The loops of the bacterial outer membrane iron transporter FepA move at different rates to adsorb and grasp the substrate ferric enterobactin before transporting it into the periplasm.
Abstract
Spectroscopic analyses of fluorophore-labeled Escherichia coli FepA described dynamic actions of its surface loops during binding and transport of ferric enterobactin (FeEnt). When FeEnt bound to fluoresceinated FepA, in living cells or outer membrane fragments, quenching of fluorophore emissions reflected conformational motion of the external vestibular loops. We reacted Cys sulfhydryls in seven surface loops (L2, L3, L4, L5, L7 L8, and L11) with fluorophore maleimides. The target residues had different accessibilities, and the labeled loops themselves showed variable extents of quenching and rates of motion during ligand binding. The vestibular loops closed around FeEnt in about a second, in the order L3 > L11 > L7 > L2 > L5 > L8 > L4. This sequence suggested that the loops bind the metal complex like the fingers of two hands closing on an object, by individually adsorbing to the iron chelate. Fluorescence from L3 followed a biphasic exponential decay as FeEnt bound, but fluorescence from all the other loops followed single exponential decay processes. After binding, the restoration of fluorescence intensity (from any of the labeled loops) mirrored cellular uptake that depleted FeEnt from solution. Fluorescence microscopic images also showed FeEnt transport, and demonstrated that ferric siderophore uptake uniformly occurs throughout outer membrane, including at the poles of the cells, despite the fact that TonB, its inner membrane transport partner, was not detectable at the poles.
INTRODUCTION
Enzymes often facilitate chemical reactions by induced fit (Koshland, 1958), which adapts the structure of the macromolecule to the form of its substrate such that their binding energy reduces the activation energy of the transition state to product formation. In induced fit, initial weak interactions between an enzyme and substrate promote conformational changes in the catalyst, usually a protein, that strengthen the binding interaction. Induced fit also encompasses ligand recognition by membrane receptors and transporters, in that conformational change is often a fundamental part of their biochemistry (Yu and Koshland, 2001; Smirnova et al., 2009). Previous findings infer induced fit in the actions of bacterial transporters (Yu and Koshland, 2001; Scott et al., 2002, Ferguson et al., 2002; James et al., 2008; Yukl et al., 2010), but this behavior is not yet understood in the context of their binding and transport activities.
The Escherichia coli cell envelope contains inner (IM) and outer (OM) membranes, separated by the periplasmic space. The selective permeability of the OM both protects Gram-negative bacteria from antagonistic molecules and facilitates the uptake of nutrients. Small (<600 D) molecules diffuse through the hydrophilic channels of general porins (Nikaido and Vaara, 1985), but larger compounds enter by other mechanisms. Siderophores (Neilands, 1995) like ferric enterobactin (FeEnt; 716 D; Pollack and Neilands, 1970) generally exceed this mass cutoff and do not penetrate porin channels. Based on their 22-stranded transmembrane β-barrels, FepA and its relatives are in the porin superfamily (Yen et al., 2002), but their channels are blocked by a 150-aa N-terminal globular domain (N-domain), their uptake reactions require the additional protein TonB, and their binding interactions with ligands potentiate their transport activity. Hence, they are called TonB-dependent transporters (TBDTs; Schauer et al., 2008) or ligand-gated porins (LGPs; Rutz et al., 1992; Newton et al., 2010). Besides TonB dependence, TBDTs accomplish proton-motive force (PMF)–driven active transport, which is unexpected in the OM bilayer because its >105 porin channels preclude the formation of a transmembrane ion gradient. Nevertheless, both radioisotopic (Bradbeer, 1993; Newton et al., 2010) and spectroscopic (Cao et al., 2003) experiments demonstrated OM active transport of metal complexes. Hence, FepA requires both PMF from the IM, and a proposed IM complex composed of TonB, ExbB, and ExbD to transport FeEnt or confer susceptibility to colicins B and D (Klebba, 2003). The corequirements for TonB and energy suggested that the former protein transfers PMF from the IM through the periplasm to the OM (Konisky, 1979). Biophysical evidence now exists supporting this idea: TonB undergoes energized, likely rotational motion, and ExbBD controls these movements (Jordan et al., 2013).
Many or all of the aforementioned activities of FepA and its homologues involve conformational change in the transporters themselves. Although the detailed mechanisms of TBDT remain uncertain, their structural characteristics are known. The architectures of the paralogous proteins in this family are equivalent to FepA, but certain residues in each protein confer individual specificities and high affinities for their different ligands. The structural congruency and biochemical similarities of the E. coli proteins BtuB (Chimento et al., 2003; Kurisu et al., 2003), FhuA (Ferguson et al., 1998; Locher et al., 1998), FecA (Ferguson et al., 2002), FepA (Buchanan et al., 1999), and Cir (Buchanan et al., 2007) denote a common mode of ligand transport in two stages: high affinity binding by surface loops, followed by energy-dependent interactions with TonB that propel the metal complex through the transmembrane pore. Both these processes require conformational motion. The movement of surface loops confines the ligand in a closed vestibule (Ferguson et al., 2002; Scott et al., 2002; Cobessi et al., 2010; Yukl et al., 2010) and propagates motion through the receptor’s tertiary structure and across the OM. This signal transduction reorients and exposes the TonB box to allow its binding by the TonB C terminus (Pawelek et al., 2006; Shultis et al., 2006), presumably initiating reorganization or displacement of the N domain (Ma et al., 2007) that allows metal transit through the pore. These proposed energy-dependent protein movements drive and regulate iron transport.
Binding of FeEnt to FepA entails hydrophobic interactions with aromatic residues in the external loops (Cao et al., 2000; Annamalai et al., 2004) and subsequent electrostatic bonds with basic residues deeper in the vestibule (Newton et al., 1997; Scott et al., 2002; Annamalai et al., 2004). At binding equilibrium, the loops enclose the metal complex atop the N domain, poised for transport. FeEnt binding is TonB-independent (Annamalai et al., 2004) and has two kinetic phases (Payne et al., 1997). We used site-directed fluorescence spectroscopy to characterize the motion of seven individual loops of FepA during its interactions with FeEnt in cells and membrane fragments. We generated Cys substitutions in these surface loops, labeled their side chains with fluorophore maleimides, and determined the extents, rates, and TonB dependency of fluorescence quenching in them during FeEnt binding and uptake. The data revealed a hierarchy of loop motion and interaction with the metal complex that translated into an elaborate process of induced fit to high-affinity binding, and further defined fluorescence spectroscopic methods of measuring membrane transport.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture conditions, and FeEnt
BN1071 (F- pro, trp, B1 entA; Klebba et al., 1982) is the parent of the plasmid host strains OKN3 (BN1071 ΔfepA; Ma et al., 2007) and OKN13 (BN1071 ΔfepA,Δ tonB; Ma et al., 2007). Plasmids pITS23 (Ma et al., 2007) and pITS47 (Smallwood et al., 2009) are derivatives of the low-copy plasmid pHSG575 (Hashimoto-Gotoh et al., 1986) that carries the wild-type fepA+ structural gene, in which we engineered mutations by QuikChange (Agilent Technologies) to introduce site-directed Cys residues (Ma et al., 2007; Smallwood et al., 2009) in the resulting FepA proteins. For spectroscopic experiments, we grew bacteria overnight at 37°C with shaking in Luria-Bertani (LB) broth (Miller, 1972) with streptomycin (100 µg/ml) and chloramphenicol (20 µg/ml), then subcultured (1%) into iron-deficient MOPS minimal media (henceforth called “MOPS media”; Neidhardt et al., 1974) with the same antibiotics. The MOPS cultures were grown with shaking at 37°C for 5.5–6 h, until cell density reached 109 cells/ml. We pelleted the cells by centrifugation at 8,000 g for 10 min at 4°C.
