A P2X4 receptor labeled with the pH-sensitive GFP superecliptic pHluorin represents a useful probe to investigate P2X4 receptor distribution, trafficking, and up-regulation.
Abstract
P2X4 receptors are adenosine triphosphate (ATP)-gated cation channels present on the plasma membrane (PM) and also within intracellular compartments such as vesicles, vacuoles, lamellar bodies (LBs), and lysosomes. P2X4 receptors in microglia are up-regulated in epilepsy and in neuropathic pain; that is to say, their total and/or PM expression levels increase. However, the mechanisms underlying up-regulation of microglial P2X4 receptors remain unclear, in part because it has not been possible to image P2X4 receptor distribution within, or trafficking between, cellular compartments. Here, we report the generation of pH-sensitive fluorescently tagged P2X4 receptors that permit evaluations of cell surface and total receptor pools. Capitalizing on information gained from zebrafish P2X4.1 crystal structures, we designed a series of mouse P2X4 constructs in which a pH-sensitive green fluorescent protein, superecliptic pHluorin (pHluorin), was inserted into nonconserved regions located within flexible loops of the P2X4 receptor extracellular domain. One of these constructs, in which pHluorin was inserted after lysine 122 (P2X4-pHluorin123), functioned like wild-type P2X4 in terms of its peak ATP-evoked responses, macroscopic kinetics, calcium flux, current–voltage relationship, and sensitivity to ATP. P2X4-pHluorin123 also showed pH-dependent fluorescence changes, and was robustly expressed on the membrane and within intracellular compartments. P2X4-pHluorin123 identified cell surface and intracellular fractions of receptors in HEK-293 cells, hippocampal neurons, C8-B4 microglia, and alveolar type II (ATII) cells. Furthermore, it showed that the subcellular fractions of P2X4-pHluorin123 receptors were cell and compartment specific, for example, being larger in hippocampal neuron somata than in C8-B4 cell somata, and larger in C8-B4 microglial processes than in their somata. In ATII cells, P2X4-pHluorin123 showed that P2X4 receptors were secreted onto the PM when LBs undergo exocytosis. Finally, the use of P2X4-pHluorin123 showed that the modulator ivermectin did not increase the PM fraction of P2X4 receptors and acted allosterically to potentiate P2X4 receptor responses. Collectively, our data suggest that P2X4-pHluorin123 represents a useful optical probe to quantitatively explore P2X4 receptor distribution, trafficking, and up-regulation.
INTRODUCTION
Extracellular ATP functions as a neurotransmitter and gliotransmitter as well as a neuromodulator in the nervous system by activating plasma membrane (PM) P2X and P2Y receptors (Burnstock, 1972; Khakh and Burnstock, 2009; Coddou et al., 2011b; Khakh and North, 2012). P2X receptors are trimeric ATP-gated cation channels (Nicke et al., 1998) comprising seven subunits (P2X1 to P2X7), six homomeric receptors, and several heteromeric assemblies (Coddou et al., 2011b; Kaczmarek-Hájek et al., 2012). Considerable progress has been made in exploring roles for P2X receptors in native systems. Important roles for P2X4 receptors in a variety of pathophysiological processes have been demonstrated based on pharmacological and genetic experiments. Thus, it has been shown that P2X4 receptors are up-regulated in microglia after neuropathic pain and epilepsy (Tsuda et al., 2003; Ulmann et al., 2013). They are also implicated in inflammatory pain (Ulmann et al., 2010), lung surfactant secretion (Miklavc et al., 2011), alcohol preference (Ostrovskaya et al., 2011; Yardley et al., 2012), cardiac function (Yang et al., 2004), and neurodevelopmental disorders (Bortolato et al., 2013; Wyatt et al., 2013).
One of the earliest and most established responses mediated by P2X4 receptors relates to neuropathic pain and microglia. Roles for P2X4 receptor–mediated responses within microglia are supported by experiments demonstrating that microglia are involved in the development of neuropathic pain (Tsuda et al., 2003; Coull et al., 2005; Inoue, 2008; Ulmann et al., 2008; Jarvis, 2010). Pharmacological and genetic strategies that reduce microglial P2X4 receptor function alleviate symptoms in neuropathic pain models (Tsuda et al., 2003). Neuropathic pain is also reduced in mice that lack P2X4 receptors (Ulmann et al., 2008). These data suggest that altered P2X4 receptor expression, trafficking, and/or function in activated microglia may be an important step in the development of neuropathic pain. As a result of these breakthroughs, P2X4 receptors are emerging as potentially important drug targets to treat pain states that are currently resistant to therapy (Jarvis, 2010; North and Jarvis, 2013). More broadly, it is probable that the mechanisms that cause P2X4 receptor up-regulation in microglia during neuropathic pain are also engaged in brain disorders that are accompanied by microglia activation (Milligan and Watkins, 2009; Graeber, 2010). If so, P2X4 receptors may be therapeutic targets for several brain diseases. To explore underlying mechanisms, there is a need to (a) develop approaches to systematically study P2X4 receptor trafficking within cells such as microglia, and (b) develop in vivo approaches to image P2X4 receptor–expressing cells in native systems. Here, we focused on the first of these tasks.
Given the known and anticipated roles for P2X4 receptors in pathophysiological processes, considerable effort has been devoted to studying P2X4 receptor pharmacology, biophysical properties, and structure–function relationships (Khakh and North, 2012). In many ways, the pinnacle of these efforts has been the availability of high resolution crystal structures of the ATP-bound and unbound forms of a zebrafish (zf) P2X4 receptor orthologue (zfP2X4.1) (Kawate et al., 2009; Hattori and Gouaux, 2012) that provide a basis to understand how the protein works at a chemical level (Browne et al., 2010; Coddou et al., 2011b; Baconguis et al., 2013; Chataigneau et al., 2013; Jiang et al., 2013; Samways et al., 2014). In addition, an extensive body of work shows that P2X4 receptors undergo dynamin-dependent endocytosis as well as PM insertion caused by lysosomal secretion (Bobanovic et al., 2002; Royle et al., 2002, 2005; Bowler et al., 2003; Royle and Murrell-Lagnado, 2003; Qureshi et al., 2007; Murrell-Lagnado and Qureshi, 2008; Stokes and Surprenant, 2009; Toulme et al., 2010). Although regulated trafficking is not unique to P2X4 among the P2X receptor family (Robinson and Murrell-Lagnado, 2013), the ability of P2X4 receptors to move onto and off the membrane is a particularly robust feature that has now been well studied by electrophysiological and biochemical methods. As a result, there is strong evidence that P2X4 receptors localize to the PM and intracellular compartments such as endosomes, lysosomes, and vacuoles. Of these, expression within lysosomes and lamellar bodies (LBs) is particularly impressive (Qureshi et al., 2007; Miklavc et al., 2011). More recently, single-molecule imaging experiments showed that P2X4 receptors display activation- and cell-dependent PM surface mobility (Richler et al., 2011; Toulme and Khakh, 2012). The physiological role of these dynamics remains to be explored, but they may accelerate recovery from desensitization in circumstances when ATP is released from a point source.
Building on the aforementioned advances, in this study we set out to engineer P2X4 receptors to carry superecliptic pHluorin (pHluorin) within the extracellular domain. PHluorin is a pH-sensitive green fluorescent protein (GFP) that is quenched at acidic pH and brightly fluorescent at neutral pH, with a pKa of ∼7.2 (Miesenböck et al., 1998; Sankaranarayanan et al., 2000). Based on past work with other ion channels and receptors (Miesenböck, 2012), we speculated that its introduction into the extracellular loop of P2X4 receptors would yield an optical reporter that could be used to quantify and track P2X4 receptor distribution within cellular compartments.
PHluorin is a 27-kD protein. Tagging it onto the N or C termini of channels is relatively simple, but attaching it within an extracellular loop without affecting function, as necessary in the case of P2X receptors, is much more challenging (Mealer et al., 2008). There was no obvious way to predict an innocuous site to insert pHluorin within the P2X4 receptor extracellular domain. Random transposon-based methods seemed like the right strategy (Sheridan et al., 2002; Giraldez et al., 2005; Mealer et al., 2008), but the recent availability of the ATP-bound and unbound zfP2X4.1 crystal structures (Kawate et al., 2009; Hattori and Gouaux, 2012) provided an opportunity to design P2X4 receptors carrying extracellularly located pHluorins.
In this technical study, we describe the design, characterization, testing, and initial deployment of pHluorin-tagged P2X4 receptors in several proof-of-concept experiments. The engineered receptors can now be used in a variety of settings to explore ATP signaling and P2X4 receptor physiology. Our experiences with P2X4 receptors also provide the rationale and basis to extend this approach to other P2X receptors (e.g., P2X1, P2X3, and P2X7) and acid-sensing ion channels that share topological features with P2X receptors (Gonzales et al., 2009; Baconguis et al., 2013).
MATERIALS AND METHODS
Sequence analysis
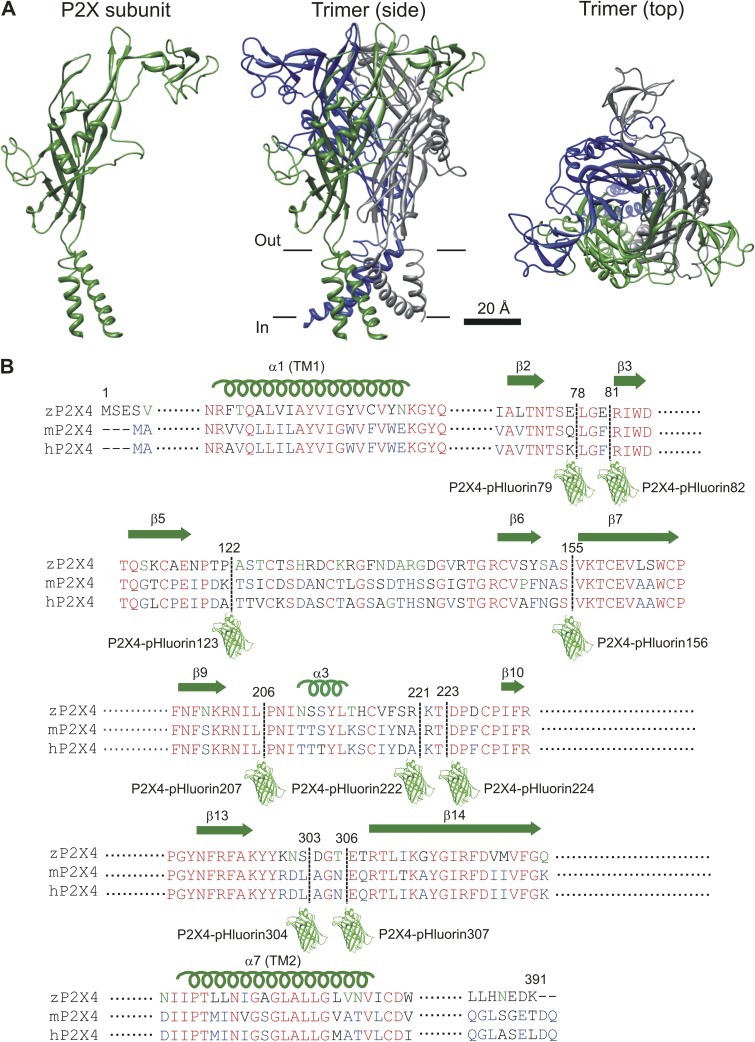
The protein sequences of zfP2X4.1 (RefSeq accession no. NP_705939.1), mouse P2X4 (RefSeq accession no. NP_035156.2), and human P2X4 (RefSeq accession no. NP_002551.2) were aligned using Vector NTI software (Life Technologies). The sequences were annotated based on conservation between the three species. In Fig. 1 B, nonsimilar regions are shown in black, conserved regions are in blue, identical regions are in red, and weakly similar regions are in green. pHluorin was inserted in stretches of five or more nonconserved residues in the extracellular domain. 10 such regions have been annotated previously based on the closed zfP2X4.1 crystal structure (Kawate et al., 2009). We avoided variable regions that were close to the ATP-binding pocket, subunit interaction surfaces, or the ion permeation pathway (Kawate et al., 2009; Hattori and Gouaux, 2012). These regions are not shown in Fig. 1 B.
Figure 1.
Design of mP2X4 receptors carrying pHluorin in the extracellular domain. (A) Cartoons illustrate the basic layout of a P2X receptor subunit and how three identical subunits are assembled into a trimeric ion channel. The location of the PM is shown approximately with black lines. (B) Sequence alignment for parts of the extracellular loop that were targeted for insertion of pHluorin. The pHluorin insertion sites are shown with a dashed line and a cartoon of GFP. Nine constructs were made and tested using approaches described in detail in the Results. They are identified in the figure.
Molecular biology
In this study, the numbering of amino acid residues is based on the mouse P2X4 protein sequence (NCBI Gene ID no. 18438, gene name P2rX4). Our goal was to generate a family of P2X4 constructs carrying pHluorin at distinct sites in the extracellular domain after amino acid residues Q78, F81, K122, S155, L206, A221, T223, L303, and N306 (see Results for how these sites were chosen). The mP2X4 receptor extracellular domain extends from the end of TM1 at position 55 to the start of TM2 at position 333 and encompasses 279 residues. The specific sites for insertion were chosen based on the ATP-bound and unbound structures of zfP2X4.1 (Kawate et al., 2009; Hattori and Gouaux, 2012), which are considered the best available representations of the closed and open forms of the channel. We named the pHluorin insertion constructs after the number of the initiating methionine in pHluorin. Thus, P2X4-pHluorin123 indicates that the initiating methionine of pHluorin was at position 123 of the engineered P2X4 receptor sequence, i.e., after the endogenous lysine at 122 (Fig. 1 B).
P2X4-pHluorin constructs were made in two steps. The first step consisted of generating a family of P2X4 constructs carrying ClaI and EcoRI sites flanked by glycine-alanine linkers at the specific insertion sites in the extracellular domain of P2X4 receptors. The DNA fragment that was inserted at these sites carried ClaI and EcoRI flanked by glycine-alanine linkers (GGAGCTGGAGCTATCGATTATGAATTCGCTGGAGCTGGA). The fragment was introduced into mP2X4 by overlapping PCR. The same basic strategy was used for all the constructs that are reported in this study. Thus, the P2X4 5′ and 3′ fragments flanking the insertion site were amplified by the primers listed in Table 1 for each specific construct. Primers P2X4 extreme 5′ and P2X4-pHluorin A-3′ were used to generate the P2X4 fragment upstream of pHluorin, and primers P2X4-pHluorin B-5′ and P2X4 extreme 3′ were used to generate the P2X4 fragment downstream of pHluorin, respectively. Primers A-3′ and B-5′ carry the overhang coding the restriction sites and glycine-alanine linker. Then, using these two fragments as template, the 5′ and 3′ fragments were combined by another round of PCR, resulting in the insertion of restriction sites and glycine-alanine linker. This “overlapping” PCR reaction used extreme 5′ and 3′ flanking primers. All of the PCR amplifications were performed with high fidelity DNA polymerase (Phusion; New England Biolabs, Inc.). PCR reactions were performed in a reaction volume of 50 µl using 50 ng of template, 0.25 µM of the primer pairs, 200 µM dNTPs, and 1 U of DNA polymerase. The extension reaction was initiated by preheating the reaction mixture to 96°C for 3 min. The PCR cycle was: 32 cycles at 96°C for 30 s, 52°C for 30 s, and 68°C for 30 s (per 1 kb). The PCR products were digested with HindIII and NotI, which flanked the entire mP2X4 cDNA and were then subcloned into pCDNA3.1. The second step consisted of PCR amplification and cloning of the pHluorin fragment between the ClaI and EcoRI sites in mP2X4. For this step, the pHluorin was PCR amplified from a parent plasmid called pCI-SEP-GluR1. All final cDNAs were propagated in DH5α Escherichia coli, and the plasmids were purified using standard techniques. All constructs were verified by DNA sequencing.
Table 1.
