Abstract
The silent mating type information regulation 2 homolog 1 gene (Sirt1) encodes an NAD-dependent deacetylase that modifies the activity of well-known transcriptional regulators affected in kidney diseases. Sirt1 is expressed in the kidney podocyte, but its function in the podocyte is not clear. Genetically engineered mice with inducible and reversible Sirt1 knockdown in widespread, podocyte-specific, or tubular-specific patterns were generated. We found that mice with 80% knockdown of renal Sirt1 expression have normal glomerular function under the basal condition. When challenged with doxorubicin (Adriamycin), these mice develop marked albuminuria, glomerulosclerosis, mitochondrial injury, and impaired autophagy of damaged mitochondria. Reversal of Sirt1 knockdown during the early phase of Adriamycin-induced nephropathy prevented the progression of glomerular injury and reduced the accumulation of dysmorphic mitochondria in podocytes but did not reverse the progression of albuminuria and glomerulosclerosis. Sirt1 knockdown mice with diabetes mellitus, which is known to cause mitochondrial dysfunction in the kidney, developed more albuminuria and mitochondrial dysfunction compared with diabetic mice without Sirt1 knockdown. In conclusion, these results demonstrate that our RNA interference–mediated Sirt1 knockdown models are valid and versatile tools for characterizing the function of Sirt1 in the kidney; Sirt1 plays a role in homeostatic maintenance of podocytes under the condition of mitochondrial stress/injury.
Sirtuin 1 (Sirt1) is a member of the sirtuin family of protein deacetylases that require NAD+ as a cofactor for enzymatic function. Sirt1 deacetylates and regulates the activity of transcription factors, including p53, FOXO, RelA, STAT3, PGC1α, and peroxisome proliferator–activated receptor.1 On the cellular level, Sirt1 functions to control autophagy,2 energetic homeostasis,3,4 mitochondrial biogenesis,5 and apoptosis.6–8 In mammals, Sirt1 participates in glucose and lipid metabolism through its action in pancreatic β cells,9,10 hepatocytes,11–13 and adipose tissue.14
Sirt1 in renal tubular cells has been shown to protect renal tubular cells from cellular stresses associated with aging, cisplatin, and hypoxia.15–17 More recently, renal tubular Sirt1 expression was reported to mitigate diabetic podocyte injury.18 Although Sirt1-mediated deacetylation is capable of modulating signaling flux through well-described pathways affected in kidney diseases, studies of Sirt1 activity in glomerular cells and glomerular biology are still in the early phases of exploration. In the past, we reported that Sirt1 expression is decreased in glomeruli of patients with diabetic nephropathy.19 More recently, we demonstrated that diabetic mice with podocyte-specific Sirt1 deletion develop accelerated diabetic nephropathy.20 Consistent with our findings, others also demonstrated a pathogenic role for Sirt1 deficiency in diabetic nephropathy.18,21–23 With the aim of developing robust and versatile murine models for evaluating Sirt1’s function in podocytes and tubular cells under the basal condition and in glomerular diseases, we engineered genetically modified mice with inducible, reversible, and tissue-specific Sirt1 knockdown.
Herein, we described the production and validation of transgenic mice with doxycycline (DOX)–inducible RNA interference (RNAi)–mediated knockdown of Sirt1 in nearly all tissues and in specific kidney cells (podocytes or tubular cells). We found that Sirt1 is dispensable for kidney and glomerular development. Under the basal condition, Sirt1 deficiency has no impact on glomerular function. However, under conditions of mitochondrial stress, due to either doxorubicin [Adriamycin (ADR)]–induced genotoxicity or diabetes-related disruption of mitochondrial dynamics, we observed that Sirt1 is necessary for autophagy of damaged mitochondria in podocytes to maintain the ultrastructure and function of the glomerular ultrafilter.
Materials and Methods
Transgenic Mouse Models
Animal studies were performed in accordance with the guidelines of and approved by the Institutional Animal Care and Use Committee at the Icahn School of Medicine at Mount Sinai (New York, NY).
In Vivo RNAi Models for Inducible and Reversible Sirt1 Knockdown
We generated transgenic mice with inducible and reversible knockdown of Sirt1 by adapting a DOX-inducible RNAi model24 to specifically target Sirt1. First, we computationally predicted four shRNA guide sequences to target Sirt1 (reference NM_019812.2). Guide sequences were embedded within the miR-30–based expression cassette of a retroviral shRNA vector (Supplemental Figure S1A). Knockdown efficiency of Sirt1 guide sequences was determined by transducing mouse podocytes with retroviral shRNA vectors. Knockdown efficiency of Sirt1-specific guide sequences was determined by comparing with a control guide sequence targeting the firefly luciferase gene24 (shLuci). Two lines of genetically engineered mice, corresponding to the Sirt1.688 and Sirt1.2191 guide sequences, were generated using the ColA1 Flp/FRT recombinase-mediated targeting system.24 A single copy of the Flp-In targeting vector was inserted downstream of the ColA1 gene in engineered KH2 ES cells, which contain an FRT-hygro-pA homing cassette. The Flp-In targeting vector, called pCol-TGM, was configured with a green fluorescent protein (GFP) spacer between a tetracycline-regulated element and the miR-30–based expression cassette. The GFP spacer sequence functions as a fluorescent reporter of shRNA expression because GFP and the shRNAmir are co-expressed as a single fusion transcript from pCol-TGM. Engineered ES cells with pCol-TGM insertion were selected and then used to generate transgenic mice, called TGM mice, by tetraploid blastocyst microinjection. Heterozygous TGM mice were bred to a line of transgenic mice that express the reverse tetracycline-controlled transactivator (rtTA) under the control of a strong synthetic promoter called CAGs (cytomegalovirus early enhancer element and chicken β-actin).24 Double-transgenic mice with the CAGs-rtTA and TGM transgenes, called CAGs;Sirt1RNAi, exhibit DOX-inducible TGM expression in a widespread manner.
Mice with Inducible Podocyte- and Tubular-Specific Sirt1 Knockdown
To generate mice with inducible Sirt1 knockdown in podocytes and tubular cells, we bred heterozygous Col-TGM mice to NPHS2-rtTA25 and Pax8-rtTA26 mice (both were purchased from Jackson Laboratory, Bar Harbor, ME). Mice with podocyte-restricted and tubular cell–restricted Sirt1 knockdown are called POD;Sirt1RNAi and TUB;Sirt1RNAi, respectively. Mice with inducible and podocyte-specific expression of shLuci targeting the firefly luciferase gene, called POD;LucRNAi mice, were generated by breeding NPHS2-rtTA mice to shLuci ColTGM mice.24 Details of construct design and transgenesis have described elsewhere.24 The genetic background of CAGS;Sirt1RNAi, POD;Sirt1RNAi, and TUB;Sirt1RNAi mice consists of contributions from C57BL/6, FVB, and 129 strains.
Induction of Sirt1 Knockdown
To induce Sirt1 knockdown, CAGs;Sirt1RNAi mice were fed 200 mg/kg DOX-supplemented chow (Bio-Serv, Frenchtown, NJ). To induce Sirt1 knockdown in POD;Sirt1RNAi and TUB;Sirt1RNAi mice, 200 mg/kg DOX-supplemented chow (Bio-Serv) and 0.2 mg/mL DOX-supplemented drinking water were given concurrently. Duration of DOX feeding is specified in the Results and figure legends.
Conditional Knockout Mice with Podocyte-Specific Sirt1 Deletion (PODΔSirt1)
Sirt1 loxP mice27 were bred to NPHS2-Cre mice28 to generate mice that are compound heterozygous for the NPHS2-Cre transgene and the Sirt1 floxed allele. Subsequent F1 × F1 crossings of these compound heterozygous mice yielded mice that were heterozygous for the NPHS2-Cre transgene and homozygous for the Sirt1 floxed allele (PODΔSirt1).
Sirt1 Gene Knockout Mice
Heterozygous Sirt1 exon 4 gene knockout mice29 have been described elsewhere.30
ADR-Induced Nephropathy
Adriamycin (Sigma-Aldrich, St. Louis, MO) was diluted in sterile 0.9% sodium chloride solution. Sensitivity to ADR nephropathy was assessed in a dose-response experiment. The i.v. administration of 12.5 mg ADR/kg of body weight did not cause proteinuria in male CAGS;Sirt1RNAi, POD;Sirt1RNAi, and TUB;Sirt1RNAi mice without DOX-induced Sirt1 knockdown. With 18.8 mg/kg of ADR, all three lines of mice without DOX-induced Sirt1 knockdown developed a significant increase in urinary albumin excretion (0.08 ± 0.006 μg/μg pre-injection versus 0.801 ± 0.081 μg/μg 10 days after injection; P < 0.05). Therefore, a dose of 18.8 mg/kg was chosen for these experiments.
Multiple Low-Dose STZ-Induced Diabetes Model
Induction of diabetes using streptozotocin (STZ; Sigma-Aldrich) has been described previously.31,32 Briefly, mice between 6 and 8 weeks of age were fasted for 4 hours before i.p. injection of 50 mg/kg of body weight STZ diluted in 50 mmol/L sodium citrate buffer (pH 5.4). Fasting and STZ injection were repeated on 5 consecutive days. Two weeks after the first day of STZ injection, a 6-hour fast blood glucose level from the tail vein was measured using the One Touch Blood Glucose Monitoring system (Lifescan, Inc., Milpitas, CA). Repeat glucose measurements were taken at least every 4 hours to verify that hyperglycemia was maintained. Successful modeling of STZ-induced diabetes was defined as sustained fasting glucose level >250 mg/dL on more than two occasions 2 to 3 weeks after STZ injection. Approximately 80% of the mice developed fasting glucose >250 mg/dL 4 weeks after STZ injection. Four of these mice were fed a DOX diet 4 weeks after STZ injection (STZ + DOX), whereas the other four mice were fed a normal chow diet (STZ) until 26 weeks of age.
Tail Cuff Noninvasive Blood Pressure
Blood pressure was measured by CODA noninvasive tail cuff blood pressure system (Kent Scientific, Torrington, CT).
