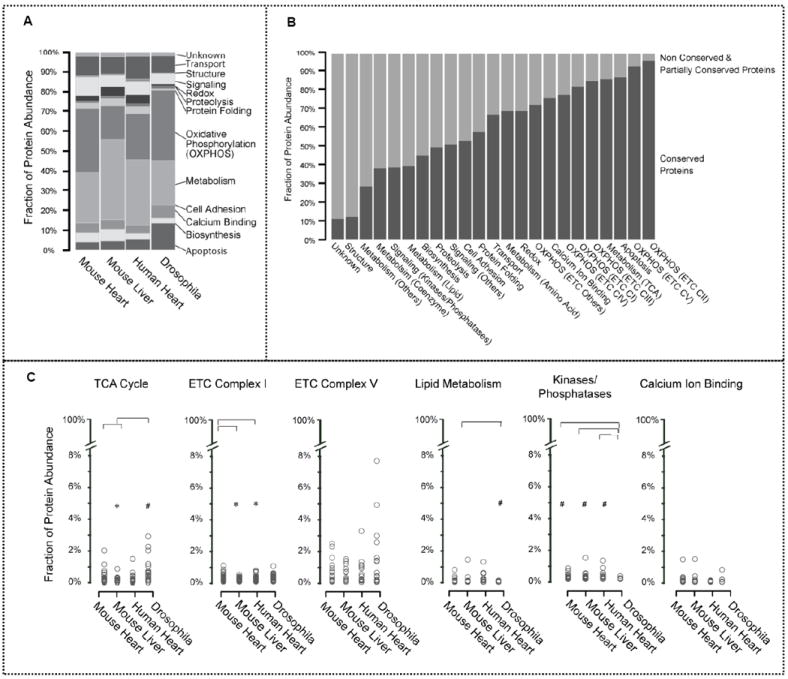
Correlation between the mitochondrial proteome and its functions. (a) Mitochondrial protein abundances were categorized by biological function. The high-abundance clusters are enriched in oxidative phosphorylation, metabolism, and transport-related proteins. In particular, mouse liver has the highest abundance values within the metabolism cluster, highlighting a metabolic specialization. Moreover, these conserved proteins are highly abundant, representing 81.0% of mitochondrial protein content in mouse heart, 59.5% in mouse liver, 79.7% in human heart, and 86.4% in drosophila. (b) Mitochondrial proteins were characterized by biological processes. Conserved proteins and non/partially conserved proteins in each category were quantified. Proteins involved in OXPHOS were concentrated in C–II and C–V. Conserved proteins were also found to be highly involved in apoptosis and TCA metabolism. Structural, signaling, and proteolytic processes have higher proportions of nonshared proteins. (c) Relative fraction of the proteome involved in specified functional pathways was determined. Mitochondrial proteins were further classified through bioinformatics analyses based on their involvement in specific functional pathways and then plotted by their NSAF values. *p < 0.05 versus mouse heart. The mouse heart, human heart, and drosophila models have high TCA cycle protein abundances, while the mouse liver shows significantly lower protein abundances, denoting the differences in TCA cycle activity between contractile and noncontractile tissues. The mouse liver mitochondria also contained the lowest abundance of C–I proteins. C–V was not found to have a significant difference in overall protein abundance among the model systems. Notably, within each Complex, differential subunit abundances spanned over three orders of magnitude. Drosophila had considerably lower lipid metabolism protein abundances in comparison with mouse liver, implicating liver mitochondria as specialized in the synthesis and degradation of lipids. In addition, drosophila mitochondria possess the lowest abundance of kinase/phosphatase proteins, suggesting that there is less phosphorylation regulation. The abundance of calcium-binding proteins was similar across the model systems; this deviates from our previous finding that shows mouse liver is more susceptible to calcium-induced swelling, indicating limitations in direct connections between proteomics data and functionality. Statistical significance was determined with a Mann–Whitney U test.
We subsequently compared mitochondrial protein abundances and their involvement in specific functional pathways using NSAF values (Figure 4c). Mouse heart mitochondria demonstrated higher TCA cycle protein concentrations compared with mouse liver mitochondria. The differences in abundance of TCA cycle proteins among mouse heart, human heart, and drosophila were indistinguishable; this illustrates the variations in TCA activity between contractile and noncontractile tissues. Similarly, liver mitochondria contained the fewest C–I proteins. However, C–V showed no significant difference in overall protein abundance among the model systems. In addition, it was found that within each Complex differential subunit abundances spanned over more than two orders of magnitude. The most abundant proteins within C–V are subunit β in mouse heart, mouse liver, and drosophila, but subunit α in human heart. The least abundant proteins within C–V are subunit α in mouse heart, subunit ε in mouse liver, and subunit β in human heart and drosophila. Moreover, we observed that proteins involved in lipid metabolism showed substantially lower abundances in drosophila when compared to mouse liver, but more than 89% of the drosophila lipid metabolism proteins were conserved among all model systems. In contrast, mouse liver possessed the highest abundance of proteins related to lipid metabolism, of which only 19% were conserved mitochondrial proteins. These results indicated that specialized pathways exist in mouse liver. Furthermore, drosophila possessed the lowest abundance of kinase/phosphatase proteins. Surprisingly, the abundance of calciumion binding proteins in liver mitochondria showed no significant difference across the model systems.
Mitochondrial-encoded proteins were further analyzed based on NSAF values. The ETC subunits appeared to be highly heterogeneous; in particular, mitochondrial-encoded subunits were found to be less abundant than nuclear-encoded subunits, primarily in mouse liver and human heart (under-detected). These subunits may serve as the limiting factors of complex assembly and may better explain the lower respiratory activities of C–I in mouse liver mitochondria and in C–V of human heart mitochondria.