We purified enterobactin from E. coli strain AN102 (Cox et al., 1970), prepared its ferric complex (Wayne and Neilands, 1975; Klebba et al., 1982), and stored the ferric siderophore on ice. When FeEnt solutions became oxidized or degraded, we repurified the authentic metal complex by chromatography on a Sephadex LH-20 (Wayne and Neilands, 1975) and determined its concentration from its absorbance at 495 nm.
Selection of sites for Cys modification and phenotypic assays
We screened existing (Liu et al., 1994; Jiang et al., 1997; Payne et al., 1997; Cao et al., 2003; Ma et al., 2007; Smallwood et al., 2009) and new Cys mutants (in L3 [S275C], L4 [G327C], L7 [S490C], L10 [G640C, T648C]) to identify accessible, modifiable sulfhydryls. The FepA Cys mutants reacted to different extents with fluorophore-maleimides. We monitored their phenotypes with siderophore nutrition (Wayne and Neilands, 1975) and colicin killing (Wayne et al., 1976) assays that qualitatively show FeEnt uptake and colicin B (ColB) susceptibility, respectively. We also performed quantitative determinations of 59FeEnt uptake (Newton et al., 1999) by the mutant FepA transporters, both before and after fluorescent modification.
In vivo fluorescence labeling and characterization by fluorescence spectroscopy
During binding the surface loops of FepA enclose FeEnt, altering the environment of covalently attached fluorophores (Scott et al., 2002; Cao et al., 2003; Smallwood et al., 2009). We measured FeEnt-mediated quenching of fluorophores attached to seven different surface loops. After growing 10-ml bacterial cultures in iron-deficient MOPS media to late log (109 cells/ml), we collected 1010 cells by centrifugation, and washed and resuspended them in 10 ml of ice-cold labeling buffer (50 mM Na2HPO4, 0.9% NaCl, pH 6.5). We exposed the cells in labeling buffer to 5 µM fluorescein-5-maleimide (FM) or Alexa Fluor 488 maleimide (A488M; Invitrogen) for 5 min at 37°C. After stopping the labeling reactions with 100 mM β-mercaptoethanol (BME), we pelleted the cells by centrifugation, and washed and resuspended them in PBS containing 0.4% glucose. We observed the fluorophore–labeled cells in a fluorescence spectrometer (SLM AMINCO 8000; SLM Instruments), upgraded with an OLIS operating system and software (OLIS SpectralWorks) to control shutters, polarizers, and data collection. For quenching experiments, we added FeEnt to 2.5 × 107 labeled cells in a 3-ml volume with stirring at 2°C. For FeEnt uptake studies in the same format, we raised the temperature to 25°C and monitored the time course of fluorescence emissions at 520 nm. For binding kinetics experiments, we measured quenching at 520 nm by rapid mixing in an OLIS stopped-flow chamber, with OM fragments containing the fluorescent FepA proteins, at 0°C.
Fluorescence quenching data were analyzed and fitted using GraFit 6.011 (Erithacus Software) by single (A(t) = A0e−kt + offset) or double (A(t) = A0(1)e−k1t + A0(2)e−k2t + offset) exponential decay equations (Payne et al., 1997). We analyzed fluorescence recovery curves that resulted from FeEnt transport according to −log (F/F0), and fitted them with a single exponential decay equation with offset (A(t) = A0e−kt + offset) that allowed calculation of the recovery half times for each FM-labeled loop.
Preparation of OM fragments
For kinetic analysis of FeEnt binding to FepA in the OM, we broke the cells and collected the cell envelope fraction. Passage of Gram-negative bacteria through a French press at 14,000 psi breaks the cells, concomitantly separating the IM and OM from one another into vesicle-like structures (Smit et al., 1975). It’s more appropriate to call them IM and OM fragments, because we did not demonstrate their physical integrity as vesicles. After a low-speed centrifugation to remove unbroken cells and debris, we directly used the lysates, containing fluoresceinated FepA in the native environment of the OM, in stopped flow studies. Only FepA was fluoresceinated in the OM fraction (Ma et al., 2007), and IM fragments and cytoplasmic proteins in the preparation did not hinder data collection (Fig. S6).
Radioisotopic binding and transport assays
59FeEnt binding and transport measurements (Newton et al., 1999) revealed the biochemical activities of FepA Cys mutants, before and after fluoresceination. Analysis of 59FeEnt binding data from cells in MOPS media at 0°C, with the bound-versus-total equation of GraFit 6.011, gave Kd and capacity values. Analysis of 59FeEnt transport data from cells in MOPS media containing 0.4% glucose at 37°C, with the enzyme kinetics equation of GraFit 6.011, gave KM and Vmax values for each sample. For convenience and uniformity in comparison of different strains, we measured these parameters in 10-ml volumes of bacterial cultures, which yields slightly inflated Kd and KM values (∼1–2 nM) relative to the affinity from determinations in 25-ml volumes (0.2 nM; Newton et al., 1999).
SDS-PAGE, fluorescence imaging, and quantitative immunoblots
Bacterial cell proteins were subjected to SDS-PAGE (Ames, 1974; Newton et al., 1999). Samples were suspended in SDS-containing sample buffer plus 3% β-mercaptoethanol, boiled for 5 min, and electrophoresed at room temperature.
