CDK4 activation/RB phosphorylation occurs in 50% of indolent but high-risk follicular lymphomas and implies susceptibility to dual CDK4 and BCL2 inhibition.
Abstract
Loss of cell cycle controls is a hallmark of cancer and has a well-established role in aggressive B cell malignancies. However, the role of such lesions in indolent follicular lymphoma (FL) is unclear and individual lesions have been observed with low frequency. By analyzing genomic data from two large cohorts of indolent FLs, we identify a pattern of mutually exclusive (P = 0.003) genomic lesions that impair the retinoblastoma (RB) pathway in nearly 50% of FLs. These alterations include homozygous and heterozygous deletions of the p16/CDKN2a/b (7%) and RB1 (12%) loci, and more frequent gains of chromosome 12 that include CDK4 (29%). These aberrations are associated with high-risk disease by the FL prognostic index (FLIPI), and studies in a murine FL model confirm their pathogenic role in indolent FL. Increased CDK4 kinase activity toward RB1 is readily measured in tumor samples and indicates an opportunity for CDK4 inhibition. We find that dual CDK4 and BCL2 inhibitor treatment is safe and effective against available models of FL. In summary, frequent RB pathway lesions in indolent, high-risk FLs indicate an untapped therapeutic opportunity.
Follicular lymphoma (FL) is an incurable B cell lymphoma that is diagnosed in 18,000 Americans and has a worldwide incidence of ∼120,000 cases per year. The clinical behavior of FLs is characterized by slow and relentless growth with inevitable relapses despite intensive chemotherapy, and eventually 50% progress toward an aggressive disease that resembles diffuse large B cell lymphoma (DLBCL). Genetically, FLs are characterized by the translocation t(14;18) that activates the anti-apoptotic BCL2 protein, and it is clear that additional lesions are required (Staudt, 2007). Accordingly, recent studies have cataloged a large number of genomic lesions in FL with increasing resolution and precision (e.g., Morin et al., 2011; Bouska et al., 2014; Okosun et al., 2014; Pasqualucci et al., 2014), and studies on serial samples have identified chromatin modifiers (e.g., EZH2 and CREBBP) as early targets followed by the acquisition of additional lesions as the disease evolves (Bödör et al., 2013; Green et al., 2013).
Loss of proliferation control is a hallmark of cancer and is also seen in aggressive B cell malignancies like mantle cell lymphoma, transformed FL, and DLBCL (Morin et al., 2011; Okosun et al., 2014; Pasqualucci et al., 2014). In contrast, in the indolent stages of FL, disruption of cell cycle checkpoints (e.g., p16 or RB1) is considered a rare event and mostly linked to disease transformation (Pinyol et al., 1998; Pasqualucci et al., 2014). This view has clinical consequences and, for example, the use of cell cycle–directed therapeutics is not typically considered at this stage (Fry et al., 2004; Relander et al., 2010; Flaherty et al., 2012).
Significantly linked—mutually exclusive or co-occurring—genetic lesions can provide insight into the genetic drivers of cancers. For example, mutual exclusivity between lesions suggests that they target either redundant or incompatible functions and this knowledge can help define the functionally relevant targets of complex aberrations. For example, in the present study we observe a mutually exclusive relation between lesions affecting the p16/CDKN2A locus, the retinoblastoma (RB) locus, and larger gains affecting chromosome 12q13. The association suggests that a cell cycle regulator may be a target of the Chr. 12q13 gain, and notably the amplicon always includes the RB1 kinase CDK4. In the present study, we examine the role of these lesions in lymphomagenesis and patient risk, and explore therapeutic implications.
RESULTS
Analysis of array-CGH data from two independent cohorts of indolent FLs
The first dataset consists of 64 FL samples collected at the Memorial Sloan-Kettering Cancer Center (MSKCC; Fig. 1 A and Table S1; data are deposited in GEO under accession no. GSE40989). The second dataset includes 198 samples collected at University of Nebraska (Bouska et al., 2014; Fig. 1 B and Table S1). Using the GISTIC algorithm (Beroukhim et al., 2010; Mermel et al., 2011), we identified 9 statistically significant amplified regions and 18 deleted regions in first dataset (Table S1), and 26 amplified and 26 deleted regions in the second dataset (Table S1). As expected, the greater number of samples in the second dataset (198 samples versus 64 samples) enhances the statistical power and enables detection of a larger number of significantly recurrent regions. Comparing the copy number analysis of the two datasets, we found that 67% of the regions in the first dataset have a match in the second dataset; notably, all significant regions from the two datasets (residual q < 1−4) are matched, indicating a remarkable similarity between these two series of indolent FL samples.
Figure 1.
Cell cycle control genes are targets of significantly mutual exclusive genomic lesions in FL. (A and B) Analysis of recurrent copy number changes with GISTIC algorithm identifies statistically significant focally amplified (red) and deleted (blue) regions in 64 FLs (MSKCC cohort; A) and 198 FLs (UNMC cohort). The height of each peak is proportional to the corresponding q-value. (C–E) mRNA expression of CDK4 (C; *, P = 0.05), RB1 (D; *, P = 0.03), and CDKN2A (E; P = 0.24) in diploid cases versus FLs harboring gains on samples from the MSK cohort. The boxplot whiskers extend from the lowest to the highest gene expression level in FL patients excluding outliers. P-values were calculated by ANOVA test. (F) Genome-wide analysis for significantly linked aberrations in 262 FLs from both cohorts reveals a significant mutually exclusive relation between lesions affecting CDKN2A/B, RB1, and CDK4 (p = 0.003) P-value was derived by constrained permutation of the alteration set.
