Abstract
Hepatocellular carcinoma (HCC), the third most common cause of cancer-related deaths worldwide, lacks effective medical therapy. Large subsets of HCC demonstrate Wnt/β-catenin activation, making this an attractive therapeutic target. We report strategy and characterization of a novel small-molecule inhibitor, ICG-001, known to affect Wnt signaling by disrupting β-catenin–CREB binding protein interactions. We queried the ZINC online database for structural similarity to ICG-001 and identified PMED-1 as the lead compound, with ≥70% similarity to ICG-001. PMED-1 significantly reduced β-catenin activity in hepatoblastoma and several HCC cells, as determined by TOPflash reporter assay, with an IC50 ranging from 4.87 to 32 μmol/L. Although no toxicity was observed in primary human hepatocytes, PMED-1 inhibited Wnt target expression in HCC cells, including those with CTNNB1 mutations, and impaired cell proliferation and viability. PMED-1 treatment decreased β-catenin–CREB binding protein interactions without affecting total β-catenin levels or activity of other common kinases. PMED-1 treatment of Tg(OTM:d2EGFP) zebrafish expressing GFP under the β-catenin/Tcf reporter led to a notable decrease in β-catenin activity. The PMED effect on β-catenin signaling lasted from 12 to 24 hours in vitro and 6 to 15 hours in vivo. Thus, using a rapid and cost-effective computational methodology, we have identified a novel and specific small-molecule inhibitor of Wnt signaling that may have implications for HCC treatment.
Hepatocellular carcinoma (HCC) is the third most common cause of cancer-related deaths worldwide; due to lack of effective treatment, HCC has a 90% fatality rate.1,2 Given the current global burden of HCC, it is paramount to develop an effective therapy against this disease. Surgical resection and liver transplantation is a desirable therapeutic approach because it may be curative and because it replaces cirrhotic liver with disease-free donor liver.3,4 However, because of shortage of donor organs, not every HCC patient can receive a transplant. Given that HCC is a vascularized tumor that derives its blood supply predominantly from the hepatic artery, transarterial chemoembolization (TACE) has been used to manage unresectable HCC.5 Because of increased expression of various receptor tyrosine kinases (RTKs), HCC has been shown to be somewhat susceptible to multi-tyrosine kinase inhibitors.6–8 Indeed, sorafenib is used to treat unresectable HCC, with some survival benefit.9 Thus, current therapeutic strategies are beginning to take advantage of specific pathways that are deregulated in HCC.
Wnt/β-catenin signaling is tightly regulated in the adult liver, and its aberrant nuclear localization, which is multifactorial, is evident in a major subset of HCCs.10 Nuclear β-catenin acts as a cofactor for the T-cell factor (TCF) family of transcription factors and recruits histone acetyl transferases such as CREB-binding protein (CBP) to regulate the expression of various target genes. Therapeutic efficacy of inhibiting β-catenin as a treatment strategy for this tumor type has been shown both in vitro and in vivo.11–15 To date, however, there are no clinically approved anti–β-catenin agents, and continued drug discovery and development are needed. The small molecule (SM) ICG-001 is one such agent that has shown to significantly reduce β-catenin activity by blocking β-catenin–CBP interactions in tumor cell nuclei.16 The next-generation derivative PRI-724 is currently in phase Ib/IIa clinical trials.17
By means of structure function similarity search to ICG-001, we identified a novel SM inhibitor of β-catenin (PMED-1) that blocks β-catenin signaling in multiple hepatoma cells, thereby reducing their viability and proliferation, but is nontoxic to primary human hepatocytes. PMED-1 lacks any off-target effects on several common kinases and, like ICG-001, it reduces β-catenin–CBP interactions. In vivo, PMED-1 treatment of transgenic zebrafish expressing GFP as a readout of β-catenin–TCF activity leads to a notable decrease in Wnt signaling. Here, we highlight the relevance of cost-effective, efficacious, and simplistic similarity searches in streamlining drug discovery and report discovery of a prospective SM that may be effective against β-catenin in HCC cells.
Materials and Methods
Similarity Search
Morphological similarity is a similarity technique dependent only on surface shape and charge characteristics of ligands.18 Morphological similarity is defined as a Gaussian function of the differences in the molecular surface distances of two molecules at weighted observation points on a uniform grid. The computed surface distances include both distances to the nearest atomic surface and distances to donor and acceptor surfaces. This function is dependent on the relative alignment of the molecules, and consequently their alignment and conformation must be optimized. The conformational optimization problem is solved by fragmentation, conformational search, alignment, and scoring, followed by incremental reconstruction from high-scoring aligned fragments. The alignment problem is addressed by exploiting the fact that two unaligned molecules or molecular fragments that have some degree of similarity will have some corresponding set of observers making the same observations. Optimization of the similarity of two unaligned molecules or molecular fragments is enabled by finding similar sets of observers of each molecule that form triangles of the same size.18
In Silico ADME and Toxicity Screening
Computational modeling tools were used to estimate absorption, distribution, metabolism, and excretion (ie, ADME) and toxicity. For hits obtained from database screening, we estimated bioavailability, aqueous solubility, and human intestinal absorption, using the ADME Suite 4.95 from Pharma Algorithms-ACD/Labs (ACD/Labs; Advanced Chemistry Development, Toronto, ON, Canada). For toxicity predictions including cytochrome P450 (ie, CYP2D6) enzyme inhibition potential, mutagenicity, and hERG inhibition of the hits obtained from the database screening, we used Tox Boxes version 2.9 (Pharma Algorithms-ACD/Labs). Lastly, ACD/Blood-Brain Barrier permeation module (ADME Suite 4.95, Pharma Algorithms-ACD/Labs) was used to estimate blood-brain barrier potential of the hits.
Cell Culture
Hep3B, HepG2, Snu-398 and Snu-449 cells were obtained from ATCC (Manassas, VA). Huh-7 cells were obtained from Japanese collection of research bioresources. HepG2 human hepatoblastoma cells (3 × 105 and 4 × 104 cells per well for 6-well and 24-well plates, respectively), Huh-7 human HCC cells (2.5 × 105 and 4 × 104 cells per well), and Hep3B human HCC cells (2.5 × 105 and 4 × 104 cells per well) from the ATCC were cultured in Eagle’s minimal essential medium (EMEM) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Flowery Branch, GA). Snu-449 or Snu-398 human HCC cells (3 × 105 and 4 × 104 cells per well) were cultured in RPMI-1640 medium (ATCC) supplemented with 10% FBS. Primary human hepatocytes for cell viability studies were obtained from Dr. Stephen Strom (then principal investigator of the Liver Tissue Cell Distribution Services, University of Pittsburgh).
Human HCC Cell Culture and Transfection with Stable β-Catenin Mutants
Hep3B (ATCC) human HCC cells were plated in 6-well plates and cultured in EMEM (ATCC) supplemented with 10% FBS (Atlanta Biologicals) at 37°C in a humidified 5% CO2 atmosphere. A constitutively active β-catenin gene resulting in a tyrosine-for-serine substitution (S33Y) was kindly provided by Dr. Jian Yu (Department of Pathology, Hillman Cancer Center, University of Pittsburgh). The cells were grown to 90% confluency, and 2 μg of wild-type and S33Y β-catenin plasmid DNA was transfected with Lipofectamine 2000 reagent (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. At 48 hours after transfection, the cells were selected by multiple passaging using 500 μg/mL geneticin (G418; Sigma-Aldrich, St. Louis, MO) to generate stable transfected cell lines. Wild-type or S33Y stably transfected Hep3B cells (3 × 105 cells per well, 6-well plates; 4 × 104 cells per well, 24-well plates) were cultured for experiments in EMEM supplemented with 10% FBS and 500 mg/mL G418.
β-Catenin–TCF Luciferase Reporter Assay
TOPflash plasmid (Upstate; EMD Millipore, Billerica, MA) has three copies of the Tcf/Lef sites upstream of a thymidine kinase (TK) promoter and the firefly luciferase gene. HCC cells were transfected simultaneously with Renilla reniformis luciferase (pRL-TK; Promega, Madison, WI) as a transfection control and with TOPflash firefly luciferase plasmids (Upstate; EMD Millipore) for β-catenin–TCF activity using FuGENE (Roche Diagnostics, Indianapolis, IN) or Lipofectamine 2000 (Life Technologies) reagent. At 24 hours after transfection, cells were treated with dimethyl sulfoxide (DMSO) (Thermo Fisher Scientific, Waltham, MA), PMED-1 (ChemBridge, San Diego, CA), PMED-2 (ChemBridge), or ICG-001 for an additional 24 or 48 hours. Lysates were harvested using the dual-luciferase reporter assay system (Promega). Firefly luciferase signals were normalized to Renilla luciferase, and the ratio between groups was compared by Student’s t-test to determine significance. P < 0.05 was considered significant and P < 0.01 was considered highly significant.