Primers used to generate mP2X4 constructs carrying pHluorin
| Primer | Sequence (5′ to 3′) |
| P2X4 extreme flanking primers | |
| P2X4 extreme 5′ | AGTTAAGCTTGGTACCGAGCTCGGATCC |
| P2X4 extreme 3′ | TATGCGGCCGCTCACTTGTCATCGTCATCCTTGTAATCCTGGTCCGTCTCTCCGGAAAG |
| P2X4 insertion site primers | |
| P2X4-pHluorin79 A-3′ | TCCAGCGAATTCATAATCGATAGCTCCAGCTCCCTGAGAAGTGTTGGTCACAGCCACACCTTTGGCTTTGG |
| P2X4-pHluorin79 B-5′ | GGAGCTGGAGCTATCGATTATGAATTCGCTGGAGCTGGACTTGGATTCCGGATCTGGGACGTGGCCGACTATGTGG |
| P2X4-pHluorin82 A-3′ | TCCAGCGAATTCATAATCGATAGCTCCAGCTCCGAATCCAAGCTGAGAAGTGTTGGTCAC |
| P2X4-pHluorin82 B-5′ | GGAGCTATCGATTATGAATTCGCTGGAGCTGGACGGATCTGGGACGTGGCCGACTATG |
| P2X4-pHluorin123 A-3′ | TCCAGCGAATTCATAATCGATAGCTCCAGCTCCCTTATCAGGAATCTCTGGACAGGTACC |
| P2X4-pHluorin123 B-5′ | GGAGCTATCGATTATGAATTCGCTGGAGCTGGAACCAGCATTTGCGATTCAGACGCCAAC |
| P2X4-pHluorin156 A-3′ | TCCAGCGAATTCATAATCGATAGCTCCAGCTCCAGACGCGTTGAATGGAACACACCTTCC |
| P2X4-pHluorin156 B-5′ | GGAGCTATCGATTATGAATTCGCTGGAGCTGGAGTGAAGACCTGTGAGGTGGCCGCATGG |
| P2X4-pHluorin207 A-3′ | TCCAGCGAATTCATAATCGATAGCTCCAGCTCCGAGGATATTCCTCTTGCTGAAATTAAAC |
| P2X4-pHluorin207 B-5′ | GGAGCTATCGATTATGAATTCGCTGGAGCTGGACCCAACATTACCACCTCCTACCTCAAG |
| P2X4-pHluorin222 A-3′ | TCCAGCGAATTCATAATCGATAGCTCCAGCTCCCGCATTATAAATGCACGACTTGAGG |
| P2X4-pHluorin222 B-5′ | GGAGCTATCGATTATGAATTCGCTGGAGCTGGACGGACGGATCCCTTCTGCCCCATATTCC |
| P2X4-pHluorin224 A-3′ | TCCAGCGAATTCATAATCGATAGCTCCAGCTCCCGTCCGCGCATTATAAATGCACGAC |
| P2X4-pHluorin224 B-5′ | GGAGCTATCGATTATGAATTCGCTGGAGCTGGAGATCCCTTCTGCCCCATATTCCGTC |
| P2X4-pHluorin304 A-3′ | TCCAGCGAATTCATAATCGATAGCTCCAGCTCCAAGGTCCCTGTAGTACTTGGCAAACC |
| P2X4-pHluorin304 B-5′ | GGAGCTATCGATTATGAATTCGCTGGAGCTGGAGCTGGCAATGAGCAACGCACACTCACC |
| P2X4-pHluorin307 A-3′ | TCCAGCGAATTCATAATCGATAGCTCCAGCTCCAGACGCGTTGAATGGAACACACCTTCC |
| P2X4-pHluorin307 B-5′ | GAGCTATCGATTATGAATTCGCTGGAGCTGGAGAGCAACGCACACTCACCAAGGCATATG |
| pHluorin amplification primers | |
| pHluorin Cla1 5′ | TATATCGATGGAGGAGCTGGAGGAGCTAGGAGTAAAGGAGAAGAACTTTTCACTGG |
| pHluorin EcoR1 3′ | ATAGAATTCAGCTCCTCCAGCTCCTCCTTTGTATAGTTCATCCATGCCATGTGTAATCC |
mP2X4 receptors were available from our past work (Toulme et al., 2010; Toulme and Khakh, 2012), and the pHluorin construct was obtained from Addgene (plasmid no. 24000). PM reporter pCS2-mcherry and ER reporter pDsRed2-ER were provided by H.A. Lester (California Institute of Technology, Pasadena, CA). Lysosome reporter Lamp1-RFP was obtained from Addgene (no. 1817).
HEK-293 cell culture and transfection
HEK-293 cells (ATCC) were maintained in 75-cm2 cell culture flasks in Dulbecco’s modified Eagle’s medium/F-12 media with Glutamax (Invitrogen) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were prepared for transfection by plating onto 6-well plates at the time of splitting, 2–3 d before transfection. They were transfected at ∼60% confluence. For transient expression in HEK-293 cells, we used ∼0.5 µg P2X4-pHluorin plasmid and the Effectene transfection reagent (QIAGEN). When P2X4-pHluorins were screened by electrophysiology, plasmids encoding control WT P2X4 or P2X4-pHluorins were cotransfected with 0.1 µg tdTomato reporter in pcDNA3.1. For the colocalization study of P2X4-pHluorin123 with organelle markers, HEK-293 cells were cotransfected with 400 ng P2X4-pHluorin123 and 25 ng PM-mcherry, 25 ng DsRed2-ER, or 50 ng Lamp1-RFP. The manufacturer’s instructions (QIAGEN) were followed for the transfections. Cells were gently dispersed and plated on poly-d-lysine–coated glass coverslips before physiological evaluations (Egan and Khakh, 2004).
C8-B4 cell culture and nucleofection
C8-B4 cells are an immortalized microglial cell line (Toulme et al., 2010). They were cultured in 25-cm2 cell culture flasks in Dulbecco’s modified Eagle’s medium (ATCC) supplemented with 10% fetal bovine serum (ATCC). The cells were split one in three when confluence reached ∼60%. C8-B4 microglia cells were transfected using a nucleofection kit (Amaxa; Lonza) with 10 µg of plasmid DNA before plating onto poly-d-lysine–coated glass coverslips. The manufacturer’s instructions were followed for the Amaxa basic nucleofection kit for mouse astrocytes (a nucleofection kit for microglia does not exist). We imaged the cells ∼1 d after transfection. We have previously published detailed methods and evaluations of P2X4 responses in C8-B4 cells before and after nucleofection (Toulme et al., 2010; Toulme and Khakh, 2012).
Hippocampal neuron culture and transfection
Hippocampal cultures were prepared as described previously (Shigetomi and Khakh, 2009). In brief, two rat pups at postnatal day 1 or 2 (Charles River) were used for hippocampal cultures. Hippocampi were dissected in Petri dishes filled with ice-cold medium. The dissected hippocampi (in medium on ice) were cut and then digested with 20 U/ml papain for 11–13 min at 37°C (Papain-022; Worthington Biochemical Corporation). After the incubation, the pieces were washed with prewarmed media and triturated with flame-polished pipettes of progressively smaller bores; 20,000 cells were used for plating onto each 12-mm coverslip. The coverslips were previously coated with 50 µg/ml poly-d-lysine (Sigma-Aldrich) and then coated overnight with 100 µl of 20 µg/ml laminin (Sigma-Aldrich) in sterile dissection medium. The cells were fed with 2 ml of prewarmed culture medium 1 h after plating. Neurons were transfected at 7–10 DIV. The neurons were in Neurobasal medium (Invitrogen) at this point. On this day, half of the media was removed (0.5 ml from each well of a 24-well plate housing one coverslip), and the neurons were fed with fresh Neurobasal medium (0.5 ml) that had been preequilibrated in the cell culture incubator (37°C; 5% CO2) for at least 2 h. The removed media was supplemented with an equal volume of new media and stored in a vented flask in the cell culture incubator (this is “the fed and conditioned media”). We used lipofectamine2000 (L2K) from Invitrogen for the transfection. For 1 well of a 24-well plate housing one 13-mm coverslip, we added 0.8 µg DNA to 50 µl MEM. In a separate tube, we added 2 µl L2K with 50 µl MEM (kept for 5 min at room temp). Then we added the 50 µl of the L2K–MEM mix to the DNA–MEM mix, flicked the tube, and incubated at room temperature for 20 min. This was added dropwise to the neurons, after which the plates were given a gentle swirl and returned to the incubator. After 6–8 h, all medium was removed from each well and replaced with 1 ml of the “fed and conditioned medium” from earlier. We imaged the cells ∼3 d after transfection.
Alveolar type II (ATII) cell isolation and electroporation
ATII cells were isolated from Sprague-Dawley rats according to published procedures (Dobbs et al., 1986) with minor modifications as described recently (Miklavc et al., 2009). In brief, rats were anesthetized with ketamin and xylazil (10 and 2%, respectively), and then injected with 400 IU/kg heparin. The lungs were perfused, removed, washed, and then incubated at 37°C with elastase and trypsin (twice for 15 min). Lungs were then immersed in a solution containing DNase and sliced into ∼1-mm3 pieces with scissors. The DNase reaction was blocked by incubating with fetal calf serum for 2 min at 37°C. The digested tissue was then filtered through gauze and nylon meshes three times, and the final filtrate was centrifuged for 8 min at 130 g. After resuspension in Dulbecco’s modified Eagle’s medium, cells were incubated on IgG-coated plastic dishes for 15 min at 37°C. Non-adherent cells were centrifuged for 8 min at 130 g, and then suspended in medium (MucilAir; Epithelix) and incubated in suspension flasks overnight at 37°C/5% CO2. The next day, 4 × 106 cells were centrifuged and resuspended in 100 µl 4D-Nucleofector Solution (Lonza), and 7 µg DNA was added. Cells were transferred to the electroporation cuvette (Nucleocuvette) and electroporated with an Amaxa 4D-Nucleofector (all from Lonza). Immediately after electroporation, 1 ml of prewarmed MucilAir medium was added and 250 µl of the resulting cell suspension were transferred to individual wells in 8-well chamber slides (ibidi GmbH). Cells were used for experiments 24 h after electroporation.
Western blot analysis of P2X4-pHluorin proteins in HEK-293 cells
Transfected HEK-293 cells were suspended in cell lysis buffer containing 50 mM Tris, 150 mM NaCl, 1 mM DTT, 1% Triton X-100, 1% IGEPAL, 0.1% SDS, and a protease inhibitor cocktail tablet (GE Healthcare). Cells were triturated with a 26-gauge syringe needle and incubated in the lysis buffer for 30 min at 4°C. This mixture was then centrifuged at 13,000 rpm for 30 min at 4°C, and the soluble proteins in the supernatant were transferred into a clean tube. Equal amounts of proteins were loaded on 10% SDS-PAGE gels and transferred to nitrocellulose membrane (GE Healthcare). After transfer, membranes were blocked by incubation with PBS containing 0.05% Tween and 5% dry milk for 2 h and incubated overnight with antibodies against GFP (1:2,000; A-11122; Invitrogen) in PBS containing 5% milk at 4°C. After washing three times for 10 min each in PBS/Tween, the membranes were incubated with anti–rabbit horseradish peroxidase secondary antibodies (1:200; Thermo Fisher Scientific) in PBS containing 5% milk for 1 h at room temperature. Membranes were washed three times for 10 min each in PBS/Tween, and the protein bands were imaged using ECL reagent (Thermo Fisher Scientific). Standard β-actin controls (anti–β actin; 1:1,000; Abcam) were included to use for normalization of the P2X band intensities.
Confocal microscopy of HEK-293 cells, hippocampal neurons, and C8-B4 cells
Imaging was performed using commercially available standard confocal microscopes. In brief, cells were imaged using a Fluoview 1000 or Fluoview 300 (Olympus) confocal microscope with a 40× water-immersion objective lens with a numerical aperture of 0.8. We used the 488-nm line of an argon laser to excite P2X4-pHluorins, with the intensity adjusted to 0.5–5% of the maximum output, which was 10 mW. The emitted light pathway consisted of an emission high pass filter (505–525 nm) before the photomultiplier tube. Red fluorescent proteins were excited by the 543-nm laser line of the HeNeG laser at 30% of the maximum output (1 mW). The emitted light pathway consisted of a dichroic mirror (SDM560) and a 560–600-nm emission filter. The time-lapse images were taken one frame per 15 s. Solution in perfusion chamber was exchanged with a local perfusion device in 5 s. Z-series images were taken every 0.5–1 µm throughout the cell’s volume. During confocal microscopy, the cells were bathed in extracellular solutions, which are listed below.
Solutions for imaging HEK-293 and C8-B4 cells comprised (mM): 150 NaCl, 1 MgCl2, 1 CaCl2, 10 HEPES, and 10 glucose, with pH adjusted to 7.4. The pH 5.4 solution comprised (mM): 150 NaCl, 1 MgCl2, 1 CaCl2, 10 HEPES, and 10 glucose, with the pH adjusted to 5.4. The NH4Cl solution comprised (mM): 125 NaCl, 1 MgCl2, 1 CaCl2, 10 HEPES, 10 glucose, and 25 NH4Cl, with pH adjusted to 7.4. Solutions for imaging hippocampal neurons comprised (mM): 110 NaCl, 0.8 MgCl2, 1.8 CaCl2, 10 HEPES, 5.4 KCl, and 10 glucose, with pH adjusted to 7.4. The pH 5.4 solution comprised (mM): 110 NaCl, 0.8 MgCl2, 1.8 CaCl2, 10 HEPES, 5.4 KCl, and 10 glucose, with pH adjusted to 5.4. The NH4Cl solution comprised (mM): 85 NaCl, 0.8 MgCl2, 1.8 CaCl2, 10 HEPES, 5.4 KCl, 10 glucose, and 25 NH4Cl, with pH adjusted to 7.4. All solutions were made fresh.
Live cell microscopy of ATII cells
Combined pHluorin and LysotrackerRed (LTR; Life Technologies) experiments were performed on an iMic digital microscope (Till Photonics) with a 488-nm excitation filter for pHluorin and a 568-nm excitation filter for LTR. Before the start of the experiment, the cells were incubated in 100 nM LTR for 10 min to detect LB fusions, washed twice in bath solution (mM: 140 NaCl, 5 KCl, 1 MgCl2, 2 CaCl2, 5 glucose, and 10 HEPES, pH 7.4), and kept in bath solution for the duration of the experiment. LB exocytosis was stimulated using 100 µM ATP and 300 nM PMA (Sigma-Aldrich). Images were analyzed using iMic Offline analysis software (Till Photonics) and ImageJ (National Institutes of Health [NIH]). Analysis of cellular distribution of P2X4 in ATII cells was performed on a Cell Observer microscope (Carl Zeiss). ATII cells were seeded in perfusion chambers (ibidi GmbH) and illuminated for 100 ms at a rate of 0.2 Hz at 480 nm. Images were acquired using MetaFluor (Molecular Devices) and analyzed using MetaFluor Analyst (Molecular Devices). Excel (Microsoft) was used for statistics and graph design. Unless otherwise stated, all data are presented as mean ± SEM.
Whole-cell voltage-clamp recording
The extracellular recording solution for HEK-293 and C8-B4 cells comprised (mM): 150 NaCl, 1 MgCl2, 1 CaCl2, 10 HEPES, and 10 glucose. The pipette solution comprised (mM): 160 KCl, 10 EGTA, and 10 HEPES. The extracellular recording solution for hippocampal neurons comprised (mM): 130 NaCl, 5.4 KCl, 0.8 MgCl2, 1.8 CaCl2, 10 HEPES, and 7.4 glucose. The pipette solution comprised (mM): 100 K-gluconate, 30 KCl, 0.5 CaCl2, 5 EGTA, 2 Mg-ATP, 0.3 Na-GTP, and 10 HEPES. Whole-cell voltage-clamp recordings were made using ∼3 MΩ borosilicate glass electrodes (World Precision Instruments) with an Axopatch 200B amplifier controlled by a computer running pCLAMP10.1 software via a Digidata 1322A interface (Molecular Devices). Data were filtered at 2 kHz and digitized at >5 kHz. Drugs were applied to single cells with a local fast perfusion device (SF-77B Perfusion Fast Step; Warner Instruments), resulting in complete solution change in ∼10 ms. The equipment used for electrophysiology has been described in detail by us (Shigetomi and Khakh, 2009; Toulme et al., 2010; Richler et al., 2011; Toulme and Khakh, 2012).
Patch-clamp photometry to measure fractional Ca2+ currents (Pf%)
We used a well-established fluorimetric flux technique (Schneggenburger et al., 1993; Egan and Khakh, 2004). In brief, the Pf% was determined by simultaneously measuring total membrane current and fluorescence in cells loaded with a high concentration (2 mM) of the calcium-sensitive dye K5FURA-2 (Invitrogen). Membrane currents were recorded from single cells held at −60 mV. The intracellular solution was as described above for voltage-clamp recording from HEK-293 cells but lacked EGTA and contained 2 mM K5FURA-2. Light emitted by FURA-2 (380-nm excitation, 510-nm emission) was gathered by a microscope objective and directed to the input of a photomultiplier detection system (model 714; Photon Technology International). To account for the day-to-day variation in the sensitivity of the microscope and photomultiplier tube, the FURA-2 signal was normalized to a “bead unit.” One bead unit equaled the average fluorescence of seven Fluoresbrite carboxy BB 4.6-µm microspheres (Polysciences, Inc.) measured one at a time on the morning of that day’s experiments (Egan and Khakh, 2004). The extracellular bath solution was as listed above for voltage-clamp recording, except that it contained 2 mM CaCl2.