Antibodies
A rabbit antibody to Wilms tumor (WT)-1 and a rabbit antibody to nephrin were purchased from Novus Biologicals (Littleton, CO). A rat antibody to podocalyxin was purchased from R&D Systems (Minneapolis, MN). A mouse antibody for Sirt1 was purchased from Millipore (Billerica, MA). A mouse antibody for GFP and a mouse antibody for TOM20 were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX). A rabbit antibody for p62/SQSTM1 and a rabbit antibody for LC3 were purchased from Cell Signaling Technology (Beverly, MA). A rabbit antibody for parkin was purchased from Abcam (Cambridge, MA). A rabbit antibody for nephrin was a gift from Dr. Larry Holzman (University of Pennsylvannia, Philadelphia, PA) Mitochondrial staining was performed using MitoTracker Red FM (Life Technologies, Carlsbad, CA).
Western Blot Analysis
Western blot analysis was performed as previously described.19 Each lane contained 60 to 100 μg of total protein. Semiquantitative assessment of band density was performed using ImageJ software version 1.43 (NIH, Bethesda, MD). Band density for the protein of interest was normalized to either glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or β-actin.
Quantitative Real-Time PCR
Real-time PCR was performed as previously described.33 For each gene target, at least two technical replicates were measured. CT values of the gene targets were normalized to GAPDH. Fold change in expression of target genes compared with the reference group was calculated using the 2−ΔΔCT method, with GAPDH as the calibrator. Primers were designed using Primer-BLAST34 to span at least one intron. Mitochondria/nuclear DNA ratios were quantified by real-time PCR and presented as a ratio of mitochondria-encoded cyclooxygenase 1/nuclear-encoded 18s rRNA. Primer sequences are in Table 1.
Table 1.
List of Real-Time PCR Primers
| Gene | Forward primer | Reverse primer |
|---|---|---|
| 18s rRNA | 5′-TGCATGGCCGTTCTTAGTTG-3′ | 5′-TAGCATGCCGAGATGCTCGTT-3′ |
| Cd2ap | 5′-TGGTGGAGTGGAACCCTGAA-3′ | 5′-GTGAGGTAGGGCCAGTCAAA-3′ |
| COX1 | 5′-TGCTAGCCGCAGGCATTACT-3′ | 5′-CGGGATCAAAGAAAGTTGTGTTT-3′ |
| Cox8a | 5′-TCCGGCTGGTTCGGCCATCT-3′ | 5′-CCAGCCCGCAGGCAGAAGAC-3′ |
| Gapdh | 5′-GCCATCAACGACCCCTTCAT-3′ | 5′-ATGATGACCCGTTTGGCTCC-3′ |
| Mpv17 | 5′-GCAGCACATCCGTGGAAAGT-3′ | 5′-CCAGCTGCTGTGAGATCATGT-3′ |
| Nd1 | 5′-TGCACCTACCCTATCACTC-3′ | 5′-ATTGTTTGGGCTACGGCT-3′ |
| Nd4 | 5′-ATAATTATAACTAGCTCAATCTGC-3′ | 5′-TCGTAGTTGGAGTTTGCTAG-3′ |
| Ndufa4 | 5′-AGCCAGGTTGCAGAAGCGGC-3′ | 5′-GCTGCTCCAGTACCCCCTGC-3′ |
| Neph1 | 5′-TCACAAGCAGGGCTTTAGGA-3′ | 5′-CTGGCTGAAGCGAGTCTGAG-3′ |
| Neph2 | 5′-AGGACCACGGTGGACTACA-3′ | 5′-TCTACCAGCAGTGACTGAGGC-3′ |
| Nphs1 | 5′-GTGCCCTGAAGGACCCTACT-3′ | 5′-CCTGTGGATCCCTTTGACAT-3′ |
| Nrf1 | 5′-AGAAACGGAAACGGCCTCAT-3′ | 5′-CATCCAACGTGGCTCTGAGT-3′ |
Immunostaining and Imaging
Staining and image acquisition were performed as previously described.33 Images of mitochondrial morphological characteristics were obtained using a Leica TCS SP5II confocal microscope (Leica Microsystems, Inc., Concord, ON, Canada). Stacked fluorescence images of TOM20 and parkin staining were acquired using a Zeiss Axioplan2 IE microscope (Carl Zeiss, Thornwood, NY). Post-processing to deconvolute stacked images was performed using the AutoQuant X2.2.1 software package (Rockville, MD).
Tissue Histological and Morphometric Data
Kidney sections were fixed and embedded in either paraffin or Tissue-Tek Optimal Cutting Temperature compound (Sakura Finetek USA, Inc. Torrance, CA), as previously described.33 For transmission electron microscopy (TEM), kidney cortex samples fixed in 2.5% glutaraldehyde were divided into sections, mounted on a copper grid, and then images were imaged using a Hitachi H7650 microscope (Hitachi, Tokyo, Japan), as previously described.31 Quantification of foot process (FP) width was performed using ImageJ software on digitized TEM images, as previously described.31 For quantification of FP width, two mice per group, five glomeruli per mouse, and >600 μm of glomerular basement membrane length were assessed per mouse.
Detection of Senescence-Associated β-Galactosidase Activity
Frozen kidney tissue embedded in OCT compound was divided into sections (4 μm thick). Tissue activity of senescence-associated β-galactosidase was determined by histochemical detection of senescent cells at pH 6.0, as previously decribed.35 Frozen kidney sections of mice with ADR-induced nephropathy, which demonstrated β-galactosidase activity in tubular cells, were used as positive controls.
Urine Albumin and Protein Analyses
Urine albumin was quantified using an ELISA kit (Bethyl Laboratory, Houston, TX). Urine creatinine was measured in the same urine sample as urine albumin using the QuantiChrom Creatinine Assay Kit (DICT-500; Bioassay Systems, Hayward, CA). Coomassie Blue staining of 5 μL of urine denatured with Laemmli sample buffer and resolved on an 8% SDS polyacrylamide gel was performed as previously described.33
Measurement of Serum Urea Nitrogen and Creatinine
Serum samples were stored at −80°C until measurement. Serum urea nitrogen content was measured using a commercially available kit (Bioassay Systems). Serum creatinine was quantified by a high-performance liquid chromatography–based method, as previously published.36 Serum (5 μL per mouse) was processed as previously described. A Shimadzu Prominence high-performance liquid chromatography unit (Shimadzu Scientific Instruments, Columbia, MD) and a Hamilton cation exchange column (model PRP-X200; Hamilton Company, Reno, NV) were used.
Renal Biopsy Specimen
Survival surgical procedures were performed to obtain a biopsy specimen of the right and left kidneys through flank incisions. A biopsy was performed of approximately 1.5 mm of the lower pole of the right kidney before the introduction of a DOX diet. Fourteen days later, a biopsy was performed of 1.5 mm of the lower pole of the left kidney. At the termination of the study, 1.5 mm of the upper pole of the right kidney was sampled. Kidney samples were stored in RNAlater (Qiagen, Valencia, CA) until RNA extraction or processed for total protein extraction, as described for Western blot analysis.
Next-Generation Sequencing
Total RNA was extracted from a biopsy specimen of kidney cortex samples using an RNAeasy mini kit (Qiagen). Next-generation sequencing of mRNA captured using polyA selection was performed by the Genomics Core Facility at the Icahn School of Medicine at Mount Sinai using the Illumina HiSeq Sequencing Platform (San Diego, CA). Raw readings from RNA sequencing were analyzed using a TopHat/Cufflinks pipeline for transcript assembly and quantification.37
Glomerular Isolation and PGEC Culture
Glomeruli from mice were isolated by perfusion of magnetic particles, as described,38 with minor modifications. Briefly, isolated glomeruli were transferred onto a 6-cm tissue culture dish coated with type I collagen and cultured in RPMI 1640 medium (Life Technologies) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. DOX was added to the culture medium to achieve a final concentration of 2 μg/mL, where specified. Glomeruli and cells were allowed to attach to the plate for 5 days in a 37°C incubator without any agitation. Five days later, glomeruli and outgrowth cells were detached from the plate using a 0.12% trypsin-EDTA solution. Trypsinized cells [primary glomerular epithelial cells (PGECs)] were strained using a 40-μm cell strainer and replated onto collagen-coated dishes. PGECs were allowed to grow to 80% confluence before passaging at a ratio of 1:3. PGECs are cells derived from decapsulated glomeruli, isolated by perfusion of magnetic particles. PGECs consist mostly of podocytes, with approximately 70% of PGECs being positive for podocalyxin staining.
Fluorescence-Activated Cell Sorting of GFP-Labeled Podocytes
To isolate GFP-positive podocytes from POD;Sirt1RNAi and POD;LucRNAi mice, we first isolated glomeruli, as previously described. Fluorescence-activated sorting of podocytes was performed as described previously.39 Briefly, isolated glomeruli were digested into a single-cell suspension using a buffered solution containing collagenase II, proteinase E, and DNase I, as described previously. Podocytes were isolated using an FACSAria II cell sorter (laser excitation, 488 nm; power, 200 MW; and sheath pressure, 20 to 30 psi using a 100-μm nozzle). DAPI staining was performed to exclude damaged cells stained with DAPI.
Cell Lines
Murine Podocyte Cell Line
A conditionally immortalized murine podocyte cell line used to test knockdown efficiency of Sirt1 shRNA guide sequences was a gift from Dr. Peter Mundel (Massachusetts General Hospital, Boston). Podocytes were cultured on collagen I–coated plates at 37°C for at least 7 days before experiments, as previously described.
Inducible Sirt1 Knockdown Podocyte Cell Line
A conditionally immortalized podocyte cell line with DOX-inducible Sirt1 knockdown from POD;Sirt1RNAi mice was established from GFP-positive podocytes by fluorescence-activated cell sorting (FACS). FACS-isolated podocytes were allowed to attach to a collage I–coated tissue culture dish for 12 hours. Isolated cells were transduced with a lentivector that expressed a temperature-sensitive SV40 T/t antigen, then maintained and propagated at 33°C permissive growth condition. Cells were thermoshifted to 37°C and maintained for 1 week before experiments. Approximately 70% of the FACS-isolated podocytes expressed GFP on DOX induction (2 ng/mL of culture medium after 3 days’ incubation). Conditionally immortalized cells used in the experiments herein were passaged less than five times.