Mitochondrial Interactome and Its Functions
Mitochondrial proteins identified in our study were queried within the IntAct database (IntAct database release 164b) for known protein–protein interactions to demonstrate associating partners among C–I, C–V, redox, and their neighboring proteins in human, mouse, and drosophila data sets. Cytoscape 3.0 Network Data Integration, Analysis, and Visualization software was then utilized to illustrate documented interactions among proteins in C–I, C–V, redox, C–I + redox, as well as C–V + redox. The resulting protein data sets focused on the differences among human, mouse, and drosophila protein connections. The size of each mitochondrial node represents the number of connections in the protein–protein interaction network, with larger nodes indicating more interactions. Interestingly, C–I proteins appeared to be centralized around NADH-ubiquinone oxidoreductase B8 subunit (NDUFA2) in human and α-synuclein in mouse (Figure 5a). In contrast, drosophila protein interactome patterns showed small scattered clusters and fragmented groupings. These results were indicative of the intergenomic protein–protein properties, which may have been influenced by the functional annotations unique to each database. Subsequently, NDUFA2 and α-synuclein were cross-analyzed for significant interactions in the mouse and human data sets, respectively. Although not a major hub, NDUFA2 was found to interact with proteins in the mouse data; however, α-synuclein was not found within the C–I human protein interactions. In the C–V interactome, subunit α (ATP5A1), β subunit (ATP5B), and subunit γ (ATP5C1) demonstrated the most protein–protein interactions (Figure 5a). Similarly, ATP5B served as the major hub in both the drosophila and the mouse network, a feature highlighting the evolutionary importance of ATP5B as a core protein. Redox proteins (Figure 5b) also portrayed a comparable pattern to C–I and C–V, with human proteins (i.e., Apoptosis Inducing Factor 1 – AIFm1) tending to serve as central hubs, whereas mouse and drosophila proteins displayed little affinity for interaction. Incorporation of both C–I + redox as well as C–V + redox revealed a lack of explicit connections between Complex proteins and redox proteins (Figure 5c). However, a single C–V protein (ATP5B) interacted with redox proteins – peroxiredoxin 1 (PRDX1), 2 (PRDX2), and 4 (PRDX4). The mouse C–I and redox protein integration serves as an important guide for the knowledge-building process regarding human protein–protein interactions. Overall, each model system showed a relatively distinct protein–protein interaction network with little overlap among the organisms for C–I, C–V, redox, C–I + redox, as well as C–V + redox proteins, indicating that the integration of protein interactions from other model systems could greatly benefit future investigations of the human interactome.
Figure 5.
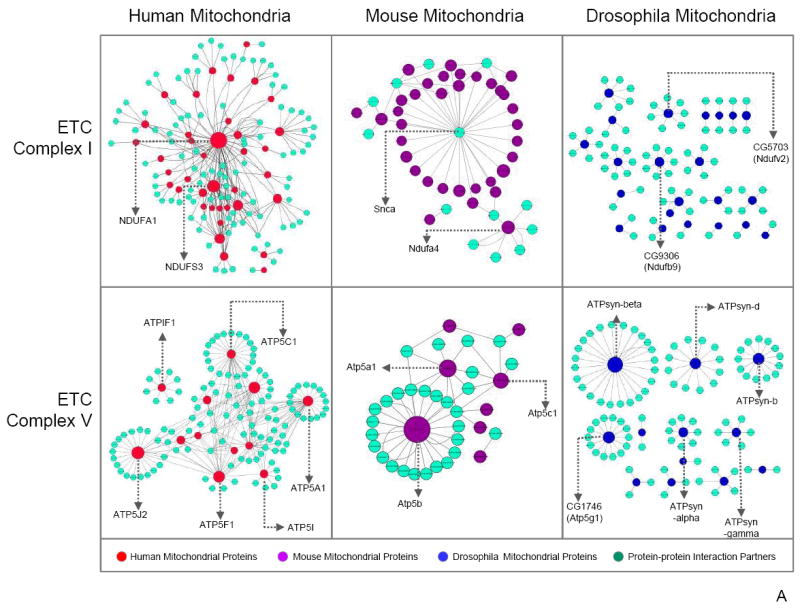
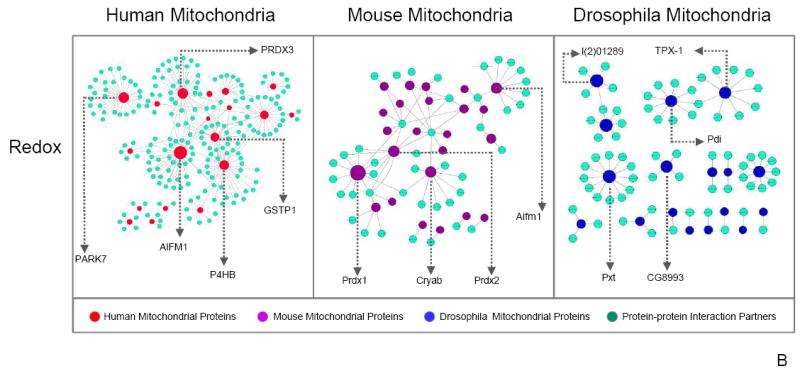
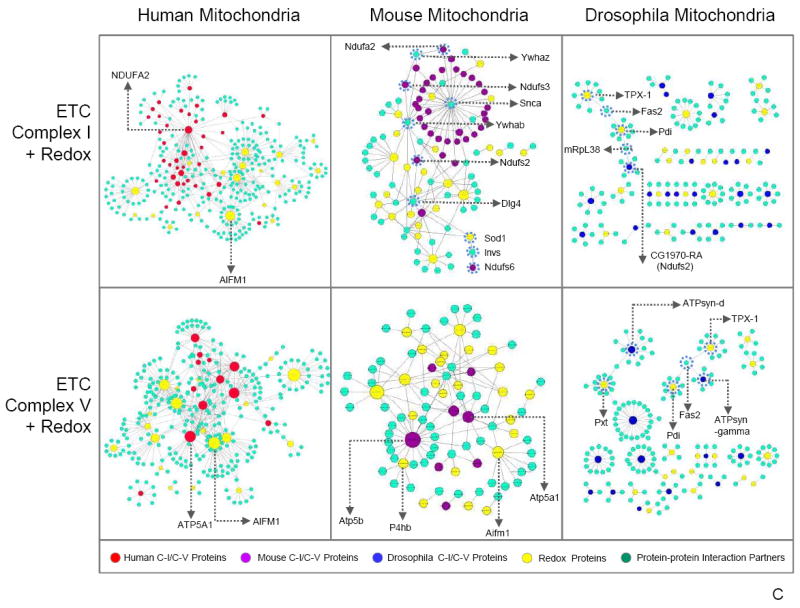
Mitochondrial protein–protein interactome analysis. An interactome analysis of C–I, C–V, redox, C–I + redox, and C–V + redox proteins was performed, and the protein–protein interactions are displayed. Major hubs have been labeled by gene name. The size of each mitochondrial node represents the number of connections in the protein–protein interaction network, with larger nodes indicating more interactions. (a) C–I showed NDUFA2 as the primary human mitochondrial protein with the most connections. Mouse mitochondrial proteins mainly centralized around one partner protein, α-synuclein. In contrast, drosophila proteins did not exhibit a clustered pattern. Three C–V proteins in human mitochondria, ATP5A1, ATP5B, and ATP5C1, were more interactive than other C–V proteins; however, only ATP5B was categorized as a major hub in both mouse and drosophila. (b) Analysis of redox protein–protein interactions demonstrated similar patterns to C–I with human and mouse proteins assembled around a single protein, whereas drosophila presented dispersed partnerships among its proteins. (c) Moreover, examination of C–I + redox and C–V + redox revealed little interaction between these complex proteins and redox proteins. An exception to the pattern, ATP5B (in C–V), interacted with redox protein PRDX1 in the mouse mitochondria analysis. Finally, the mouse C–I + redox interactome revealed an intertwining C–I and redox protein interaction network, a feature unique to the mouse mitochondrial proteome. The series of interlocking C–I and redox proteins found in the mouse C–I + redox interactome have been circled by a dotted line.