Epifluorescence and total internal reflection fluorescence (TIRF) microscopy
In brief, we used a microscope (IX71; Olympus) equipped with an IXON DV887DCS EMCCD camera (Andor Technology), epifluorescence condenser, control unit, and monochromator (Polychrome V; TILL Photonics). P2X4-pHluorin was illuminated by 488-nm light from the monochromator. The control of excitation and image acquisition was achieved using TILLVision software (TILL Photonics). The beams of argon (100 mW) and 442-nm solid-state (45 mW) lasers were combined and controlled with a polyline (TILL Photonics) laser combiner, TIRF dual-port condenser, and acoustoptical tunable filter and controller (TILL Photonics) and fed into a broadband fiber for entry into the TIRF condenser. We used a 60× 1.45 numerical aperture lens (Olympus) to achieve TIRF. The time-lapse images were taken at one frame per second using 2 × 2 binning on the EMCCD chip. The epifluorescence and TIRF rig have been described in detail by us (Richler et al., 2008; Shigetomi and Khakh, 2009).
Data analysis
Electrophysiological analysis was performed with Clampfit 10.1 (Molecular Devices), Igor Pro (Wavemetrics, Inc.), Origin 6.1 or 7.5 (OriginLab Corporation), and GraphPad Instat (GraphPad Software). Image analysis was performed with iMic Offline analysis software (Till Photonics) and ImageJ (NIH). The figures were assembled in CorelDraw12 (Corel Corporation). The Pf% was calculated as
where QT (the integral of the ATP-gated ionic current) and QCa are given by
FMax is the calibration constant used to relate ΔQCa to ΔF380. It was calculated using procedures described in a separate series of experiments under conditions where QT is expected to equal QCa (Egan and Khakh, 2004). Pf% was often measured multiple times in a single cell (these measures were then averaged to give the Pf% for the individual cell).
We performed a series of experiments in cells expressing P2X4-pHluorin to calculate the contribution of fluorescence from receptors on the cell surface, within acidic compartments and within other intracellular compartments. The rationale and logic of these experiments are well established. In brief, we exploited the pH sensitivity of the pHluorin with the knowledge that intracellular compartments have acidic pH that can be alkalized with brief bath applications of NH4Cl (Miesenböck et al., 1998; Sankaranarayanan et al., 2000). The cells were initially bathed in extracellular solutions at pH 7.4. Under these settings, the total cellular fluorescence (Ftotal) consists of channels on the surface (Fsurface) with the pHluorin facing the extracellular pH 7.4 buffer, channels within acidic compartments (Facidic) with the pHluorin quenched, and receptors/fluorescence in other compartments (Fother) of unknown pH environment. Thus,
where
Here, Fbasal is the basal fluorescence of the cells at pH 7.4, and FNH4Cl is the fluorescence of the cells in NH4Cl, when all acidic compartments are also at a pH greater than ∼7.4. Fsurface was measured by applying extracellular buffers at pH 5.4, which quenches surface receptors. Facidic was measured by applying NH4Cl, which alkalizes acidic compartments to a pH of ∼7.4 (Miesenböck et al., 1998; Sankaranarayanan et al., 2000). Thus,
The TIRF experiments were analyzed in the same way.
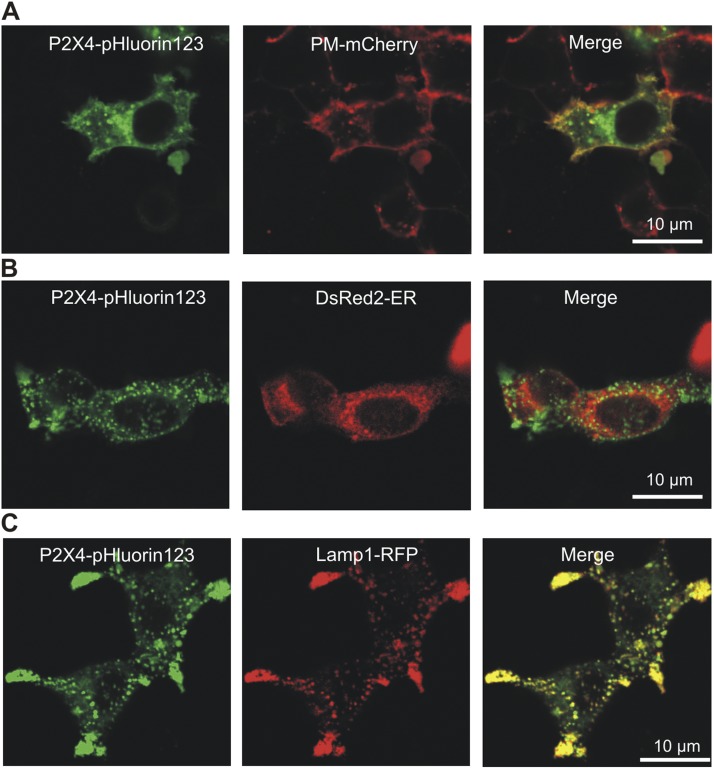
Colocalization was assessed using dual-color confocal images of P2X4-pHluorin and PM-mCherry, DsRed2-ER (Srinivasan et al., 2012), and Lamp1-RFP using the using the Coloc_2 plugin in the Fiji image-processing package for ImageJ. We unambiguously analyzed colocalization across many cells (n numbers are provided in the text) by measuring the Pearson’s correlation coefficient (r) and comparing these values statistically (Fig. 7). In analyzing and interpreting the data, we followed published guidelines (Zinchuk and Zinchuk, 2008). In brief, we interpreted Pearson’s coefficient (which can range from −1 to 1) in the following way: −1, perfect negative correlation; 0, no correlation; 1, perfect positive correlation, i.e., colocalization. Pearson’s correlation coefficients of greater than ∼0.5 are interpreted as indicative of reliable colocalization between two spectrally separated fluorophores (Zinchuk and Zinchuk, 2008). Data are presented as the mean ± SEM from n experiments as indicated in the text. Statistical tests were run in Origin 8.2 (OriginLab Corporation) or GraphPad Instat (version 3.10, 32 bit for Windows; GraphPad Software). Significance was declared at a p-value of <0.05.
Figure 7.
Cellular distribution of P2X4-pHluorin123 receptors in HEK-293 cells. (A) Representative images of HEK-293 cells expressing P2X4-pHluorin123 (green) and PM-mCherry. (B and C) As in A, but for P2X4-pHlurin and DsRed2-ER (B), and P2X4-pHluorin123 and Lamp1-RFP (C). Average data for Pearson’s correlation coefficients are shown in the Results.
Temperature
All imaging and electrophysiology experiments were performed at room temperature (21–23°C).
Chemicals
All of the reagents were from VWR International, Sigma-Aldrich, Tocris Bioscience, Thermo Fisher Scientific, or Ascent Scientific. The names of several molecules have been abbreviated.
Online supplemental material
Video 1 shows confocal Z-series images through a HEK-293 cell expressing P2X4-YFP; Video 2 shows confocal Z-series images through a HEK-293 cell expressing P2X4-pHluorin123; and Video 3 shows confocal Z-series images through a HEK-293 cell expressing P2X4-pHluorin123 at pH 7.4, pH 5.4, and in NH4Cl. The online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201411169/DC1.
RESULTS
Selection of extracellular sites in mP2X4 receptor subunits for pHluorin insertions
We begin with a necessary brief introduction to the structural organization of P2X receptors. Our current understanding of the P2X4 receptor is based on the zfP2X4.1 receptor crystal structure (Kawate et al., 2009; Hattori and Gouaux, 2012) combined with nearly two decades of structure–function studies of mammalian subunits (Coddou et al., 2011b; Kaczmarek-Hájek et al., 2012; Khakh and North, 2012). Each P2X subunit has two transmembrane domains and an extracellular loop (Fig. 1 A). The P2X receptor is assembled as a trimer (Nicke et al., 1998) with a large extracellular domain, the tip of which stands ∼70 Å above the PM (Fig. 1 A). Here, the three subunits embrace each other, with a narrow central pore along their axis of symmetry. In the juxtamembrane regions, the extracellular domain contains three lateral portals that form access portals for ion flow into an extracellular vestibule above the transmembrane pore. The ATP-binding site is located ∼45 Å above the PM. In seeking to identify sites to insert pHluorin within the P2X4 receptor extracellular domain, we avoided regions of known function, such as the ATP-binding site; the lateral portals; and the upper, central, and extracellular vestibules (Khakh and North, 2012). We also avoided the pore and the connecting rods thought to couple ATP binding to pore opening (Browne et al., 2010). We then identified regions within the extracellular domain that correspond to flexible loops or unfolded regions within the zfP2X4.1 structure. Finally, we compared the amino acid sequences of these regions to those of regions representing nonconserved areas based on sequence analysis (Fig. 1 B). The combination of these approaches suggested nine sites for pHluorin insertion. These were immediately after Q78, F81, K122, S155, L206, A221, T223, L303, and N306 (mP2X4 numbering; Fig. 1 B). We inserted pHluorin at each of these sites independently using PCR and named the cognate receptors P2X4-pHluorin79, P2X4-pHluorin82, P2X4-pHluorin123, P2X4-pHluorin156, P2X4-pHluorin207, P2X4-pHluorin222, P2X4-pHluorin224, P2X4-pHluorin304, and P2X4-pHluorin307, respectively (Fig. 1 B). All of these constructs are available from us. The most useful construct, P2X4-pHluorin123, has been deposited at Addgene for distribution. In each case, the pHluorin was flanked by flexible linkers. Thus, the insertion cassette was -GAGAILGGAGGA-pHluorin-GGAGGAEFAGAG-. The flanking amino acid sequences were chosen to give rise to linkers that are likely to be flexible, hence the choice of glycine and alanine residues. The amino acids IL and EF correspond to the 5′ and 3′ restriction sites we used during the cloning step (ClaI and EcoRI). We did not systematically vary the linker sequences, and this is something that could be explored in follow-up studies, perhaps most usefully for insertion sites that were nonfunctional. As reported below, P2X4-pHluorin123 was functional and thus there was no immediate need to vary the flanking sequences.
Screening of nine mP2X4-pHluorin receptors for function and expression
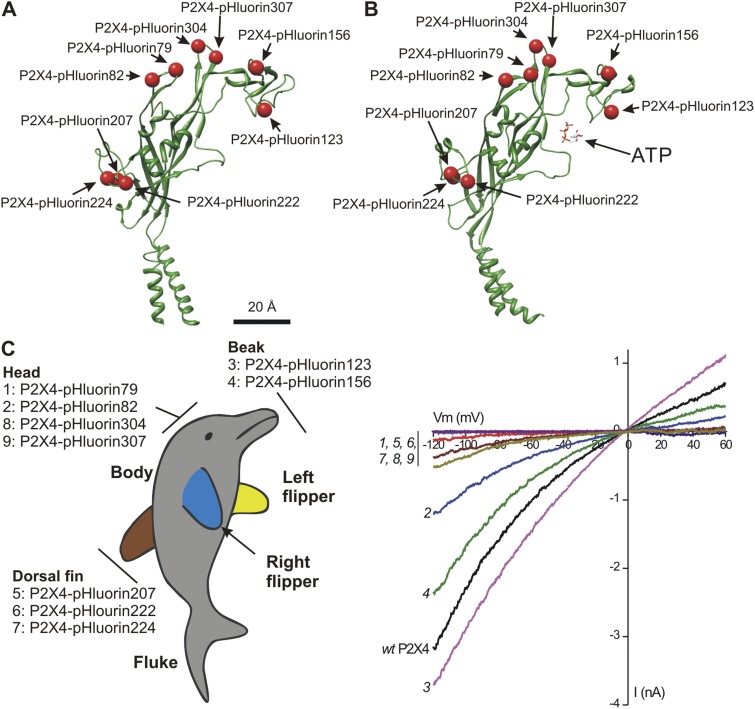
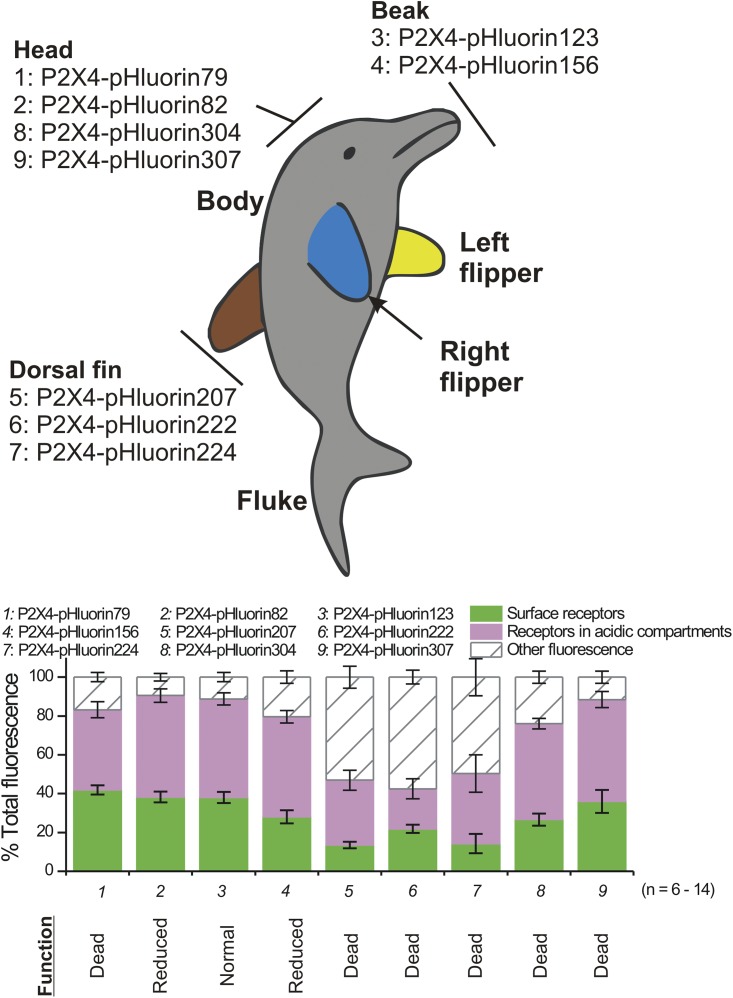
Fig. 2 illustrates the sites chosen for pHluorin insertion as red balls on the structure of a single P2X4 subunit in the unbound (Fig. 2 A) and ATP-bound states (Fig. 2 B). Most of the sites chosen were within loops. The domains within P2X subunits were named on the basis of the zfP2X4.1 crystal structure by viewing it as a dolphin rising from the water, with its fluke representing the transmembrane segments and parts of its body representing distinct regions of the P2X subunit (Kawate et al., 2009). In this view, the body of a P2X subunit consists of the beak, head, body, dorsal fin, fluke, and right and left flippers (Fig. 2 C). The pHluorin sites we used map to the beak, head, and dorsal fin domains (Fig. 2 C). We avoided the left and right flippers because these are thought to be involved in stabilizing subunit–subunit interactions.
Figure 2.
Anatomy of P2X4-pHluorin receptors and initial screening. (A) The cartoon shows a single P2X4 subunit with the insertion sites for pHluorin indicated as red balls. (B) As in A, but for a P2X4 subunit bound to ATP. (C) The sites for pHluorin insertion mapped onto a dolphin model of a P2X4 subunit. Hence, the constructs we made map to the beak, head, and dorsal fin domains. The graph shows leak-subtracted current–voltage relations for 100 µM of ATP-activated whole-cell currents from HEK-293 cells. The numbers next to each trace correspond to the individual pHluorin constructs, as shown in the dolphin model on the left. From this initial screen, it was apparent that P2X4-pHluorin123 was most similar to WT P2X4. The average data for experiments such as these are shown in Table 1.
We used whole-cell voltage-clamp electrophysiology to screen the pHluorin-tagged P2X4 receptors expressed in HEK-293 cells and made parallel measurements from WT P2X4 receptors. We applied 100 µM ATP and measured current–voltage relations (Fig. 2 C; n = 3–17). Several of the pHluorin-tagged receptors produced functional responses with macroscopic current–voltage relations that were similar to WT P2X4 (Fig. 2 C and Table 2). From this initial analysis, P2X4-pHluorin123 was the most similar to WT P2X4. Average data and statistics are shown in Table 2. Indeed, P2X4-pHluorin123 displayed ATP-evoked currents that were not significantly different to WT P2X4 (Fig. 3 A and Table 2; P > 0.05 using ANOVA). Interestingly, epitope tags (e.g., AU5, HA) are well tolerated in rP2X4 receptors at amino acid positions 76 and 78 (Bobanovic et al., 2002; Toulme and Khakh, 2012), whereas pHluorin insertion at Q78 of mP2X4 was detrimental to function. This empirical result speaks to the difficulty/unpredictability of inserting GFP-like proteins into the middle of receptors and channels (Mealer et al., 2008), and implies the necessity for random insertion methods (Giraldez et al., 2005) or structure-based design as in the case here.
Table 2.