For time-course analysis, S33Y β-catenin stably transfected Hep3B cells were cultured in serum-free EMEM containing 500 μg/mL G418 for 4 hours before Lipofectamine 2000 (Life Technologies) cotransfection using Renilla and TOPflash luciferase plasmids for 4 hours. Cells were then exposed to EMEM containing 2% FBS and 500 μg/mL G418 overnight. On the next morning, cells were serum-starved again for 4 hours to synchronize the cell cycle and then were treated with either 0.05% DMSO or 25 μmol/L PMED-1 in EMEM containing 10% FBS and 500 μg/mL G418 for 3, 6, 9, 12, 18, 24, 36, or 48 hours before harvest and analysis of samples for luciferase activity as described above.
To determine the half-maximal inhibitory concentration (IC50) of the drug required to reduce luciferase activity, TOPflash reporter assay was performed. Snu-398, Snu-449, Hep3B, and Huh-7 cells were plated in 96-well plates at 2 × 104 cells/well at 80% confluence were transfected using Lipofectamine 2000 with 100 ng of TOPflash firefly luciferase and 10 ng of Renilla luciferase. Cells were treated without or with serial dilutions in triplicate from 50 μmol/L to 1.5 μmol/L of PMED-1 in RPMI-1640 or EMEM medium supplemented with FBS. PMED-1, dissolved in DMSO (0.2% final concentration), was administered in one dose at 4 hours after transfection. At 24 hours after treatment, cells were harvested and lysed with passive lysis buffer (Promega). Luciferase assay was performed as described above, and relative luciferase activity (ratio) was reported as fold induction after normalization for transfection efficiency.
Western Blot Analysis
Whole-cell lysates were prepared using radioimmunoprecipitation assay buffer containing 1% IgePAL CA-630, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate in 1× PBS after 24 hours of treatment with DMSO, PMED-1, or ICG-001 SM at 25 μmol/L. The lysate (9 to 30 ng) was run on a 7.5% polyacrylamide gel (Bio-Rad Laboratories, Hercules, CA) at 60 V for 0.5 hours and then at 100 V for 1.5 hours. Gels were transferred onto a polyvinylidene fluoride membrane (Millipore) for 1 hour at 4°C. Membranes were blocked in 5% nonfat milk (LabScientific, Livingston, NJ) in Blotto blocking reagent [0.15 mol/L NaCl, 0.02 mol/L Tris at pH 7.5, 0.1% Tween in deionized water] for 1 hour at room temperature. Primary antibodies for β-catenin (BD Transduction Laboratories; BD Biosciences, San Jose, CA), β-actin (Chemicon International; EMD Millipore), c-Myc (Santa Cruz Biotechnology, Dallas, TX), cyclin D1 (Thermo Fisher Scientific), GAPDH (Santa Cruz Biotechnology), glutamine synthetase (Santa Cruz Biotechnology), SMP30/regucalcin (Santa Cruz Biotechnology), and VEGF-A (Santa Cruz Biotechnology) were diluted 1:1000, 1:2500, 1:200, 1:200, 1:800, 1:200, 1:200, and 1:200, respectively, in 5% nonfat milk and Blotto reagent and were incubated on membranes overnight at 4°C. Membranes were then washed in Blotto reagent for 1 hour at room temperature before incubation of membranes with rabbit (1:10,000), mouse (1:25,000), or goat (1:10,000) secondary antibodies (EMD Millipore) in 1% nonfat milk and Blotto reagent for 1 hour. Membranes were again washed in Blotto reagent for 1 hour at room temperature before exposure to SuperSignal West Pico or Femto chemiluminescent substrate (Pierce; Thermo Fisher Scientific) for 1 to 2 minutes at room temperature. The bands reflective of target proteins were visualized by autoradiography.
Immunoprecipitation
HepG2 cells treated with DMSO, 25 μmol/L PMED-1, or 25 μmol/L ICG-001 for 24 hours in 10-cm dishes. Whole-cell lysates were prepared as described above, and 0.25 μg of control mouse IgG (Santa Cruz Biotechnology) was used to preclear lysates. β-Catenin monoclonal antibody (2 μg) (BD Transduction Laboratories) was added to 500 μg of each sample, which was incubated for 1 hour at 4°C on a rocking platform. Protein A/G PLUS agarose beads (20 mL) (Santa Cruz Biotechnology) were added to each sample, which was incubated overnight at 4°C on a rocking platform. Samples were then centrifuged at 10,000 × g for 5 minutes and the supernatant was removed. The bead pellet was then washed with radioimmunoprecipitation assay buffer, followed by three additional centrifugations and washes. After the final wash, bead pellets of each sample were resuspended in 40 mL of 2× SDS buffer (Bio-Rad Laboratories) and boiled at 95°C for 5 minutes. Samples were centrifuged one last time for 1 minute, and 20 μL of each sample was run on a 5% polyacrylamide gel (Bio-Rad Laboratories). Gel was run and transferred as described above. The membrane was probed as described above with primary antibody for CBP (A-22; Santa Cruz Biotechnology) diluted 1:200 in 5% nonfat milk and Blotto reagent and secondary anti-rabbit antibody diluted 1:10,000 in 1% nonfat milk and Blotto reagent. SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific) was used to develop the membrane as described above. Densitometry was performed using ImageJ software version 1.47 (NIH, Bethesda, MD).
MTT Assay for Toxicity
HCC cells were plated in 6-well plates and primary hepatocytes were plated in 12-well plates for 24 hours. Cells were then treated for 24 hours with DMSO, PMED-1, or PMED-2. After incubation, cultures were changed into 1% MTT (w/v) (Sigma-Aldrich) in PBS for 0.5 hours at 37°C. Cells were then lysed using room-temperature isopropanol. Samples were read at 570 nm for colorimetric assessment.
Thymidine Incorporation Proliferation Assay
HepG2 or Snu-398 cells were plated in 6-well plates for 24 hours. Cells were then treated with DMSO or PMED-1 for 24 or 72 hours in the presence of 2.5 μCi of 5′-[3H]thymidine (PerkinElmer). Medium was aspirated, and cells were washed with 1× PBS and incubated with 5% trichloroacetic acid (Sigma-Aldrich) for 15 minutes at 4°C. Next, plates were washed in running water and placed to dry at 37°C. Cells were then lysed in 0.33 N NaOH for 20 minutes at room temperature. Samples were combined with scintillation fluid and measured in a Beckman LS 6000 IC scintillation counter (Beckman Coulter, Brea, CA).
Live Cell Imaging
HepG2 cells plated in 6-well plates were maintained in medium supplemented with DMSO or 200 μmol/L PMED-1 under live-cell bright-field imaging for 24 hours. Cells were imaged using a Nikon (Tokyo, Japan) Eclipse Ti live-cell imaging system.
Immunofluorescent Staining
HepG2 cells were plated 2 × 103 in 4-well chamber slides for 24 hours, followed by 24 hours of DMSO or 25 μmol/L PMED-1 treatment. For detection of cells in active cell cycle (late G1, S, G2, and M phases), immunofluorescence for Ki-67 was performed. After DMSO or PMED-1 treatment of HepG2 cells grown on chamber slides, cells were fixed in 4% paraformaldehyde. Wells were washed with 1× PBS and cells were permeabilized with 0.1% Triton-X surfactant in PBS, followed by washes in PBS alone and then with 0.5% bovine serum albumin in PBS (PBB). Samples were blocked in 2% PBB for 1 hour, washed with 0.5% PBB, and incubated in 0.5% PBB containing primary antibody for Ki-67 (Santa Cruz Biotechnology) at 1:100 for 1 hour. After a washing in 0.5% PBB, secondary antibody anti-goat Alexa Fluor 555, diluted 1:1000 in 0.5% PBB, was applied to each well for 1 hour. Wells were washed with 0.5% PBB, then with 1× PBS, and finally were stained with Hoechst dye 33342 for 30 seconds. Samples were coverslipped and imaged using a Nikon Eclipse Ti live-cell imaging system.