Mitochondrial Proteome and Protein Translocation
To explore the mechanisms promoting nuclear-encoded protein translocation into mitochondria, we determined the number of mitochondrial proteins with an N-terminal mitochondrial targeting sequence (MTS) using TargetP and the UniProt KnowledgeBase. Interestingly, only a small subpopulation of mitochondrial proteins possessed an MTS, ranging from 9.7% in drosophila to 19.9, 20.7, and 22.4% in mouse liver, human heart, and mouse heart, respectively (Figure 6a). Analysis of the core proteins demonstrated a similar pattern (17.4% in drosophila, 37.9% in human heart, 48.7% in mouse liver, and 49.7% in mouse heart) as exemplified in Figure 6b. Further examination illustrated that mitochondrial proteins with and without an MTS had similar functional distributions. A functional perspective demonstrated that proteins with an MTS were found to be fundamental in metabolism and biosynthesis and were localized within the general mitochondrion. Conversely, mitochondrial proteins essential in metabolism and OXPHOS without an MTS were primarily localized in nonmitochondrial domains (Figure 6c). However, the subcellular localization distribution of protein abundance showed that mitochondrial proteins with an MTS were predominantly concentrated in both the inner mitochondrial membrane (IMM) and the matrix (Figure 6d).
Figure 6.
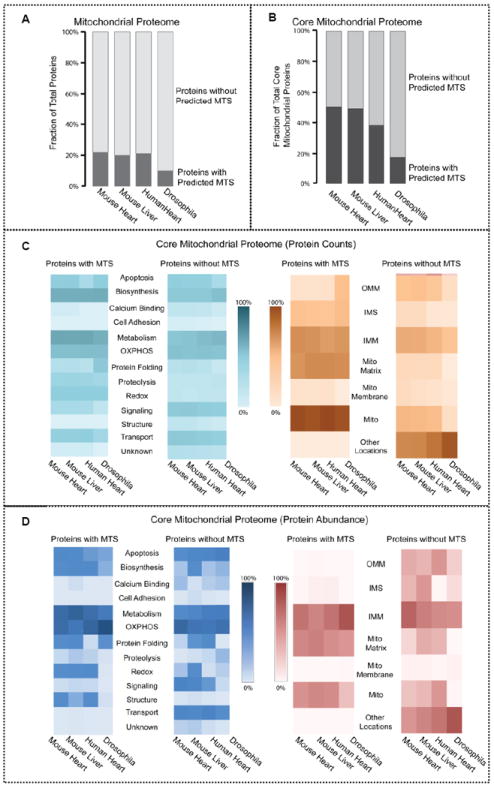
Distribution of proteins with mitochondrial targeting sequences in the mitochondrial proteome. (a) The number of mitochondrial proteins with an N-terminal mitochondrial targeting sequence (MTS) within the mitochondrial proteome was examined. Only a small percentage of mitochondrial proteins carried an MTS. Mitochondrial proteins with an MTS accounted for 9.7% of the total mitochondrial proteome in drosophila, 19.9% in mouse heart, 20.7% in human heart, and 22.4% in mouse liver. (b) Analysis of the core proteins demonstrated a similar pattern of mitochondrial proteins with an MTS – 17.4% in drosophila, 37.9% in human heart, 48.7% in mouse liver, and 49.7% in mouse heart. This suggests that the mitochondrial proteins without N-terminal targeting sequences are utilizing alternative methods to localize in the mitochondria. (c) Number of core mitochondrial proteins with an MTS and without an MTS was determined by their different functionalities and subcellular localizations. A majority of the mitochondrial proteins with an MTS were involved in metabolism and biosynthesis and were found throughout the mitochondrion rather than a specific mitochondrial compartment. In contrast, mitochondrial proteins without an MTS were predominantly integral in metabolism and OXPHOS and were localized in other cellular regions. (d) While the abundance of core mitochondrial proteins with an MTS and core mitochondrial proteins without an MTS were both highly involved in metabolism, OXPHOS, apoptosis, and biosynthesis, the core mitochondrial proteins with an MTS were most abundant in the outer and inner mitochondrial membrane, whereas the core mitochondrial proteins without an MTS were most abundant in the inner mitochondrial membrane, intermembrane space, matrix, and other cellular locations.
Mitochondrial Proteome and Disease
Mitochondrial dysfunction has been shown to be associated with the onset of disease. Literature curation using predefined terms (e.g., diabetes, obesity, muscular dystrophy, and neurodegeneration) suggested that human disease-associated genes are enriched in the conserved mitochondrial proteome, especially in conserved respiratory chain proteins (Figure 7). The diversified mitochondrial proteome contains fewer known disease associations than the conserved mitochondrial proteome but still has a higher occurrence of disease gene associations than did the whole human genome (~15%). In addition, a significant portion of the conserved respiratory proteins (~27%) and core mitochondrial proteome (~17%) plays a role in known cardiac diseases. Other conserved proteins are prime candidates for disease association as well. These data highlight the central importance of the core mitochondrial proteome to normal physiological functions.
Figure 7.