Properties of P2X4-pHluorin receptors expressed in HEK-293 cells
| Construct | Position region | Fluorescence intensity | GFP/β actin ratio | Erev | Rectification index | IATP | Surface fraction | Acidic fraction | Other fluorescence |
| a.u. | mV | −pA | % | % | % | ||||
| WT P2X4 | — | — | — | −0.1 ± 0.8 | 0.79 ± 0.08 | 2,351 ± 315 | — | — | — |
| (n = 14) | (n = 14) | (n = 17) | |||||||
| P2X4-YFP | C tail | 738 ± 62 | 1.0 ± 0.05 | — | — | — | — | — | — |
| (n = 9) | (n = 3) | ||||||||
| P2X4-pHluorin79 | Head | 163 ± 17 | 0.9 ± 0.1 | ND | ND | 24 ± 11 | 42 ± 2 | 41.2 ± 4.1 | 16.8 ± 2.3 |
| (n = 16) | (n = 3) | (n = 8) | (n = 11) | (n = 11) | (n = 11) | ||||
| P2X4-pHluorin82 | Head | 188 ± 28 | 1.8 ± 0.03 | 2.4 ± 1.2 | 0.53 ± 0.03 | 587 ± 155 | 38 ± 3 | 52.2 ± 3.4 | 9.5 ± 1.9 |
| (n = 13) | (n = 3) | (n = 7) | (n = 7) | (n = 8) | (n = 11) | (n = 11) | (n = 11) | ||
| P2X4-pHluorin123 | Beak | 359 ± 25 | 2.0 ± 0.2 | 0.4 ± 0.7 | 0.67 ± 0.09 | 1,853 ± 271 | 38 ± 3 | 50.8 ± 3.1 | 11.2 ± 2.4 |
| (n = 14) | (n = 3) | (n = 9) | (n = 9) | (n = 10) | (n = 14) | (n = 14) | (n = 14) | ||
| P2X4-pHluorin156 | Beak | 343 ± 31 | 1.9 ± 0.2 | −0.03 ± 0.90 | 0.47 ± 0.04 | 472 ± 81 | 28 ± 3 | 51.6 ± 3.2 | 20.4 ± 3.2 |
| (n = 16) | (n = 3) | (n = 7) | (n = 8) | (n = 9) | (n = 12) | (n = 12) | (n = 12) | ||
| P2X4-pHluorin207 | Dorsal fin | 240 ± 9 | 1.5 ± 0.1 | ND | ND | 0.4 ± 0.4 | 13 ± 2 | 33.5 ± 5.1 | 53.1 ± 5.6 |
| (n = 7) | (n = 3) | (n = 3) | (n = 6) | (n = 6) | (n = 6) | ||||
| P2X4-pHluorin222 | Dorsal fin | 120 ± 10 | 0.9 ± 0.1 | ND | ND | 0.9 ± 1.1 | 22 ± 2 | 20.7 ± 5.2 | 57.5 ± 3.5 |
| (n = 6) | (n = 3) | (n = 5) | (n = 5) | (n = 5) | (n = 5) | ||||
| P2X4-pHluorin224 | Dorsal fin | 199 ± 25 | 0.9 ± 0.1 | ND | ND | 5 ± 4 | 14 ± 5 | 36.0 ± 9.6 | 49.7 ± 9.5 |
| (n = 9) | (n = 3) | (n = 5) | (n = 6) | (n = 6) | (n = 6) | ||||
| P2X4-pHluorin304 | Head | 177 ± 19 | 1.9 ± 0.2 | −3.9 ± 1.3 | 0.70 ± 0.09 | 58 ± 15 | 27 ± 3 | 49.4 ± 2.7 | 24.0 ± 3.1 |
| (n = 12) | (n = 3) | (n = 8) | (n = 8) | (n = 8) | (n = 12) | (n = 12) | (n = 12) | ||
| P2X4-pHluorin307 | Head | 185 ± 24 | 1.6 ± 0.3 | −1.7 ± 0.8 | 0.54 ± 0.09 | 88 ± 20 | 36 ± 6 | 52.4 ± 4.1 | 11.6 ± 3.1 |
| (n = 12) | (n = 3) | (n = 8) | (n = 8) | (n = 9) | (n = 10) | (n = 10) | (n = 10) |
Fluorescence intensity documents the average fluorescence intensity of HEK-293 cells expressing the indicated constructs. The rectification index is the ratio of current + and −40 of the ATP-evoked current reversal potential (Erev). The surface, acidic, and nonacidic fractions of receptors were calculated as described in Materials and methods and Results.
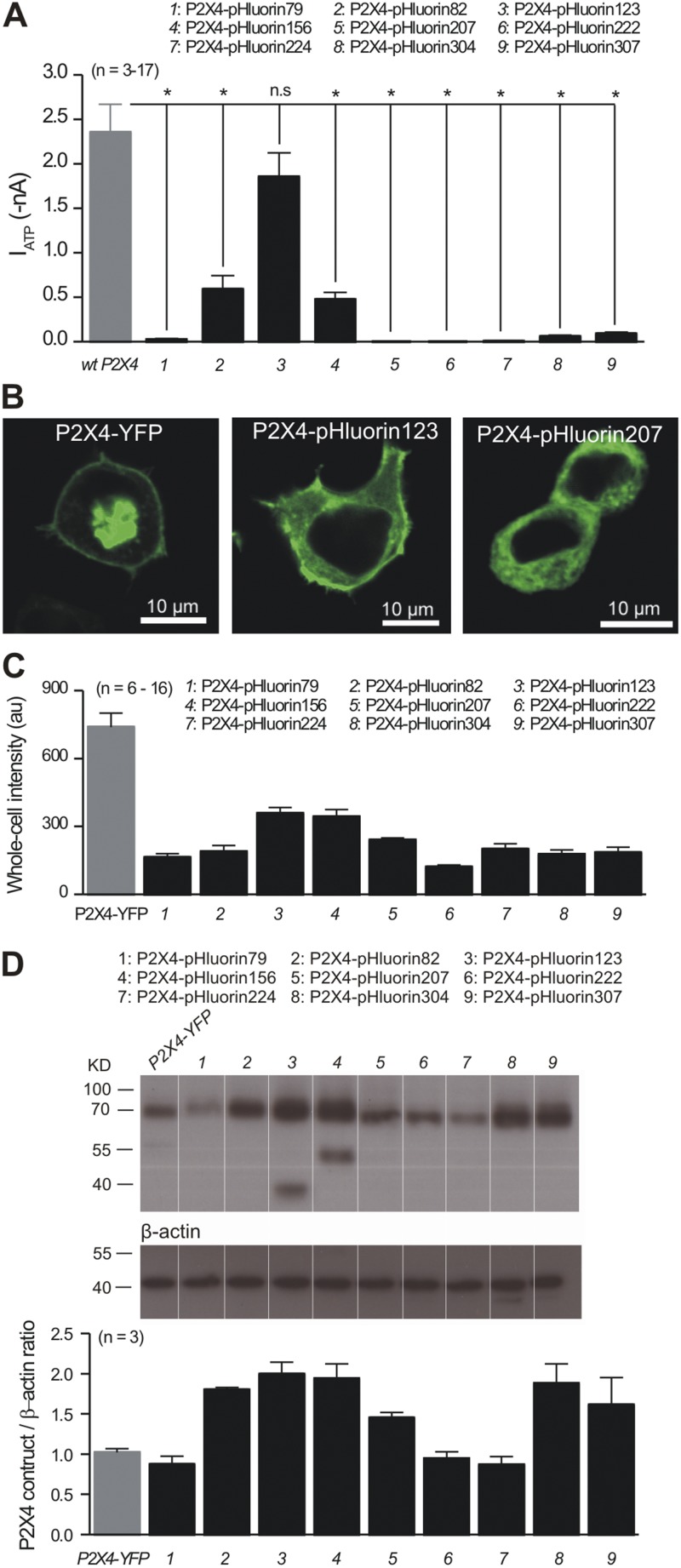
Figure 3.
Characterization of P2X4-pHluorin constructs in HEK-293 cells. (A) The bar graph summarizes the average data for peak 100-µM ATP-evoked currents at −60 mV for HEK-293 cells expressing each of the indicated constructs. All of the pHluorin constructs were compared with WT P2X4 using a Dunnett one-way ANOVA test. With the exception of P2X4-pHluorin123, they all displayed reduced currents. (B) Representative confocal images of HEK-293 cells expressing the three indicated constructs. Note that no membrane localization was observed for P2X4-pHluorin207. (C) Bar graph summarizes the fluorescence intensity of HEK-293 cells expressing the indicated constructs; all of the data were acquired using identical conditions for transfection and imaging. (D) The top panels show representative Western blots for the indicated constructs, whereas the bottom bar graph summarizes the data for P2X4 band intensities relative to β actin. Note that we observed additional bands in the case of P2X4-pHluorin123 and P2X4-pHluorin156. These likely correspond to partial degradation of the protein. Table 1 summarizes statistical data from experiments such as those shown in Fig. 3. Error bars represent mean ± SEM.
We next examined expression of the pHluorin-tagged receptors in HEK-293 cells by confocal microscopy. For these experiments, we used P2X4 receptors tagged on the C-terminal tail with YFP as the control; past work shows that receptors carrying C-terminal YFP or GFP fusions function like WT P2X4 (Bobanovic et al., 2002; Khakh and Egan, 2005; Toulme and Khakh, 2012). All of the P2X4-pHluorin constructs were expressed within HEK-293 cells as judged by marked bright green fluorescence (Fig. 3, B and C; n = 6–16), as well as by Western blot analysis of HEK-293 cell lysates (Fig. 3 D; n = 3 in each case; P2X4-pHluorin is expected to run at 70.5 kD). Thus, P2X4-pHluorin79, P2X4-pHourin207, P2X4-pHluorin222, and P2X4-pHluorin224 were robustly expressed (Fig. 3, B–D) but failed to give functional responses (Figs. 2 C and 3 A). Confocal microscopy revealed that these constructs were trapped inside the cells and failed to reach the surface (e.g., Fig. 3 B for P2X4-pHluorin207). In contrast, P2X4-pHluorin 82, P2X4-pHluorin123, and P2X4-pHluorin156 were functional and brightly fluorescent, and reached the cell surface as judged by bright fluorescence at the cell perimeter (e.g., Fig. 3 B for P2X4-pHluorin123). Of the P2X4-pHluorin constructs made and tested, P2X4-pHluorin123 revealed the highest fluorescence intensity (Fig. 3 C), recalling the large ATP-evoked currents measured with this receptor (Figs. 2 B and 3 A, and Table 2).
We comment briefly on the fact that the fluorescence intensity of P2X4-YFP and P2X4-pHluorin123 expressed in HEK-293 cells is not identical (Fig. 3, B and C). What factors contribute to this difference? Recall that the YFP moiety in P2X4-YFP is on the cytosolic domain and is therefore always exposed to the neutral intracellular pH. In contrast, the pHluorin moiety in P2X4-pHluorin123 is in the extracellular domain and is exposed to the acidic lumen of vesicles within cells. These facts imply that the overall fluorescence intensity of cells expressing P2X4-pHluorin123 is expected to be lower than that of cells expressing P2X4-YFP, which recalls the empirical data (Fig. 3 C). More precise intensity comparisons are vitiated by the fact that values for quantum yields of YFP and pHluorin at equivalent pH values are not available. Interestingly, past work and data shown herein (Fig. 3 A) show that P2X4-YFP and P2X4-pHluorin123 receptors both give rise to whole-cell ATP-evoked currents that are indiscernible from WT P2X4 (Bobanovic et al., 2002; Khakh and Egan, 2005). Collectively, these data provide strong evidence that P2X4-pHluorin123 is expressed at levels comparable to P2X4-YFP and WT P2X4. Moreover, the difference in fluorescence intensity between P2X4-YFP and P2X4-pHluorin123 receptors implies a significant acidic pool of receptors that is not revealed by the P2X4-YFP fusion construct but is revealed by P2X4-pHluorin123 (as being quenched). We explored this possibility in subsequent experiments.
From Fig. 3 B it is also clear that P2X4-pHluorin123 displays more fluorescence in the cytosol as compared with P2X4-YFP, which likely reflects partial localization to the ER. Further work is needed to understand this difference, but one possibility is that the YFP tag on the C-terminal tail of P2X4 in P2X4-YFP receptors affects ER localization. Nonetheless, P2X4-pHluorin123 and P2X4-YFP both give rise to functional responses indiscernible from WT P2X4 receptors (Figs. 2 C and 3 A) (Toulme and Khakh, 2012).
Properties of mP2X4-pHluorin123 receptors in relation to WT P2X4
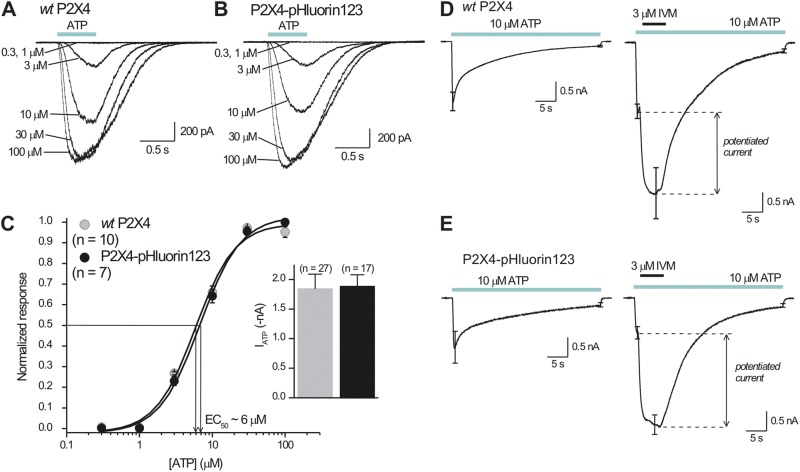
We studied the properties of P2X4-pHluorin123 receptors in more detail because this receptor most resembled WT P2X4 in our initial screening experiments (Figs. 1–3). ATP evoked concentration-dependent inward currents that were almost identical in HEK-293 cells expressing WT P2X4 and P2X4-pHluorin123 receptors (Fig. 4, A–C; n = 7 and 10). Thus, the concentration–effect curves for WT P2X4 and P2X4-pHluorin123 overlapped with similar EC50 values at ∼6 µM and similar Hill slopes of ∼2, which are both consistent with past work on mP2X4 receptors (Jones et al., 2000). Moreover, the peak 30-µM ATP-evoked inward currents were not significantly different (Fig. 4 C; n = 27 and 17). Next, we applied 10 µM ATP for 30 s and examined the extent of desensitization for WT P2X4 and P2X4-pHluorin123 receptors and found subtle but statistically significant differences between them (Fig. 4, D and E; n = 5 and 6). Thus, WT P2X4 and P2X4-pHluorin123 receptors desensitized by 92 ± 1 and 86 ± 1%, respectively, over 30 s of 10-µM ATP applications (P = 0.0006 when assessed with an unpaired Student’s t test). The subtle effect of pHluorin insertion on desensitization may be explained by the fact that the head domain of P2X receptors is proposed to move during activation and desensitization (Lörinczi et al., 2012). We also note that ∼90% desensitization during 30-s applications of 10 µM ATP is consistent with past studies on P2X4 receptors (Toulme et al., 2010; Toulme and Khakh, 2012). Moreover, the allosteric modulator ivermectin (IVM) (Khakh et al., 1999; Priel and Silberberg, 2004) potentiated ATP-evoked currents mediated by WT P2X4 and P2X4-pHluorin123 receptors equally (Fig. 4, D and E; n = 6 and 6; P = 0.46 when compared with an unpaired Student’s t test). Collectively, these experiments suggest that the basic pharmacological properties of P2X4 receptors are not altered by the addition of pHluorin after K122 in the extracellular domain. From a structural perspective, this is in accord with the knowledge that both ATP and IVM bind to regions of the protein distanced from the loop within the beak carrying pHluorin. The best available data suggest that ATP binds within an intersubunit pocket lined with positive charges cradled by the head domain, upper body, lower body, left flipper, and dorsal fin (Hattori and Gouaux, 2012), whereas IVM binds within the transmembrane domains (Silberberg et al., 2007; Jelínkova et al., 2008; Coddou et al., 2011a).
Figure 4.
Functional properties of P2X4-pHluorin123 receptors expressed in HEK-293 cells. (A and B) Representative traces for concentration-dependent ATP-evoked currents for WT P2X4 and P2X4-pHluorin123 receptors expressed in HEK-293 cells. Note the close similarities in the waveforms. (C) Average concentration–effect curves for ATP-evoked currents for WT P2X4 and P2X4-pHluorin123. The inset graph shows the peak 100-µM ATP-evoked currents from multiple cells gathered over a 3-mo period. The graphs for WT P2X4 and P2X4-pHluorin123 were identical, with the receptors displaying similar EC50 values, Hill slopes, and peak currents. (D and E) Current waveforms for WT P2X4 and P2X4-pHluorin123 to assess the extent of desensitization during ∼30-s, 10-µM ATP applications to assess the degree of current potentiation during applications of 3 µM IVM. The data for WT P2X4 and P2X4-pHluorin123 were indiscernible (n = 6 for WT P2X4 and n = 6 for P2X4-pHluorin123). Error bars represent mean ± SEM.