We determined the efficiency of fluoresceination of individual FepA Cys substitutions in live cells by measuring the extent of FM labeling relative to the level of FepA expression. After growth in MOPS media to late log and labeling with FM or AM, we spectrophotometrically measured cell density at 600 nM, collected 5 × 108 cells of each strain by centrifugation, resuspended the cells in 150 µl of SDS-PAGE sample buffer, boiled samples for 5 min, briefly centrifuged the samples to remove debris, and subjected 30 µl (the lysate of 108 cells) to SDS-PAGE. We processed the gels in two stages. For quantification of fluorescence we briefly rinsed them with water, scanned them with a Typhoon imager (GE Healthcare) to record emissions at 520 nm, and analyzed the results with ImageQuant software (version 5.0; Molecular Dynamics). For determination of FepA expression levels, we performed quantitative Western immunoblots. We transferred the proteins from the same gels to nitrocellulose paper, blocked the papers with 50 mM Tris chloride, pH 7.5, containing 0.9% NaCl and 1% gelatin (TBSG), incubated them with mouse anti-FepA mAbs 41 and 45 (0.5%; Murphy et al., 1990) in TBSG for 1 h, washed them five times with TBS containing 0.05% Tween 20 (TBST), and exposed the nitrocellulose to [125I]protein A (Newton et al., 1999) in TBSG for 1 h. We discarded the [125I]protein A, washed the paper three times with tap water, dried the nitrocellulose, exposed it to a phosphorescent screen, and analyzed the screen on a Typhoon phosphoimager (GE Healthcare). The 125I decay from the bands in the images provided a direct measurement of the amount of FepA in each sample. We quantified the amounts (using ImageQuant) relative to internal standards of purified FepA included in the same gels/immunoblots.
Confocal microscopy
Fluorescence microscopy experiments were performed on a confocal laser scanning microscope (LSM 700; Carl Zeiss) in the Confocal Microfluorometry and Microscopy Core Facility at Kansas State University. OKN13/pGT/pfepAS271C (Jordan et al., 2013), which encodes GFP-TonB, was grown in MOPS media for 5–6 h, labeled with A546M at an OD600 nm = 1.0, and resuspended in PBS, pH 7.4. The bacterial cells were adhered to poly-l-lysine–coated, 8-well microslides (Ibidi Inc.) and microscopically observed for changes in fluorescence after introduction of FeEnt. 5 × 108 cells in 350 µl of PBS were observed before and after the addition of 5 nM FeEnt. GFP and A546M were sequentially excited with 488-nm and 555-nm laser light, respectively, with a total scan time of 20 s. Images were collected at 100× magnification over a 23-min time course.
Online supplemental material
Table S1 and Fig. S1 show additional data regarding optimization of fluorescence labeling. Fig. S2 shows the concentration dependence of FeEnt quenching. Table S2 shows the efficiency of FepA function before and after fluoresceination. Figs. S3–S5 show FepA fluoresceination and expression in tonB+ and ΔtonB bacteria. Fig. S6 shows raw data from stopped-flow fluorescence quenching measurements. Online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201311159/DC1.
RESULTS
Site-directed fluorescence labeling of Cys substitutions in the surface loops of FepA
We determined the reactivity of single Cys sulfhydryls at various positions in FepA to modification by fluorophore maleimides (FM, A546M, A555M, and A680M). Certain sulfhydryls in the surface loops and periplasmic regions of the N-terminal globule and the C-terminal β-barrel were well labeled by the fluorophores (Table S1), including stoichiometric labeling of several Cys substitutions in the outer vestibule by FM or the Alexa Fluors. We used FM to optimize the labeling reactions by assessing modification of surface residue S271C. The results showed best selectivity and satisfactory reactivity at pH 6.5, in as little as 1–5 min (Fig. S1). This pH minimizes maleimide reactivity with Lys. Temperature variations had little impact: we achieved comparable results at 0 or 37°C.
The procedure specifically labeled cell surface sulfhydryls with the maleimide fluorophores (Figs. 1 and S1). Efficiency of Cys modification on the periplasmic surface of FepA depended on the mass of the fluorophore. FM (427 D) penetrated the OM (through OmpF/C channels; Ma et al., 2007) and labeled Cys substitutions on the internal rim of the FepA β-barrel (at S150, T666), and on the underside of the N domain (I14, T30, T32, and A33). However, the larger reagent A546M (1,040 D) was size-excluded from this route and did not label periplasmic Cys residues (unpublished data). FM is within the size limit of porin channels, but at the low external levels (i.e., 1–5 µM) used for modification of surface sulfhydryls it only weakly labeled periplasmic targets. Higher concentrations of FM (300 µM) quantitatively labeled the periplasmic sites T30C, T32C, A33C, and T666C (Table S1), but without exclusivity for FepA: other cellular proteins were also modified at the higher FM levels (unpublished data).
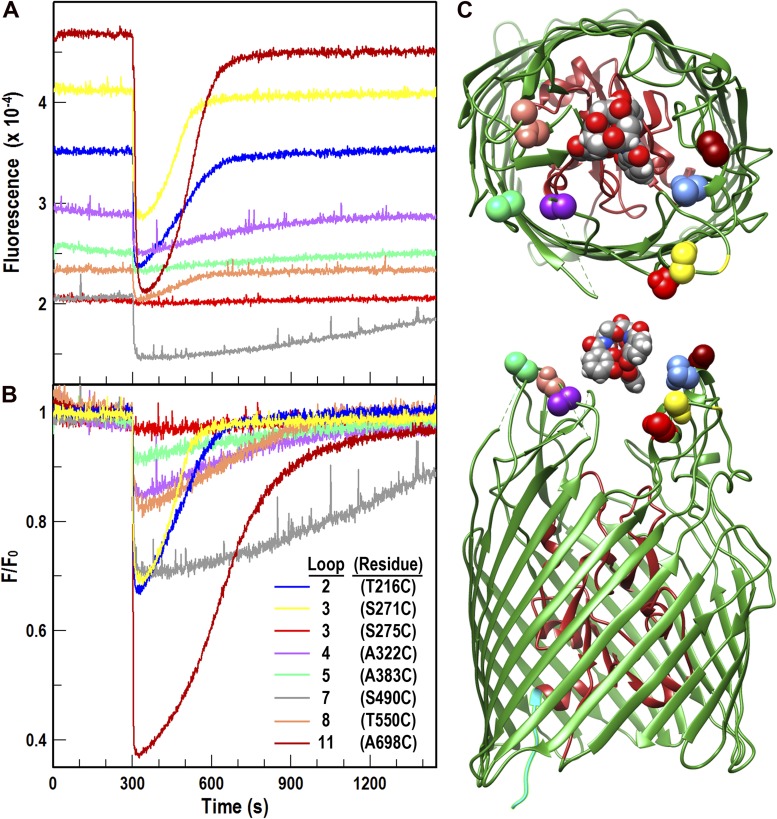
Figure 1.
FeEnt-induced quenching of FepA surface loop–FM fluorescence, and recovery from FeEnt transport. (A) Raw data. Bacteria expressing FepA Cys substitution mutants were grown in MOPS media, and each individual site-directed Cys mutant was modified by FM (see Materials and methods). We then added 10 nM FeEnt to 2.5 × 107 cells/ml in MOPS media plus 0.4% glucose at t = 300 s, and aligned the raw fluorescence time courses. Fluorescence intensity readings showed characteristic extents of quenching and subsequent recovery, as a result of FeEnt uptake from the media. Data from T216C (L2, blue), S271C (L3, yellow), S275C (L3, red), A322C (L4, purple), A383C, (L5, green), S490C (L7, gray), T550C (L8, flesh), and A698C (L11, dark red) were analyzed and plotted in GraFit 6.011. (B) Normalized data. We normalized results from A to the initial fluorescence of the cells before FeEnt addition, and replotted the quenching and recovery curves. F/F0 revealed different relative extents of quenching and rates of recovery (Table S2). The data were analyzed and plotted in GraFit 6.011. (C) Structural models. The structural illustrations are models. The precise location of FeEnt was not solved within the original crystal structure of FepA (Buchanan et al., 1999), and the pictures depict the size of FeEnt relative to that of FepA, and the positions of the target residues relative to the predicted path of FeEnt into the outer vestibule.