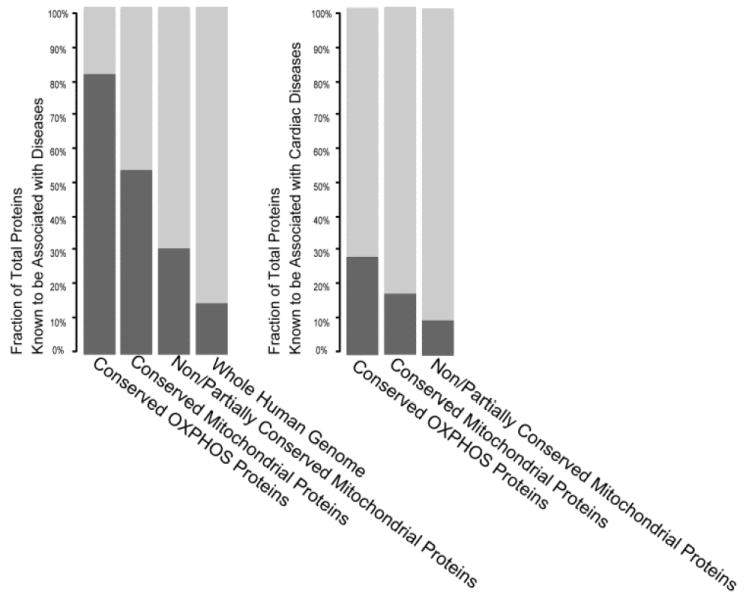
Pathological phenotypes of the mitochondrial proteomes. The involvement of mitochondrial proteins as well as the OXPHOS subproteome in disease phenotypes (left) and cardiac disease phenotypes (right) was determined by literature curation using predefined search terms. Human disease-associated genes are more prominent within the conserved mitochondrial proteome, and dysfunction of conserved respiratory chain proteins is particularly detrimental with 54% of these proteins implicated in disease. In contrast, only 15% of the human genome is correlated with causing disorders. Among these, 27% of conserved OXPHOS proteins and 17% of conserved mitochondrial proteins are directly associated with cardiac disease. These results illustrate the importance of the core mitochondrial proteome to physiological functions.
DISCUSSION
In summary, we combined biochemistry assays, quantitative proteomics profiling, and interactome analysis to define four mitochondrial populations. Our analysis significantly expands the existing knowledge on the mitochondrial proteome with approximately 3350 unique proteins identified, including a core conserved mitochondrial proteome comprised of 419 proteins. These data portrayed inter- and intraspecies heterogeneities of mitochondria from both expressional and functional perspectives. Our data suggest that proteome–function correlations are subject to complex organizations with protein–protein interactions and that concomitant molecular function assessments are essential to contextualize global proteomics data and locate decisive perturbations.
Mitochondrial Core Proteome Properties
Comparative genomics predicted that ~100 human mitochondrial genes have homologues in nine diverse eukaryotes and are mainly enriched in biological processes related to transport, metabolism, and signal transduction.57 Indeed, a series of reports supports the existence of a highly specified, conserved, and heterogeneous mitochondrial proteome.23,24,31,32 Our experimental data sets redefined the core mitochondrial proteome and provided additional evidence of the connection between conserved proteins and disease phenotypes. The data suggest a two-tiered architecture of the mitochondrial proteome, with the 419 conserved proteins in the core mitochondrial proteome representing essential hubs of protein networks involved in basal processes.
The Approach to Assess Protein Abundance
Existing MS approaches to assess protein abundance include label-free qualification and labeled quantification. In this particular study, we elected to quantify relative protein abundance through a label-free approach using NSAF values; this allowed us to overcome the challenge associated with synthesizing tens of thousands of stable isotope labeled peptides.56 However, we are aware of the limitations of label-free approaches.55 Specifically, the accuracy of such quantifications is affected by LC–MS run-to-run variability and the weight of shared peptides among different proteins. We implemented specific approaches to address these challenges. Multiple biological and technical replicates (from 18 replicates of the mouse heart to 29 replicates of the human heart) were performed to gain a reliable evaluation of protein abundance. Furthermore, an algorithm was developed to proportionally allocate shared peptides to their corresponding proteins based on the ratio of unique peptides identified among these proteins. Within the current technological capabilities, limiting factors are certainly evident; however, we believe that our approach to systematically assess relative protein abundance adequately addresses these shortcomings.
Links Between Mitochondrial Protein Composition and Function
Although the relationship between protein expression and functional differences may be affected by multiple factors, protein abundances are thought to be a selectable trait that reflects functional attributes, especially in closely related species.58 The recent demonstration of transcript-level clustering recapitulates the phylogeny of mammalian lineages and supports the notability of expression levels in adaptation.59 Interestingly, protein abundance, as calculated using NSAF values, varied according to their primary functions across the model systems. The two most abundant proteins in mouse heart, ADP/ATP translocase 1 and ATP synthase subunit α, are involved in the generation and transport of bioenergetics. Similarly, the most abundant proteins in human heart, ATP synthase subunit α and ATP synthase subunit β, as well as the two most abundant proteins in drosophila, ATP synthase subunit β and ADP/ATP translocase 1, are similarly involved in bioenergetics. In contrast, carbamoyl phosphate synthase 1 and 3-oxoacyl-CoA thiolase are the two most abundant proteins in mouse liver and are mainly involved in the urea cycle and β-oxidation of fatty acids, respectively. The relationship between the most abundant proteins for each model system and their respective function implies that abundance is strongly related to tissue type and mitochondrial location.
Distinct Mitochondrial Metabolic Function among the Model Systems
Previous studies have reported that mitochondrial respiratory function is heavily connected to many pathological processes, including heart failure, ischemic injury, and aging.2,17,60 In the current study, variations in mitochondrial respiratory capacity among the examined mitochondria were observed in ETC complex activity and oxygen consumption. Mitochondria in different species/tissues dramatically differ in their ability to consume oxygen and make ATP. The higher C–I, C–V, and oxygen consumption activity seen in drosophila and mouse heart tissue, respectively, indicates that these model systems have a high NADH delivery and ATP generation rate. Consistent with our functional data, we found that C–I subunits in mouse heart are significantly more abundant compared with mouse liver and human heart. However, C–V showed no significant differences in protein abundance, signifying the existence of potential regulatory mechanisms such as post-translational modifications and complex assembly, other than protein expression level. C–I activity and respiratory rates of liver mitochondria are not particularly reflected in the overall complex subunit abundance but instead correlate with those of the mitochondrial-encoded subunits (Figure 4d). Similarly, lower C–V activity in human heart mitochondria is consistent with the lower abundance of mitochondrial-encoded C–V proteins. These results suggest that complex assembly may be significantly restricted by the abundance of mitochondrial-encoded proteins. Furthermore, our data emphasize that mitochondria in different species/tissues are specialized for diverse metabolic duties. The metabolism functional group not only contains the most proteins but also possesses the most nonconserved and conserved proteins.