Kinase Assays
Kinase assays were performed to assess activity of PMED-1 against AKT, protein kinase D (PKD), polo-like kinases 1 and 2 (Plk1 and Plk2), and cyclin-dependent kinase-activating kinase (CAK). Opaque Small Volume plates from Greiner Bio-One (Monroe, NC) were used: black for fluorescence polarization (FP) assay and white for time-resolved fluorescence energy transfer (TR-Fret) assay. IMAP binding reagent, binding buffer, terbium, peptide substrates (AKT: 5FAM-GRPRTSSFAEG-COOH; PKD: 5FAM-KKLNRTLSVA-COOH; Plk1 and CAK: 5FAM-KKRNRRLSVA-OH; Plk2: 5FAM-LKKLTRRPSFSAQ-COOH), and kinase reaction buffers [10 mmol/L Tris-HCl at pH 7.2, 10 mmol/L MgCl2, 0.05% NaN3, 1 mmol/L dithiothreitol, and carrier (PKD, AKT: 0.1% bovine serum albumin; Plk1, Plk2, CAK: 0.01% Tween 20)] were purchased from Molecular Devices (Sunnyvale, CA). Active Plk1 and AKT were purchased from Cell Signaling Technology (Danvers, MA), PKD and CAK from Upstate (EMD Millipore), and Plk2 from Life Technologies. ATP was purchased from GE Healthcare (Little Chalfont, UK) and H89 from EMD Millipore.
Compounds were solubilized in DMSO to stock 10 mmol/L and then were diluted to 3× starting concentration 300 μmol/L (3% DMSO) and serially diluted in 3% DMSO–water. In brief, 2 μL of 3× concentration substrate and ATP in 2× complete reaction buffer was added to each well of a microtiter plate. After 2 μL of 3× concentration compound was added, 2 μL of 3× concentration enzyme in 1× complete reaction buffer was added to start the 6 μL total volume kinase reaction. At the designated time, the reaction was stopped by addition of binding solution containing the binding reagent (FP assay) or binding reagent and terbium (TR-Fret assay). Plates were read at excitation/emission wavelengths Ex485/Em525 for FP or at Ex330/Em490, 520 for TR-Fret on a Molecular Devices SpectraMax M5. Specific final reaction conditions were as follows.
IMAP-based Plk2 TR-Fret assay: 0.34 mU/μL Plk2; 550 nmol/L substrate peptide; 35 μmol/L ATP; 1% DMSO; 2.5 hours kinase reaction; 15 μL binding solution (70% A, 30% B + 1/600 binding reagent + 1/400 terbium); 2 hours binding incubation.
IMAP-based PKD FP assay: 0.06 mU/μL PKD; 100 nmol/L substrate peptide; 20 μmol/L ATP; 1% DMSO; 90 minutes kinase reaction; 18 μL binding solution (100% A + 1/400 binding reagent); 2 hours binding incubation.
IMAP-based Plk1 TR-Fret assay: 0.034 mU/μL Plk1; 750 nmol/L substrate peptide; 25 μmol/L ATP; 1% DMSO; 3 hours kinase reaction; 18 μL binding solution (70% A, 30% B + 1/600 binding reagent + 1/400 terbium); 5 hours binding incubation.
IMAP-based CAK TR-Fret assay: 0.1 mU/μL CAK; 500 nmol/L substrate peptide; 25 μmol/L ATP; 1% DMSO; 3 hours kinase reaction; 12 μL binding solution (70% A, 30% B + 1/600 binding reagent + 1/400 terbium).
IMAP-based AKT FP assay: 0.05 mU/μL AKT; 300 nmol/L substrate peptide; 10 μmol/L ATP; 1% DMSO; 90 minutes kinase reaction; 18 μL binding solution (75% A, 25% B + 1/400 binding reagent); 45 minutes binding incubation.
Zebrafish Experiments
Embryos and adult fish were raised and maintained under standard laboratory conditions.19 We used the Tg(OTM:d2EGFP) line to assess Wnt/β-catenin signaling in vivo.20 Five millimolar stock of PMED-1 and 10 mmol/L stock of XAV939 (Cayman Chemical, Ann Arbor, MI) were prepared in 1× DMSO and diluted to 15 and 10 μmol/L, respectively, with egg water supplemented with 0.2 mmol/L 1-phenyl-2-thiourea (Sigma-Aldrich). As a control, 0.5% DMSO solution in the egg water was used. For imaging, embryos were anesthetized with 0.16 mg/mL tricaine (Sigma-Aldrich) and mounted in egg water containing 3% methyl cellulose (Sigma-Aldrich); a Leica M205 FA epifluorescence microscope (Leica Microsystems, Wetzlar, Germany) was used to obtain bright-field and GFP fluorescence images.
Results
Similarity Search Identifies SMs with ≥70% Structural Similarity to ICG-001
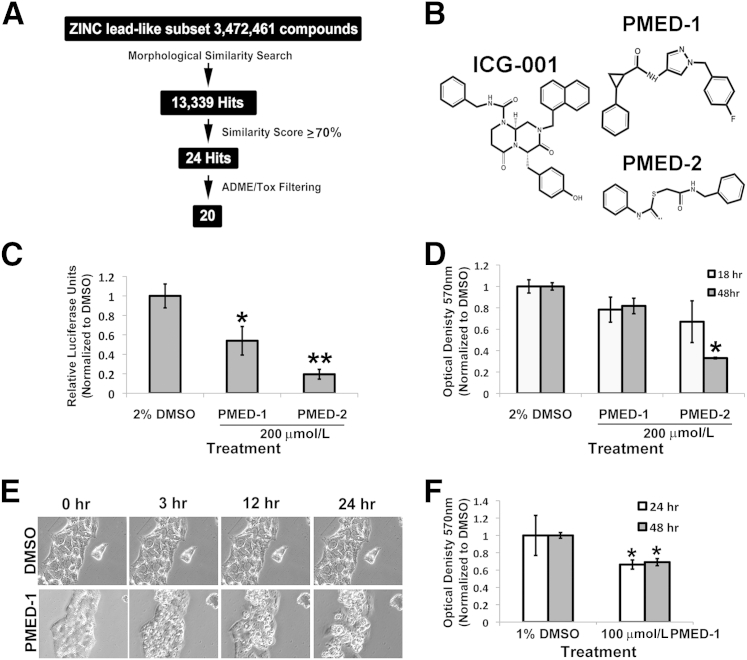
Recognition of SMs by proteins is mediated largely by molecular surface complementarities. Structure-based approaches to drug design use this as the fundamental guiding principle: that is, closely related molecules will elicit similar activity in a biological assay.21,22 Our starting point was to identify compounds similar to ICG-00116 that could serve as structural analogs by screening the lead-like subset of the ZINC 10 database (http://zinc.docking.org/subsets/lead-like, last accessed February 15, 2010) (Figure 1A).23 This search yielded 13,339 compounds, which were sorted by similarity score. We retained 24 compounds with a similarity score of 70% or greater.
Figure 1.
Computational approach to identifying SMs that inhibit Wnt signaling to affect primary human hepatocytes and hepatic tumor cells. A: A search of the ZINC version 8.0 database lead-like subset identified 13,399 SMs structurally similar to ICG-001. A similarity score of ≥70% restricted the hits to 24 molecules, of which 20 passed in silico ADME/tox filtering. B: Chemical structure of ICG-001, PMED-1, and PMED-2. C: HepG2 cells treated with 200 μmol/L PMED-1 or PMED-2 for 24 hours exhibited a significant decrease in TOPflash luciferase reporter activity, compared with 2% DMSO. D: Primary hepatocytes treated with 2% DMSO or with 200 μmol/L PMED-1 or PMED-2 for 18 or 48 hours exhibited a significant decrease in viability by MTT assay only after PMED-2 treatment. E: HepG2 cells treated with 200 μmol/L PMED-1 for 24 hours monitored by live-cell imaging exhibit notable morphological changes reminiscent of cell death as early as 3 hours after treatment, compared with the 2% DMSO-treated controls. F: HepG2 cells treated with 100 μmol/L PMED-1 exhibited a significant decrease in cell viability as assessed by MTT assay, compared with 1% DMSO control. Data are expressed as means ± SD. ∗P < 0.05, ∗∗P < 0.01. Original magnification, ×200 (E).