Single Cys substitution mutations usually did not impair expression of FepA, nor its ability to bind and transport ferric enterobactin, but in individual cases we observed some variations in these results. Fluorescent modification did not occur at numerous sites in the protein interior (Ma et al., 2007; Smallwood et al., 2009), and labeling of a few residues inhibited 59FeEnt binding and transport (Table S1). The cell surface labeling sites, nevertheless, were usually free of such detrimental effects. Based on these and other data, we chose seven Cys substitutions for study of loop motion: T216 (Loop 2), S271 (L3), A322 (L4), A383 (L5), S490 (L7), T550 (L8), and A698 (L11; Fig. 1). These had comparable functionality to wild-type FepA before modification, and they bound and transported FeEnt after modification (Table S1), although fluoresceination sometimes reduced their transport rate. We assessed both FM and A488M for modification of the target sites. The pH insensitivity and photostability of A488M are advantageous, but we generally found better labeling efficiency with FM, less impairment of FepA functionality, and no significant differences in experimental outcomes between the two reagents (unpublished data). Hence we used the more cost-effective reagent FM, and after the labeling reactions we performed the biochemical experiments in phosphate-buffered solutions at pH 7.4, where FM has a stable, high quantum yield.
Loop motion during FeEnt binding and transport by FepA
FeEnt binding to FepA-FM quenches fluorescence from loop motion that changes the environment of the probe (Payne et al., 1997; Cao et al., 2003). Subsequent uptake of the ferric siderophore by the bacteria depletes it from solution, allowing fluorescence to rebound to its original level. We characterized seven fluoresceinated loops during this FeEnt binding and transport time course.
Light emissions from the FM-labeled Cys residues occurred to different extents (presumably determined by their local environments), yielding about a fivefold range of initial fluorescence intensity of the bacterial cells (Fig. 1 A). Second, FM attached to individual loops showed different degrees of quenching during FeEnt binding. We recorded emissions from FM-labeled FepA mutants T216C, S271C, S275C, A322C, A383C, S490C, T550C, and A698C (2.5 × 107 cells/ml in a 3-ml cuvette) before and after the addition of 10 nM FeEnt (at 300 s; Fig. 1, A and B). Binding of the ferric siderophore quenched 10–60% of the original intensity of FM attached to most loops (Fig. 1 B). The iron complex quenched FM more in L2, L3, and L11, and less in L4, L5, L7, and L8. The different reductions reflected the locations of the fluorophore in the loops, as well as individual motions of the loops themselves (see the following paragraph and the Discussion section). A few targets were too poorly labeled to analyze (G327C [L4], G640C and T648C [L10]), whereas one (S275C [L3]; Fig. 1) was fluoresceinated but unaffected by FeEnt. We did not further study these mutants.
At physiological temperatures the bacteria transported FeEnt and depleted it from solution, reverting fluorescence to its original level. This recovery reaction had several attributes. (1) Its energy and TonB dependence (for all the mutants; unpublished data) indicated that it derived from FeEnt transport. (2) The elapsed time to recovery depended on the initial FeEnt concentration (Fig. S2). (3) The individual rates of FeEnt uptake, estimated by the progress of fluorescence recovery, varied as much as 10-fold among the FM-labeled Cys loop mutants (Fig. 1 and Table S2). Recovery half-times ranged from 70 to 700 s at 25°C, which indicates that mutagenesis or fluoresceination at some sites impaired the ability of FepA to transport FeEnt (A322C-FM, A383C-FM, S490C-FM, and T550C-FM; Table S2). We further studied these effects with 59FeEnt.
Correspondence between fluorescence spectroscopic and radioisotopic measurements of FeEnt uptake
To assess the effects of fluoresceinated Cys residues in the loops of FepA, we measured 59FeEnt binding affinity and transport rates, and related these data to the half-times of fluorescence recovery after FeEnt-mediated quenching (Table S2). The single Cys substitutions themselves did not much change the binding or transport properties of FepA. The affinity for 59FeEnt remained comparable to that of wild-type FepA (Kd = KM = 1–3 nM). The exceptions were A383C (Kd = 6.7 nM, KM = 3 nM) and S490C (Kd = 15 nM, KM = 9 nM). Before fluoresceination most mutants had 59FeEnt transport rates comparable to wild-type FepA (Vmax = ∼100 pMol/109 cells/min). The exceptions were A322C, A383C, and S490C, which transported at about half that rate. Fluorescence modification did not generally degrade affinity for 59FeEnt (Kd = KM = 2–6 nM) relative to wild-type FepA (Kd = KM = 1–2 nM), except for S490C-FM (Kd = 90 nM, KM = 57 nM). But, fluoresceination usually reduced the rate of 59FeEnt uptake: Vmax was 3–10-fold lower for A322C-FM, A383C-FM, S490C-FM, and T550C-FM. The spectroscopic transport rates derived from half-times of recovery of fluorescence intensity (Table S2) reiterated these decreases in the FeEnt uptake rate. Modification of S490C, T550C, and A383C increased the recovery time 10-fold compared with S271C-FM, the protein least affected by fluoresceination. Hence, all the FM-labeled Cys mutants actively transported FeEnt, but some of them transported more slowly, likely from the bulkiness of the fluorophore in mechanistically important regions of protein structure (see Discussion).
Effects of ΔtonB on ligand adsorption to FepA in vivo
James et al. (2008), reported loop motion in FhuA as TonB-dependent, which led us to examine the behavior of FepA loops in ΔtonB cells. We compared FeEnt binding in the host strains OKN3 (tonB+, ΔfepA) and OKN13 (ΔtonB, ΔfepA), expressing the seven fluoresceinated FepA Cys-substitution mutants (e.g., FepAS271C-FM) from pHSG575 plasmids. The magnitude of FeEnt-induced quenching was always less in the ΔtonB strain than in the tonB+ background. Thus, the extent of fluorescence quenching during FeEnt binding appeared to be TonB-dependent (Fig. S3), which suggests that TonB conferred greater loop motion. However, closer study found that the lesser quenching in ΔtonB bacteria arose from lower expression of the FepA mutant derivatives in that strain, which led to decreased fluoresceination of FepA relative to background, nonspecific labeling (Figs. S4 and S5). Hence, the appearance of TonB-dependent loop motion from differences in the extent of FeEnt-mediated fluorescence quenching was an artifact that originated from compromised expression of FepA in the ΔtonB cells.