Evolved Mitochondrial Phosphorylation Events among the Model Systems
Phosphorylation signaling regulated by kinases and phosphatases plays a critical role in regulating biological processes in all living organisms. However, identifying the entire phosphoproteome is a daunting task. Due to its unique evolutionary origin and enriched functionality, mitochondrial phosphoproteome studies have begun to draw increasing attention.43,61-64 Gnad et al.63 compared the phosphoproteomes of different organisms and found that serine/threonine phosphorylation in prokaryotes has occurred relatively recently in evolution. Another study carried out by Beltrao and colleagues65 showed that kinase–substrate interactions change more slowly than transcription factor–promoter interactions and that protein kinases are an important source of phenotypic diversity. Accordingly, we observed that the distribution of kinase/phosphatase in drosophila is scarce in terms of number and abundance of proteins compared with other model systems. These findings are consistent with the notion that phosphorylation rapidly expanded in higher metazoan lineages as a general mechanism for increased cellular signaling and complexity.63
Differential Mitochondrial Calcium Regulation among the Model Systems
Functional assays on calcium-induced swelling show that liver mitochondria are the most susceptible to calcium damage. This agrees with previous data66 showing that rat liver mitochondria have larger uptake of calcium than heart mitochondria and is probably a reflection of the heart being an excitable organ under constant calcium flux. At face value, one might reasonably posit that liver mitochondria, being in a nonexcitable tissue, would contain fewer calcium binding proteins. On the contrary, there was no significant difference in the total expression of calcium-binding proteins between liver mitochondria and other mitochondria. Although we cannot differentiate the effect of differing affinities of calcium influx, efflux, and buffering proteins across organisms, this nevertheless indicates that protein calcium regulation mechanisms besides protein expression levels should be taken into account. An attempt to correlate individual protein abundance with functional differences proved more promising, as we observed that liver has a relatively high abundance of calcium uniporters (MCUs) but almost no mitochondrial sodium/calcium exchanger (NCX), which taken together would suggest increased calcium influx and decreased efflux. A recently published study suggests that the activity and abundance of the mitochondrial calcium uniporter varies greatly between tissues of different species; mouse heart and drosophila flight muscle showed low mitochondrial calcium uniporter activity in comparison with that of mouse liver, kidney, and brown fat.67 This result is consistent with the higher susceptibility of liver mitochondria to calcium overload compared with mitochondria from mouse heart, human heart, and drosophila. In our opinion, these examples demonstrate the limitations of reducing proteomics data to simplistic functional categories and the values of functional assays in augmenting proteomics data.
Broadening the Mitochondrial Interactome
There is a need to bridge the translational divide by determining the protein–protein interactions within the interactomes of nonhuman organisms as well as highlighting similar structures in the human interactome. While animal models and computational insights remain indispensable, the success of translational discoveries is fostered by our understanding of protein connectivity in human cellular networks.
Each of the model systems characterized in our investigation demonstrated a distinct protein–protein interaction pattern; ostensibly these molecular interactions may be limited by the present knowledge of the scientific community as well as the IntAct database. A more accurate and comprehensive data set collecting multiple model systems has yet to be available due to investigator-specific foci of research, targeting a specific organ, tissue, disease, or pathway. An integrated framework addressing protein interactions from different organs, tissues, diseases, and pathways may elucidate mechanistic insights that play a fundamental role in targeting disease origins in humans, as many diseases affect protein partnerships in multiple organ systems. Proteins whose interactions have not been extensively studied in humans such as α-synuclein may act as future areas of research in translational medicine.
Although it was evident that a distinguished pattern arose in each model system, one major mouse hub emerged among the C–I protein–protein interactions. Although not a Complex protein, α-synuclein was found to be central for C–I mitochondrial proteins in mouse; this differed greatly in the human C–I interactome, as it was not seen to interact with human C–I proteins. As a protein heavily involved in signaling, α-synuclein has been studied extensively in humans and is a key protein associated with Parkinson’s and other neurodegenerative diseases.68 The interaction of α-synuclein with multiple C–I proteins in the mouse may implicate its important regulatory role in energy metabolism and therefore be relevant to human molecular pathways.
In addition, examination of C–I + redox and C–V + redox protein networks revealed little interaction between C–I and redox proteins as well as C–V and redox proteins, with the exception of ATP5B in C–V. This evolutionarily conserved C– V protein serves as a major hub in human, mouse, and drosophila. However, in the mouse interactome, ATP5B interacts with several peroxiredoxin (redox) proteins. PRDX1 and ATP5B have emerged as key proteins in this C–V + redox protein interaction network. As seen in previous studies, the centrality of both proteins predisposes them to be highly associated with the onset of disease. The intermingled network of connections between C–I and redox proteins in the mouse interactome serves as a proponent for the necessity of analyzing human protein–protein interactions that have yet to be investigated. Investigation of proteins highly involved in C–I, C–V, and redox in nonhuman organisms could facilitate the identification of new target proteins for translational medicine and scientific research.
Mitochondrial-Targeted Protein Localization
Mitochondrial targeting sequences facilitate the translocation of mitochondrial proteins to their designated location. In this study, we identified mitochondrial proteins with an N-terminal MTS, specifically focusing on the core conserved proteins. Our results suggest that many mitochondrial proteins without an MTS are utilizing alternative ways of translocating into the mitochondria, although a sophisticated method of predicting proteins possessing internal mitochondrial targeting sequences has yet to be developed. We postulate that these proteins lacking an MTS may have been imported through various mechanisms or attracted by biochemical signals (i.e., cardiolipin, ΔΨ, motifs, positively charged amino acids residues, etc.). These results indicate that some mitochondrial proteins may either share multiple cellular localizations or only transiently reside in the mitochondria. Identification of currently unknown sequence patterns may aid in the clarification of this issue and provide clinical relevance to the field of medicine.69
Involvement of Mitochondrial Proteins in Diseases
As the center of metabolism and energy production, mitochondrial protein dysfunction, caused by either genetic or environmental alterations, has been shown to be directly associated with diseases, specifically cardiomyopathies, neurodegenerative disorders, and diabetes.11,60,70,71 Through literature curation, we found that the 419 conserved mitochondrial core proteins are heavily involved in disease phenotypes (~54%). In particular, ~82% of conserved OXPHOS proteins are related to the development of disease. These data highlight the evolutionary prominence of these proteins.
The involvement of mitochondrial proteins in cardiovascular diseases, including ischemic injury, heart failure, and congenital heart disease, has also been analyzed. Corroborating prior studies, the dysfunction of a significant portion of core mitochondrial proteins (>17%) and conserved OXPHOS proteins (>27%), compared with shared and nonconserved proteins among the model systems, plays a role in cardiac diseases. In addition, OXPHOS proteins implicated in these pathologies have high abundance values and can be considered a requirement for normal physiological OXPHOS function. A mutation of one gene-encoded OXPHOS protein might show multiple clinical symptoms, whereas the mutation of various OXPHOS proteome genes may cause the same disease; our data mark the OXPHOS system as an injury-prone subproteome.