ADME/Tox Predictions Identify 20 SMs That Pass in Silico Screening
Because most failures in clinical trials result from issues related to drug pharmacodynamics and pharmacokinetics, all state-of-the-art drug-discovery strategies and programs now incorporate the evaluation of absorption, distribution, metabolism, and excretion (ADME) and toxicological (Tox) properties early in the discovery process, to mitigate this risk. We therefore applied in silico ADME/Tox filtering to prioritize compounds for purchase and further characterization. Of the 24 compounds, 20 passed all of the ADME/Tox filters. Based on the highest structural similarity to ICG-001, the top two of these molecules, which we designated PMED-1 and PMED-2, were selected for further characterization. The structures of PMED-1, PMED-2, and ICG-001 are shown in Figure 1B.
Identification of an SM That Inhibits β-Catenin Activity Without Toxicity to Primary Human Hepatocytes
To characterize the effectiveness and safety of the top two SMs in inhibiting β-catenin–TCF activity, we treated HepG2 cells (a human hepatoblastoma cell line that harbors constitutively active β-catenin because of a monoallelic CTNNB1 exon-3 deletion) with a relatively high dose of PMED-1 or PMED-2 (200 μmol/L) or with 2% DMSO for 24 hours.24 TOPflash luciferase reporter assay revealed a significant decrease in β-catenin–TCF activity after PMED-1 or PMED-2 treatment (Figure 1C). However, primary human hepatocytes treated with the same high concentration showed significant toxicity as assessed by MTT assay only to PMED-2 (200 μmol/L) after 48 hours (Figure 1D). Because PMED-1 was nontoxic to human hepatocytes, we next treated HepG2 cells with the same high concentration (200 μmol/L) for durations up to 24 hours. This treatment led to notable cell death, observed as clumping and rounding, compared with normally spread viable cells in DMSO-treated cultures (Figure 1E). Furthermore, even at 100 μmol/L the PMED-1 treatment of these cells for 24 and 48 hours led to approximately 30% to 35% reduction in viability as assessed by MTT assay (Figure 1F). These results indicate that PMED-1, but not PMED-2, is selectively toxic to tumor cells by affecting β-catenin activity.
PMED-1 Treatment Blocks Cell-Cycle Progression, Proliferation, and Downstream Wnt Signaling in HepG2 Cells
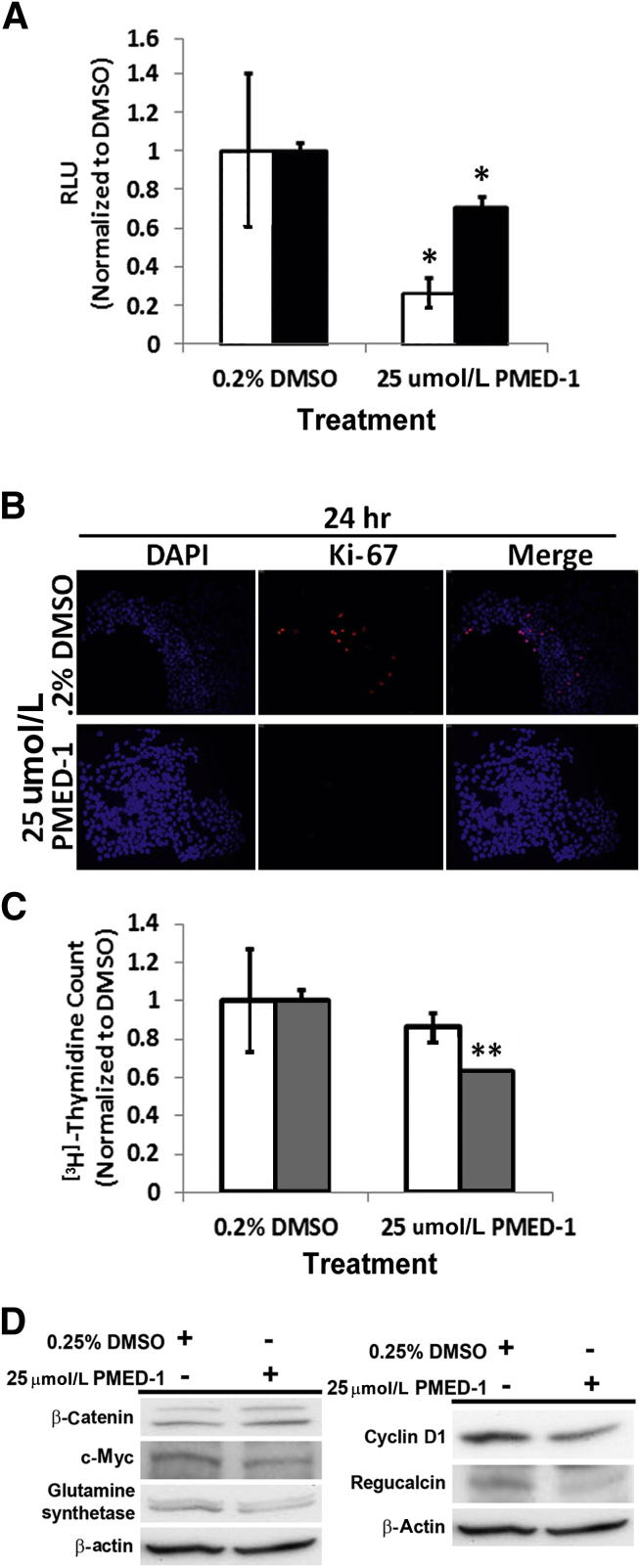
Next, IC50 was assessed for PMED-1 in reducing TOPflash reporter activity in HepG2 cells after 24 hours of treatment with concentrations ranging from 200 μmol/L to 500 nmol/L. A significant decrease in TOPflash reporter activity was observed only at a concentration of ≥25 μmol/L in this human hepatoblastoma cell line (data not shown). Interestingly, HepG2 cells treated with PMED-1 at 25 μmol/L showed maximal decrease in TOPflash reporter activity at 24 hours, with some rebound at 48 hours (Figure 2A). To determine the effect of blocking β-catenin activity on cell proliferation, we first assessed cells in active cell cycle by Ki-67 immunofluorescence. A notable decrease in the number of Ki-67+ HepG2 cells was observed at 24 hours after PMED-1 treatment (Figure 2B). To further address this decrease in DNA synthesis over time, we performed a [3H]thymidine incorporation assay, which revealed a persistent decrease over 72 hours after PMED-1 treatment (Figure 2C). We next examined protein expression of β-catenin and some of its targets in HepG2 cells at 24 hours after 25 μmol/L PMED-1 treatment. Although no change in total β-catenin was evident, a notable decrease in glutamine synthetase, cyclin D1, c-Myc, and regucalcin protein levels was evident (Figure 2D). Thus, PMED-1 treatment impedes β-catenin signaling in HepG2 cells.
Figure 2.
PMED-1 targets β-catenin signaling to affect cell proliferation in HepG2 cells. A: HepG2 cells treated with 25 μmol/L PMED-1 for 24 hours (white bars) exhibited a significant decrease in TOPflash reporter activity, but less so at 48 hours (black bars). B: Decrease in Ki-67+ cells (red) was observed after treatment of HepG2 cells with 25 μmol/L PMED-1 for 24 hours, compared with 0.2% DMSO. C: DNA synthesis in HepG2 cells as measured by [3H]thymidine incorporation exhibited a decrease in response to 24 (white bars) or 72 (gray bars) hours of treatment with 25 μmol/L of PMED-1, compared with 0.2% DMSO. D: Western blots using HepG2 cell lysates reveal decreased β-catenin targets after 25 μmol/L PMED-1 treatment. Data are expressed as means ± SD. ∗P < 0.05, ∗∗P < 0.01. Original magnification, ×100 (B).