Rates of FM quenching in different loops of FepA
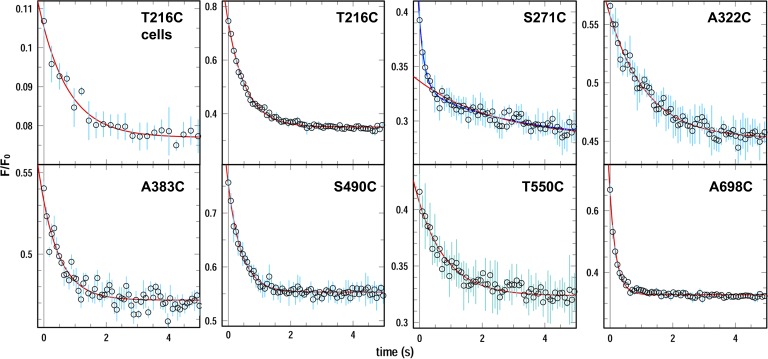
When monitored with purified FepAE280C-FM (in L3) in detergent solution, FeEnt binding caused a biphasic decay of fluorescence (Payne et al., 1997). Fits of the quenching data yielded two rates, which is consistent with two stages of conformational motion. In that context we observed the quenching of FM attached to the seven individual surface loops of FepA during FeEnt binding. We attempted to measure these rates in live bacteria by stirring cells in a cuvette during the addition of FeEnt, and also by rapidly mixing FM-labeled cells with FeEnt in a stopped-flow device. The former technique was of little value for kinetic studies because reagent mixing took 3–5 s. Thus, the individual quenching rates were too rapid to differentiate in the 2-ml format. The latter method was occasionally successful, but despite instances of highly labeled cells that allowed stopped-flow measurements (see T216C in Figs. 2 and S6), this approach was usually unachievable because of insufficient fluorescence intensity to overcome the turbidity of the bacterial suspension in the small volume of the stopped-flow chamber. To circumvent these difficulties we concentrated the bacteria and generated OM fragments by French press lysis, which gave clarified solutions that permitted stopped-flow measurements of FeEnt-FepA binding in the native environment of the OM. In these rapid mixing experiments the observed rate of ligand binding to FepA was much faster (45–350-fold; k = 0.8–6.4 s−1) than ever previously measured (1.8 × 10−2 s−1 and 2.1 × 10−3 s−1; Payne et al., 1997). In addition, the rates of FM quenching differed in the individual loops: S271C (L3) > A698C (L11) > S490C (L7) > T216C (L2) > T550C (L8) > A383C (L5) > S322C (L4). These data described a sequence of surface loop motion, with L3 and L11 moving about twice as fast as the other loops. Finally, measurements of motion with S271C-FM reiterated the biphasic kinetics of L3 (originally shown for FepAE280C-FM; Payne et al., 1997). However, FeEnt quenching of FM attached to any of the other loops fit well with single exponential decays (Fig. 2). The half-times of the quenching reactions (Table 1) showed that the conformational changes in FepA during FeEnt binding completed within about a second.
Figure 2.
Rapid mixing stopped-flow measurements of FeEnt binding to FepA. 1010 cells of OKN3 harboring pHSG575 derivatives with fepA Cys substitutions were grown, fluoresceinated, washed, resuspended in PBS, and lysed by two passages in a French pressure cell at 14,000 psi (see Materials and methods). We collected the cell envelope fractions, containing OM fragments (Smit et al., 1975), by centrifugation, resuspended them in 2 ml of PBS, and performed rapid-mixing stopped-flow experiments with an equal volume of 200 nM FeEnt in an OLIS-SLM8000 fluorometer. We collected FeEnt quenching time courses and averaged and analyzed the data with GraFit 6.011. The top left panel shows rapid-mixing stopped-flow data for intact cells expressing FepAT216C-FM; other panels derive from analysis of OM fragments from the indicated FM-labeled FepA Cys mutants. We performed the experiment three times for each mutant and averaged the results (open circles); the standard deviations of the mean values are shown as cyan error bars; red curves derive from fits to single exponential relaxations; the blue curve for S271C-FM derives from a fit to a double exponential decay process. See Fig. S6 for the raw data, and Table 1 for the resulting rate constants from these measurements.
Table 1.
Rates of FM quenching in different surface loops of FepA during FeEnt binding
| Mutant | Loop | k (SE) | t1/2 | Rank |
| s−1 | s | |||
| T216a | 2 | 1.08 (0.08) | 0.64 | 7 |
| T216 | 2 | 1.73 (0.04) | 0.40 | 4 |
| S271Cb | 3 | 5.72 (0.81); | 0.12; | 1 |
| 0.31 (0.04) | 2.23 | |||
| A322C | 4 | 0.84 (0.05) | 0.83 | 8 |
| A383C | 5 | 1.64 (0.15) | 0.43 | 5 |
| S490C | 7 | 2.32 (0.13) | 0.30 | 3 |
| T550C | 8 | 1.09 (0.06) | 0.64 | 6 |
| A698C | 11 | 4.90 (0.18) | 0.14 | 2 |
We measured quenching by rapid mixing of FM-labeled FepA Cys substitution mutants (in OM fragments) with an equal volume of 200 nM FeEnt. Rates for the exponential decay processes (with parenthetic standard errors) were derived from fitting the data with GraFit 6.011; decay half times were calculated from t1/2 = −ln 0.5/k.
Determined in live bacteria expressing FepAT216C-FM.
FepAS271C-FM was the only construct that manifested biphasic decay kinetics during FeEnt binding. The amplitudes of the first and second kinetic phases were 0.063 and 0.047, respectively. The offset was 0.28.
Bulk observations of FeEnt transport in living cells
In iron-deficient conditions, Gram-negative bacteria contain 25,000–50,000 FepA proteins in the OM (Newton et al., 1999), but only 1,000–3,000 TonB proteins in the IM (Higgs et al., 2002). Yet, uptake of FeEnt through FepA involves direct interactions between the receptor and TonB (Cadieux et al., 2000; Shultis et al., 2006; Pawelek et al., 2006), so this numerical discrepancy creates a mechanistic puzzle in their biochemical relationship. The exclusion of TonB, but not FepA, from the poles of the cell (Jordan et al., 2013; Fig. 3) posed the question: are polar-localized FepA proteins biochemically inactive, as a result of the apparent absence of TonB from those locations? The spectroscopic iron uptake assays suggested fluorescence microscopy as a means to observe the activity of FepA proteins in different locations around the cell surface (Fig. 3). After growth of OKN13/pfepAS271C/pGT (Jordan et al., 2013) in MOPS media, we fluoresceinated FepAS271C, observed the iron-deficient cells by confocal fluorescence microscopy, and exposed them to uptake of 5 nM FeEnt. Close scrutiny of the images showed the discrepancy in number between FepA and GFP-TonB, as well as the absence of TonB from the poles and central areas underlying the OM (Fig. 3 B). The time course again portrayed the iron uptake process as the diminution of fluorescence when FeEnt bound, and its return to original levels as transport depleted the ferric siderophore from solution. Fluorescence uniformly recovered at the same rate all over the bacterial cell surface, including at the poles, despite the absence of GFP-TonB from polar locations. These data indicated that all FepA proteins in the cell actively transported FeEnt at the same rate, even when TonB was not in obvious proximity as a result of its lower abundance, and restriction to central locations of the cell envelope.