Statistical Analysis
Data are expressed as means ± SEM. Statistical analyses were performed using GraphPad Prism version 5.0a (La Jolla, CA). For comparison of means between groups, one-way analysis of variance was performed, followed by a Tukey post-test using a significance cutoff of P < 0.05.
Results
A Murine Model of Inducible and Reversible Sirt1 Knockdown
We adapted a DOX-inducible RNAi-mediated knockdown model24 to target Sirt1. First, we computationally predicted four shRNA guide sequences targeting Sirt1 (Supplemental Figure S1A). Two of the four guide sequences, Sirt1.688 and Sirt1.2191, exhibited >50% Sirt1 knockdown in a murine podocyte cell line (Supplemental Figure S1, B and C). Two lines of genetically engineered mice with either the Sirt1.688 or Sirt1.2191 guide sequence were generated. To generate mice with robust and widespread knockdown of Sirt1, we produced double-transgenic CAGs;Sirt1RNAi mice in which DOX-induced TGM expression is regulated by the strong, ubiquitous CAGs promoter (Figure 1A). CAGs;Sirt1RNAi mice expressing either the Sirt1.688 or Sirt1.2191 guide sequence exhibit robust DOX-inducible knockdown of Sirt1 protein and mRNA transcript levels (Figure 1, B and C). DOX treatment also induced GFP expression from the TGM transgene, which allows tracking of cells with Sirt1 knockdown. GFP expression was stronger in CAGs;Sirt1RNAi mice expressing the Sirt1.688 guide sequence (Figure 1B). Therefore, the Sirt1.688 line was used in all subsequent experiments. This discrepancy in GFP expression between the Sirt1.688 and Sirt1.2191 lines was likely due to improved shRNAmir processing of the Sirt1.2191 guide sequence.
Figure 1.
In vivo RNAi model with inducible Sirt1 knockdown. A: Double-transgenic CAGs. Sirt1RNAi mice harbor two transgenes: TGM and CAGs-rtTA. B: Representative Western blot analyses of Sirt1, GFP, and GAPDH on kidney protein lysates from two different lines of CAGs;Sirt1RNAi mice, with or without 5 days of DOX feeding, starting at 8 weeks of age. Each lane represents one mouse. C: Normalized Sirt1 band density from Western blot analyses in B. D: Immunostaining of WT-1 on kidney sections of CAGs;Sirt1RNAi mice, with and without DOX feeding. Dashed line in the lowest panel outlines WT-1–positive cells. Representative Western blot analyses of Sirt1 in PGECs isolated from CAGs-only control mice and CAGs;Sirt1RNAi mice. E: PGECs were cultured with and without 2 μg/mL DOX for 3 days. F: Normalized Sirt1 mRNA transcript levels of PGECs cultured with and without DOX. ∗P < 0.05. Scale bar = 50 μm (D). CAGG, CMV-enhanced chicken β-actin promoter; Hygro, hygromycin resistance gene; I, interstitial cells; P, WT-1–positive podocytes; PGK, phosphoglycerate kinase 1 promoter; rtTA3, third-generation reverse tet-transactivator; SAdpA (stop), splicing acceptor donor polyA sequence; T, tubular cells; TRE, tetracycline-regulated element.
The pattern of GFP expression in the kidney indicates that nearly all of the cells within the glomerular and tubulointerstitial compartments of CAGs;Sirt1RNAi kidney were targeted for Sirt1 knockdown (Figure 1D). GFP expression was absent from renal interstitial cells and nonpodocyte glomerular cells (Figure 1D). Colocalization of GFP with WT-1, which is a marker of podocytes, confirmed that podocytes were targeted for Sirt1 knockdown (Figure 1D). To assess the extent of Sirt1 knockdown in podocytes, we isolated PGECs from CAGs;Sirt1RNAi mice and cultured these PGECs in the presence or absence of DOX. PGECs were also isolated from control mice with only the CAGs-rtTA transgene (ie, CAGs-only mice). DOX treatment significantly reduced the protein and mRNA expression of Sirt1 in CAGs;Sirt1RNAi PGECs (Figure 1, E and F). As expected, Sirt1 expression was reduced and GFP expression was present in multiple organs of DOX-fed CAGs;Sirt1RNAi mice (Supplemental Figure S1D).
To demonstrate that shRNA-mediated knockdown of Sirt1 is reversible on withdrawal of DOX, we performed transcriptome-wide assessment of renal gene expression by next-generation sequencing of RNA transcripts extracted from kidneys of mice before DOX (D0), after 14 days of DOX treatment (D14), and 14 days after discontinuing DOX treatment (D28) (Supplemental Figure S1E). The transcript and protein levels of Sirt1 on D14 were significantly lower than those on D0 and D28 (Supplemental Figure S1, F and G). Of all of the mammalian sirtuins (Sirt1 to Sirt7), only the Sirt1 mRNA transcript level was significantly lower on D14 compared with both on D0 and D28 (Supplemental Figure S1H), suggesting that Sirt1 knockdown in our model was not associated with a compensatory change in the expression of other members of the sirtuin family.
Functional Role of Sirt1 in the Kidney under the Basal Condition
First, we examined the impact of acute Sirt1 knockdown (5 days of DOX) on the renal function of male CAGs;Sirt1RNAi mice. We found that urinary albumin/creatinine ratio (UACR) and serum levels of urea nitrogen and creatinine were not significantly different between mice with and without DOX feeding to knockdown Sirt1 (Supplemental Figure S2, A–C). Renal histological features were also indistinguishable between the two groups of mice (data not shown). Next, we examined the impact of chronic Sirt1 knockdown (11 weeks of DOX) on renal function of CAGs;Sirt1RNAi mice. Chronic DOX feeding significantly decreased protein and mRNA expression of Sirt1 compared with age-matched mice without DOX feeding (Figure 2, A and B). Urinary albumin excretion (ie, UACR) and serum levels of urea nitrogen and creatinine were also not significantly different between mice with and without chronic Sirt1 knockdown (Figure 2, C–E). Because Sirt1 participates in the regulation of cellular senescence in response to cellular stress,40,41 we evaluated the impact of chronic Sirt1 knockdown on cellular senescence of renal cells. Markers of cellular senescence, including senescence-associated β-galactosidase activity (Figure 2F) and mRNA transcript levels of the Cdkn2a locus (INK4a and ARF) (Figure 2B), were not markedly different between mice with and without Sirt1 knockdown. Furthermore, the renal expression of podocyte genes—Nphs1, Synpo, CD2AP, WT-1, Neph1, and Neph2—and histological features of glomeruli, podocytes, podocyte foot processes, mesangial cells, glomerular endothelium, and tubular cells were also unaffected by chronic Sirt1 knockdown (Figure 2, B and G–L).
Figure 2.
Renal gene expression, function, and histological features of mice with chronic Sirt1 knockdown. A: Representative Western blot analyses of Sirt1, GFP, and β-actin of age-matched CAGs;Sirt1RNAi mice with and without 11 weeks of DOX feeding. The arrow indicates the Western blot band corresponding to Sirt1. B: Glomerular expressions of Sirt1, markers of cellular senescence (ARF and p16INK4a), and podocyte-specific genes (Nphs1, Synpo, CD2AP, WT-1, Neph1, and Neph2) in mice with and without 11 weeks of DOX feeding. N = 4 mice per group. Black bars, DOX; white bars, no DOX. C: UACR of CAGs;Sirt1RNAi mice before and after 11 weeks of DOX feeding. Serum urea nitrogen (D) and serum creatinine (E) of CAGs;Sirt1RNAi mice after 11 weeks of DOX compared with those not fed with DOX. N = 4 to 5 mice per group. F: Staining for β-galactosidase activity assessed at a pH of 6.0 (known as senescence-associated β-galactosidase activity) on frozen kidney sections of CAGs;Sirt1RNAi mice with and without 11 weeks of DOX feeding. Staining of a section of mice with ADR nephropathy (ADR-N) served as a positive control. Arrowheads indicate tubular cells with positive staining. G–L: Representative images of kidney glomeruli, podocyte, podocyte foot processes, mesangial cell, glomerular endothelium, and tubular cell of CAGs;Sirt1RNAi mice after 11 weeks of DOX feeding. ∗P < 0.05. Scale bars: 50 μm (F and G); 2 μm (H–L). Pos., positive.
To further confirm that Sirt1 is dispensable for glomerular function under the basal condition, we examined the renal phenotype in two other Sirt1 gene deletion models: heterozygous Sirt1-null mice (Sirt1+/−)30 and podocyte-conditional Sirt1 deletion mice (PODΔSirt1). Sirt1 was approximately 45% lower in the renal cortex of 22-week-old Sirt1+/− mice and approximately 80% lower in PGECs of PODΔSirt1 mice compared with corresponding age-matched control mice (Sirt1+/+ mice and NPHS2-Cre mice) (Supplemental Figure S3, A–D). Similar to CAGs;Sirt1RNAi mice with chronic Sirt1 knockdown, Sirt1+/− mice and PODΔSirt1 mice did not develop any abnormality in renal histological characteristics or urinary protein excretion when followed up to 40 and 50 weeks of age, respectively (data not shown). Taken together, these data strongly suggest that up to an 80% reduction of Sirt1 in the podocyte and kidney, either acutely or chronically, does not interfere with glomerular function and development under the basal condition.
Mice with Inducible Podocyte- and Tubule-Specific Sirt1 Knockdown
To study Sirt1 function in podocytes and tubular cells, we generated double-transgenic mice with either podocyte- or tubular-restricted Sirt1 knockdown (eg, POD;Sirt1RNAi and TUB;Sirt1RNAi mice) (Figure 3A). Podocyte-specific Sirt1 knockdown in POD;Sirt1RNAi mice was confirmed by colocalization of GFP with a podocyte cell surface marker, podocalyxin (Supplemental Figure S4A). GFP expression was also observed in PGECs of POD;Sirt1RNAi mice that displayed highly arborized morphological characteristics, but not in PGECs with a cobblestone appearance (Figure 3B). These findings suggest that GFP expression and Sirt1 knockdown are restricted to the podocytes of POD;Sirt1RNAi mice. To determine the magnitude of Sirt1 knockdown in the podocytes, we isolated the GFP-positive podocytes by FACS from POD;Sirt1RNAi mice and control POD;LuciRNAi mice that have been fed DOX for 7 days (Supplemental Figure S4B). We found that the Sirt1 transcript level of GFP-positive podocytes, isolated from POD;Sirt1RNAi mice, was 31.3% ± 1.27% of that from POD;LucRNAi mice (Figure 3C). To determine the specificity of Sirt1 knockdown in TUB;Sirt1RNAi mice, we evaluated the pattern of GFP expression in kidney sections and found that GFP was expressed in the tubular compartment and absent from the glomeruli of DOX-fed TUB;Sirt1RNAi mice (Supplemental Figure S4C). Sirt1 expression in the kidney cortex of DOX-fed TUB;Sirt1RNAi mice was approximately 70% lower than TUB;Sirt1RNAi mice without DOX feeding (Figure 3D).