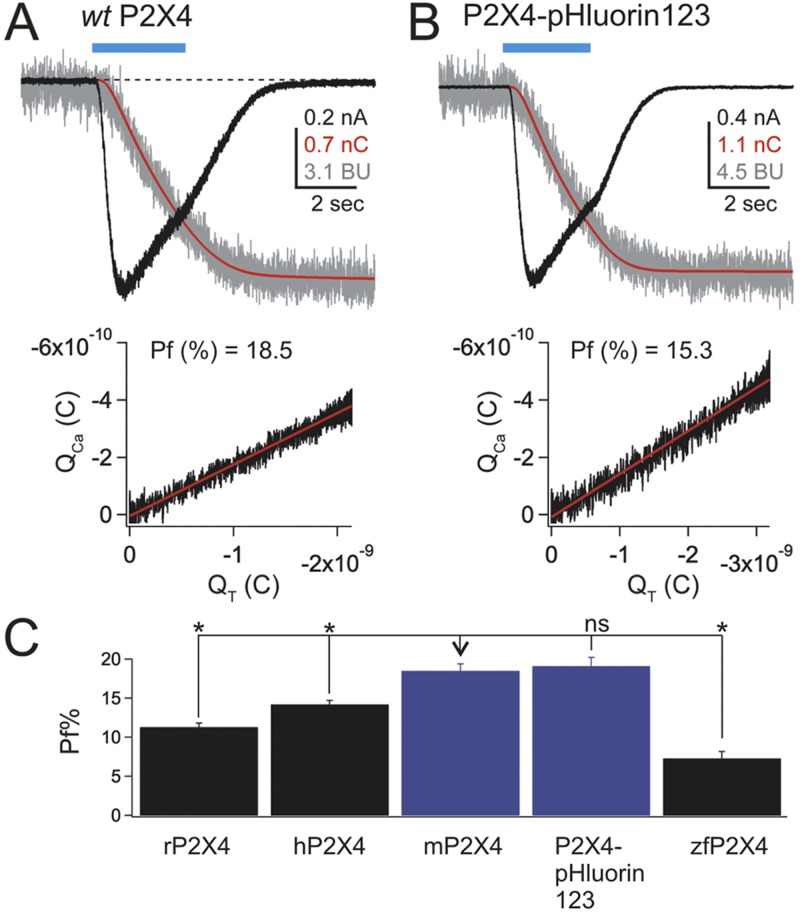
Native and recombinant P2X4 receptors carry significant Ca2+ currents (Egan and Khakh, 2004; Toulme et al., 2010). The Pf% of rP2X4 receptors is ∼12% and is considered a physiologically important attribute of this protein, although the Pf% value for mP2X4 has not hitherto been reported. In light of this, we used the dye overload method to determine and compare the Pf% values for mP2X4 and P2X4-pHluorin123 in relation to other mammalian P2X receptors (Fig. 5; n = 20, 78, 9, 8, and 10 for rP2X4, hP2X4, mP2X4, P2X4-pHluorin123, and zfP2X4, respectively). We made several observations. First, the Pf% value for mP2X4 and P2X4-pHluorin123 was not significantly different at ∼18.5 and 19%, respectively, but it was greater than zfP2X4, rP2X4, and hP2X4 when assessed using ANOVA between the multiple groups (Fig. 5 C). Collectively, these findings provide strong evidence that the Pf% value of mP2X4 is not altered by the pHluorin tag at K122. In accord, Ca2+ permeability and flux in P2X receptors are known to be regulated by residues that line the lateral portals and the pore domain, both of which are distanced from K122 (Migita et al., 2001; Egan and Khakh, 2004; Samways and Egan, 2007; Samways et al., 2011, 2012).
Figure 5.
Pf% for P2X4-pHluorin123 receptors in relation to other P2X receptors. (A and B) The top panels show waveforms for ATP-evoked currents (black), charge (red), and changes in the fluorescence of FURA-2 (red) for WT P2X4 and P2X4-pHluorin123 receptors expressed in HEK-293 cells. The bottom graphs plot the change in fluorescence of FURA-2 against charge transfer. The slope of this graph provides the Pf(%) value. In these graphs, the change in fluorescence intensity of FURA-2 is shown in “bead units” (see Materials and methods). (C) Summary bar graph showing the average Pf(%) value for WT mP2X4 and mP2X4-pHluorin123 in relation to other P2X receptors. The values were compared using a Tukey–Kramer multiple comparisons ANOVA test. rP2X4, hP2X4, and zfP2X4 were significantly different to mP2X4 (*, P < 0.05), but P2X4-pHluorin123 was not (P > 0.05). Error bars represent mean ± SEM.
mP2X4-pHluorin123 receptors display pH-dependent fluorescence changes
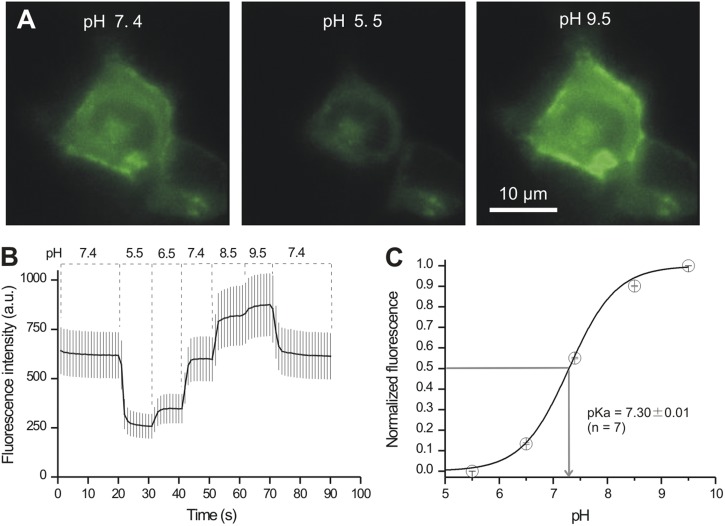
The preceding sections show that P2X4-pHluorin123 receptors function like WT P2X4, prompting us to evaluate if the pHluorin moiety within these receptors also displays the expected pH-sensitive fluorescence changes. We tested this possibility by applying buffered pH solutions to HEK-293 cells expressing P2X4-pHluorin123 (Fig. 6). We measured robust pH-dependent alterations in the intensity of HEK-293 cells (Fig. 6; n = 7). The apparent pKa was ∼7.30, which is close to the reported value of ∼7.2 for pHluorin (Sankaranarayanan et al., 2000). We conclude that insertion of pHluorin into mP2X4 receptors at K122 does not affect the pH sensitivity of the fluorescent protein.
Figure 6.
P2X4-pHluorin123 receptors display pH-dependent fluorescence changes. (A) Representative images of a HEK-293 cell expressing P2X4-pHluorin123 at the indicated pH values. (B) Average traces for pH-dependent changes in the fluorescence of HEK-293 cells expressing P2X4-pHluorin123. (C) Summary data for pH-dependent changes in the fluorescence of P2X4-pHluorin123. The pKa value was ∼7.3, which is similar to past characterization work on pHluorin (see Results).
Distribution of mP2X4-pHluorin123 receptors expressed in HEK-293 cells
We used live cell imaging to study the distribution of P2X4-pHluorin123 receptors expressed in HEK-293 cells (Fig. 7). To examine expression within the PM regions, we explored colocalization between P2X4-pHluorin with mCherry targeted to the PM (PM-mCherry is from a pCS2-mCherry plasmid containing the mCherry fluorescent protein tagged to a lyn-kinase PM localization signal). To assess colocalization in a systematic way, we used Pearson’s correlation coefficient analysis: a value >0.5 represents significant colocalization (Zinchuk and Zinchuk, 2008). In the case of PM-mCherry and P2X4-pHluorin, the Pearson’s correlation coefficient for whole-cell regions of interest (ROIs) was 0.6 ± 0.04, whereas it was 0.9 ± 0.01 for ROIs selected to be near the PM (Fig. 7 A; n = 6). In contrast, we found little colocalization between P2X4-pHluorin and DsRed2 targeted to the ER (DsRed2-ER). The Pearson’s correlation coefficient was 0.5 ± 0.10 (Fig. 7 B; n = 6). Finally, we assessed expression within lysosomes by using Lamp1-RFP as a marker. The Pearson’s correlation coefficient was 0.9 ± 0.03 (Fig. 7 C; n = 6). Collectively, these data indicate that P2X4-pHluorin123 was expressed predominantly in PM and lysosomal compartments (Bobanovic et al., 2002; Royle et al., 2002; Royle and Murrell-Lagnado, 2003; Qureshi et al., 2007).
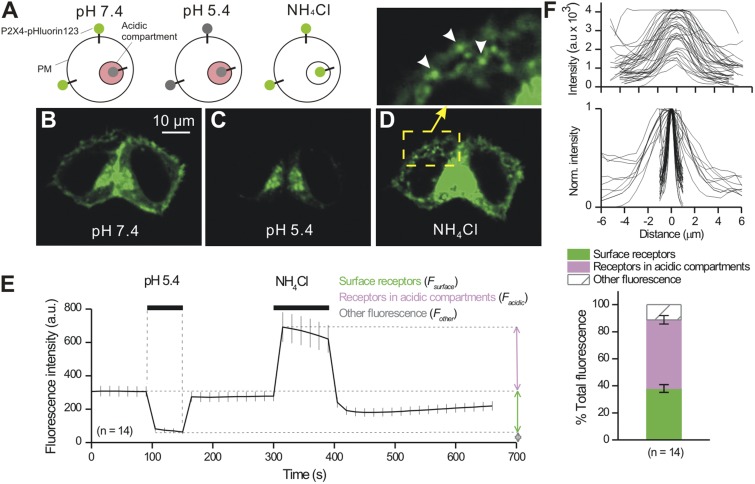
Next, we exploited the pH dependence of pHluorin to quantify the fraction of P2X4-pHluorin123 receptors on the PM and in acidic compartments within HEK-293 cells (Fig. 8). To this end, we applied acidic buffers (pH 5.4) to measure cell surface fractions followed by 25 mM NH4Cl to alkalize acidic compartments and measure the total receptor fraction (Fig. 8, A–F; n = 14). In this context, NH4Cl is used because it alkalizes receptors throughout the cells, hence relieving them from being quenched within acidic compartments by protons. From these measurements, we could arithmetically calculate the proportion of the total receptors that were found on the cell surface and within acidic compartments at ∼40 and ∼50%, with a further ∼10% residing in other compartments (Fig. 8 F). These additional intracellular compartments may represent the slight degradation of P2X4-pHluorin123 that was apparent from the Western blot analysis in Fig. 3. During applications of NH4Cl, numerous intracellular vesicle-like structures became visible, displaying a range of intensities and apparent sizes ranging from ∼1 to ∼6 µm full width at half maximum (FWHM; Fig. 8 F). Based on the observations from Fig. 7, these vesicle-like structures are likely endosomes and lysosomes carrying variable amounts of P2X4-pHluorin123 cargo. During control-imaging experiments that also lasted ∼700 s, cells expressing P2X4-pHluorin123 receptors bleached minimally by 4.0 ± 1.7% (n = 19), highlighting P2X4-pHluorin123’s utility for imaging studies without obvious loss of signal.
Figure 8.
Evaluation of total, surface, and intracellular fractions of P2X4-pHluorin123 receptors in HEK-293 cells. (A) The schematic shows P2X4-pHluorin123 receptors expressed on the PM and within vesicles. The P2X4-pHluorin123 receptors were fluorescent in neutral environments, quenched in the acidic compartments of intracellular vesicles, and unquenched in all compartments when the cells were bathed in NH4Cl. (B–D) Representative images of HEK-293 cells shown under the indicated conditions. Note, as shown in the cartoon in A, that the fluorescence intensity was quenched in acidic buffers and increased in NH4Cl. The inset shows a zoomed-in version of the cell in NH4Cl. Under this condition, numerous vesicles became visible. (E) Average graph showing how the fluorescence of HEK-293 cells expressing P2X4-pHluorin123 changes under the indicated conditions. (F) The top two panels show the FWHM of vesicles that became visible in the presence of NH4Cl. The graphs reveal a range of vesicle sizes (FWHM) and intensities, perhaps representing different loads of P2X4-pHluorin123. The bottom bar graph shows the proportion of P2X4-pHluorin123 receptors on the cell surface, within intracellular compartments, and within other compartments of HEK-293 cells. To generate this graph, the data were analyzed as described in Materials and methods using data from experiments such as those shown in E. Error bars represent mean ± SEM.
In the preceding section, we focused on P2X4-pHluorin123 because this receptor was indiscernible from WT mP2X4 using several measures of function (Figs. 2–5). For comparative purposes, in Fig. 9 we also report cell surface and intracellular fractions for all of the P2X4-pHluorin123 receptors we made (Figs. 1–2). In Fig. 9, P2X4-pHluorin123 is replotted from Fig. 8 F for comparison, and the bar graph lists the function of each receptor relative to WT P2X4 based on functional evaluations (Figs. 2–3 and Table 2). From this analysis, it emerges that P2X4-pHluorin79 and P2X4-pHluorin123 are almost identical in their cell surface and intracellular distribution patterns, despite the fact that P2X4-pHluorin79 is electrophysiologically silent (i.e., dead). This result suggests two things. First, the distribution of P2X4 receptors within cellular compartments may not be regulated by P2X4 channel function, and second, P2X4-pHluorin79 (and P2X4-pHluorin307) emerges as a useful control/comparative construct for future studies to evaluate the contributions of channel function to P2X4 receptor distributions within cells.
Figure 9.
Summary data for all pHluorin-tagged P2X4 receptors. The top panel represents a dolphin model of P2X4 receptors with the locations of the pHluorin insertion sites shown. The bottom bar graph shows data for each of these constructs and reports the fraction of the total receptors on the cell surface and within intracellular compartments. The data for P2X4-pHluorin123 in this graph are replotted from Fig. 8 for comparison. Of the indicated constructs, only P2X4-pHluorin123 and P2X4-pHluorin79 are interesting because both of these show the same overall distribution in the cells, but P2X4-pHluorin79 is nonfunctional, whereas P2X4-pHluorin123 receptors function like WT P2X4. Error bars represent mean ± SEM.
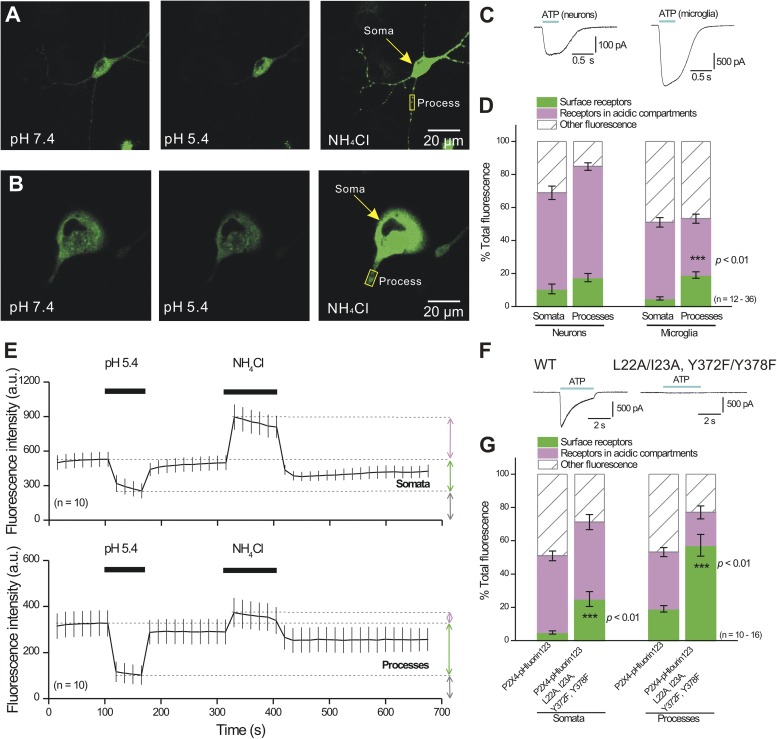
Distribution of mP2X4-pHluorin123 receptors in neurons and C8-B4 microglial cells
As proof-of-concept, we set out to evaluate the utility of P2X4-pHluorin123 in native cell types. Using protocols and analysis methods tested on HEK-293 cells (Fig. 8), we next evaluated P2X4-pHluorin123 expression in cultured hippocampal neurons and the C8-B4 microglial cell line (Toulme et al., 2010; Samways et al., 2012; Toulme and Khakh, 2012). In so doing, we analyzed fluorescence signals from the somatic regions as well as in regions from processes (n = 12–36), and we also measured whole-cell ATP-evoked currents (Fig. 10). The currents evoked by 100 µM ATP were −11 ± 4 (n = 6) and −56 ± 1 pA/pF (n = 7; Fig. 10 C) for neurons and C8-B4 microglial cells, respectively. There were no differences in the relative proportions of cell surface and intracellular P2X4-pHluorin123 receptors in the somata or processes of neurons (Fig. 10, A and D; n = 16 somata and n = 37 processes). However, for C8-B4 microglial cells, the cell surface fraction was significantly larger in the processes as compared with somata (Fig. 10 D; P < 0.01, n = 12 somata and n = 16 processes). This was clear from the average data shown in Fig. 10 D as well as from the traces for somatic and process regions (Fig. 10 E; n = 10). Moreover, a quadruple P2X4 mutant (L22A, I23A, Y372F, and Y378F; Fig. 10 F) that lacks endocytic and lysosomal targeting (Qureshi et al., 2007) significantly increased the fraction of receptors on the cell surface for C8-B4 microglial processes and somata (Fig. 10 G; P < 0.01; n = 16). These data support the findings that the distribution of P2X4-pHluorin123 receptors within cellular compartments is determined by specific amino acid residues in the cytosolic N and C termini (Qureshi et al., 2007), whereas our data with “dead” P2X4-pHluorin79 receptors (Fig. 9) imply little detectable role for ion channel function in these phenomena. Interestingly, we found that the quadruple mutant was functionally compromised: the ATP-evoked currents were −45 ± 5 (n = 5) and −1.2 ± 0.3 pA/pF (n = 7) for P2X4-pHluorin123 and P2X4-pHluorin123L22A/I23A/Y372F/Y378F, respectively. As far as we know, the quadruple mutant was not evaluated using electrophysiology in past studies (Qureshi et al., 2007), but it is not surprising that the four mutations affect function because they occur in regions known to be important for normal functioning of P2X receptors (North, 2002; Coddou et al., 2011b; Kaczmarek-Hájek et al., 2012).