Figure 3.
Fluorescence microscopic observation of FeEnt binding and transport in single cells. OKN13 (ΔtonB,ΔfepA)/pGT/pfepAS271C was grown in MOPS media and labeled with A546M (see Materials and methods). We adhered the cells, which contained GFP-TonB in the IM (Jordan et al., 2013) and FepAS271C-A546M in the OM, to poly-l-lysine–coated chamber-slides. (A) The images show the response of GFP-TonB and FepAS271C-FM (in the top and bottom series, respectively), to the addition of 10 nM FeEnt at t = 0 min. The fluorescence of GFP-TonB (at 520 nm) was unaffected, whereas that of FepAS271C-A546M (at 585 nm) was immediately quenched by binding of the ferric siderophore, but rebounded over the next 6 min during transport. (B) The image superimposes the two individual images from the 6-min time point, and designates polar (p) and median (m) regions of the cells from which we performed a fluorescence intensity quantification (ImageQuant). (C) The analyzed data show that TonB-dependent iron transport by FepA occurs uniformly with the same kinetics throughout the OM, despite the almost complete absence of TonB from the poles of the cells.
DISCUSSION
Conformational motion that regulates OM active transport of metal complexes is a defining attribute of TBDT. FepA is unusually conformationally active, and several findings (Scott et al., 2002; Ferguson et al., 2002; Smallwood et al., 2009), including the individual loop motion reported herein, infer that its surface loops achieve high affinity for FeEnt by induced fit (Koshland, 1958). In the ligand-free state the receptor’s loops open to optimize solute collection from the environment (Scott, 2000; Smallwood et al., 2009). The protein progresses to a closed state as it tightly associates with FeEnt at binding equilibrium (Buchanan et al., 1999; Ferguson et al., 2002; Scott et al., 2002). The subnanomolar affinity of TBDT for their metal complexes allows selection among potentially hundreds of ferric siderophores in the microbial environment. Our data show that the loops of FepA move at different rates to capture FeEnt before transport, creating a kinetic hierarchy in the context of the transporter’s architecture. Crystallography shows the loops of FepA grouped on opposite sides of the external vestibule (Fig. 1), and these elements move at different rates during FeEnt adsorption, like the fingers of two hands catching an object. These data imply loop dynamics that create rapid, high-affinity binding and hold the metal complex in the FepA vestibule until TonB facilitates its transport.
Site-directed fluorescence labeling procedures combined the high quantum yield of fluorescein and Alexa Fluors with the precision, kinetic capabilities and analytical sophistication of spectroscopic instrumentation, creating a sensitive technology for study of OM protein biochemistry. The methodology is generally applicable to cell surface proteins of Gram-negative or -positive (unpublished data) bacterial cells. Nevertheless, for E. coli it required preliminary experiments to optimize labeling procedures, maximize specificity, and allow quantification. OM proteins contain few cysteines, and when present they predominantly form disulfide bonds (FepA, a protein of 724 aa, has a single disulfide: C487-C494 in L7) that are unreactive unless reduced (Liu et al., 1994). Specific labeling of FepA was essential, and we adjusted conditions to minimize reactivity with other proteins.
Consistent with the idea that labeling efficiency derives from a combination of FepA’s abundance and the accessibility of Cys targets in its tertiary structure, different sites within different loops fluoresceinated to different levels. Mutants A698C and T216C (surface-exposed, on the exteriors of L11 and L2, respectively), were expressed at the highest levels and labeled with the highest efficiency. FeEnt binding quenched the emissions of FM >60% at both sites, and similarly quenched A322C-FM (L4) that was labeled at much lower levels (only 50% of A698C). Other Cys substitutions were less well expressed, less well fluoresceinated, and less quenched: FeEnt binding only reduced emissions of S490C-FM (L7) 30%. The variations in FeEnt-mediated quenching of FepA in a TonB-deficient strain illustrated some caveats of the procedure. As labeling time increased, specificity for FepA decreased (i.e., nonspecific background labeling increased), and the ultimate level of quenching by FeEnt depended on the degree of nonspecific labeling of the cells. FepA was expressed and labeled less in the ΔtonB strain, so background labeling became a more significant component of the total cellular fluorescence. Consequently, we saw less overall quenching in the ΔtonB host relative to its ΔtonB+ parent strain. The bottom line was that rapid labeling (<5 min) with low concentrations of FM (1–5 µM) and moderate conditions (pH 6.5–7, at 0 or 37°C) was effective and selective for Cys side chains in the loops of FepA. Additionally, it was best to compare mutant strains that were simultaneously grown, prepared, and labeled with the same FM solution, for the same time, etc. Comparing data from different strains labeled on different days, especially tonB+ versus ΔtonB strains analyzed on different days, led to variability and potential artifacts.
The rapid-mixing, stopped-flow analyses of quenching, from multiple fluoresceinated loops in the native environment of the OM, provided a detailed view of surface loop motion. These experiments, never before accomplished for a bacterial OM protein, showed rapid rates of loop conformational change. Most loops followed simple closing mechanisms, well fit by single exponential relaxation. However, the results verified the two-state closing process of L3. It’s not obvious why L3 closes in two stages, but interactions with adjacent loops that affect its local environment are a conceivable explanation. The slower second component may not represent L3 motion per se, but instead its reactions to the movements of other loops, especially L2 beside and L7 across the vestibule, which both close at a slower rates; the second phase of L3 motion temporally coincides with the slower actions of L2 and L7. The second kinetic phase may represent adjustments in the final configuration of L3 as FeEnt reaches binding equilibrium in the vestibule. The upshot of the kinetic studies is that the ferric siderophore binds within a second or two, by interacting with loops that close at different rates, and may interact with one another.
Radioisotopic experiments revealed that the attachment of FM to the surface loops usually only slightly affected the affinity of the ligand–receptor interaction (two- to three-fold), which imparts confidence in the rate measurements of loop motion that lead to equilibrium binding. Both radioisotopic and spectroscopic assays showed that fluoresceination may impair the FeEnt transport rate (as much as 10-fold). But it was noteworthy that all the covalently modified FepA mutant derivatives accomplished TonB-dependent uptake, and that certain among them transported at rates that were comparable to that of wild-type FepA (S271C-FM, 65%; A698C-FM, 90%). FM only adds 427 D, the mass of 2–3 amino acids, to its target, so its inhibitory effects likely arise from steric factors as the transporter undergoes conformational motion during ligand internalization.