In summary, we conducted a comprehensive analysis to integrate information from biochemical, proteomic, and genomic data sets to characterize mitochondrial biology from a multifaceted perspective. Among the published reports describing the mitochondrial proteome, our current analysis uniquely and significantly expands the mitochondrial proteome pools, highlighting a core mitochondrial proteome from four model systems. These analyses simultaneously portray the inter- and intraspecies heterogeneity of mitochondria from both expressional and functional perspectives. Our study of the proteome–function correlation confirms previous observations of mitochondrial biology and contributes new evidence of the expediency of diagnosing mitochondrial disease through proteomic parameters. This investigation bridges the knowledge gap between molecular compositions and their accompanying functions, which ultimately aids in the translation of mitochondrial proteomics data to a contextualized understanding of complex mitochondrial biology as well as allows for the acquisition of a prospective source for not only the diagnosis of mitochondrial pathologies but also the procurement of mitochondrial therapeutic targets.
Supplementary Material
Acknowledgments
We thank the members of our laboratory for their helpful discussions. We are supported by the NHLBI Proteomics Center Award (HHSN268201000035C) to Dr. Peipei Ping; NIH Award HL-63901 to Dr. Peipei Ping; and AHA predoctoral fellowship 12PRE11610024 to Edward Lau.
Footnotes
Supporting Information
Materials, isolation, and purification of functionally viable mitochondria from mouse heart, mouse liver, human heart, and drosophila; assessment of mitochondrial function; proteomic profiling of mitochondria from mouse heart, mouse liver, human heart, and drosophila; mitochondrial proteome and properties; approach to assess protein abundance; schematic diagram of experimental workflow; functionally-enabled recovery of the individuals; summary of mass spectrometry experiments; biochemical features of mitochondrial proteins; proteins identified from mitochondria across organs and organisms; and mitochondrial proteome annotation and inter/intra-genomic comparison across organs and organisms. Additional supporting raw data can be found at http://149.142.212.48/ (user name: pinglab; password: proteomics). This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
References
- 1.Johnson DT, Harris RA, Blair PV, Balaban RS. Functional consequences of mitochondrial proteome heterogeneity. Am J Physiol : Cell Physiol. 2007;292:C698–707. doi: 10.1152/ajpcell.00109.2006. [DOI] [PubMed] [Google Scholar]
- 2.Balaban RS. Regulation of oxidative phosphorylation in the mammalian cell. Am J Physiol. 1990:377–389. doi: 10.1152/ajpcell.1990.258.3.C377. [DOI] [PubMed] [Google Scholar]
- 3.Shao D, Oka S, Brady CD, Haendeler J, Eaton P, Sadoshima J. Redox modification of cell signaling in the cardiovascular system. J Mol Cell Cardiol. 2012;52:550–558. doi: 10.1016/j.yjmcc.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Lin A, Powers J, Lam MP, Lotz C, Liem D, Lau E, Wang D, Deng N, Korge P, Zong NC, Cai H, Weiss J, Ping P. Perspectives on: Sgp symposium on mitochondrial physiology and medicine: Mitochondrial proteome design: From molecular identity to pathophysiological regulation. J Gen Physiol. 2012;139:395–406. doi: 10.1085/jgp.201210797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gucek M, Murphy E. What can we learn about cardioprotection from the cardiac mitochondrial proteome? Cardiovasc Res. 2010;88:211–218. doi: 10.1093/cvr/cvq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balaban RS. The mitochondrial proteome: A dynamic functional program in tissues and disease states. Environ Mol Mutagen. 2010;51:352–359. doi: 10.1002/em.20574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollander JM, Baseler WA, Dabkowski ER. Proteomic remodeling of mitochondria in heart failure. Congestive Heart Failure. 2011;17:262–268. doi: 10.1111/j.1751-7133.2011.00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karamanlidis G, Bautista-Hernandez V, Fynn-Thompson F, Del Nido P, Tian R. Impaired mitochondrial biogenesis precedes heart failure in right ventricular hypertrophy in congenital heart disease. Circ : Heart Failure. 2011;4:707–713. doi: 10.1161/CIRCHEARTFAILURE.111.961474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neubauer S. The failing heart–an engine out of fuel. N Engl J Med. 2007;356:1140–1151. doi: 10.1056/NEJMra063052. [DOI] [PubMed] [Google Scholar]
- 10.Rosca M, Popescu BA, Beladan CC, Calin A, Muraru D, Popa EC, Lancellotti P, Enache R, Coman IM, Jurcut R, Ghionea M, Ginghina C. Left atrial dysfunction as a correlate of heart failure symptoms in hypertrophic cardiomyopathy. J Am Soc Echocardiography. 2010;23:1090–1098. doi: 10.1016/j.echo.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 11.Wallace DC. Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen. 2010:440–450. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- 12.Yoshioka J, Chutkow WA, Lee S, Kim JB, Yan J, Tian R, Lindsey ML, Feener EP, Seidman CE, Seidman JG, Lee RT. Deletion of thioredoxin-interacting protein in mice impairs mitochondrial function but protects the myocardium from ischemiareperfusion injury. J Clin Invest. 2012;122:267–279. doi: 10.1172/JCI44927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez KA, Wywial E, Perez VI, Lambert AJ, Edrey YH, Lewis KN, Grimes K, Lindsey ML, Brand MD, Buffenstein R. Walking the oxidative stress tightrope: A perspective from the naked mole-rat, the longest-living rodent. Curr Pharm Des. 2011;17:2290–2307. doi: 10.2174/138161211797052457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ago T, Liu T, Zhai P, Chen W, Li H, Molkentin JD, Vatner SF, Sadoshima J. A redox-dependent pathway for regulating class ii hdacs and cardiac hypertrophy. Cell. 2008;133:978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 15.Chen EI, Hewel J, Krueger JS, Tiraby C, Weber MR, Kralli A, Becker K, Yates JR, 3rd, Felding-Habermann B. Adaptation of energy metabolism in breast cancer brain metastases. Cancer Res. 2007;67:1472–1486. doi: 10.1158/0008-5472.CAN-06-3137. [DOI] [PubMed] [Google Scholar]