Figure 3.
Genetically engineered mice with inducible and conditional Sirt1 knockdown in podocytes and tubular cells. A: A scheme depicting two lines of double-transgenic mice with inducible Sirt1 knockdown in podocytes (POD;Sirt1RNAi) or tubular cells (TUB;Sirt1RNAi). Podocyte-specific knockdown was driven by a 2.5-kb NPHS2 promoter, and tubular-specific knockdown was driven by a 5.6-kb Pax8 promoter. B: A representative fluorescein isothiocyanate (FITC) fluorescence image overlapped with a phase-contrast image of glomeruli (arrowheads) isolated from POD;Sirt1RNAi mice. Highly arborized PGECs emanating from glomeruli are marked with arrows. The asterisk indicates GFP-negative PGECs outgrown from the glomeruli. C: Normalized Sirt1 expression of GFP-positive podocytes isolated by fluorescence-activated cell sorting from POD;Sirt1RNAi mice and control POD;LucRNAi mice (N = 4 mice in each group). D: Normalized Sirt1 mRNA expression in renal cortex of TUB;Sirt1RNAi mice with and without DOX feeding for 5 days. N = 3 mice per group. ∗P < 0.05.
Sirt1 Knockdown Enhances Susceptibility to ADR Nephropathy
Because Sirt1 knockdown has no impact on glomerular function under the basal condition, next we investigated whether Sirt1 is required for the maintenance of glomerular function in mice with ADR-induced nephropathy. ADR is an anthracycline antibiotic. When injected into the tail vein of mice, ADR causes podocyte damage and glomerular injury. Proteinuria and glomerulosclerosis manifest as early as 10 and 28 days after ADR injection, respectively.42 Mitochondrial function43 and autophagy response44 of podocytes are key determinants of strain-dependent susceptibility to ADR. Because Sirt1 is known to regulate mitochondrial function and autophagy, we examined whether CAGs;Sirt1RNAi mice with Sirt1 knockdown are more susceptible to ADR-induced glomerular injury. DOX was given to male CAGs;Sirt1RNAi mice 5 days before ADR injection and continued until 10 days after ADR injection (Figure 4A). CAGs;Sirt1RNAi mice with and without DOX feeding were injected with 0.9% saline as a control. DOX-fed and ADR-injected mice (ie, ADR + DOX) lose more body weight compared with all other groups of mice (Supplemental Figure S5A). Urinary albumin excretion, as assessed by Coomasie Blue staining of urinary proteins and UACR, was significantly more in the ADR + DOX group compared with the ADR-only group (1.860 ± 0.42 versus 0.8 ± 0.081 μg/μg; P < 0.05) (Figure 4, B and C). Serum urea nitrogen of the ADR + DOX group was also higher than the ADR-only group (Supplemental Figure S5B), whereas no significant difference in serum creatinine was observed (data not shown). Dilated renal tubules with protein casts were present diffusely in the ADR + DOX kidneys (>50% of the tubular area), infrequently in the ADR-only kidneys (<25% of the tubular area), and absent in the saline-only and saline + DOX kidneys (Figure 4D). No obvious disruption of glomerular structure was observed by light microscopy. Ultrastructural examination of the podocyte foot processes (FPs), however, revealed that FP width was significantly wider in the ADR + DOX group compared with the ADR-only group (1240 ± 41 nm versus 641 ± 30 nm) (Figure 4, E and F). These data indicate that Sirt1 deficiency exacerbates ADR-induced disruption of podocyte FP organization and glomerular ultrafilter function during the early phase of ADR-induced nephropathy.
Figure 4.
Glomerular function and structure of CAGs;Sirt1RNAi mice with ADR-induced nephropathy. A: A scheme of study design to determine the susceptibility of CAGs;Sirt1RNAi mice to ADR. B: A Coomasie Blue–stained gel with urinary proteins and albumin standards resolved by SDS-PAGE. (Albumin standards: 0.05, 0.1, 0.5, 1.0, and 5.0 μg per lane). C: A graph of UACR on D0 and D10 after ADR or normal saline (NS) injection. N = 5 in the ADR-only group, N = 6 in the ADR + DOX group, N = 3 in the NS-only group, and N = 3 in the NS + DOX group. D: Representative images of PAS-stained kidney sections. Dilated tubules filled with protein casts are present in the ADR + DOX group. E: Representative TEM images of podocytes (P). Red arrows indicate areas of foot process effacement. F: A bar graph representing podocyte FP width. ∗P < 0.05, ∗∗∗P < 0.001. Std, standard.
Sirt1 in the Podocytes, But Not Tubules, Maintains Glomerular Function in ADR Nephropathy
Because Sirt1 knockdown is widespread in CAGs;Sirt1RNAi mice, it was not clear whether their susceptibility to ADR nephropathy was due to Sirt1 knockdown in podocytes and/or other renal cell types. To specifically address podocyte Sirt1’s role in ADR nephropathy, we compared the progression of ADR nephropathy between POD;Sirt1RNAi and TUB;Sirt1RNAi mice. The length of the experiment was extended to 28 days after ADR injection to capture the development of ADR-induced glomerulosclerosis. Simultaneously, we also investigated whether reversal of Sirt1 knockdown modifies the progression of nephropathy. Male POD;Sirt1RNAi and TUB;Sirt1RNAi mice were injected with ADR and then fed DOX for either 14 or 28 days (Figure 5A). Mice with a shorter duration of Sirt1 knockdown were fed DOX for 14 days before withdrawal of DOX [ie, the ADR + DOX_WD (doxycycline withdraw) group], and mice with a longer duration of knockdown (ie, the ADR + DOX group) received DOX for all 28 days of the experiment. Mice were observed for 28 days after ADR injection. On day 14, the level of UACR was significantly higher in both of the POD;Sirt1RNAi groups (ADR + DOX and ADR + DOX_WD) compared with the corresponding TUB;Sirt1RNAi groups (Figure 5B). Between days 14 and 28, UACR continued to increase for the ADR + DOX group of POD;Sirt1RNAi mice, but remained unchanged for the ADR + DOX_WD group of POD;Sirt1RNAi mice as well as both groups of TUB;Sirt1RNAi mice. Although serum creatinine levels were not different between the four groups on day 14, the creatinine level for the ADR + DOX group of POD;Sirt1RNAi mice on day 28 was significantly higher than all other groups (Figure 5C). Segmental glomerulosclerosis was observed in >75% of the glomeruli in the ADR + DOX group, in <25% of the glomeruli in the ADR + DOX_WD group, and in <10% of the glomeruli in either groups of TUB;Sir1RNAi mice (Figure 5D). For the POD;Sirt1RNAi mice, tubular dilation and tubular protein casts were observed in >50% of the tubular lumen in the ADR + DOX group and in <10% of the ADR + DOX_WD group. Tubular casts were absent in both groups of TUB;Sirt1RNAi mice. Assessment of podocyte FPs revealed focal areas of effacement in both groups of POD;Sirt1RNAi mice (Figure 5E). The width of FPs was significantly higher in the ADR + DOX group compared with the ADR + DOX_WD group (Figure 5, E and F). These findings suggest that Sirt1’s function in the podocyte, and not tubular cells, contributes to CAGs;Sirt1RNAi mice’s susceptibility to ADR. In addition, our data suggest that reversal of Sirt1 knockdown 2 weeks after ADR injection prevented the progression, but did not completely reverse the development, of ADR-induced glomerulopathy.
Figure 5.
Progression and reversibility of ADR nephropathy in POD;Sirt1RNAi and TUB;Sir1RNAi mice. A: A scheme of study design. B: UACR of mice 0, 14, and 28 days after ADR injection. ∗P < 0.05 versus UACR on D0. (N = 7 POD;Sirt1RNAi mice in both ADR + DOX and ADR + DOX_WD groups, and N = 8 TUB;Sirt1RNAi mice in both ADR + DOX and ADR + DOX_WD groups). C: Serum creatinine levels of mice after ADR injection. D: Representative images of PAS-stained kidney sections showing global glomerulosclerosis (black arrows), segmental glomerulosclerosis (black arrowheads), normal glomeruli (gray arrowheads), and dilated tubules with protein casts. E: Representative electron microscopy images for POD;Sirt1RNAi mice showing focal areas of effacement (red arrows) with areas of normal FPs (black arrows). F: A bar graph of FP width for POD;Sirt1RNAi mice. (N = 3 mice in each group of ADR + DOX and ADR + DOX_WD.) ∗P < 0.05, ∗∗∗P < 0.001. Scale bar = 50 μm (D). P, podocytes.
Sirt1 Knockdown Enhances ADR-Induced Mitochondrial Damage in Podocytes
To delineate the cellular mechanism that drives ADR-induced renal injury in Sirt1-deficient mice, we focused on ADR-induced mitochondrial damage. First, we confirmed that 0.4 μg/mL ADR exposure for 6 hours significantly reduced the mitochondrial DNA (mtDNA) content of primary PGECs isolated from mice without Sirt1 knockdown (Figure 6A). ADR exposure was also found to significantly up-regulate the expression of genes encoding essential mitochondrial proteins, including those involved in mitochondrial respiration (Nrf1, Ndufa4, and Sco2), mitochondrial reactive oxygen species metabolism (Mpv17 and Sod2), and mtDNA transcription (Polrmt, Tfb1m, and Tfb2m) (Figure 6B). Interestingly, the expression of Prkdc, which encodes a nuclear DNA double-stranded break repair protein that prevents ADR-induced mtDNA damage and nephropathy,43 was significantly increased by ADR. Sirt1 was also significantly increased by ADR treatment.