PMED-1 Affects β-Catenin Signaling to Impair Viability and Proliferation in Multiple HCC Cell Lines
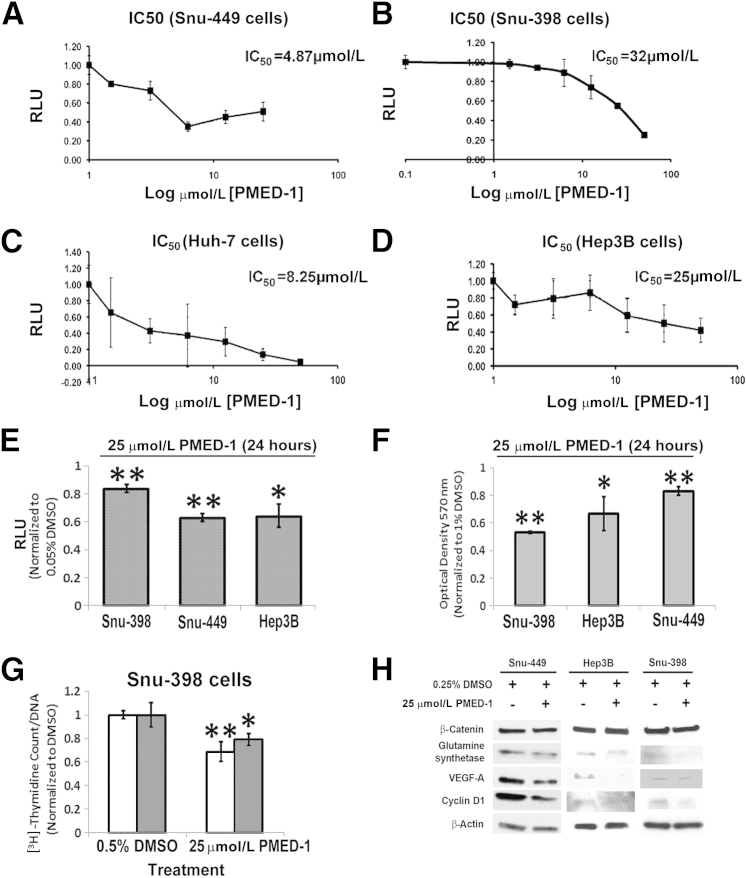
To determine IC50 of PMED-1 in reducing β-catenin–TCF activity in multiple HCC cells, we treated Snu-449, Huh-7, and Hep3B cells [which contain the wild-type β-catenin gene (CTNNB1)] and Snu-398 cells (which harbor the S37C mutation in CTNNB1) with a range of PMED-1 doses, as described under Materials and Methods. Based on this analysis, PMED-1 had an IC50 of 4.87 μmol/L for Snu-449 cells, 8.25 μmol/L for Huh-7 cells, 25 μmol/L for Hep3B cells, and 32 μmol/L for Snu-398 cells (Figure 3, A–D). Next, to compare the effect on β-catenin activity of PMED-1 at the single dose of 25 μmol/L, we treated Snu-449, Hep3B, and Snu-398 cells for 24 hours. A significant decrease in TOPflash reporter activity was evident in all cells, compared with DMSO, albeit to varying extent in accord with the respective IC50 values (Figure 3E). Intriguingly, the maximum effect of PMED-1–mediated decrease in cell viability by MTT assay was evident in β-catenin–mutated Snu-398 cells, demonstrating their addiction to β-catenin signaling (Figure 3F); however, decreased survival was evident in other HCC cells as well (Figure 3F). A significant decrease in [3H]thymidine incorporation was also noted in Snu-398 cells treated for 24 or 72 hours with PMED-1 at 25 μmol/L (Figure 3G).
Figure 3.
IC50 of PMED-1 in various HCC cells and the effect of PMED-1 on β-catenin signaling and biology. A–D: IC50 was determined by response of TOPflash reporter activity to PMED-1 treatment for 24 hours in Snu-449 (A), Snu-398 (B), Huh-7 (C), and Hep3B (D) cells. E: Relative decrease in TOPflash reporter activity after 25 μmol/L PMED-1 treatment of various HCC cells for 24 hours. F: The most robust decrease in viability as measured by MTT assay was observed in Snu-398 cells after 24 hours of 25 μmol/L PMED-1 treatment. G: DNA synthesis as measured by [3H]thymidine incorporation was significantly reduced in Snu-398 cells treated for 24 (white bars) or 72 (gray bars) hours with 25 μmol/L PMED-1, compared with DMSO. H: Western blots using whole-cell lysates from Snu-398, Snu-449, and Hep3B cells treated with 25 μmol/L PMED-1 reveal notable decreases in β-catenin downstream targets. Data are expressed as means ± SD. ∗P < 0.05, ∗∗P < 0.01.
Whole-cell lysates from three HCC cell lines treated for 24 hours with 25 μmol/L PMED-1 were also assessed for protein expression of β-catenin and its targets. A notable but varying decrease in glutamine synthetase, cyclin D1, and VEGF-A expression was observed in all cells, without any influence on total β-catenin levels (Figure 3H). Thus, PMED-1 effectively targets β-catenin activity in multiple HCC cells to impair their viability and proliferation.
PMED-1 Inhibits β-Catenin–CBP Interactions
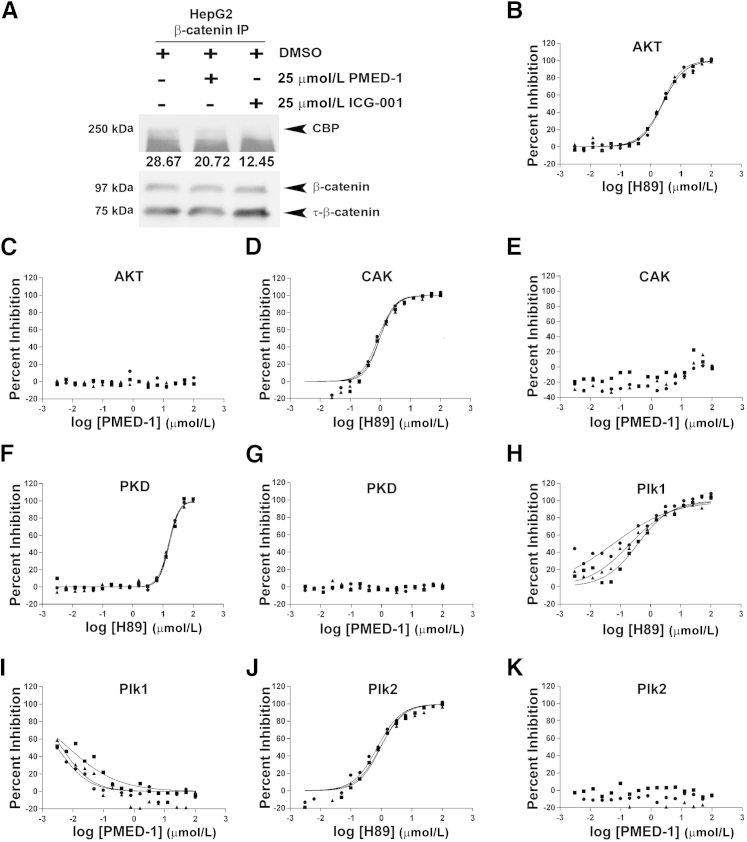
To address the molecular basis of β-catenin inhibition by PMED-1, we assessed β-catenin–CBP interactions, because their disruption is reported to be the major mechanism of β-catenin inhibition by ICG-001.16 Indeed, treatment of HepG2 cells with PMED-1 led to a notable decrease in CBP-β-catenin as assessed by immunoprecipitation studies, compared with DMSO treatment, although the effect was less robust than with ICG-001 (Figure 4A). Similar modest decreases in β-catenin–CBP interactions were observed in response to PMED-1 treatment of other cells, such as Snu-398 cells (data not shown).
Figure 4.
PMED-1 affects β-catenin–CBP interactions but does not affect activity of other common kinases. A: Western blot analysis of whole-cell lysates from untreated HepG2 cells or HepG2 cells treated for 24 hours with either 2% DMSO, 25 μmol/L PMED-1, or 25 μmol/L ICG-001 used for immunoprecipitation revealed significantly less CBP pull-down with anti–β-catenin antibody in cells treated with PMED-1 or ICG-001, compared with DMSO. Densitometry values (arbitrary units) for CBP are provided under the bands. B–K: Effect of H89 and PMED-1 on activity of kinases as assessed by IMAP-based FP assay (AKT and PKD) or TR-Fret assay (CAK, Plk1, and Plk2). Three replicates are shown. B: H89 decreases CAK activity in a dose-dependent manner. C: PMED-1 does not affect AKT activity. D: H89 decreases CAK activity. E: PMED-1 does not affect CAK activity. F: H89 decreases PKD activity at higher concentrations. G: PMED-1 does not affect PKD activity. H: H89 decreases Plk1 activity in a largely dose-dependent manner. I: PMED-1 had an inhibitory effect on Plk1 up to −1 μmol/L on the log scale (0.097 μmol/L); however, no decrease was evident at higher concentrations. J: H89 decreases Plk2 activity in a dose-dependent manner. K: PMED-1 does not affect Plk2 activity. IP, immunoprecipitation.