Physical interactions occur between FepA and TonB during FeEnt transport that involve recruitment of the TonB box at the FepA N terminus by the TonB C terminus (Pawelek et al., 2006). The fluorescence microscopic transport assay method revealed that despite uniform distribution of the FepA throughout the OM, and restriction of GFP-TonB to central regions of the cell, FeEnt uptake occurred at the same rate in all regions of the cell envelope, including in the polar regions devoid of TonB. This unexpected finding illustrates that much remains unknown about the bioenergetic phenomena that drive OM metal transport. The two proteins must find each in the cell envelope more rapidly than the FeEnt turnover number (∼0.03 s−1; Newton et al., 1999; Annamalai et al., 2004), but the apparent exclusion of TonB from the poles, combined with the low fluidity of the OM (Nikaido and Vaara, 1985), raises questions about how that occurs.
Supplementary Material
Acknowledgments
We are grateful to the Confocal Microfluorometry and Microscopy Core Facility in the College of Veterinary Medicine at Kansas State University.
The research reported herein was supported by National Institutes of Health grant GM53836 and National Science Foundation grant MCB0952299 (to P.E. Klebba and S.M.C. Newton).
The authors declare no competing financial interests.
Sharona E. Gordon served as editor.
Footnotes
Abbreviations used in this paper:
- FeEnt
- ferric enterobactin
- FM
- fluorescein-5-maleimide
- IM
- inner membrane
- OM
- outer membrane
- PMF
- proton-motive force
- TBDT
- TonB-dependent transporter
References
- Ames G.F. 1974. Resolution of bacterial proteins by polyacrylamide gel electrophoresis on slabs. Membrane, soluble, and periplasmic fractions. J. Biol. Chem. 249:634–644 [PubMed] [Google Scholar]
- Annamalai R., Jin B., Cao Z., Newton S.M., Klebba P.E. 2004. Recognition of ferric catecholates by FepA. J. Bacteriol. 186:3578–3589 10.1128/JB.186.11.3578-3589.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbeer C. 1993. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J. Bacteriol. 175:3146–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan S.K., Smith B.S., Venkatramani L., Xia D., Esser L., Palnitkar M., Chakraborty R., van der Helm D., Deisenhofer J. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56–63 10.1038/4931 [DOI] [PubMed] [Google Scholar]
- Buchanan S.K., Lukacik P., Grizot S., Ghirlando R., Ali M.M., Barnard T.J., Jakes K.S., Kienker P.K., Esser L. 2007. Structure of colicin I receptor bound to the R-domain of colicin Ia: implications for protein import. EMBO J. 26:2594–2604 10.1038/sj.emboj.7601693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadieux N., Bradbeer C., Kadner R.J. 2000. Sequence changes in the ton box region of BtuB affect its transport activities and interaction with TonB protein. J. Bacteriol. 182:5954–5961 10.1128/JB.182.21.5954-5961.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., Qi Z., Sprencel C., Newton S.M., Klebba P.E. 2000. Aromatic components of two ferric enterobactin binding sites in Escherichia coli FepA. Mol. Microbiol. 37:1306–1317 10.1046/j.1365-2958.2000.02093.x [DOI] [PubMed] [Google Scholar]
- Cao Z., Warfel P., Newton S.M., Klebba P.E. 2003. Spectroscopic observations of ferric enterobactin transport. J. Biol. Chem. 278:1022–1028 10.1074/jbc.M210360200 [DOI] [PubMed] [Google Scholar]
- Chimento D.P., Mohanty A.K., Kadner R.J., Wiener M.C. 2003. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat. Struct. Biol. 10:394–401 10.1038/nsb914 [DOI] [PubMed] [Google Scholar]
- Cobessi D., Meksem A., Brillet K. 2010. Structure of the heme/hemoglobin outer membrane receptor ShuA from Shigella dysenteriae: heme binding by an induced fit mechanism. Proteins. 78:286–294 10.1002/prot.22539 [DOI] [PubMed] [Google Scholar]
- Cox G.B., Gibson F., Luke R.K., Newton N.A., O’Brien I.G., Rosenberg H. 1970. Mutations affecting iron transport in Escherichia coli. J. Bacteriol. 104:219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson A.D., Hofmann E., Coulton J.W., Diederichs K., Welte W. 1998. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science. 282:2215–2220 10.1126/science.282.5397.2215 [DOI] [PubMed] [Google Scholar]
- Ferguson A.D., Chakraborty R., Smith B.S., Esser L., van der Helm D., Deisenhofer J. 2002. Structural basis of gating by the outer membrane transporter FecA. Science. 295:1715–1719 10.1126/science.1067313 [DOI] [PubMed] [Google Scholar]
- Hashimoto-Gotoh T., Kume A., Masahashi W., Takeshita S., Fukuda A. 1986. Improved vector, pHSG664, for direct streptomycin-resistance selection: cDNA cloning with G:C-tailing procedure and subcloning of double-digest DNA fragments. Gene. 41:125–128 10.1016/0378-1119(86)90275-1 [DOI] [PubMed] [Google Scholar]
- Higgs P.I., Larsen R.A., Postle K. 2002. Quantification of known components of the Escherichia coli TonB energy transduction system: TonB, ExbB, ExbD and FepA. Mol. Microbiol. 44:271–281 10.1046/j.1365-2958.2002.02880.x [DOI] [PubMed] [Google Scholar]
- James K.J., Hancock M.A., Moreau V., Molina F., Coulton J.W. 2008. TonB induces conformational changes in surface-exposed loops of FhuA, outer membrane receptor of Escherichia coli. Protein Sci. 17:1679–1688 10.1110/ps.036244.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Payne M.A., Cao Z., Foster S.B., Feix J.B., Newton S.M., Klebba P.E. 1997. Ligand-specific opening of a gated-porin channel in the outer membrane of living bacteria. Science. 276:1261–1264 10.1126/science.276.5316.1261 [DOI] [PubMed] [Google Scholar]
- Jordan L.D., Zhou Y., Smallwood C.R., Lill Y., Ritchie K., Yip W.T., Newton S.M., Klebba P.E. 2013. Energy-dependent motion of TonB in the Gram-negative bacterial inner membrane. Proc. Natl. Acad. Sci. USA. 110:11553–11558 10.1073/pnas.1304243110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba P.E. 2003. Three paradoxes of ferric enterobactin uptake. Front. Biosci. 8:s1422–s1436 10.2741/1233 [DOI] [PubMed] [Google Scholar]
- Klebba P.E., McIntosh M.A., Neilands J.B. 1982. Kinetics of biosynthesis of iron-regulated membrane proteins in Escherichia coli. J. Bacteriol. 149:880–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konisky J. 1979. Specific transport systems and receptors for colicins and phages. Bacterial Outer Membranes: Biogenesis and Function. Inouye M., editor John Wiley and Sons, New York: 319–359 pp. [Google Scholar]