Figure 6.
Mitochondrial response of podocytes with Sirt1 knockdown to ADR treatment. A: mtDNA/nuclear DNA (nuDNA) ratio of PGECs with and without 0.4 μg/mL ADR treatment for 6 hours. Results from four independent experiments. B: Relative expression of mitochondrial genes and other genes of interest for PGECs with and without ADR treatment. Results from at least five independent experiments. C: Mitochondria staining of POD;Sirt1RNAi podocytes using a mitochondrial membrane–dependent MTR probe. D: Co-immunostaining of mitochondria and a marker of mitophagy by anti-TOM20 (red) and anti-parkin (pseudocolored: white) antibodies, respectively. Inset, right panel: High-power images of the area circumscribed by the dashed-line squares in the larger images. ADR, 0.4 μg/mL ADR for 6 hours; ADR + DOX, dose and duration of treatment same as previously described; Control, no treatment; DOX, 2 μg/mL DOX for 48 hours. ∗P < 0.05, ∗∗∗P < 0.001. Scale bar = 20 μm (C and D); 5 μm (D, insets).
To examine the impact of ADR treatment on mitochondria of podocytes with Sirt1 knockdown, we performed mitochondrial staining using a mitochondrial membrane potential-dependent probe, called MitoTracker (MTR; Life Technologies). Cytoplasmic MTR staining was markedly reduced by ADR treatment in podocytes without DOX-induced Sirt1 knockdown (ADR group) and in podocytes with DOX-induced Sirt1 knockdown (ADR + DOX group) (Figure 6C). Sirt1 knockdown alone (DOX), however, did not reduce MTR staining. To determine whether the ADR-induced decrease in MTR staining was due to a loss of mitochondrial membrane potential or a reduction in mitochondrial number, we performed immunostaining for TOM20, which is a mitochondrial outer membrane translocase protein. We found that cytoplasmic staining of TOM20 was not affected by ADR or DOX, either separately or in combination (Figure 6D), suggesting that reduced MTR staining in ADR-treated cells was more likely caused by a loss of mitochondrial membrane potential rather than a reduction in mitochondrial number (Figure 6D). A perinuclear pattern of TOM20 and MTR labeling was observed in the ADR and ADR + DOX groups.
Autophagy is a key cellular mechanism for removal of damaged mitochondria. Selective mitochondrial elimination by autophagy is mediated by an E3 ubiquitin ligase called parkin. Parkin translocates to the mitochondrial outer membrane on dissipation of the mitochondrial membrane potential in response to oxidant stress and labels damaged mitochondria for removal by autophagy.45 Because ADR treatment induces mtDNA depletion and depolarization, we examined whether ADR promotes autophagy of damaged mitochondria and whether Sirt1 deficiency prevents ADR-induced autophagy by studying the accumulation of parkin on TOM20-labeled mitochondria. We found that ADR increased intracellular parkin-labeled puncti and the extent of parkin and TOM20 colocalization (Figure 6D). ADR-induced colocalization of parkin and TOM20, however, was markedly reduced in Sirt1 knockdown cells (ie, the ADR + DOX group), suggesting that ADR-induced parkin recruitment is an Sirt1-dependent process. To examine Sirt1’s role on autophagy flux in podocytes, we studied the accumulation of lipidated LC3 (ie, LC3-II) and p62 in the presence and absence of bafilomycin A1, which inhibits degradation of autolysosome content. We observed that podocytes have a high LC3-II/LC3-I ratio under the basal condition (lane 1) (Figure 7), which is consistent with the previous observation that podocytes in vivo exhibit a high autophagy flux.44 ADR treatment increased the accumulation of both LC3-II and p62/SQSTM1 in the presence of bafilomycin A1 compared with those without ADR treatment (lane 3 versus 1) (Figure 7), which was abrogated in DOX-treated cells with Sirt1 knockdown (lane 3 versus 4). Taken together, these findings suggest that Sirt1 is required to facilitate the ADR-triggered autophagy response. Interestingly, ADR treatment alone did not markedly alter p62 expression in cells not treated with bafilomycin (lane 2 versus 1).
Figure 7.
Markers of autophagy in POD;Sirt1RNAi podocytes treated with ADR. A: Representative Western blot analyses of LC3, p62/SQSTM1, and β-actin of POD;Sirt1RNAi podocytes with and without DOX-induced Sirt1 knockdown treated with ADR in the absence or presence of an autophagy inhibitor, bafilomycin A. B: Band densitometric values of Western blot analyses for LC3-II and p62 relative to β-actin and normalized to lane 1. Values are averages from two sets of experiments. Black bars, p62/actin; white bars, LC3-II/actin.
Podocyte Sirt1 Facilitates Autophagy without Increasing the Glomerular Expression of Mitochondrial Genes
To validate the in vitro observations, we compared the expression of mitochondrial genes and the morphological characteristics of mitochondria in ADR-injected POD;Sirt1RNAi mice with Sirt1 knockdown. The mitochondria-to-nuclear DNA content in glomeruli of POD;Sirt1RNAi mice with a longer duration of Sirt1 knockdown (ADR + DOX) was significantly lower than that of POD;Sirt1RNAi mice with a shorter duration of Sirt1 knockdown (ADR + DOX_WD) (Figure 8A). Mitochondria of podocytes in the ADR + DOX group exhibited loss of internal cristae and appeared more fragmented and swollen (Figure 8B). Early autophagosomes containing heterogeneous cytoplasmic material were observed in both ADR + DOX and ADR + DOX_WD podocytes, but appeared less abundant in the ADR + DOX group. Taken together, these findings suggest that prolonged knockdown of Sirt1 reduces the glomerular mtDNA content, increases the accumulation of dysmorphic mitochondria, and prevents formation of autophagosomes in ADR-treated mice.
Figure 8.
Mitochondrial gene expression and morphological characteristics in POD;Sirt1RNAi mice with podocyte Sirt1 knockdown. A: A bar graph depicting mtDNA content normalized to nuDNA in glomeruli of POD;Sirt1RNAi mice with (ADR + DOX_WD) and without (ADR + DOX) reversal of Sirt1 knockdown. N = 4 samples per group. B: Electron microscopy images of mitochondria in podocytes of POD;Sirt1RNAi mice. Green arrows indicate fragmented mitochondria with loss of internal cristae structure; red arrows, normal-appearing mitochondria with intact cristae; green arrowheads, early autophagosomes containing mitochondrial profile similar to an adjacent mitochondrion in the cytoplasm; red arrowheads, autophagosomes containing heterogeneous cytoplasmic material. C: Normalized expressions of selected mitochondrial genes in isolated glomeruli of POD;Sirt1RNAi mice with or without reversal of Sirt1 knockdown. N = 4 samples per group. ∗∗P < 0.01. Black bars, ADR+DOX_WD; blue bars, ADR+DOX; FP, foot processes.
Because Sirt1 also activates PGC1α, which is a master regulator of mitochondrial gene transcription and mitochondrial biogenesis,46,47 we compared the glomerular expression of mitochondrial genes in ADR-injected POD;Sirt1RNAi mice. Glomerular expression of nuclear DNA-encoded mitochondrial genes in podocytes (Nrf1, Ndufa4, Sco2, Mpv17, Sod3, Polrmt, Tfb1m, and Tfb2m) that were up-regulated and in podocytes treated with ADR in vitro were not significantly different between ADR + DOX_WD and ADR + DOX groups (Figure 8C). Specifically, the glomerular expression of mitochondrial genes important for mitochondrial biogenesis—Nrf1, Tfb1m, and Tfb2m—was not different between ADR + DOX_WD and ADR + DOX.
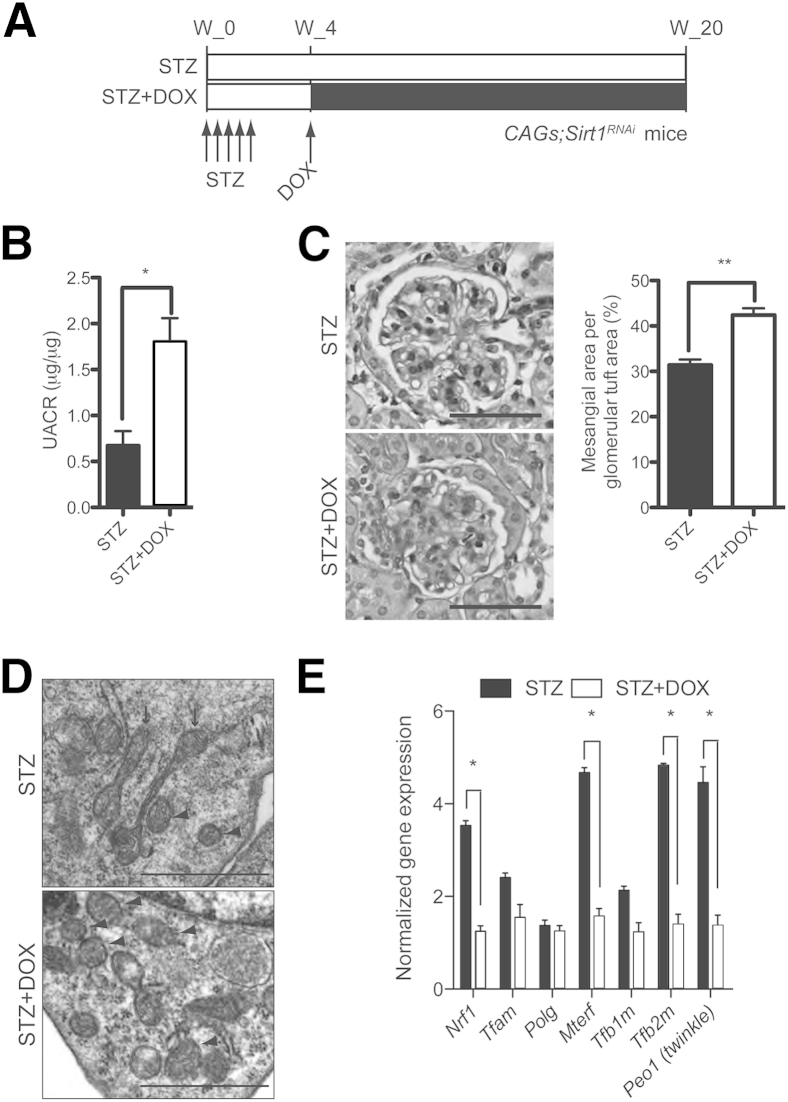
Sirt1 Deficiency Promotes Mitochondrial Injury of Glomerular Cells in Diabetic Mice
Because a pathogenic role for Sirt1 in diabetic nephropathy has been reported by us and others18–20,22 and mitochondrial dysfunction is involved in the pathogenesis of diabetic kidney injury,48,49 we evaluated the progression of diabetic nephropathy in CAGs;Sirt1RNAi mice with and without Sirt1 knockdown (STZ + DOX and STZ, respectively). Diabetes mellitus was induced in these mice between 6 and 7 weeks of age by multiple low-dose STZ injections (Figure 9). Mice were followed up for 20 weeks after the start of STZ injections (Figure 9A). Four-hour fasting serum glucose levels, body weights, kidney/body weight ratio, systolic blood pressures, and serum creatinine concentration were measured at the end of 20 weeks of observation and were not significantly different between STZ and STZ + DOX groups (Table 2). However, urinary albumin excretion was significantly higher in the STZ + DOX mice compared with STZ mice (1.99 ± 0.22 versus 0.74 ± 0.11 μg albumin/μg creatinine; P = 0.0063) (Figure 9B). In addition, mesangial matrix area of the STZ + DOX group was significantly higher than the STZ group (mesangial area per glomerular tuft area, 42.4% ± 1.5% versus 31.4% ± 1.2%; P = 0.0004) (Figure 9C). Ultrastructural evaluation of the mitochondria in podocytes revealed numerous mitochondria with a smaller aspect ratio and loss of internal cristae (Figure 9D), which are suggestive of mitochondrial fragmentation, in the STZ + DOX group, whereas the long filamentous mitochondria were observed in the STZ group (Figure 9D). Glomerular mRNA expression of genes involved in mitochondrial gene transcription (Nrf1, Mterf, and Tfb2m) and mtDNA replication (Peo1/twinkle) was significantly lower in the STZ + DOX group (Figure 9D). Together, these findings provide further validation that Sirt1 deficiency plays a pathogenic role in another experimental model, in which mitochondrial injury is critically involved in the pathogenesis of glomerular disease.