PMED-1 Does Not Inhibit Common Kinases
Next, we determined whether PMED-1 affects activity of common kinases that could be the basis of some of the observed biological effects. We compared the efficacy of PMED-1 to the multikinase inhibitor H89, which at high doses can inhibit AKT, PKD, CAK, Plk1, and Plk2.25 Although dose-dependent inhibition of AKT (Figure 4B), CAK (Figure 4D), PKD (Figure 4F), Plk1 (Figure 4H) and Plk2 (Figure 4J) in response to H89 was observed, no inhibition was evident after PMED-1 treatment, especially at 25 μmol/L, which equates to 1.39 log μmol/L (Figure 4, C, E, G, I, and K), a dose at which significant decreases in β-catenin–TCF activity were observed. Intriguingly, low doses of PMED-1 (−2.5 to −1.0 log μmol/L, equating to 0.003 μmol/L to 0.09 μmol/L) led to an inverse decrease in Plk1 kinase; at higher doses, however, this effect was lost (Figure 4I). Thus, PMED-1 does not appear to have any significant off-target effects on these major kinases, especially at doses that effectively blocked β-catenin activity.
PMED-1 Impairs Wnt/β-Catenin Signaling in Vivo in Zebrafish Embryos
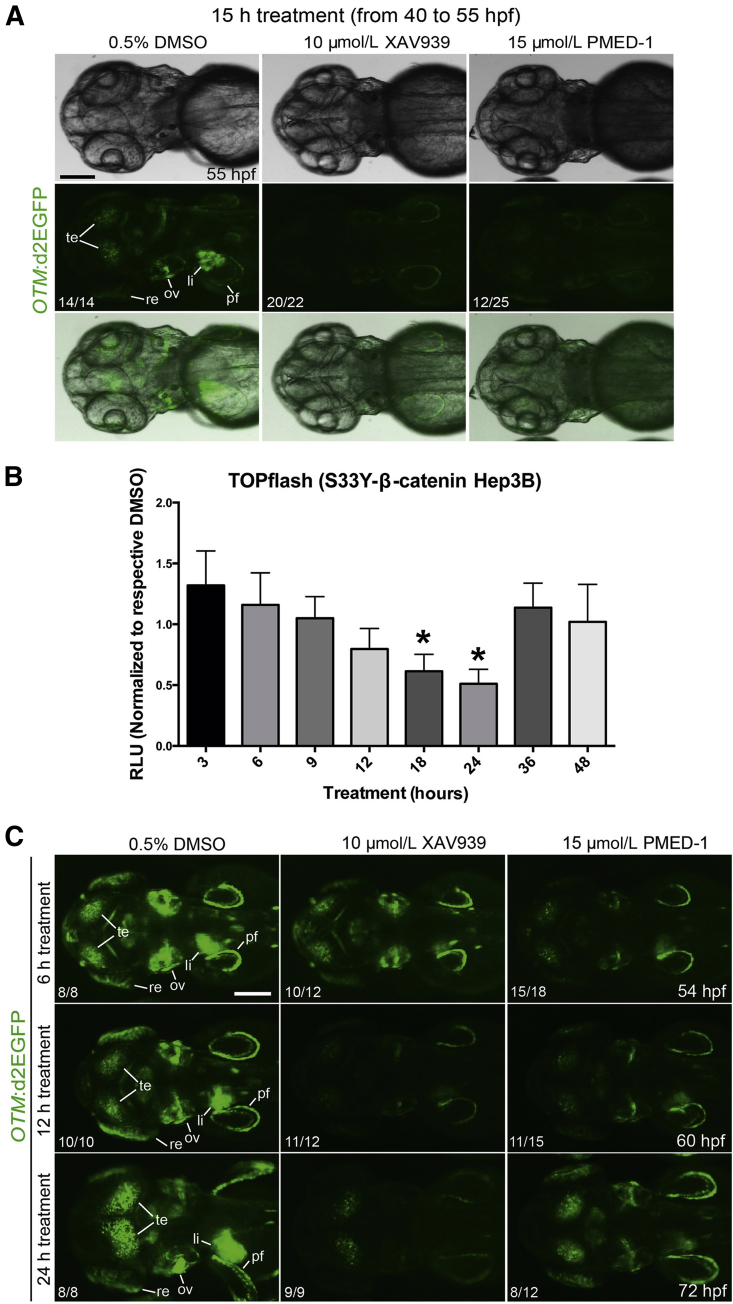
To determine whether PMED-1 can inhibit Wnt/β-catenin signaling in vivo, we used the Tg(OTM:d2EGFP) zebrafish line, which expresses destabilized GFP under the promoter containing six copies of Tcf/Lef binding sites.20 XAV939, a known β-catenin inhibitor in zebrafish, was used as a positive control.13 GFP expression was greatly reduced in 20/22 (91%) of the Tg(OTM:d2EGFP) embryos treated with XAV939 for 15 hours, from 40 to 55 hours after fertilization (hpf), and was weakly reduced in the remaining embryos, compared with DMSO-treated controls (Figure 5A). Importantly, GFP expression was greatly reduced in 12/25 (48%) of the embryos treated with PMED-1, compared with the controls (Figure 5A). These data indicate the in vivo efficacy of PMED-1 as a Wnt/β-catenin signaling inhibitor.
Figure 5.
Efficacy and duration of the effect of PMED-1 effect on β-catenin activity in vivo and in vitro. A: GFP+ embryos obtained from outcrossing Tg(OTM:d2EGFP) zebrafish were treated with DMSO, XAV939, or PMED-1 for 15 hours (40 to 55 hpf), and GFP expression was assessed under a fluorescence microscope. Shown are bright-field images (top row), GFP fluorescence images (middle row), and merged images (bottom row). B: Stably transfected Hep3B cells with S33Y-β-catenin, treated with a single dose of PMED-1 at 25 μmol/L, exhibited a gradual decrease in TOPflash reporter activity; the decrease reached significance at 18 and 24 hours after treatment, with rebound at 36 and 48 hours. C: Epifluorescence images of GFP expression in Tg(OTM:d2EGFP) embryos. Embryos strongly GFP+ [obtained from incrossing Tg(OTM:d2EGFP) zebrafish] were treated with DMSO, XAV939, or PMED-1 from 48 hpf. Embryos were harvested 6, 12, or 24 hours later, and GFP expression was assessed under a fluorescence microscope. ∗P < 0.05. The proportion of embryos (n/N) exhibiting the GFP expression is indicated at the lower left of each image. All images are dorsal view, with anterior to the left. Scale bars: 100 μm (A and C). li, liver; ov, otic vesicle; pf, pectoral fin; re, retina; te, tectum.
Duration of Action of PMED-1 in Cell Culture and in Vivo
To address the duration of the effect of PMED-1 on β-catenin activity in cell culture and in vivo, we treated Hep3B cells that were stably transfected with constitutively active S33Y-β-catenin mutant. After a single-dose treatment of PMED-1 at 25 μmol/L, the cells were harvested at times ranging from 3 hours to 48 hours for TOPflash reporter activity. As early as 6 hours after PMED-1 treatment, a downward trend in TOPflash reporter activity was evident, compared with DMSO (Figure 5B). Statistically significant inhibition of β-catenin activity was evident at 18 to 24 hours after PMED-1 treatment. Intriguingly, at 36 hours TOPflash reporter activity was restored. Thus, a single treatment with 25 μmol/L PMED-1 progressively decreases β-catenin activity from 6 to 24 hours in cell culture.