Figure 10.
Distribution of P2X4-pHluorin123 receptors in cultured hippocampal neurons and C8-B4 microglia. (A and B) Representative images of P2X4-pHluorin123 receptors expressed in cultured hippocampal neurons and C8-B4 microglia under the conditions indicated (pH 7.4, pH 5.4, and 50 mM NH4Cl). (C) Representative traces for ATP-evoked currents recorded from neurons and C8-B4 microglia (average data are reported in the Results). (D) Distribution of P2X4-pHluorin123 receptors in cellular compartments of neurons and C8-B4 microglia (somata and processes are separated). (E) Graphs show average data for C8-B4 microglia ROIs in somata and processes. (F) Representative traces for ATP-evoked currents for WT P2X4-pHluorin123 and for P2X4-pHluorin123 carrying quadruple mutations that block lysosomal targeting and endocytosis. (G) Summary data showing that proportions of P2X4-pHluorin123 receptors in cellular compartments of C8-B4 microglial somata and process ROIs are altered in quadruple mutant channels; the cell surface fraction increases significantly. Error bars represent mean ± SEM.
Distribution of P2X4-pHluorin receptors in primary ATII cells
Similar to the experiments performed in cultured hippocampal neurons and the C8-B4 microglial cell line, we next evaluated expression of P2X4-pHluorin123 in rat primary ATII cells. It has been reported recently that P2X4 is expressed on LBs, which are large lysosome-related organelles storing pulmonary surfactant; P2X4 receptors readily become part of the apical membrane upon exocytosis of LBs (Miklavc et al., 2011). Activation of vesicular P2X4 receptors upon exocytosis of LBs is essential for surfactant release and contributes to “activation” of surfactant in the extracellular space (Miklavc et al., 2011, 2013; Thompson et al., 2013).
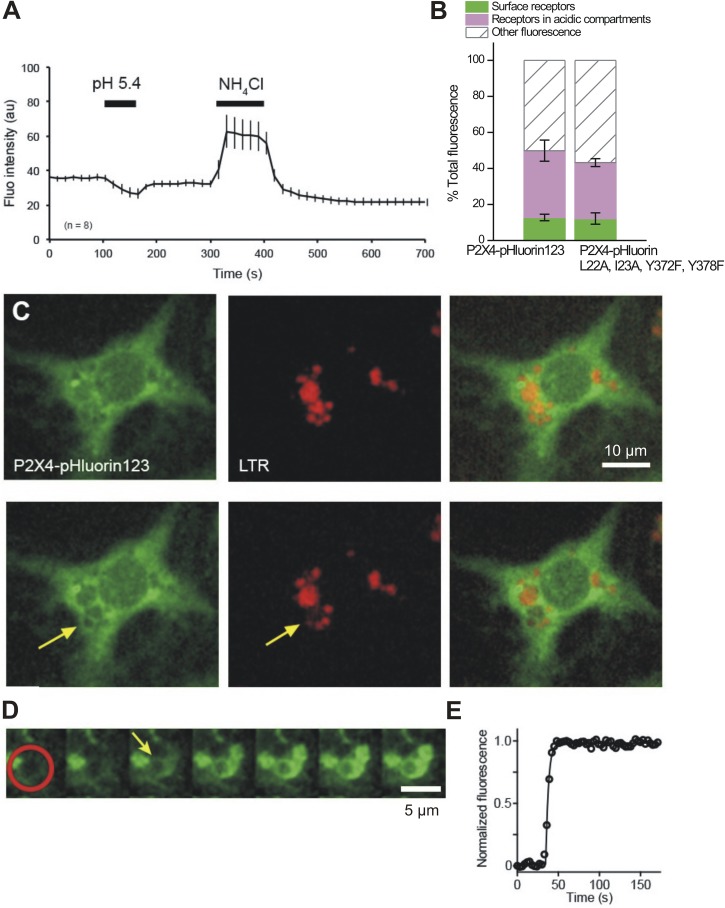
We first analyzed the relative proportions of cell surface and intracellular P2X4-pHluorin receptors in ATII cells expressing either WT P2X4-pHluorin123 or the P2X4 mutant (L22A, I23A, Y372F, and Y378F; Fig. 11, A and B). In contrast to neurons and microglia, the distribution of P2X4 among cell surface, receptors in acidic compartments, and other compartments was not different in cells expressing WT or mutant P2X4 (Fig. 11 B). Little surface expression of P2X4 (12.8 ± 1.8%, n = 8, and 12.2 ± 3.1%, n = 14, in cells expressing WT and mutant P2X4, respectively) is in line with previous reports (Miklavc et al., 2011) and could reflect that a large proportion of PM P2X4 results from exocytosis of LBs. Such fusion events are sparse under nonstimulated resting conditions (Frick et al., 2001). In addition, the lack of difference between WT and mutant P2X4 surface expression likely results from little membrane turnover and endocytic activity in alveolar epithelial cells kept in standard cell culture conditions (Huh et al., 2010). Under such conditions, WT P2X4 receptors are expected to be trapped in the PM once inserted. However, a detailed understanding of P2X4 trafficking in ATII cells is still elusive.
Figure 11.
Distribution of P2X4-pHluorin123 receptors in primary ATII cells. (A) Graph illustrating how average fluorescence of primary ATII cells expressing P2X4-pHluorin changes under the indicated conditions. (B) Summary data showing the proportions of P2X4-pHluorin receptors in cellular compartments of ATII cells expressing either WT P2X4-pHluorin or P2X4-pHluorin carrying quadruple mutations that block lysosomal targeting and endocytosis. (C) Simultaneous imaging of P2X4-pHluorin123 (green) and LTR (red) before (top row) and after (bottom images) exocytic fusion of an LB with the PM. Arrow indicates fused LB, revealed by a decrease in vesicular LTR fluorescence caused by diffusion of LTR from the vesicle lumen. (D) Time-lapse image series of an increase in P2X4-pHluorin123 fluorescence on the membrane of an LB after fusion caused by the increase in luminal pH of the fused LB. The red circle indicates the region where the increase in P2X4-pHluorin123 fluorescence was measured after fusion of LB with the PM. Images were acquired at 0.3 Hz; the arrow indicates time of fusion. (E) Time course of P2X4-pHluorin123 fluorescence within the area of the red circle in D. Error bars represent mean ± SEM.
Similar to the observations in HEK-293 cells, neurons, and microglia, a substantial proportion of P2X4 resides within acidic compartments (37.1 ± 5.9%, n = 8, and 31.0 ± 2.2%, n = 14, in cells expressing WT and mutant P2X4, respectively). To test whether P2X4 is expressed in LBs and whether fusion of LBs results in the insertion of P2X4 in the cell surface, we performed experiments to directly visualize an increase of vesicular pHluorin fluorescence upon fusion of LBs with the PM. LB fusions with PM were detected by selective loss of LTR fluorescence from individual LBs after fusion pore opening (Haller et al., 1998). After fusion of LBs with the PM, a selective ring-like increase in pHluorin fluorescence at the site of vesicle fusion was observed, most likely resembling the LB membrane (Fig. 11 C). Detailed analysis of the fluorescence increase revealed that P2X4-pHluorin123 fluorescence does increase immediately after LB fusion and is restricted to the LB membrane of the fused vesicle (Fig. 11 D). Moreover, mean P2X4-pHluorin123 fluorescence does not significantly decrease within fused LBs for prolonged times (98.5 ± 1.1%, n = 5, and 85.6 ± 8.8%, n = 5, of maximum pHluorin123 fluorescence 30 and 60 s after fusion, respectively). This suggests that P2X4-pHluorin123 is restricted to the LB membrane after LB fusion with the PM; P2X4-pHluorin123 does not rapidly diffuse into the PM or appear to be recaptured via endocytosis over these time scales.
Collectively, our experiences with P2X4-pHluorin123 receptors expressed in HEK-293 cells (Fig. 8), hippocampal neurons and C8-B4 microglial cells (Fig. 10), and ATII cells (Fig. 11) emphasize cell- and compartment-specific expression patterns of this protein and in so doing provide an optical assay to study these differences and their molecular basis in future work.
IVM potentiates mP2X4-pHluorin123 functional responses without altering the surface fraction of receptors
Having shown that P2X4-pHluorin123 is useful for studies within native cell types (Figs. 10 and 11), we next used it to address a specific mechanistic question related to IVM actions at P2X4 receptors. The application of micromolar amounts of IVM to cells expressing P2X4 receptors dramatically potentiates and prolongs ATP-evoked currents in a manner that is largely selective for P2X4 receptors over other P2X receptors (Jarvis and Khakh, 2009). When this property was discovered, it was suggested that IVM was an allosteric regulator of P2X4 receptors (Khakh et al., 1999). This conclusion gained support from subsequent electrophysiology and structure–function work (Priel and Silberberg, 2004; Silberberg et al., 2007; Jelínkova et al., 2008; Coddou et al., 2011a), and studies of Ca2+ flux (Samways et al., 2012). However, another study using biochemical methods suggested that IVM potentiated P2X4 receptor responses by reducing endocytosis (Toulmé et al., 2006), which in turn increased the pool of functional P2X4 receptors on the PM.
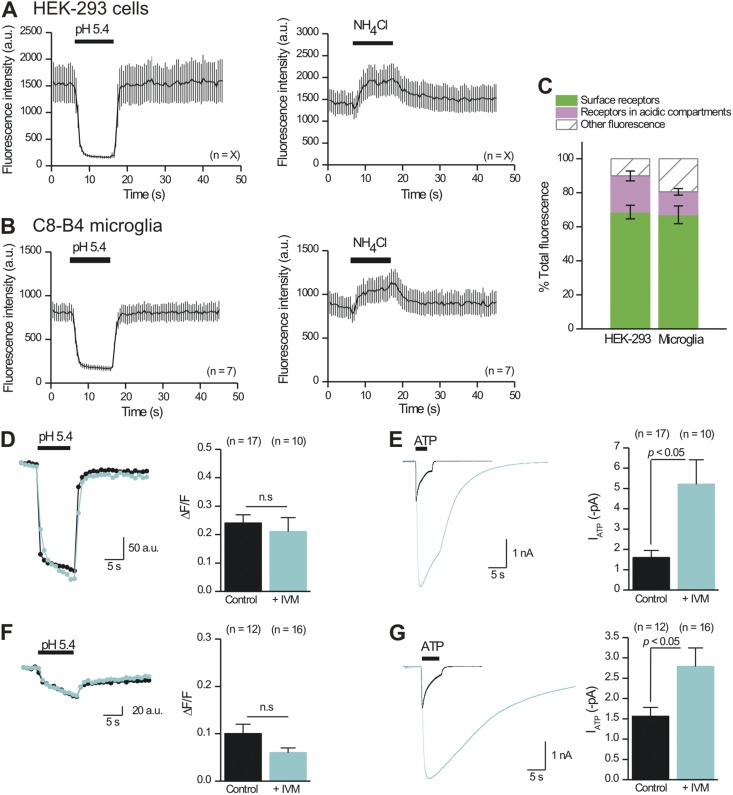
Because P2X4-pHluorin123 receptors are potentiated by IVM (Fig. 4), we exploited them to further evaluate if IVM produced its potentiating effects on these receptors (Fig. 4) by increasing the surface pool of receptors, which we could estimate directly before and during IVM applications using brief application of pH 5.4 solutions (Figs. 8–10). We started by determining if an intracellular pool of receptors existed near the PM of HEK-293 cells and C8-B4 microglia using TIRF microscopy and applications of buffers at pH 5.4 and containing NH4Cl (Fig. 12, A–C; n = 7 and 9). We did not use hippocampal neurons for these experiments because they sit on a layer of astrocytes (Shigetomi and Khakh, 2009), making TIRF problematic because it requires that the cells be attached directly to glass. Using TIRF microscopy, we found evidence for a large pool of cell surface P2X4-pHluorin123 receptors that represented ∼80% of the total within the evanescent field (Fig. 12 C). We also found clear evidence for P2X4-pHluorin123 receptors residing within ∼100 nm of the PM within acidic compartments that could be alkalized with NH4Cl; these represented ∼10% of the total pool within the evanescent field (Fig. 12 C). Collectively, these data clearly show that an intracellular pool of P2X4 receptors exists near the PM.
Figure 12.
IVM does not detectably increase the cell surface fraction of P2X4-pHluorin123 receptors in HEK-293 cells or C8-B4 microglia. (A) The graphs plot the effect of acidic and alkalinizing solutions on the intensity of P2X4-pHluorin123 receptors expressed in HEK-293 cells and studied by TIRF microscopy, which reports only on receptors within ∼100 nm of the PM. (B) As in A, but for P2X4-pHluorin123 receptors expressed in C8-B4 microglial cells. (C) Summary data for the fraction of P2X4-pHluorin123 receptors on the cell surface and within acidic compartments located within ∼100 nm of the PM for HEK-293 cells and C8-B4 microglia. Only cell bodies were imaged reliably with TIRF, likely because the interactions between processes and glass were minimal. (D) Representative traces and average data for the acid-quenched component of P2X4-pHluorin123 receptors in HEK-293 cells before and during 3-µM IVM applications. (E) Representative traces and average data for 100-µM ATP-evoked currents mediated by P2X4-pHluorin123 receptors in HEK-293 cells before and during 3-µM IVM applications. (F and G) As in D and E, but for P2X4-pHluorin123 receptors expressed in C8-B4 microglia. Error bars represent mean ± SEM.
If IVM produces its potentiating effects on P2X4 responses by increasing the numbers of channels in the PM, we would predict that IVM would increase the magnitude of the fluorescence signal quenched by pH 5.4 buffers. We tested this possibility by applying pH 5.4 buffers to single HEK-293 cells and C8-B4 microglia expressing P2X4-pHluorin123 before and during ∼1-min applications of 3 µM IVM. We then performed whole-cell recordings on the same cells to determine if a subsequent application of IVM potentiated the ATP-evoked currents. To permit comparisons with whole-cell electrophysiology, the imaging experiments were performed with epifluorescence imaging. For both HEK-293 cells and C8-B4 microglia, we found that IVM did not significantly change the surface fraction of receptors (Fig. 12, D and F), but in both cases, IVM did significantly potentiate and prolong the ATP-evoked currents (Fig. 12, E and G). Collectively, our experiments support the view that IVM acts allosterically to potentiate ATP-evoked responses mediated by P2X4 receptors (Khakh et al., 1999; Priel and Silberberg, 2004; Silberberg et al., 2007; Samways et al., 2012). We found no evidence to indicate that IVM increases the cell surface fraction of P2X4-pHluorin123 receptors.