Figure 9.
Diabetic CAGs;Sirt1RNAi mice with Sirt1 knockdown. A: A scheme depicting the experimental design for determining the impact of STZ-induced diabetes on the development of nephropathy in CAGs;Sirt1RNAi mice with and without DOX-induced Sirt1 knockdown (STZ + DOX and STZ, respectively). B: Urinary albumin/creatinine ratio of mice 20 weeks after the start of STZ injections. C: PAS-stained kidney sections. Adjacent bar graph depicts the percentage of mesangial area/glomerular tuft area in the two groups of mice. D: TEMs of mitochondria within podocytes. Arrowheads indicate round, fragmented mitochondria with absence of internal cristae structure; arrows, long filamentous mitochondria. E: Glomerular expression of mitochondrial genes. ∗P < 0.05, ∗∗P < 0.01. Scale bars: 50 μm (C); 1 μm (D).
Table 2.
Physiological and Biochemical Parameters of Diabetic CAGs;Sirt1RNAi Mice 20 Weeks after STZ Injection
| Parameter | STZ (n = 4) | STZ + DOX (n = 6) |
|---|---|---|
| 4-Hour fasting serum glucose (mg/dL) | 409 ± 71 | 416 ± 59 |
| Body weight (g) | 35.3 ± 4.0 | 35.7 ± 1.9 |
| Kidney weight/body weight (mg/g) | 9.6 ± 0.95 | 10.8 ± 0.10 |
| Systolic BP (mmHg) | 134 ± 5 | 141 ± 9 |
| Serum creatinine (mg/dL) | 0.13 ± 0.01 | 0.14 ± 0.01 |
Values are expressed as means ± SEM. No statistically significant differences were observed between the STZ and STZ + DOX groups of mice at the time of sacrifice (20 weeks after STZ injection).
Discussion
Herein, we generated and validated several lines of genetically engineered mice exhibiting tissue-specific, DOX-inducible knockdown of Sirt1 expression in a reversible manner. We demonstrated that post-natal Sirt1 reduction, either acutely or chronically, widespread or restricted to the kidneys, does not interfere with glomerular function. However, we determined that Sirt1 plays a key role in autophagy of mitochondria in podocytes of mice with ADR nephropathy and diabetic nephropathy.
Conditional and inducible gene deletion models for the podocytes have been described previously.50,51 However, these inducible gene reduction models rely on Cre-mediated recombination, which involves irreversible and direct modification of the gene of interest. Gene knockdown by RNAi, on the other hand, is reversible and does not involve direct modification to the gene of interest. Thus, this model is appropriate for rescue experiments in which the investigator can examine the impact that reversal of gene knockdown in mice with established pathological features has on the course of disease. Others have proposed the application of an in vivo RNAi approach in renal research,52,53 but we are not aware of any successful attempts in the published literature. Our model is uniquely suited for studying kidney podocyte- and tubule- specific roles of Sirt1 in disease processes, as we demonstrated herein with ADR nephropathy. Herein, we observed that diabetic mice with widespread Sirt1 knockdown manifested early features of diabetic nephropathy after 20 weeks of hyperglycemia, including mesangial matrix accumulation and albuminuria, as well as alterations in the mitochondrial morphological characteristics and gene expression. These findings in mice with widespread Sirt1 knockdown are consistent with a recent study showing that Sirt1 deficiency in the tubular cells promotes diabetic albuminuria18 and our own observation that podocyte-specific Sirt1 deletion aggravates albuminuria in diabetic db/db mice.20 Although overexpression of Sirt1 in the proximal tubules of transgenic mice prevents the development of albuminuria in diabetic mice,18 it remains to be determined whether reversal of Sirt1 knockdown in our model and overexpression of Sirt1 in podocytes can prevent or reverse the development of diabetic nephropathy.
A podocyte is a post-mitotic, terminally differentiated cell with a limited capacity for cell division. As such, dependence on homeostatic mechanisms to cope with cellular stress, such as ADR-induced mitochondrial stress, is critical to prevent amplification of the initial stressor, which otherwise would further impair the respiratory chain or promote mutations in mitochondrial DNA54 and could ultimately contribute to cell death. The essential role of mitochondrial quality control in podocytes is highlighted by our study. Herein, we elicited the critical role served by Sirt1 in podocytes by imposing ADR-induced genotoxic stress on the mitochondrial genome. Knocking down Sirt1 in podocytes markedly diminished the podocyte’s ability to cope with ADR-induced and diabetes-related mitochondrial damage. Although Sirt1 is known to protect against tubular injury,15–17 mice with tubule-restricted Sirt1 knockdown (TUB;Sirt1RNAi mice) were not susceptible to ADR nephropathy, which was not unexpected, especially because ADR is only known to affect podocytes55 and glomerular endothelial cells,56 but not tubular cells.
Mitochondrial biogenesis is a mechanism to replenish mitochondrial number. Sirt1 controls activation of PGC1α, which is a key regulator of mitochondrial biogenesis. Although POD-Sirt1RNAi mice with a shorter duration of Sirt1 knockdown developed less glomerular injury compared with mice with a longer duration of knockdown, we did not observe a significant difference in the expression of mitochondrial genes between these two groups of mice. The lack of a difference in glomerular expression of mitochondrial genes, however, does not exclude Sirt1/PGC1α-mediated mitochondrial biogenesis as a contributing mechanism for the difference in glomerular injury between mice with shorter and longer duration of Sirt1 knockdown: First, isolated glomeruli were used to determine mitochondrial gene expression, not isolated podocytes. Because podocytes constitute only a small fraction of the cellular mass in the glomerulus, isolated glomeruli—which are an aggregate of at least three different cell types (podocytes, endothelial cells, and mesangial cells)—might not reflect gene expression on the level of the podocytes. Second, samples were obtained from mice at the termination of the experiment (ie, 28 days after ADR injection) when considerable glomerulosclerosis and reduction in glomerular cells have developed in the ADR + DOX group. The pattern of gene expression 28 days after ADR injection might not reflect acute cellular response to ADR-induced mitochondrial damage. It is possible that mitochondrial biogenesis was present during the acute, and not the chronic, phase of ADR-induced injury. Because of these methodological limitations, we are unable to completely exclude that podocyte Sirt1 regulates mitochondrial biogenesis.
Mitochondrial fission is another mechanism to generate new mitochondria. Fission is critical for dividing cells to populate them with an adequate number of mitochondria. However, the role of fission in non-proliferating cells, such as podocytes, is less clear. Fission and fusion appear to be closely linked to autophagic removal of damaged mitochondria.57,58 Herein, we observed an accumulation of fragmented mitochondria in ADR-treated mice with Sirt1 knockdown. Although mitochondrial fission is observed under conditions of cellular stress and exaggerated fission can facilitate apoptosis during high levels of cellular stress, mitochondrial fission per se is not detrimental to cell survival. Accumulation of fragmented mitochondria could be consistent with either increased fission or a lack of removal of fragmented mitochondria by autophagy. In our model, the lack of parkin recruitment to mitochondria of podocytes with Sirt1 knockdown suggests that accumulation of fragmented mitochondria was likely due to impaired autophagy.
In conclusion, we generated and validated in vivo RNAi models with spatial and temporal control of Sirt1 expression. We found that Sirt1 is dispensable for podocyte and glomerular function under the basal condition. We also identified that podocyte Sirt1 is critical for coping with ADR-induced and diabetes-related damage by facilitating the removal of damaged mitochondria.
Acknowledgments
We thank Drs. Peter Mundel (Massachusetts General Hospital, Boston, MA) for sharing cultured conditionally immortalized podocytes and Larry Holzman (University of Pennsylvannia, Philadelphia, PA) for providing rabbit antibody for nephrin.
P.Y.C. and J.C.H. contributed to the experimental design; P.Y.C., J.X., Y.D., S.K.M., R.Y., L.G., and P.K.P. performed the experiments; and P.Y.C. wrote the manuscript.