To determine how long PMED-1 can inhibit Wnt/β-catenin signaling in vivo, we examined GFP expression in Tg(OTM:d2EGFP) zebrafish embryos at multiple time points after a single PMED-1 treatment. To better reveal GFP expression in the embryos, we used homozygous Tg(OTM:d2EGFP) embryos, in which GFP expression is stronger than in hemizygous embryos. GFP+ Tg(OTM:d2EGFP) embryos were treated with DMSO, XAV939, or PMED-1 at 48 hpf and GFP expression was examined at 6, 12, or 24 hours later. At 54 hpf (6 hours after treatment), most embryos treated with XAV939 (10/12) or PMED-1 (15/18) exhibited reduced GFP expression, compared with controls (Figure 5C). At 60 hpf (12 hours after treatment), GFP expression was further reduced in XAV939-treated embryos, and it continued to be reduced in PMED-1–treated embryos (Figure 5C). Interestingly, at 72 hpf (24 hours after treatment), GFP expression was significantly restored in most PMED-1–treated embryos (8/12), but it continued to be suppressed in XAV939-treated embryos (Figure 5C). These data indicate that the Wnt/β-catenin inhibitory activity of PMED-1 lasts a shorter time than that of XAV939.
Discussion
Currently, there is a lack of an effective medical therapy for HCC. The multikinase inhibitor sorafenib has been shown to improve overall survival of patients with advanced HCC.9,26 A distinct pathway active in significant subsets of HCC patients is the Wnt/β-catenin signaling pathway.27 Activation of β-catenin in HCC may lead to increased expression of genes encoding for cyclin D1, glutamine synthetase, survivin, c-Myc, EpCAM, and VEGF-A, which contribute to HCC by regulating cell proliferation, survival, metabolism, cancer stem cell expansion, and angiogenesis.12,28–33 Thus, inhibition of β-catenin in a subset of HCC is expected to be of significant therapeutic relevance. However, there are no effective inhibitors of β-catenin approved for clinical use, although its inhibition in HCC is being increasingly discussed,10,34 thus making it an interesting target for drug design.
Developing a drug from bench to bedside is an arduous and slow process. High-throughput screens assess SMs for potential efficacy against a desired target; however, because there is little to no selectivity or specificity in these screens, the eventual effect may be due to an off-target effect.35 The intention in such studies is to discover a molecule that will cause cell death in culture (although with little to no knowledge of the mechanism), which can then be tested for its efficacy in vivo. We took a novel approach by basing our drug discovery on ICG-001, a previously established SM inhibitor of β-catenin–CBP interactions. Based on the premise of structure–function similarity, we found that PMED-1, a SM resembling the structure of ICG-001, shows similar function in inhibiting β-catenin in multiple HCC cell lines, although at a higher concentration. Like ICG-001, PMED-1 inhibits β-catenin–CBP interactions and thus impedes downstream signaling. CBP is a histone acetyl transferase and interacts with β-catenin in the nucleus once it has been activated and has been shown to be critical for β-catenin–dependent target gene expression.16 Importantly, PMED-1 did not show any off-target kinase inhibition. We also checked the quality of PMED-1 by high-performance liquid chromatography and nuclear magnetic resonance, to determine whether any contaminant or degradation product could be responsible for inhibiting β-catenin–TCF activity. The results showed that the PMED-1 used in our studies was >95% pure (data not shown).
PMED-1 successfully targeted β-catenin signaling in a hepatoblastoma cell line with exon-3 deletion in β-catenin gene (HepG2) and a HCC cell line with β-catenin point mutation (Snu-398) with an IC50 of 25 μmol/L and 32 μmol/L, respectively. Several downstream targets of β-catenin signaling were suppressed, which led to impaired tumor cell proliferation and survival. Additional HCC cells with wild-type β-catenin gene were more responsive to inhibitory effect of PMED-1, although these cells showed less addiction to Wnt signaling for their growth and survival. Indeed, Snu-398 cells showed a robust decrease in tumor cell survival and proliferation on PMED-1 treatment. An in vitro time course showed that the anti–β-catenin effect of a single dose of PMED-1 lasts up to 24 hours. This was in contrast to ICG-001, which has a prolonged effect lasting several days (unpublished data; M.K.). Interestingly, the effect of a single dose of PMED-1 on cell proliferation or viability was evident at 72 hours, indicating that even transient interruption of β-catenin signaling cascade may impair downstream biological processes for a longer duration. Indeed, similar prolonged biological effects of transient β-catenin suppression by other modalities such as peptide nucleic acid antisense on cell proliferation and survival have been reported.12 How PMED-1 is metabolized and inactivated remains a topic of ongoing investigation.
A major limitation of the present study is the issue of solubility of PMED-1. As a hydrophobic SM, PMED-1 could be dissolved only in DMSO, which limited our ability to perform in vivo studies in orthotopic or xenograft models of HCC in mice. However, we demonstrated its in vivo efficacy in a zebrafish model. In this model, similar to cell culture, PMED-1 exhibited peak anti–β-catenin activity from 5 to 15 hours after treatment, further evidence of a relatively short half-life. This short half-life most likely explains the restoration of β-catenin–TCF-driven EGFP expression in approximately 50% of the zebrafish embryos at 15 hours after PMED-1 treatment and in 67% of the embryos at 24 hours. Although we did not directly investigate the mechanism of PMED-1 in vivo, it likely also involves blockade of β-catenin–CBP interactions. Additional studies will be necessary to address whether chemical or structural modifications can alter the solubility and metabolism of PMED-1 for improved drug delivery.
Ultimately, the present study demonstrates the practical application of in silico screening in effective identification of a specific molecule against β-catenin–driven HCC, with future therapeutic implications. This technique led us to identify PMED-1, which is similar to its parent compound ICG-001 in some ways and different in others. PMED-1 is especially different from ICG-001 in having a less robust β-catenin inhibitory effect and in having a shorter duration of action. However, these seemingly negative attributes of PMED-1 could make this agent more suitable for use in certain specific applications, such as locoregional therapies. This may be especially relevant, because PMED-1 is quite hydrophobic, a property that may allow for it to be retained in the tumor microenvironment. Similarly, a shorter half-life of the agent in vivo theoretically lessens the systemic adverse effects. Further studies will be critical to address such important questions relevant to PMED-1.
Acknowledgments
We thank Drs. Peter Wipf and Erin Skoda (University of Pittsburgh) for help with assessing the quality of PMED-1 by high-performance liquid chromatography and nuclear magnetic resonance.
Footnotes
Supported by NIH grants 1R01DK062277, 1R01DK100287, and 1R01DK095498 (S.P.M.) and the University of Pittsburgh Endowed Chair for Experimental Pathology (S.P.M.).
Disclosures: None declared.
Contributor Information
Gabriela Mustata Wilson, Email: gmwilson@usi.edu.
Satdarshan P. Monga, Email: smonga@pitt.edu.