- 16.Calvo SE, Mootha VK. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosca MG, Hoppel CL. Mitochondria in heart failure. Cardiovasc Res. 2010;88:40–50. doi: 10.1093/cvr/cvq240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindsey ML, Goshorn DK, Comte-Walters S, Hendrick JW, Hapke E, Zile MR, Schey K. A multidimensional proteomic approach to identify hypertrophy-associated proteins. Proteomics. 2006;6:2225–2235. doi: 10.1002/pmic.200500013. [DOI] [PubMed] [Google Scholar]
- 19.Wu F, Zhang J, Beard DA. Experimentally observed phenomena on cardiac energetics in heart failure emerge from simulations of cardiac metabolism. Proc Natl Acad Sci U S A. 2009;106:7143–7148. doi: 10.1073/pnas.0812768106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Merkle H, Hendrich K, Garwood M, From AH, Ugurbil K, Bache RJ. Bioenergetic abnormalities associated with severe left ventricular hypertrophy. J Clin Invest. 1993;92:993–1003. doi: 10.1172/JCI116676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pflieger D, Le Caer JP, Lemaire C, Bernard BA, Dujardin G, Rossier J. Systematic identification of mitochondrial proteins by lc-ms/ms. Anal Chem. 2002;74:2400–2406. doi: 10.1021/ac011295h. [DOI] [PubMed] [Google Scholar]
- 22.Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. The proteome of saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, Rossant J, Hughes TR, Frey B, Emili A. Global survey of organ and organelle protein expression in mouse: Combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 24.Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 25.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex i disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Li X, Mueller M, Wang Y, Zong C, Deng N, Vondriska TM, Liem DA, Yang JI, Korge P, Honda H, Weiss JN, Apweiler R, Ping P. Systematic characterization of the murine mitochondrial proteome using functionally validated cardiac mitochondria. Proteomics. 2008;8:1564–1575. doi: 10.1002/pmic.200700851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaucher SP, Taylor SW, Fahy E, Zhang B, Warnock DE, Ghosh SS, Gibson BW. Expanded coverage of the human heart mitochondrial proteome using multidimensional liquid chromatography coupled with tandem mass spectrometry. J Proteome Res. 2004;3:495–505. doi: 10.1021/pr034102a. [DOI] [PubMed] [Google Scholar]
- 28.Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, Ghosh SS. Characterization of the human heart mitochondrial proteome. Nat Biotechnol. 2003;21:281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- 29.Reifschneider NH, Goto S, Nakamoto H, Takahashi R, Sugawa M, Dencher NA, Krause F. Defining the mitochondrial proteomes from five rat organs in a physiologically significant context using 2d blue-native/sds-page. J Proteome Res. 2006;5:1117–1132. doi: 10.1021/pr0504440. [DOI] [PubMed] [Google Scholar]
- 30.Forner F, Foster LJ, Campanaro S, Valle G, Mann M. Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol Cell Proteomics. 2006;5:608–619. doi: 10.1074/mcp.M500298-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Phillips D, Covian R, Aponte AM, Glancy B, Taylor JF, Chess D, Balaban RS. Regulation of oxidative phosphorylation complex activity: Effects of tissue-specific metabolic stress within an allometric series and acute changes in workload. Am J Physiol : Regul Integr Comp Physiol. 2012;302:R1034–1048. doi: 10.1152/ajpregu.00596.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White MY, Brown DA, Sheng S, Cole RN, O’Rourke B, Van Eyk JE. Parallel proteomics to improve coverage and confidence in the partially annotated oryctolagus cuniculus mitochondrial proteome. Mol Cell Proteomics. 2011;10:M110–004291. doi: 10.1074/mcp.M110.004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alonso J, Rodriguez JM, Baena-Lopez LA, Santaren JF. Characterization of the drosophila melanogaster mitochondrial proteome. J Proteome Res. 2005;4:1636–1645. doi: 10.1021/pr050130c. [DOI] [PubMed] [Google Scholar]
- 34.Brunner E, Ahrens CH, Mohanty S, Baetschmann H, Loevenich S, Potthast F, Deutsch EW, Panse C, de Lichtenberg U, Rinner O, Lee H, Pedrioli PG, Malmstrom J, Koehler K, Schrimpf S, Krijgsveld J, Kregenow F, Heck AJ, Hafen E, Schlapbach R, Aebersold R. A high-quality catalog of the drosophila melanogaster proteome. Nat Biotechnol. 2007;25:576–583. doi: 10.1038/nbt1300. [DOI] [PubMed] [Google Scholar]
- 35.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 36.Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP. Quantitative proteomic analysis of distinct mammalian mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci U S A. 2006;103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mosley AL, Florens L, Wen Z, Washburn MP. A label free quantitative proteomic analysis of the saccharomyces cerevisiae nucleus. J Proteomics. 2009;72:110–120. doi: 10.1016/j.jprot.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bier E, Bodmer R. Drosophila, an emerging model for cardiac disease. Gene. 2004;342:1–11. doi: 10.1016/j.gene.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 39.Taghli-Lamallem O, Akasaka T, Hogg G, Nudel U, Yaffe D, Chamberlain JS, Ocorr K, Bodmer R. Dystrophin deficiency in drosophila reduces lifespan and causes a dilated cardiomyopathy phenotype. Aging Cell. 2008;7:237–249. doi: 10.1111/j.1474-9726.2008.00367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukhida K, Kobayashi NR, Mendez I. A novel role for parkin in trauma-induced central nervous system secondary injury. Med Hypotheses. 2005;64:1120–1123. doi: 10.1016/j.mehy.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Liem DA, Mueller M, Wang Y, Zong C, Deng N, Vondriska TM, Korge P, Drews O, Maclellan WR, Honda H, Weiss JN, Apweiler R, Ping P. Altered proteome biology of cardiac mitochondria under stress conditions. J Proteome Res. 2008;7:2204–2214. doi: 10.1021/pr070371f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lau E, Wang D, Zhang J, Yu H, Lam MP, Liang X, Zong N, Kim TY, Ping P. Substrate- and isoform-specific proteome stability in normal and stressed cardiac mitochondria. Circ Res. 2012;110:1174–1178. doi: 10.1161/CIRCRESAHA.112.268359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng N, Zhang J, Zong C, Wang Y, Lu H, Yang P, Wang W, Young GW, Wang Y, Korge P, Lotz C, Doran P, Liem DA, Apweiler R, Weiss JN, Duan H, Ping P. Phosphoproteome analysis reveals regulatory sites in major pathways of cardiac mitochondria. Mol Cell Proteomics. 2011;10:M110.000117. doi: 10.1074/mcp.M110.000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirby DM, Thorburn DR, Turnbull DM, Taylor RW. Biochemical assays of respiratory chain complex activity. Methods Cell Biol. 2007;80:93–119. doi: 10.1016/S0091-679X(06)80004-X. [DOI] [PubMed] [Google Scholar]