Footnotes
Supported by NIH grants 1R01DK078897 and 1R01DK088541-01A1 (P.Y.C.) and 5K08DK082760 (J.H.C.) and Veterans Affairs Merit Review Award1I01BX000345 (J.C.H.).
Disclosures: P.K.P. is the Chief Executive Officer and a shareholder of Mirimus, Inc., which specializes in engineering customized mouse models with reversible gene silencing.
Supplemental Data
A: A schematic figure representing the guide sequence embedded within the stem-loop structure of shRNAmir and the four guide sequences targeting Sirt1. B: Western blot analysis of Sirt1 in cultured podocytes transduced with different viral constructs that expressed shRNAmirs of different Sirt1 guide sequences. C: A bar graph of band densitometry for different guide sequences. D: Western blot analyses of Sirt1, GFP, and two different housekeeping proteins (GAPDH and β-actin) for lysates of different organs from CAGS;Sirt1RNAi mice with (+) or without (0) DOX feeding. E: A scheme of the experiment to confirm reversibility of Sirt1 knockdown. F: Normalized Sirt1 expression in the renal cortex of mice on days 0, 14, and 28 of the experiment. Gene expression quantified by next-generation sequencing of mRNA. Gene expression is expressed as fragments per kilobase of transcript per million mapped reads (FPKM). G: Western blot analyses of Sirt1 and GFP in kidney cortex of mice. H: A heat map of log2 normalized gene expression for sirtuins (Sirt1 to Sirt7) in kidney cortex of CAGS;Sirt1RNAi mice. Yellow box: P = 7.26 × 10−5. ∗P < 0.05. Hrt, heart; Lg, lung; Lv, liver; Ms, muscle; Spl, spleen.
Glomerular function of CAGs;Sirt1RNAi mice with acute Sirt1 knockdown. A: UACR of mice with and without DOX feeding (N = 8 mice without DOX feeding, and N = 10 mice with DOX feeding). B and C: Levels of serum urea nitrogen (B) and creatinine (C) for four mice without DOX feeding and four mice with DOX feeding.
Comparisons of renal Sirt1 protein expression in different Sirt1 gene reduction models (Sirt1−/− versus PODΔSirt1 versus CAGs;Sirt1RNAi). A: Western blot analyses of Sirt1 and GAPDH in kidney cortices of Sirt1+/+ and Sirt1−/− mice, as well as CAGs;Sirt1RNAi mice with and without 5 days of DOX feeding. B: A graph of normalized Sirt1/GAPDH band densitometric values for Western blot analyses in A. C: Western blot analyses of Sirt1 and GAPDH for primary glomerular epithelial cells of PODΔSirt1 mice compared with control NPHS2-Cre mice. D: A graph of normalized Sirt1/GAPDH band densitometric values for C.
Renal expression of GFP in kidneys of POD;Sirt1RNAi and TUB;Sirt1RNAi mice. A: Representative images of kidney sections from DOX-fed POD;Sirt1RNAi mice stained with an antibody against podocalyxin. B: Representative plots of side versus forward scatter (inset) and TRITC versus fluorescein isothiocyanate (FITC) fluorescence of single-cell suspension of glomerular cells used for FACS. High-FITC cells (green box) were isolated by FACS. C: Representative images of kidney sections from DOX-fed TUB;Sirt1RNAi mice stained with an antibody against nephrin.
Body weight (A) and serum urea nitrogen (B) levels of CAGs;Sirt1RNAi mice with or without DOX feeding when injected with saline or 18 mg/kg ADR. ∗P < 0.05.
References
- 1.Nakagawa T., Guarente L. Sirtuins at a glance. J Cell Sci. 2011;124:833–838. doi: 10.1242/jcs.081067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salminen A., Kaarniranta K. SIRT1: regulation of longevity via autophagy. Cell Signal. 2009;21:1356–1360. doi: 10.1016/j.cellsig.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 3.Rodgers J.T., Lerin C., Haas W., Gygi S.P., Spiegelman B.M., Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 4.Nemoto S., Fergusson M.M., Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 5.Scarpulla R.C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta. 2011;1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo J., Nikolaev A.Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 7.Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Hu L.S., Cheng H.L., Jedrychowski M.P., Gygi S.P., Sinclair D.A., Alt F.W., Greenberg M.E. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 8.van der Horst A., Tertoolen L.G., de Vries-Smits L.M., Frye R.A., Medema R.H., Burgering B.M. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J Biol Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 9.Moynihan K.A., Grimm A.A., Plueger M.M., Bernal-Mizrachi E., Ford E., Cras-Meneur C., Permutt M.A., Imai S. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Bordone L., Motta M.C., Picard F., Robinson A., Jhala U.S., Apfeld J., McDonagh T., Lemieux M., McBurney M., Szilvasi A., Easlon E.J., Lin S.J., Guarente L. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodgers J.T., Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X., Zhang S., Blander G., Tse J.G., Krieger M., Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 13.Hou X., Xu S., Maitland-Toolan K.A., Sato K., Jiang B., Ido Y., Lan F., Walsh K., Wierzbicki M., Verbeuren T.J., Cohen R.A., Zang M. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J Biol Chem. 2008;283:20015–20026. doi: 10.1074/jbc.M802187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De Oliveira R., Leid M., McBurney M.W., Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He W., Wang Y., Zhang M.Z., You L., Davis L.S., Fan H., Yang H.C., Fogo A.B., Zent R., Harris R.C., Breyer M.D., Hao C.M. Sirt1 activation protects the mouse renal medulla from oxidative injury. J Clin Invest. 2010;120:1056–1068. doi: 10.1172/JCI41563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasegawa K., Wakino S., Yoshioka K., Tatematsu S., Hara Y., Minakuchi H., Sueyasu K., Washida N., Tokuyama H., Tzukerman M., Skorecki K., Hayashi K., Itoh H. Kidney-specific overexpression of Sirt1 protects against acute kidney injury by retaining peroxisome function. J Biol Chem. 2010;285:13045–13056. doi: 10.1074/jbc.M109.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kume S., Uzu T., Horiike K., Chin-Kanasaki M., Isshiki K., Araki S., Sugimoto T., Haneda M., Kashiwagi A., Koya D. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest. 2010;120:1043–1055. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa K., Wakino S., Simic P., Sakamaki Y., Minakuchi H., Fujimura K., Hosoya K., Komatsu M., Kaneko Y., Kanda T., Kubota E., Tokuyama H., Hayashi K., Guarente L., Itoh H. Renal tubular Sirt1 attenuates diabetic albuminuria by epigenetically suppressing Claudin-1 overexpression in podocytes. Nat Med. 2013;19:1496–1504. doi: 10.1038/nm.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuang P.Y., Dai Y., Liu R., He H., Kretzler M., Jim B., Cohen C.D., He J.C. Alteration of forkhead box O (foxo4) acetylation mediates apoptosis of podocytes in diabetes mellitus. PLoS One. 2011;6:e23566. doi: 10.1371/journal.pone.0023566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu R., Zhong Y., Li X., Chen H., Jim B., Zhou M.M., Chuang P.Y., He J.C. Role of transcription factor acetylation in diabetic kidney disease. Diabetes. 2014 doi: 10.2337/db13-1810. http://dx.doi.org/10.2337/db13-1810 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitada M., Kume S., Imaizumi N., Koya D. Resveratrol improves oxidative stress and protects against diabetic nephropathy through normalization of Mn-SOD dysfunction in AMPK/SIRT1-independent pathway. Diabetes. 2011;60:634–643. doi: 10.2337/db10-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tikoo K., Singh K., Kabra D., Sharma V., Gaikwad A. Change in histone H3 phosphorylation, MAP kinase p38, SIR 2 and p53 expression by resveratrol in preventing streptozotocin induced type I diabetic nephropathy. Free Radic Res. 2008;42:397–404. doi: 10.1080/10715760801998646. [DOI] [PubMed] [Google Scholar]
- 23.Kitada M., Takeda A., Nagai T., Ito H., Kanasaki K., Koya D. Dietary restriction ameliorates diabetic nephropathy through anti-inflammatory effects and regulation of the autophagy via restoration of Sirt1 in diabetic Wistar fatty (fa/fa) rats: a model of type 2 diabetes. Exp Diabetes Res. 2011;2011:908185. doi: 10.1155/2011/908185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Premsrirut P.K., Dow L.E., Kim S.Y., Camiolo M., Malone C.D., Miething C., Scuoppo C., Zuber J., Dickins R.A., Kogan S.C., Shroyer K.R., Sordella R., Hannon G.J., Lowe S.W. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145:145–158. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shigehara T., Zaragoza C., Kitiyakara C., Takahashi H., Lu H., Moeller M., Holzman L.B., Kopp J.B. Inducible podocyte-specific gene expression in transgenic mice. J Am Soc Nephrol. 2003;14:1998–2003. doi: 10.1681/ASN.V1481998. [DOI] [PubMed] [Google Scholar]
- 26.Traykova-Brauch M., Schonig K., Greiner O., Miloud T., Jauch A., Bode M., Felsher D.W., Glick A.B., Kwiatkowski D.J., Bujard H., Horst J., von Knebel Doeberitz M., Niggli F.K., Kriz W., Grone H.J., Koesters R. An efficient and versatile system for acute and chronic modulation of renal tubular function in transgenic mice. Nat Med. 2008;14:979–984. doi: 10.1038/nm.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H., Rajendran G.K., Liu N., Ware C., Rubin B.P., Gu Y. SirT1 modulates the estrogen-insulin-like growth factor-1 signaling for postnatal development of mammary gland in mice. Breast Cancer Res. 2007;9:R1. doi: 10.1186/bcr1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moeller M.J., Sanden S.K., Soofi A., Wiggins R.C., Holzman L.B. Podocyte-specific expression of cre recombinase in transgenic mice. Genesis. 2003;35:39–42. doi: 10.1002/gene.10164. [DOI] [PubMed] [Google Scholar]