References
- 1.Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Yang J.D., Roberts L.R. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fortune B.E., Umman V., Gilliland T., Emre S. Liver transplantation for hepatocellular carcinoma: a surgical perspective. J Clin Gastroenterol. 2013;47(Suppl):S37–S42. doi: 10.1097/MCG.0b013e318286ff8e. [DOI] [PubMed] [Google Scholar]
- 4.Imamura H., Matsuyama Y., Tanaka E., Ohkubo T., Hasegawa K., Miyagawa S., Sugawara Y., Minagawa M., Takayama T., Kawasaki S., Makuuchi M. Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J Hepatol. 2003;38:200–207. doi: 10.1016/s0168-8278(02)00360-4. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki K., Kono N., Ono A., Osuga Y., Kiyokawa H., Mineo I., Matsuda Y., Miyoshi S., Kawata S., Minami Y., Moriwaki K., Tarui S. Transcatheter arterial chemo-embolization for humoral hypercalcemia of hepatocellular carcinoma. Gastroenterol Jpn. 1988;23:29–36. doi: 10.1007/BF02918853. [DOI] [PubMed] [Google Scholar]
- 6.Stock P., Monga D., Tan X., Micsenyi A., Loizos N., Monga S.P. Platelet-derived growth factor receptor-alpha: a novel therapeutic target in human hepatocellular cancer. Mol Cancer Ther. 2007;6:1932–1941. doi: 10.1158/1535-7163.MCT-06-0720. [DOI] [PubMed] [Google Scholar]
- 7.Ng I.O., Poon R.T., Lee J.M., Fan S.T., Ng M., Tso W.K. Microvessel density, vascular endothelial growth factor and its receptors Flt-1 and Flk-1/KDR in hepatocellular carcinoma. Am J Clin Pathol. 2001;116:838–845. doi: 10.1309/FXNL-QTN1-94FH-AB3A. [DOI] [PubMed] [Google Scholar]
- 8.Dhar D.K., Naora H., Yamanoi A., Ono T., Kohno H., Otani H., Nagasue N. Requisite role of VEGF receptors in angiogenesis of hepatocellular carcinoma: a comparison with angiopoietin/Tie pathway. Anticancer Res. 2002;22:379–386. [PubMed] [Google Scholar]
- 9.Bronte F., Bronte G., Cusenza S., Fiorentino E., Rolfo C., Cicero G., Bronte E., Di Marco V., Firenze A., Angarano G., Fontana T., Russo A. Targeted therapies in hepatocellular carcinoma. Curr Med Chem. 2014;21:966–974. doi: 10.2174/09298673113209990234. [DOI] [PubMed] [Google Scholar]
- 10.Nejak-Bowen K.N., Monga S.P. Beta-catenin signaling, liver regeneration and hepatocellular cancer: sorting the good from the bad. Semin Cancer Biol. 2011;21:44–58. doi: 10.1016/j.semcancer.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behari J., Zeng G., Otruba W., Thompson M.D., Muller P., Micsenyi A., Sekhon S.S., Leoni L., Monga S.P. R-Etodolac decreases beta-catenin levels along with survival and proliferation of hepatoma cells. J Hepatol. 2007;46:849–857. doi: 10.1016/j.jhep.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delgado E., Bahal R., Yang J., Lee J.M., Ly D.H., Monga S.P. β-Catenin knockdown in liver tumor cells by a cell permeable gamma guanidine-based peptide nucleic acid. Curr Cancer Drug Targets. 2013;13:867–878. doi: 10.2174/15680096113139990081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S.M., Mishina Y.M., Liu S., Cheung A., Stegmeier F., Michaud G.A. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 14.Thompson M.D., Dar M.J., Monga S.P. Pegylated interferon alpha targets Wnt signaling by inducing nuclear export of beta-catenin. J Hepatol. 2011;54:506–512. doi: 10.1016/j.jhep.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng G., Apte U., Cieply B., Singh S., Monga S.P. siRNA-mediated beta-catenin knockdown in human hepatoma cells results in decreased growth and survival. Neoplasia. 2007;9:951–959. doi: 10.1593/neo.07469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emami K.H., Nguyen C., Ma H., Kim D.H., Jeong K.W., Eguchi M., Moon R.T., Teo J.L., Kim H.Y., Moon S.H., Ha J.R., Kahn M. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription. Proc Natl Acad Sci USA. 2004;101:12682–12687. doi: 10.1073/pnas.0404875101. [Erratum appeared in Proc Natl Acad Sci USA 2004, 101:16707] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi-Yanaga F., Kahn M. Targeting Wnt signaling: can we safely eradicate cancer stem cells? Clin Cancer Res. 2010;16:3153–3162. doi: 10.1158/1078-0432.CCR-09-2943. [DOI] [PubMed] [Google Scholar]
- 18.Jain A.N. Morphological similarity: a 3D molecular similarity method correlated with protein-ligand recognition. J Comput Aided Mol Des. 2000;14:199–213. doi: 10.1023/a:1008100132405. [DOI] [PubMed] [Google Scholar]
- 19.Westerfield M: The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio), ed 4. University of Oregon Press, Eugene, 2000
- 20.Shimizu N., Kawakami K., Ishitani T. Visualization and exploration of Tcf/Lef function using a highly responsive Wnt/beta-catenin signaling-reporter transgenic zebrafish. Dev Biol. 2012;370:71–85. doi: 10.1016/j.ydbio.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 21.Martin Y.C., Kofron J.L., Traphagen L.M. Do structurally similar molecules have similar biological activity? J Med Chem. 2002;45:4350–4358. doi: 10.1021/jm020155c. [DOI] [PubMed] [Google Scholar]
- 22.Wallqvist A., Huang R., Thanki N., Covell D.G. Evaluating chemical structure similarity as an indicator of cellular growth inhibition. J Chem Inf Model. 2006;46:430–437. doi: 10.1021/ci0501544. [DOI] [PubMed] [Google Scholar]
- 23.Irwin J.J., Sterling T., Mysinger M.M., Bolstad E.S., Coleman R.G. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model. 2012;52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carruba G., Cervello M., Miceli M.D., Farruggio R., Notarbartolo M., Virruso L., Giannitrapani L., Gambino R., Montalto G., Castagnetta L. Truncated form of beta-catenin and reduced expression of wild-type catenins feature HepG2 human liver cancer cells. Ann N Y Acad Sci. 1999;886:212–216. doi: 10.1111/j.1749-6632.1999.tb09419.x. [DOI] [PubMed] [Google Scholar]
- 25.Lochner A., Moolman J.A. The many faces of H89: a review. Cardiovasc Drug Rev. 2006;24:261–274. doi: 10.1111/j.1527-3466.2006.00261.x. [DOI] [PubMed] [Google Scholar]
- 26.Ibrahim N., Yu Y., Walsh W.R., Yang J.L. Molecular targeted therapies for cancer: sorafenib mono-therapy and its combination with other therapies (review) Oncol Rep. 2012;27:1303–1311. doi: 10.3892/or.2012.1675. [DOI] [PubMed] [Google Scholar]
- 27.Monga S.P. Role of Wnt/β-catenin signaling in liver metabolism and cancer. Int J Biochem Cell Biol. 2011;43:1021–1029. doi: 10.1016/j.biocel.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cadoret A., Ovejero C., Terris B., Souil E., Lévy L., Lamers W.H., Kitajewski J., Kahn A., Perret C. New targets of beta-catenin signaling in the liver are involved in the glutamine metabolism. Oncogene. 2002;21:8293–8301. doi: 10.1038/sj.onc.1206118. [DOI] [PubMed] [Google Scholar]
- 29.Loeppen S., Schneider D., Gaunitz F., Gebhardt R., Kurek R., Buchmann A., Schwarz M. Overexpression of glutamine synthetase is associated with beta-catenin-mutations in mouse liver tumors during promotion of hepatocarcinogenesis by phenobarbital. Cancer Res. 2002;62:5685–5688. [PubMed] [Google Scholar]
- 30.Ma H., Nguyen C., Lee K.S., Kahn M. Differential roles for the coactivators CBP and p300 on TCF/beta-catenin-mediated survivin gene expression. Oncogene. 2005;24:3619–3631. doi: 10.1038/sj.onc.1208433. [DOI] [PubMed] [Google Scholar]
- 31.Nejak-Bowen K.N., Thompson M.D., Singh S., Bowen W.C., Jr., Dar M.J., Khillan J., Dai C., Monga S.P. Accelerated liver regeneration and hepatocarcinogenesis in mice overexpressing serine-45 mutant beta-catenin. Hepatology. 2010;51:1603–1613. doi: 10.1002/hep.23538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan X., Behari J., Cieply B., Michalopoulos G.K., Monga S.P. Conditional deletion of beta-catenin reveals its role in liver growth and regeneration. Gastroenterology. 2006;131:1561–1572. doi: 10.1053/j.gastro.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 33.Yao H., Ashihara E., Strovel J.W., Nakagawa Y., Kuroda J., Nagao R., Tanaka R., Yokota A., Takeuchi M., Hayashi Y., Shimazaki C., Taniwaki M., Strand K., Padia J., Hirai H., Kimura S., Maekawa T. AV-65, a novel Wnt/β-catenin signal inhibitor, successfully suppresses progression of multiple myeloma in a mouse model. Blood Cancer J. 2011;1:e43. doi: 10.1038/bcj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoshida Y., Toffanin S., Lachenmayer A., Villanueva A., Minguez B., Llovet J.M. Molecular classification and novel targets in hepatocellular carcinoma: recent advancements. Semin Liver Dis. 2010;30:35–51. doi: 10.1055/s-0030-1247131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teriete P., Pinkerton A.B., Cosford N.D. Inhibitors of tissue-nonspecific alkaline phosphatase (TNAP): from hits to leads. Methods Mol Biol. 2013;1053:85–101. doi: 10.1007/978-1-62703-562-0_5. [DOI] [PubMed] [Google Scholar]