- Koshland D.E. 1958. Application of a Theory of Enzyme Specificity to Protein Synthesis. Proc. Natl. Acad. Sci. USA. 44:98–104 10.1073/pnas.44.2.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisu G., Zakharov S.D., Zhalnina M.V., Bano S., Eroukova V.Y., Rokitskaya T.I., Antonenko Y.N., Wiener M.C., Cramer W.A. 2003. The structure of BtuB with bound colicin E3 R-domain implies a translocon. Nat. Struct. Biol. 10:948–954 10.1038/nsb997 [DOI] [PubMed] [Google Scholar]
- Liu J., Rutz J.M., Klebba P.E., Feix J.B. 1994. A site-directed spin-labeling study of ligand-induced conformational change in the ferric enterobactin receptor, FepA. Biochemistry. 33:13274–13283 10.1021/bi00249a014 [DOI] [PubMed] [Google Scholar]
- Locher K.P., Rees B., Koebnik R., Mitschler A., Moulinier L., Rosenbusch J.P., Moras D. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell. 95:771–778 10.1016/S0092-8674(00)81700-6 [DOI] [PubMed] [Google Scholar]
- Ma L., Kaserer W., Annamalai R., Scott D.C., Jin B., Jiang X., Xiao Q., Maymani H., Massis L.M., Ferreira L.C., et al. 2007. Evidence of ball-and-chain transport of ferric enterobactin through FepA. J. Biol. Chem. 282:397–406 10.1074/jbc.M605333200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 466 pp [Google Scholar]
- Murphy C.K., Kalve V.I., Klebba P.E. 1990. Surface topology of the Escherichia coli K-12 ferric enterobactin receptor. J. Bacteriol. 172:2736–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F.C., Bloch P.L., Smith D.F. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilands J.B. 1995. Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270:26723–26726 10.1074/jbc.270.45.26723 [DOI] [PubMed] [Google Scholar]
- Newton S.M., Allen J.S., Cao Z., Qi Z., Jiang X., Sprencel C., Igo J.D., Foster S.B., Payne M.A., Klebba P.E. 1997. Double mutagenesis of a positive charge cluster in the ligand-binding site of the ferric enterobactin receptor, FepA. Proc. Natl. Acad. Sci. USA. 94:4560–4565 10.1073/pnas.94.9.4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton S.M., Igo J.D., Scott D.C., Klebba P.E. 1999. Effect of loop deletions on the binding and transport of ferric enterobactin by FepA. Mol. Microbiol. 32:1153–1165 10.1046/j.1365-2958.1999.01424.x [DOI] [PubMed] [Google Scholar]
- Newton S.M., Trinh V., Pi H., Klebba P.E. 2010. Direct measurements of the outer membrane stage of ferric enterobactin transport: postuptake binding. J. Biol. Chem. 285:17488–17497 10.1074/jbc.M109.100206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H., Vaara M. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelek P.D., Croteau N., Ng-Thow-Hing C., Khursigara C.M., Moiseeva N., Allaire M., Coulton J.W. 2006. Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science. 312:1399–1402 10.1126/science.1128057 [DOI] [PubMed] [Google Scholar]
- Payne M.A., Igo J.D., Cao Z., Foster S.B., Newton S.M., Klebba P.E. 1997. Biphasic binding kinetics between FepA and its ligands. J. Biol. Chem. 272:21950–21955 10.1074/jbc.272.35.21950 [DOI] [PubMed] [Google Scholar]
- Pollack J.R., Neilands J.B. 1970. Enterobactin, an iron transport compound from Salmonella typhimurium. Biochem. Biophys. Res. Commun. 38:989–992 10.1016/0006-291X(70)90819-3 [DOI] [PubMed] [Google Scholar]
- Rutz J.M., Liu J., Lyons J.A., Goranson J., Armstrong S.K., McIntosh M.A., Feix J.B., Klebba P.E. 1992. Formation of a gated channel by a ligand-specific transport protein in the bacterial outer membrane. Science. 258:471–475 10.1126/science.1411544 [DOI] [PubMed] [Google Scholar]
- Schauer K., Rodionov D.A., de Reuse H. 2008. New substrates for TonB-dependent transport: do we only see the ‘tip of the iceberg’? Trends Biochem. Sci. 33:330–338 10.1016/j.tibs.2008.04.012 [DOI] [PubMed] [Google Scholar]
- Scott D.C. 2000. Mechanism of ferric enterobactin transport through Escherichia coli FepA: evolution of a bacterial venus fly trap. Dissertation. The University of Oklahoma. https://shareok.org/handle/11244/403 (accessed June 13, 2014)
- Scott D.C., Newton S.M., Klebba P.E. 2002. Surface loop motion in FepA. J. Bacteriol. 184:4906–4911 10.1128/JB.184.17.4906-4911.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultis D.D., Purdy M.D., Banchs C.N., Wiener M.C. 2006. Outer membrane active transport: structure of the BtuB:TonB complex. Science. 312:1396–1399 10.1126/science.1127694 [DOI] [PubMed] [Google Scholar]
- Smallwood C.R., Marco A.G., Xiao Q., Trinh V., Newton S.M., Klebba P.E. 2009. Fluoresceination of FepA during colicin B killing: effects of temperature, toxin and TonB. Mol. Microbiol. 72:1171–1180 10.1111/j.1365-2958.2009.06715.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova I., Kasho V., Sugihara J., Kaback H.R. 2009. Probing of the rates of alternating access in LacY with Trp fluorescence. Proc. Natl. Acad. Sci. USA. 106:21561–21566 10.1073/pnas.0911434106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit J., Kamio Y., Nikaido H. 1975. Outer membrane of Salmonella typhimurium: chemical analysis and freeze-fracture studies with lipopolysaccharide mutants. J. Bacteriol. 124:942–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne R., Neilands J.B. 1975. Evidence for common binding sites for ferrichrome compounds and bacteriophage phi 80 in the cell envelope of Escherichia coli. J. Bacteriol. 121:497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne R., Frick K., Neilands J.B. 1976. Siderophore protection against colicins M, B, V, and Ia in Escherichia coli. J. Bacteriol. 126:7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen M.R., Peabody C.R., Partovi S.M., Zhai Y., Tseng Y.H., Saier M.H. 2002. Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim. Biophys. Acta. 1562:6–31 10.1016/S0005-2736(02)00359-0 [DOI] [PubMed] [Google Scholar]
- Yu E.W., Koshland D.E., Jr 2001. Propagating conformational changes over long (and short) distances in proteins. Proc. Natl. Acad. Sci. USA. 98:9517–9520 10.1073/pnas.161239298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukl E.T., Jepkorir G., Alontaga A.Y., Pautsch L., Rodriguez J.C., Rivera M., Moënne-Loccoz P. 2010. Kinetic and spectroscopic studies of hemin acquisition in the hemophore HasAp from Pseudomonas aeruginosa. Biochemistry. 49:6646–6654 10.1021/bi100692f [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.