- 29.Wang R.H., Li C., Deng C.X. Liver steatosis and increased ChREBP expression in mice carrying a liver specific SIRT1 null mutation under a normal feeding condition. Int J Biol Sci. 2010;6:682–690. doi: 10.7150/ijbs.6.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng H.L., Mostoslavsky R., Saito S., Manis J.P., Gu Y., Patel P., Bronson R., Appella E., Alt F.W., Chua K.F. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallipattu S.K., Liu R., Zhong Y., Chen E.Y., D’Agati V., Kaufman L., Ma’ayan A., Klotman P.E., Chuang P.Y., He J.C. Expression of HIV transgene aggravates kidney injury in diabetic mice. Kidney Int. 2013;83:626–634. doi: 10.1038/ki.2012.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu K.K., Huan Y. Streptozotocin-induced diabetic models in mice and rats. Curr Protoc Pharmacol. 2008;Chapter 5 doi: 10.1002/0471141755.ph0547s40. Unit 5.47. [DOI] [PubMed] [Google Scholar]
- 33.Gu L., Dai Y., Xu J., Mallipattu S., Kaufman L., Klotman P.E., He J.C., Chuang P.Y. Deletion of podocyte STAT3 mitigates the entire spectrum of HIV-1-associated nephropathy. AIDS. 2013;27:1091–1098. doi: 10.1097/QAD.0b013e32835f1ea1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Debacq-Chainiaux F., Erusalimsky J.D., Campisi J., Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protoc. 2009;4:1798–1806. doi: 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- 36.Yuen P.S., Dunn S.R., Miyaji T., Yasuda H., Sharma K., Star R.A. A simplified method for HPLC determination of creatinine in mouse serum. Am J Physiol Renal Physiol. 2004;286:F1116–F1119. doi: 10.1152/ajprenal.00366.2003. [DOI] [PubMed] [Google Scholar]
- 37.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takemoto M., Asker N., Gerhardt H., Lundkvist A., Johansson B.R., Saito Y., Betsholtz C. A new method for large scale isolation of kidney glomeruli from mice. Am J Pathol. 2002;161:799–805. doi: 10.1016/S0002-9440(10)64239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boerries M., Grahammer F., Eiselein S., Buck M., Meyer C., Goedel M., Bechtel W., Zschiedrich S., Pfeifer D., Laloe D., Arrondel C., Goncalves S., Kruger M., Harvey S.J., Busch H., Dengjel J., Huber T.B. Molecular fingerprinting of the podocyte reveals novel gene and protein regulatory networks. Kidney Int. 2013;83:1052–1064. doi: 10.1038/ki.2012.487. [DOI] [PubMed] [Google Scholar]
- 40.Chua K.F., Mostoslavsky R., Lombard D.B., Pang W.W., Saito S., Franco S., Kaushal D., Cheng H.L., Fischer M.R., Stokes N., Murphy M.M., Appella E., Alt F.W. Mammalian SIRT1 limits replicative life span in response to chronic genotoxic stress. Cell Metab. 2005;2:67–76. doi: 10.1016/j.cmet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 41.Langley E., Pearson M., Faretta M., Bauer U.M., Frye R.A., Minucci S., Pelicci P.G., Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayashi K., Sasamura H., Ishiguro K., Sakamaki Y., Azegami T., Itoh H. Regression of glomerulosclerosis in response to transient treatment with angiotensin II blockers is attenuated by blockade of matrix metalloproteinase-2. Kidney Int. 2010;78:69–78. doi: 10.1038/ki.2010.81. [DOI] [PubMed] [Google Scholar]
- 43.Papeta N., Zheng Z., Schon E.A., Brosel S., Altintas M.M., Nasr S.H., Reiser J., D’Agati V.D., Gharavi A.G. Prkdc participates in mitochondrial genome maintenance and prevents Adriamycin-induced nephropathy in mice. J Clin Invest. 2010;120:4055–4064. doi: 10.1172/JCI43721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartleben B., Godel M., Meyer-Schwesinger C., Liu S., Ulrich T., Kobler S., Wiech T., Grahammer F., Arnold S.J., Lindenmeyer M.T., Cohen C.D., Pavenstadt H., Kerjaschki D., Mizushima N., Shaw A.S., Walz G., Huber T.B. Autophagy influences glomerular disease susceptibility and maintains podocyte homeostasis in aging mice. J Clin Invest. 2010;120:1084–1096. doi: 10.1172/JCI39492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Austin S., St-Pierre J. PGC1alpha and mitochondrial metabolism: emerging concepts and relevance in ageing and neurodegenerative disorders. J Cell Sci. 2012;125:4963–4971. doi: 10.1242/jcs.113662. [DOI] [PubMed] [Google Scholar]
- 47.Scarpulla R.C., Vega R.B., Kelly D.P. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol Metab. 2012;23:459–466. doi: 10.1016/j.tem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang W., Wang Y., Long J., Wang J., Haudek S.B., Overbeek P., Chang B.H., Schumacker P.T., Danesh F.R. Mitochondrial fission triggered by hyperglycemia is mediated by ROCK1 activation in podocytes and endothelial cells. Cell Metab. 2012;15:186–200. doi: 10.1016/j.cmet.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu T., Robotham J.L., Yoon Y. Increased production of reactive oxygen species in hyperglycemic conditions requires dynamic change of mitochondrial morphology. Proc Natl Acad Sci U S A. 2006;103:2653–2658. doi: 10.1073/pnas.0511154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Juhila J., Roozendaal R., Lassila M., Verbeek S.J., Holthofer H. Podocyte cell-specific expression of doxycycline inducible Cre recombinase in mice. J Am Soc Nephrol. 2006;17:648–654. doi: 10.1681/ASN.2005050547. [DOI] [PubMed] [Google Scholar]
- 51.Wang J., Wang Y., Long J., Chang B.H., Wilson M.H., Overbeek P., Danesh F.R. Tamoxifen-inducible podocyte-specific iCre recombinase transgenic mouse provides a simple approach for modulation of podocytes in vivo. Genesis. 2010;48:446–451. doi: 10.1002/dvg.20635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kohan D.E. Progress in gene targeting: using mutant mice to study renal function and disease. Kidney Int. 2008;74:427–437. doi: 10.1038/ki.2008.146. [DOI] [PubMed] [Google Scholar]
- 53.Gawlik A., Quaggin S.E. Deciphering the renal code: advances in conditional gene targeting. Physiology (Bethesda) 2004;19:245–252. doi: 10.1152/physiol.00009.2004. [DOI] [PubMed] [Google Scholar]
- 54.Hill B.G., Benavides G.A., Lancaster J.R., Jr., Ballinger S., Dell’Italia L., Jianhua Z., Darley-Usmar V.M. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol Chem. 2012;393:1485–1512. doi: 10.1515/hsz-2012-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee V.W., Harris D.C. Adriamycin nephropathy: a model of focal segmental glomerulosclerosis. Nephrology (Carlton) 2011;16:30–38. doi: 10.1111/j.1440-1797.2010.01383.x. [DOI] [PubMed] [Google Scholar]
- 56.Jeansson M., Bjorck K., Tenstad O., Haraldsson B. Adriamycin alters glomerular endothelium to induce proteinuria. J Am Soc Nephrol. 2009;20:114–122. doi: 10.1681/ASN.2007111205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., Alroy J., Wu M., Py B.F., Yuan J., Deeney J.T., Corkey B.E., Shirihai O.S. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim I., Lemasters J.J. Mitophagy selectively degrades individual damaged mitochondria after photoirradiation. Antioxid Redox Signal. 2011;14:1919–1928. doi: 10.1089/ars.2010.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: A schematic figure representing the guide sequence embedded within the stem-loop structure of shRNAmir and the four guide sequences targeting Sirt1. B: Western blot analysis of Sirt1 in cultured podocytes transduced with different viral constructs that expressed shRNAmirs of different Sirt1 guide sequences. C: A bar graph of band densitometry for different guide sequences. D: Western blot analyses of Sirt1, GFP, and two different housekeeping proteins (GAPDH and β-actin) for lysates of different organs from CAGS;Sirt1RNAi mice with (+) or without (0) DOX feeding. E: A scheme of the experiment to confirm reversibility of Sirt1 knockdown. F: Normalized Sirt1 expression in the renal cortex of mice on days 0, 14, and 28 of the experiment. Gene expression quantified by next-generation sequencing of mRNA. Gene expression is expressed as fragments per kilobase of transcript per million mapped reads (FPKM). G: Western blot analyses of Sirt1 and GFP in kidney cortex of mice. H: A heat map of log2 normalized gene expression for sirtuins (Sirt1 to Sirt7) in kidney cortex of CAGS;Sirt1RNAi mice. Yellow box: P = 7.26 × 10−5. ∗P < 0.05. Hrt, heart; Lg, lung; Lv, liver; Ms, muscle; Spl, spleen.
Glomerular function of CAGs;Sirt1RNAi mice with acute Sirt1 knockdown. A: UACR of mice with and without DOX feeding (N = 8 mice without DOX feeding, and N = 10 mice with DOX feeding). B and C: Levels of serum urea nitrogen (B) and creatinine (C) for four mice without DOX feeding and four mice with DOX feeding.
Comparisons of renal Sirt1 protein expression in different Sirt1 gene reduction models (Sirt1−/− versus PODΔSirt1 versus CAGs;Sirt1RNAi). A: Western blot analyses of Sirt1 and GAPDH in kidney cortices of Sirt1+/+ and Sirt1−/− mice, as well as CAGs;Sirt1RNAi mice with and without 5 days of DOX feeding. B: A graph of normalized Sirt1/GAPDH band densitometric values for Western blot analyses in A. C: Western blot analyses of Sirt1 and GAPDH for primary glomerular epithelial cells of PODΔSirt1 mice compared with control NPHS2-Cre mice. D: A graph of normalized Sirt1/GAPDH band densitometric values for C.
Renal expression of GFP in kidneys of POD;Sirt1RNAi and TUB;Sirt1RNAi mice. A: Representative images of kidney sections from DOX-fed POD;Sirt1RNAi mice stained with an antibody against podocalyxin. B: Representative plots of side versus forward scatter (inset) and TRITC versus fluorescein isothiocyanate (FITC) fluorescence of single-cell suspension of glomerular cells used for FACS. High-FITC cells (green box) were isolated by FACS. C: Representative images of kidney sections from DOX-fed TUB;Sirt1RNAi mice stained with an antibody against nephrin.
Body weight (A) and serum urea nitrogen (B) levels of CAGs;Sirt1RNAi mice with or without DOX feeding when injected with saline or 18 mg/kg ADR. ∗P < 0.05.