DISCUSSION
Main findings from this study
There are seven main findings from this study. (1) We generated a series of mP2X4 receptors carrying pHluorin tags in the extracellular domain and report that one of these functions like WT P2X4 as judged by several measures of function and expression. (2) P2X4-pHluorin123 can be used to explore the cell surface and intracellular fractions of receptors in living cells. (3) The cell surface and intracellular fractions of P2X4-pHluorin123 receptors are cell and compartment specific, as they are of different magnitudes between C8-B4 microglia and neurons, and between microglial processes and somata. (4) The fraction of P2X4-pHluorin123 receptors within cell surface and intracellular pools in C8-B4 microglia is controlled by specific amino acid residues in the N and C termini of P2X4 receptors, but seemingly independently of a functional P2X4 channel pore. (5) By measuring functional responses as well as cell surface fractions of mP2X4-pHluorin123 receptors in HEK-293 cells and C8-B4 microglia, we found no evidence that IVM affects the surface fraction, adding further support to the view that IVM acts as an allosteric modulator. (6) Our experiences and studies of P2X4 receptors provide the rational and specific insertion sites to generate similar P2X receptors carrying pHluorin tags (e.g., for P2X1, P2X3, and P2X7). The sites may also be predictive for generating similar constructs for acid-sensing channels that share structural and topological similarities with P2X receptors (Gonzales et al., 2009; Baconguis et al., 2013). The utility of P2X4-pHluorin123 could be extended by generating further insertion sites for different colored fluorescent proteins using random transposon-based methods (Sheridan et al., 2002; Giraldez et al., 2005; Mealer et al., 2008). (7) mP2X4-pHluorin123 receptors can now be used to image and quantify the cell surface and intracellular fractions of these receptors in live cells, thus permitting exploration of the molecular mechanisms that lead to up-regulation of P2X4 receptors. The same optical approach could be used to screen for inhibitors of P2X4-pHluorin123 up-regulation, which may be of value therapeutically for epilepsy and neuropathic pain.
As expected, we detected P2X4-pHluorin receptors on the cell surface and within acidic intracellular compartments by using acidic pH jumps and ammonium chloride to alkalize intracellular acidic compartments. However, we also measured fluorescence that we called other fluorescence in Figs. 8–12 that cannot be obviously accounted for by P2X4-pHluorin cell surface expression or expression within acidic compartments (e.g., Fig. 8 E). What might this other fluorescence represent? It is possible that P2X4-pHluorin123 receptors reside in compartments that are not neatly classified by acidic pH jumps and ammonium chloride. Such compartments could conceivably be synthetic or degradation pathways. In accord with the latter possibility, P2X4 (Qureshi et al., 2007) and P2X4-pHluorin123 receptors are known to traffic to lysosomes (Fig. 7). It is also notable from Fig. 9 that receptors that were nonfunctional and intracellularly retained (Table 2) also displayed large fractions for other fluorescence. Hence, it is possible that P2X4-pHluorin123 could be used to measure receptors within cell surface, acidic, and degradation compartments. Further studies are needed to systematically explore this possibility, but at this juncture, the use of the general term “other fluorescence” seems the most parsimonious as it implies no specific mechanism and encompasses the possibility that the fluorescence may be from partially synthesized or partially degraded proteins.
Other approaches and further future developments
We show that our optical reporter, P2X4-pHluorin123, causes minimal disruption of receptor function and allows pH-dependent measurements of P2X4 receptors in the cellular secretory and lysosomal pathways. When complemented with classical biochemical methods, P2X4-pHluorin123 is a powerful tool to study P2X4 receptor biology.
Classical biochemical methods such as surface biotinylation and antibody labeling of P2X receptors (Bobanovic et al., 2002; Toulmé et al., 2006, 2010; Qureshi et al., 2007) require cell fixation and consequently they cannot report PM receptor insertion and/or internalization in live cells. Furthermore, if P2X4 receptor trafficking occurs within minutes, the extended incubation needed for biotinylation and antibody labeling of receptors may complicate the measurement of receptor trafficking kinetics. The penetration of antibody or labeling reagents would also be a factor to consider if receptors within intact tissues such as brain slices need to be studied. In contrast, P2X4-pHluorin123 allows the measurement of receptor insertion, turnover, and perhaps degradation in real time and at a much higher resolution than that achieved with classical biochemistry. Of key importance is that P2X4-pHluorin123 receptors can be imaged in different areas of the cell with ease (e.g., dendrites, somata, axons). However, the use of P2X4-pHluorin123 requires the protein to be tagged with a pH-sensitive form of GFP, appropriate optical instrumentation, and cellular expression at levels that can be observed with light microscopy.
Starting with initial studies, fusion of GFP to the C terminus of P2X4 subunits has been used with great success to study receptor surface expression and trafficking (Bobanovic et al., 2002; Royle et al., 2002). However, because the C-terminal tail of P2X4 receptors is always exposed to the cytosol, assigning PM localization of P2X4-GFP receptors is inaccurate, as the GFP would be in the same pH environment whether it was in the PM or within near-PM vesicles. This specific issue is avoided in P2X4-pHluorin123, which carries a pH-sensitive GFP variant in the extracellular domain of P2X4 receptors. The same feature permits study of P2X4-pHluorin123–laden vesicle fusion because the pH of the vesicle lumen changes during fusion with the PM. However, the use of P2X4-pHluorin123 could be improved further. First, it would be interesting to engineer P2X4-pHluorin receptors to also carry 13–amino acid α-bungarotoxin–binding sites in the extracellular domain, and then subsequently label these with red fluorescent bungarotoxin (Sekine-Aizawa and Huganir, 2004). Such approaches should allow one to image receptor endocytosis directly and in real time, because only cell surface receptors will be labeled. Hence, the fate of cell surface receptors and how they mingle with those in lysosomes could be studied directly. Second, in future studies, the use of P2X4-pHluorin123 could be expanded by combining it with subunits that carry pH-sensitive Tomato proteins (pHTomato) within appropriate extracellular locations (Li and Tsien, 2012). In such settings, the spectral differences between pHluorin and pHTomato would permit the study of the trafficking of two different ion channel populations simultaneously, a feature that could be used to explore pairs of interacting receptors such as P2X4 and P2X7 (Murrell-Lagnado, 2009).
By necessity, all of our studies used standard plasmid-based mammalian cell transfection methods that are frequently used for cells in culture. These methods have been used based on past experiences with HEK-293, C8-B4, and ATII cells, and hippocampal neurons. In particular, we note that for C8-B4 cells, we used nucleofection because past studies showed that this method did not cause detectable microglial cell activation (Toulme et al., 2010; Toulme and Khakh, 2012). We clarify that we did not use the same transfection method across all experiments and used methods that we found most suitable for the cells we studied on a case-by-case basis. Hence, our comparisons for P2X4-pHluorin in different cell types should be interpreted with this consideration in mind. In future studies it will be important to build on these experiments by expressing P2X4-pHluorin123 in vivo using viruses, transgenic mice, or knock-in gene-targeting methods. Of these, the generation and use of knock-in mice would provide the most faithful representation of native P2X4 expression levels. Once such mice are made and tested, it will be useful to directly compare the results to those reported here with transient transfection. Such analyses may reveal how different methods used to deliver P2X4-pHluorin123 can affect the cell surface and intracellular pools of receptors. This is something that the studies reported herein were not designed to assess.
The preceding section discusses several advantages and potential future developments to P2X4-pHluorin123 to experimentally explore P2X4 receptor trafficking. Collectively, we suggest that P2X4-pHluorin123 receptors represent a straightforward and useful complementary approach to study P2X4 receptor cell biology and biophysics.
Proof-of-concept experiments
All of the constructs reported herein are available from us for unrestricted distribution, and their use may shed light on several important biological questions. Based on our initial proof-of-concept experiments, we provide three examples of how P2X4-pHluorin123 receptors could be used in follow-up studies of outstanding questions in the field.
First, P2X4-pHluorin123 could be used to image up-regulation of P2X4 receptors in microglial cells and other cells during pathological processes such as the development of neuropathic pain and epilepsy. So far, the evidence to implicate P2X4 receptors in these complex disorders has come from pharmacological and genetic studies (see Introduction). Through the use of in vivo expression via knock-in mice or viral transduction, P2X4-pHlouirn would build upon and extend these insights by providing real-time measures of P2X4 receptor trafficking before and during the development of pain. Conversely, drugs that treat neuropathic pain could be assessed for their ability to reverse P2X4 up-regulation, thus permitting causative correlations that are required to further explore the roles of P2X4 receptors in complex disease states such as neuropathic pain.
Second, P2X4-pHluorin123 receptors could be used to image LB exocytosis from lung ATII cells and thus shed light on the process of fusion-activated Ca2+ entry (Miklavc et al., 2011). Specifically, P2X4-pHluorin123 will allow one to image P2X4-laden vesicle fusion with the PM, and it will permit imaging of the subsequent fate of P2X4 receptors and determine whether they are recaptured by the same vesicle during endocytosis, left stranded at the site of insertion, or relinquished to diffuse into the PM bulk. As such, the use of P2X4-pHluorin123 would permit direct study of the biophysical and molecular mechanisms that determine receptor insertion into the PM and their relationships to fusion activated Ca2+ entry (Miklavc et al., 2011).
Third, the proposed roles of P2X4 receptors in neuropathic pain and epilepsy has prompted the search for P2X4 receptor antagonists for pain therapeutics by high throughput screening of functional P2X4 responses in clonal cell lines such as HEK-293. However, an alternative therapeutic strategy would be to prevent P2X4 receptor up-regulation rather than block the receptors after they have been up-regulated. Progress on this front has been limited because of the lack of available methods to image P2X4 up-regulation in a manner that could be adapted for high throughput screening. From this perspective, P2X4-pHluorin123 represents an ideal tool because up-regulation can be measured simply as an increase in fluorescence intensity of cells expressing P2X4-pHluorin123. If so, P2X4-pHluorin123 receptors represent a designer and novel assay for screening small compound libraries for drug-like molecules to target P2X4 receptor up-regulation and their use in neuropathic pain and epilepsy therapy. Use of P2X4-pHluorin in high throughout assays could occur quite rapidly and may also benefit from knowledge of the excitation and emission spectra of pHluorin in a range of pH values. Finally, it is possible that fluorescence anisotropy of the fluorophore in P2X4-pHluorin123 could be used to relate receptor functions such as activation and desensitization to trafficking and endocytosis, as the domain of P2X receptors (that carries pHluorin within the beak region) is proposed to undergo conformational changes associated with activation and desensitization (Lörinczi et al., 2012).
More generally, the last few years have witnessed the development of several genetically encoded optical approaches to measure P2X receptor properties, including location within cells (Khakh et al., 2001; Bobanovic et al., 2002), conformational changes (Fisher et al., 2004; Chaumont and Khakh, 2008), proximity (Khakh et al., 2005), dimensions (Young et al., 2008), protein interactions (Chaumont et al., 2008), mobility (Richler et al., 2011; Shrivastava et al., 2011, 2013; Toulme and Khakh, 2012), and activation (Richler et al., 2008; Parkinson et al., 2014), as well as approaches to optically trigger P2X channel opening (Lemoine et al., 2013; Browne et al., 2014). This study adds to this toolkit by providing an imaging approach to track P2X receptor subcellular distribution and trafficking. The paucity of selective pharmacological tools remains a hindrance to understanding P2X receptor cell biology and physiological roles. From this perspective, the combination of the aforementioned genetic and optical approaches provides exciting opportunities to explore the physiological functions of P2X receptors with light.
Supplementary Material
Acknowledgments
This study was supported partly by the NIH National Institute of Neurological Disorders and Stroke (grant NS073980) and partly by unrestricted funds (to B.S. Khakh). Some of the work was supported by the NIH Heart, Lung and Blood Institute (grant HL56236 to T.M. Egan) and the Deutsche Forschungsgemeinschaft (grant DI 1402/3-1) and a grant from the Ministry of Science, Research and the Arts of Baden-Wurttemberg (Az: 32-7533.-6-10/15/5) to M. Frick, respectively.
The authors declare no competing financial interests.
Sharona E. Gordon served as editor.
Footnotes
Abbreviations used in this paper:
- ATII
- alveolar type II
- FWHM
- full width at half maximum
- GFP
- green fluorescent protein
- IVM
- ivermectin
- LB
- lamellar body
- LTR
- LysotrackerRed
- Pf%
- fractional Ca2+ current(s)
- PM
- plasma membrane
- ROI
- region of interest
- TIRF
- total internal reflection fluorescence
References
- Baconguis I., Hattori M., Gouaux E. 2013. Unanticipated parallels in architecture and mechanism between ATP-gated P2X receptors and acid sensing ion channels. Curr. Opin. Struct. Biol. 23:277–284 10.1016/j.sbi.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobanovic L.K., Royle S.J., Murrell-Lagnado R.D. 2002. P2X receptor trafficking in neurons is subunit specific. J. Neurosci. 22:4814–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolato M., Yardley M.M., Khoja S., Godar S.C., Asatryan L., Finn D.A., Alkana R.L., Louie S.G., Davies D.L. 2013. Pharmacological insights into the role of P2X4 receptors in behavioural regulation: lessons from ivermectin. Int. J. Neuropsychopharmacol. 16:1059–1070 10.1017/S1461145712000909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler J.W., Bailey R.J., North R.A., Surprenant A. 2003. P2X4, P2Y1 and P2Y2 receptors on rat alveolar macrophages. Br. J. Pharmacol. 140:567–575 10.1038/sj.bjp.0705459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne L.E., Jiang L.H., North R.A. 2010. New structure enlivens interest in P2X receptors. Trends Pharmacol. Sci. 31:229–237 10.1016/j.tips.2010.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne L.E., Nunes J.P., Sim J.A., Chudasama V., Bragg L., Caddick S., Alan North R. 2014. Optical control of trimeric P2X receptors and acid-sensing ion channels. Proc. Natl. Acad. Sci. USA. 111:521–526 10.1073/pnas.1318582111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnstock G. 1972. Purinergic nerves. Pharmacol. Rev. 24:509–581 [PubMed] [Google Scholar]
- Chataigneau T., Lemoine D., Grutter T. 2013. Exploring the ATP-binding site of P2X receptors. Front. Cell. Neurosci. 7:273 10.3389/fncel.2013.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont S., Khakh B.S. 2008. Patch-clamp coordinated spectroscopy shows P2X2 receptor permeability dynamics require cytosolic domain rearrangements but not Panx-1 channels. Proc. Natl. Acad. Sci. USA. 105:12063–12068 10.1073/pnas.0803008105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont S., Compan V., Toulme E., Richler E., Housley G.D., Rassendren F., Khakh B.S. 2008. Regulation of P2X2 receptors by the neuronal calcium sensor VILIP1. Sci. Signal. 1:ra8 10.1126/scisignal.1162329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coddou C., Stojilkovic S.S., Huidobro-Toro J.P. 2011a. Allosteric modulation of ATP-gated P2X receptor channels. Rev. Neurosci. 22:335–354 10.1515/rns.2011.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coddou C., Yan Z., Obsil T., Huidobro-Toro J.P., Stojilkovic S.S. 2011b. Activation and regulation of purinergic P2X receptor channels. Pharmacol. Rev. 63:641–683 10.1124/pr.110.003129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coull J.A., Beggs S., Boudreau D., Boivin D., Tsuda M., Inoue K., Gravel C., Salter M.W., De Koninck Y. 2005. BDNF from microglia causes the shift in neuronal anion gradient underlying neuropathic pain. Nature. 438:1017–1021 10.1038/nature04223 [DOI] [PubMed] [Google Scholar]
- Dobbs L.G., Gonzalez R., Williams M.C. 1986. An improved method for isolating type II cells in high yield and purity. Am. Rev. Respir. Dis. 134:141–145 [DOI] [PubMed] [Google Scholar]