- 45.Kramarova TV, Shabalina IG, Andersson U, Westerberg R, Carlberg I, Houstek J, Nedergaard J, Cannon B. Mitochondrial atp synthase levels in brown adipose tissue are governed by the c-fo subunit p1 isoform. FASEB J. 2008;22:55–63. doi: 10.1096/fj.07-8581com. [DOI] [PubMed] [Google Scholar]
- 46.Wittig I, Braun HP, Schagger H. Blue native page. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 47.Magrane M, Consortium U. Uniprot knowledgebase: A hub of integrated protein data. Database. 2011;2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their n-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 49.Kasprzyk A. Biomart: Driving a paradigm change in biological data management. Database. 2011;2011:bar049. doi: 10.1093/database/bar049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Auston I, Cahn MA, Selden CR. Literature Search Methods for the Development of Clinical Practice Guidelines; Report No PB92-221175. National Library of Medicine; Bethesda, MD: 1992. [Google Scholar]
- 51.Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, Jandrasits C, Jimenez RC, Khadake J, Mahadevan U, Masson P, Pedruzzi I, Pfeiffenberger E, Porras P, Raghunath A, Roechert B, Orchard S, Hermjakob H. The intact molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. A travel guide to cytoscape plugins. Nat Methods. 2012;9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wallenstein S, Zucker CL, Fleiss JL. Some statistical methods useful in circulation research. Circ Res. 1980;47:1–9. doi: 10.1161/01.res.47.1.1. [DOI] [PubMed] [Google Scholar]
- 54.Zong NC, Li H, Li H, Lam MP, Jimenez RC, Kim CS, Deng N, Kim AK, Choi JH, Zelaya I, Liem D, Meyer D, Odeberg J, Fang C, Lu HJ, Xu T, Weiss J, Duan H, Uhlen M, Yates JR, III, Apweiler R, Ge J, Hermjakob H, Ping P. Integration of cardiac proteome biology and medicine by a specialized knowledgebase. Circ Res. 2013;113(9):1043–1053. doi: 10.1161/CIRCRESAHA.113.301151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilm M. Quantitative proteomics in biological research. Proteomics. 2009;9:4590–4605. doi: 10.1002/pmic.200900299. [DOI] [PubMed] [Google Scholar]
- 56.Neilson KA, Ali NA, Muralidharan S, Mirzaei M, Mariani M, Assadourian G, Lee A, van Sluyter SC, Haynes PA. Less label, more free: Approaches in label-free quantitative mass spectrometry. Proteomics. 2011;11:535–553. doi: 10.1002/pmic.201000553. [DOI] [PubMed] [Google Scholar]
- 57.Richly E, Chinnery PF, Leister D. Evolutionary diversification of mitochondrial proteomes: Implications for human disease. Trends Genet. 2003;19:356–362. doi: 10.1016/S0168-9525(03)00137-9. [DOI] [PubMed] [Google Scholar]
- 58.Agnetti G, Husberg C, Van Eyk JE. Divide and conquer: The application of organelle proteomics to heart failure. Circ Res. 2011;108:512–526. doi: 10.1161/CIRCRESAHA.110.226910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brawand D, Soumillon M, Necsulea A, Julien P, Csardi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, Albert FW, Zeller U, Khaitovich P, Grutzner F, Bergmann S, Nielsen R, Paabo S, Kaessmann H. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 60.Fosslien E. Mitochondrial medicine - cardiomyopathy caused by defective oxidative phosphorylation. Ann Clin Lab Sci. 2003;33:371–395. [PubMed] [Google Scholar]
- 61.Lee J, Xu Y, Chen Y, Sprung R, Kim SC, Xie S, Zhao Y. Mitochondrial phosphoproteome revealed by an improved imac method and ms/ms/ms. Mol Cell Proteomics. 2007;6:669–676. doi: 10.1074/mcp.M600218-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reinders J, Wagner K, Zahedi RP, Stojanovski D, Eyrich B, van der Laan M, Rehling P, Sickmann A, Pfanner N, Meisinger C. Profiling phosphoproteins of yeast mitochondria reveals a role of phosphorylation in assembly of the atp synthase. Mol Cell Proteomics. 2007;6:1896–1906. doi: 10.1074/mcp.M700098-MCP200. [DOI] [PubMed] [Google Scholar]
- 63.Gnad F, Forner F, Zielinska DF, Birney E, Gunawardena J, Mann M. Evolutionary constraints of phosphorylation in eukaryotes, prokaryotes, and mitochondria. Mol Cell Proteomics. 2010;9:2642–2653. doi: 10.1074/mcp.M110.001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lam MP, Lau E, Scruggs SB, Wang D, Kim TY, Liem DA, Zhang J, Ryan CM, Faull KF, Ping P. Site-specific quantitative analysis of cardiac mitochondrial protein phosphorylation. J Proteomics. 2013;81:15–23. doi: 10.1016/j.jprot.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beltrao P, Trinidad JC, Fiedler D, Roguev A, Lim WA, Shokat KM, Burlingame AL, Krogan NJ. Evolution of phosphoregulation: Comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009;7:e1000134. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Favaron M, Bernardi P. Tissue-specific modulation of the mitochondrial calcium uniporter by magnesium ions. FEBS Lett. 1985;183:260–264. doi: 10.1016/0014-5793(85)80789-4. [DOI] [PubMed] [Google Scholar]
- 67.Fieni F, Lee SB, Jan YN, Kirichok Y. Activity of the mitochondrial calcium uniporter varies greatly between tissues. Nat Commun. 2012;3:1317. doi: 10.1038/ncomms2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O’Connor K, Magne J, Rosca M, Pierard LA, Lancellotti P. Impact of aortic valve stenosis on left atrial phasic function. Am J Cardiol. 2010;106:1157–1162. doi: 10.1016/j.amjcard.2010.06.029. [DOI] [PubMed] [Google Scholar]
- 69.Yamada Y, Harashima H. Mitochondrial drug delivery systems for macromolecule and their therapeutic application to mitochondrial diseases. Adv Drug Delivery Rev. 2008;60:1439–1462. doi: 10.1016/j.addr.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 70.Cwerman-Thibault H, Sahel JA, Corral-Debrinski M. Mitochondrial medicine: To a new era of gene therapy for mitochondrial DNA mutations. J Inherited Metab Dis. 2011;34:327–344. doi: 10.1007/s10545-010-9131-5. [DOI] [PubMed] [Google Scholar]
- 71.Lesnefsky EJ, Hoppel CL. Ischemia-reperfusion injury in the aged heart: Role of mitochondria. Arch Biochem Biophys. 2003;420:287–297. doi: 10.1016/j.abb.2003.09.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