- Egan T.M., Khakh B.S. 2004. Contribution of calcium ions to P2X channel responses. J. Neurosci. 24:3413–3420 10.1523/JNEUROSCI.5429-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J.A., Girdler G., Khakh B.S. 2004. Time-resolved measurement of state-specific P2X2 ion channel cytosolic gating motions. J. Neurosci. 24:10475–10487 10.1523/JNEUROSCI.3250-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick M., Eschertzhuber S., Haller T., Mair N., Dietl P. 2001. Secretion in alveolar type II cells at the interface of constitutive and regulated exocytosis. Am. J. Respir. Cell Mol. Biol. 25:306–315 10.1165/ajrcmb.25.3.4493 [DOI] [PubMed] [Google Scholar]
- Giraldez T., Hughes T.E., Sigworth F.J. 2005. Generation of functional fluorescent BK channels by random insertion of GFP variants. J. Gen. Physiol. 126:429–438 10.1085/jgp.200509368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales E.B., Kawate T., Gouaux E. 2009. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature. 460:599–604 10.1038/nature08218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber M.B. 2010. Changing face of microglia. Science. 330:783–788 10.1126/science.1190929 [DOI] [PubMed] [Google Scholar]
- Haller T., Ortmayr J., Friedrich F., Völkl H., Dietl P. 1998. Dynamics of surfactant release in alveolar type II cells. Proc. Natl. Acad. Sci. USA. 95:1579–1584 10.1073/pnas.95.4.1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Gouaux E. 2012. Molecular mechanism of ATP binding and ion channel activation in P2X receptors. Nature. 485:207–212 10.1038/nature11010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D., Matthews B.D., Mammoto A., Montoya-Zavala M., Hsin H.Y., Ingber D.E. 2010. Reconstituting organ-level lung functions on a chip. Science. 328:1662–1668 10.1126/science.1188302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K. 2008. Purinergic systems in microglia. Cell. Mol. Life Sci. 65:3074–3080 10.1007/s00018-008-8210-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis M.F. 2010. The neural-glial purinergic receptor ensemble in chronic pain states. Trends Neurosci. 33:48–57 10.1016/j.tins.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Jarvis M.F., Khakh B.S. 2009. ATP-gated P2X cation-channels. Neuropharmacology. 56:208–215 10.1016/j.neuropharm.2008.06.067 [DOI] [PubMed] [Google Scholar]
- Jelínkova I., Vávra V., Jindrichova M., Obsil T., Zemkova H.W., Zemkova H., Stojilkovic S.S. 2008. Identification of P2X4 receptor transmembrane residues contributing to channel gating and interaction with ivermectin. Pflugers Arch. 456:939–950 10.1007/s00424-008-0450-4 [DOI] [PubMed] [Google Scholar]
- Jiang R., Taly A., Grutter T. 2013. Moving through the gate in ATP-activated P2X receptors. Trends Biochem. Sci. 38:20–29 10.1016/j.tibs.2012.10.006 [DOI] [PubMed] [Google Scholar]
- Jones C.A., Chessell I.P., Simon J., Barnard E.A., Miller K.J., Michel A.D., Humphrey P.P.A. 2000. Functional characterization of the P2X4 receptor orthologues. Br. J. Pharmacol. 129:388–394 10.1038/sj.bjp.0703059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek-Hájek K., Lörinczi E., Hausmann R., Nicke A. 2012. Molecular and functional properties of P2X receptors—recent progress and persisting challenges. Purinergic Signal. 8:375–417 10.1007/s11302-012-9314-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawate T., Michel J.C., Birdsong W.T., Gouaux E. 2009. Crystal structure of the ATP-gated P2X4 ion channel in the closed state. Nature. 460:592–598 10.1038/nature08198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh B.S., Burnstock G. 2009. The double life of ATP. Sci. Am. 301:84–92 10.1038/scientificamerican1209-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh B.S., Egan T.M. 2005. Contribution of transmembrane regions to ATP-gated P2X2 channel permeability dynamics. J. Biol. Chem. 280:6118–6129 10.1074/jbc.M411324200 [DOI] [PubMed] [Google Scholar]
- Khakh B.S., North R.A. 2012. Neuromodulation by extracellular ATP and P2X receptors in the CNS. Neuron. 76:51–69 10.1016/j.neuron.2012.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh B.S., Proctor W.R., Dunwiddie T.V., Labarca C., Lester H.A. 1999. Allosteric control of gating and kinetics at P2X4 receptor channels. J. Neurosci. 19:7289–7299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh B.S., Smith W.B., Chiu C.S., Ju D., Davidson N., Lester H.A. 2001. Activation-dependent changes in receptor distribution and dendritic morphology in hippocampal neurons expressing P2X2-green fluorescent protein receptors. Proc. Natl. Acad. Sci. USA. 98:5288–5293 10.1073/pnas.081089198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh B.S., Fisher J.A., Nashmi R., Bowser D.N., Lester H.A. 2005. An angstrom scale interaction between plasma membrane ATP-gated P2X2 and α4β2 nicotinic channels measured with fluorescence resonance energy transfer and total internal reflection fluorescence microscopy. J. Neurosci. 25:6911–6920 10.1523/JNEUROSCI.0561-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine D., Habermacher C., Martz A., Méry P.F., Bouquier N., Diverchy F., Taly A., Rassendren F., Specht A., Grutter T. 2013. Optical control of an ion channel gate. Proc. Natl. Acad. Sci. USA. 110:20813–20818 10.1073/pnas.1318715110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Tsien R.W. 2012. pHTomato, a red, genetically encoded indicator that enables multiplex interrogation of synaptic activity. Nat. Neurosci. 15:1047–1053 10.1038/nn.3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lörinczi É., Bhargava Y., Marino S.F., Taly A., Kaczmarek-Hájek K., Barrantes-Freer A., Dutertre S., Grutter T., Rettinger J., Nicke A. 2012. Involvement of the cysteine-rich head domain in activation and desensitization of the P2X1 receptor. Proc. Natl. Acad. Sci. USA. 109:11396–11401 10.1073/pnas.1118759109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mealer R., Butler H., Hughes T. 2008. Functional fusion proteins by random transposon-based GFP insertion. Methods Cell Biol. 85:23–44 10.1016/S0091-679X(08)85002-9 [DOI] [PubMed] [Google Scholar]
- Miesenböck G. 2012. Synapto-pHluorins: Genetically encoded reporters of synaptic transmission. Cold Spring Harb. Protoc. 2012:213–217 10.1101/pdb.ip067827 [DOI] [PubMed] [Google Scholar]
- Miesenböck G., De Angelis D.A., Rothman J.E. 1998. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 394:192–195 10.1038/28190 [DOI] [PubMed] [Google Scholar]
- Migita K., Haines W.R., Voigt M.M., Egan T.M. 2001. Polar residues of the second transmembrane domain influence cation permeability of the ATP-gated P2X2 receptor. J. Biol. Chem. 276:30934–30941 10.1074/jbc.M103366200 [DOI] [PubMed] [Google Scholar]
- Miklavc P., Wittekindt O.H., Felder E., Dietl P. 2009. Ca2+-dependent actin coating of lamellar bodies after exocytotic fusion: A prerequisite for content release or kiss-and-run. Ann. NY Acad. Sci. 1152:43–52 10.1111/j.1749-6632.2008.03989.x [DOI] [PubMed] [Google Scholar]
- Miklavc P., Mair N., Wittekindt O.H., Haller T., Dietl P., Felder E., Timmler M., Frick M. 2011. Fusion-activated Ca2+ entry via vesicular P2X4 receptors promotes fusion pore opening and exocytotic content release in pneumocytes. Proc. Natl. Acad. Sci. USA. 108:14503–14508 10.1073/pnas.1101039108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklavc P., Thompson K.E., Frick M. 2013. A new role for P2X4 receptors as modulators of lung surfactant secretion. Front. Cell. Neurosci. 7:171 10.3389/fncel.2013.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan E.D., Watkins L.R. 2009. Pathological and protective roles of glia in chronic pain. Nat. Rev. Neurosci. 10:23–36 10.1038/nrn2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell-Lagnado R. 2009. More cross-talk between purinergic receptors. J. Physiol. 587:2713–2714 10.1113/jphysiol.2009.174961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell-Lagnado R.D., Qureshi O.S. 2008. Assembly and trafficking of P2X purinergic receptors (Review). Mol. Membr. Biol. 25:321–331 10.1080/09687680802050385 [DOI] [PubMed] [Google Scholar]
- Nicke A., Bäumert H.G., Rettinger J., Eichele A., Lambrecht G., Mutschler E., Schmalzing G. 1998. P2X1 and P2X3 receptors form stable trimers: a novel structural motif of ligand-gated ion channels. EMBO J. 17:3016–3028 10.1093/emboj/17.11.3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North R.A. 2002. Molecular physiology of P2X receptors. Physiol. Rev. 82:1013–1067 [DOI] [PubMed] [Google Scholar]
- North R.A., Jarvis M.F. 2013. P2X receptors as drug targets. Mol. Pharmacol. 83:759–769 10.1124/mol.112.083758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrovskaya O., Asatryan L., Wyatt L., Popova M., Li K., Peoples R.W., Alkana R.L., Davies D.L. 2011. Ethanol is a fast channel inhibitor of P2X4 receptors. J. Pharmacol. Exp. Ther. 337:171–179 10.1124/jpet.110.176990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson K., Baines A.E., Keller T., Gruenheit N., Bragg L., North R.A., Thompson C.R. 2014. Calcium-dependent regulation of Rab activation and vesicle fusion by an intracellular P2X ion channel. Nat. Cell Biol. 16:87–98 10.1038/ncb2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priel A., Silberberg S.D. 2004. Mechanism of ivermectin facilitation of human P2X4 receptor channels. J. Gen. Physiol. 123:281–293 10.1085/jgp.200308986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi O.S., Paramasivam A., Yu J.C., Murrell-Lagnado R.D. 2007. Regulation of P2X4 receptors by lysosomal targeting, glycan protection and exocytosis. J. Cell Sci. 120:3838–3849 10.1242/jcs.010348 [DOI] [PubMed] [Google Scholar]
- Richler E., Chaumont S., Shigetomi E., Sagasti A., Khakh B.S. 2008. Tracking transmitter-gated P2X cation channel activation in vitro and in vivo. Nat. Methods. 5:87–93 10.1038/nmeth1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richler E., Shigetomi E., Khakh B.S. 2011. Neuronal P2X2 receptors are mobile ATP sensors that explore the plasma membrane when activated. J. Neurosci. 31:16716–16730 10.1523/JNEUROSCI.3362-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson L.E., Murrell-Lagnado R.D. 2013. The trafficking and targeting of P2X receptors. Front. Cell. Neurosci. 7:233 10.3389/fncel.2013.00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle S.J., Murrell-Lagnado R.D. 2003. Constitutive cycling: A general mechanism to regulate cell surface proteins. Bioessays. 25:39–46 10.1002/bies.10200 [DOI] [PubMed] [Google Scholar]
- Royle S.J., Bobanović L.K., Murrell-Lagnado R.D. 2002. Identification of a non-canonical tyrosine-based endocytic motif in an ionotropic receptor. J. Biol. Chem. 277:35378–35385 10.1074/jbc.M204844200 [DOI] [PubMed] [Google Scholar]
- Royle S.J., Qureshi O.S., Bobanović L.K., Evans P.R., Owen D.J., Murrell-Lagnado R.D. 2005. Non-canonical YXXGΦ endocytic motifs: recognition by AP2 and preferential utilization in P2X4 receptors. J. Cell Sci. 118:3073–3080 10.1242/jcs.02451 [DOI] [PubMed] [Google Scholar]
- Samways D.S., Egan T.M. 2007. Acidic amino acids impart enhanced Ca2+ permeability and flux in two members of the ATP-gated P2X receptor family. J. Gen. Physiol. 129:245–256 10.1085/jgp.200609677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samways D.S., Khakh B.S., Dutertre S., Egan T.M. 2011. Preferential use of unobstructed lateral portals as the access route to the pore of human ATP-gated ion channels (P2X receptors). Proc. Natl. Acad. Sci. USA. 108:13800–13805 10.1073/pnas.1017550108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samways D.S., Khakh B.S., Egan T.M. 2012. Allosteric modulation of Ca2+ flux in ligand-gated cation channel (P2X4) by actions on lateral portals. J. Biol. Chem. 287:7594–7602 10.1074/jbc.M111.322461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samways D.S., Li Z., Egan T.M. 2014. Principles and properties of ion flow in P2X receptors. Front. Cell. Neurosci. 8:6 10.3389/fncel.2014.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan S., De Angelis D., Rothman J.E., Ryan T.A. 2000. The use of pHluorins for optical measurements of presynaptic activity. Biophys. J. 79:2199–2208 10.1016/S0006-3495(00)76468-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneggenburger R., Zhou Z., Konnerth A., Neher E. 1993. Fractional contribution of calcium to the cation current through glutamate receptor channels. Neuron. 11:133–143 10.1016/0896-6273(93)90277-X [DOI] [PubMed] [Google Scholar]
- Sekine-Aizawa Y., Huganir R.L. 2004. Imaging of receptor trafficking by using α-bungarotoxin-binding-site-tagged receptors. Proc. Natl. Acad. Sci. USA. 101:17114–17119 10.1073/pnas.0407563101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan D.L., Berlot C.H., Robert A., Inglis F.M., Jakobsdottir K.B., Howe J.R., Hughes T.E. 2002. A new way to rapidly create functional, fluorescent fusion proteins: random insertion of GFP with an in vitro transposition reaction. BMC Neurosci. 3:7 10.1186/1471-2202-3-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigetomi E., Khakh B.S. 2009. Measuring near plasma membrane and global intracellular calcium dynamics in astrocytes. J. Vis. Exp. pii:1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava A.N., Triller A., Sieghart W., Sarto-Jackson I. 2011. Regulation of GABAA receptor dynamics by interaction with purinergic P2X2 receptors. J. Biol. Chem. 286:14455–14468 10.1074/jbc.M110.165282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava A.N., Rodriguez P.C., Triller A., Renner M. 2013. Dynamic micro-organization of P2X7 receptors revealed by PALM based single particle tracking. Front. Cell. Neurosci. 7:232 10.3389/fncel.2013.00232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberberg S.D., Li M., Swartz K.J. 2007. Ivermectin interaction with transmembrane helices reveals widespread rearrangements during opening of P2X receptor channels. Neuron. 54:263–274 10.1016/j.neuron.2007.03.020 [DOI] [PubMed] [Google Scholar]
- Srinivasan R., Richards C.I., Xiao C., Rhee D., Pantoja R., Dougherty D.A., Miwa J.M., Lester H.A. 2012. Pharmacological chaperoning of nicotinic acetylcholine receptors reduces the endoplasmic reticulum stress response. Mol. Pharmacol. 81:759–769 10.1124/mol.112.077792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes L., Surprenant A. 2009. Dynamic regulation of the P2X4 receptor in alveolar macrophages by phagocytosis and classical activation. Eur. J. Immunol. 39:986–995 10.1002/eji.200838818 [DOI] [PubMed] [Google Scholar]
- Thompson K.E., Korbmacher J.P., Hecht E., Hobi N., Wittekindt O.H., Dietl P., Kranz C., Frick M. 2013. Fusion-activated cation entry (FACE) via P2X4 couples surfactant secretion and alveolar fluid transport. FASEB J. 27:1772–1783 10.1096/fj.12-220533 [DOI] [PubMed] [Google Scholar]
- Toulme E., Khakh B.S. 2012. Imaging P2X4 receptor lateral mobility in microglia: Regulation by calcium and p38 MAPK. J. Biol. Chem. 287:14734–14748 10.1074/jbc.M111.329334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmé E., Soto F., Garret M., Boué-Grabot E. 2006. Functional properties of internalization-deficient P2X4 receptors reveal a novel mechanism of ligand-gated channel facilitation by ivermectin. Mol. Pharmacol. 69:576–587 10.1124/mol.105.018812 [DOI] [PubMed] [Google Scholar]
- Toulme E., Garcia A., Samways D., Egan T.M., Carson M.J., Khakh B.S. 2010. P2X4 receptors in activated C8-B4 cells of cerebellar microglial origin. J. Gen. Physiol. 135:333–353 10.1085/jgp.200910336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M., Shigemoto-Mogami Y., Koizumi S., Mizokoshi A., Kohsaka S., Salter M.W., Inoue K. 2003. P2X4 receptors induced in spinal microglia gate tactile allodynia after nerve injury. Nature. 424:778–783 10.1038/nature01786 [DOI] [PubMed] [Google Scholar]
- Ulmann L., Hatcher J.P., Hughes J.P., Chaumont S., Green P.J., Conquet F., Buell G.N., Reeve A.J., Chessell I.P., Rassendren F. 2008. Up-regulation of P2X4 receptors in spinal microglia after peripheral nerve injury mediates BDNF release and neuropathic pain. J. Neurosci. 28:11263–11268 10.1523/JNEUROSCI.2308-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmann L., Hirbec H., Rassendren F. 2010. P2X4 receptors mediate PGE2 release by tissue-resident macrophages and initiate inflammatory pain. EMBO J. 29:2290–2300 10.1038/emboj.2010.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmann L., Levavasseur F., Avignone E., Peyroutou R., Hirbec H., Audinat E., Rassendren F. 2013. Involvement of P2X4 receptors in hippocampal microglial activation after status epilepticus. Glia. 61:1306–1319 10.1002/glia.22516 [DOI] [PubMed] [Google Scholar]
- Wyatt L.R., Godar S.C., Khoja S., Jakowec M.W., Alkana R.L., Bortolato M., Davies D.L. 2013. Sociocommunicative and sensorimotor impairments in male P2X4-deficient mice. Neuropsychopharmacology. 38:1993–2002 10.1038/npp.2013.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A., Sonin D., Jones L., Barry W.H., Liang B.T. 2004. A beneficial role of cardiac P2X4 receptors in heart failure: rescue of the calsequestrin overexpression model of cardiomyopathy. Am. J. Physiol. Heart Circ. Physiol. 287:H1096–H1103 10.1152/ajpheart.00079.2004 [DOI] [PubMed] [Google Scholar]
- Yardley M.M., Wyatt L., Khoja S., Asatryan L., Ramaker M.J., Finn D.A., Alkana R.L., Huynh N., Louie S.G., Petasis N.A., et al. 2012. Ivermectin reduces alcohol intake and preference in mice. Neuropharmacology. 63:190–201 10.1016/j.neuropharm.2012.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.T., Fisher J.A., Fountain S.J., Ford R.C., North R.A., Khakh B.S. 2008. Molecular shape, architecture, and size of P2X4 receptors determined using fluorescence resonance energy transfer and electron microscopy. J. Biol. Chem. 283:26241–26251 10.1074/jbc.M804458200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinchuk V., Zinchuk O. 2008. Quantitative colocalization analysis of confocal fluorescence microscopy images. Curr. Protoc. Cell Biol. 4:4.19 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.