Next, we used an unbiased and genome-wide approach to identify significant relationships between chromosomal changes. The analysis is based on a mutual exclusivity module (MEMo; Ciriello et al., 2012) and described in detail in the Materials and methods. We merged the list of copy number altered regions and the total now includes all 262 patient samples. Strikingly, among the most significantly linked mutually exclusive events were three chromosomal changes affecting key cell cycle control loci (FDR corrected p-value = 0.003) in 109/262 (42%) of the samples (Fig. 1 F and Table S2). In detail, the mutually exclusive lesions were: first, an amplified peak (seen in 29%) that affected chromosome 12, and within the peak, CDK4 as the most frequently altered gene (Fig. 1, A and B); second, a highly focal loss centered on the CDKN2a/b locus at chromosome 9p21.3 (19 samples: 5%; homozygous: 3%, heterozygous: 2%; Fig. 1, A and B); and third, a loss of 13q14 that included the RB1 locus (32 samples: homozygous, 16 [6%]; heterozygous, 16 [6%]; Fig. 1, A and B). Quantitative RT-PCR confirmed corresponding changes in the mRNA levels of CDK4 (P = 0.05), RB1 (P = 0.03), and CDKN2A (P = 0.24; Fig. 1, C–E). Hence, a genome-wide analysis of array-CGH data representing >250 indolent FLs identifies a mutually exclusive relation between focal lesions affecting RB and CDKN2A and gains of chromosome 12 that include the RB kinase CDK4.
The CDK4 kinase requires a D-type cyclin for its activity toward RB (Matsushime et al., 1992). In our FL cohort, the CCND3 expression level is significantly higher than CCND1 and CCND2 (Fig. 2 A; data deposited in GEO under accession no. GSE37088), and Sanger sequencing of 69 samples confirmed CCND3 mutations in 6% of FLs (Fig. 2, B and C; and Table S3). As reported by Morin et al. (2011), these mutations clustered in the PEST domain, they were typically heterozygous, and only one case harbored a homozygous T283A mutation (#107-2124). Similar mutations in the CCND3 PEST domain occur in aggressive lymphomas and other cancers (Morin et al., 2010, 2011; Musgrove et al., 2011; Pasqualucci et al., 2011). The CCND3 mutations co-occur with gains in CDK4 in two of four cases (unpublished data), which is consistent with its cofactor function for CDK4/6. Functionally, these mutations increase the stability of the CCND3 protein; for example, the T283A mutation affects a phosphorylation site that is required for proteasomal degradation (Naderi et al., 2004). Accordingly, cycloheximide treatment of 293T and immortalized B cells engineered to express mutant and wild-type CCND3 confirms increased stability of the mutant CCND3 proteins (T283A, V287D, and Q276*; Fig. 2, D–F).
Figure 2.
Cyclin D3 is mutated in 6% of indolent FL. (A) mRNA levels of CCND1, CCND2, and CCND3 in 72 FL samples. The boxplot whiskers extend from the lowest to the highest gene expression level in FL patients excluding outliers. (B) Graphical representation of CCND3 mutations in FL. (C) Representative sequencing traces of homozygous (#I07-2124) and heterozygous (#1125P-I06-1301) CCND3 mutations in FL. (D–F) Immunoblot on lysates of 293T (D) or FL5-12 pre–B cells (E) expressing the indicated wild-type or mutant forms of CYCLIN D3 and either untreated or treated for the indicated times with cycloheximide (CHX). A quantification of the immunoblot results is shown in F. (G) Comparison of the frequency of CDKN2A, RB, and CDK4 lesions in indolent (FL) and transformed FLs (tFL).
Cell cycle control loci have been implicated in FL transformation (e.g., Elenitoba-Johnson et al., 1998; Pinyol et al., 1998; Pasqualucci et al., 2014). The UMCN series also includes 67 transformed FL cases and detailed analysis confirms lesions at the CDKN2A locus (18 samples, 17.9% homozygous, 9% heterozygous), the RB locus (10 samples, 4.5% homozygous, 10.4% heterozygous), and gains of CDK4 in 22 samples (Fig. 2 G). This confirms a further increase of RB pathway lesions upon transformation.
RB pathway lesions are associated with high-risk disease
The FL international prognostic index (FLIPI; Solal-Céligny et al., 2004) provides a prognostic index that reflects parameters of disease spread (e.g., stage, number of involved lymph nodes, serum lactate dehydrogenase, hemoglobin, and patient age) and is predictive of outcome (Solal-Céligny et al., 2004). In the MSKCC cohort, 45% of patients were low risk by FLIPI, 26% intermediate, and 29% high-risk (FLIPI was not available for the UNMC cohort). FLIPI was highly predictive of poor outcome in our cohort (P = 0.05). We find that genomic lesions at cell cycle loci were significantly associated with high risk by FLIPI (P = 0.002), and most high-risk tumors by FLIPI harbored cell cycle activating lesions (13/17; Table 1, A). The MSKCC cohort also provides information on histological grade and all grades of indolent FL are present. Consistent with prior studies (Martin et al., 1995; Hans et al., 2003; Hsi et al., 2004), we find no association between grade and outcomes (P = 0.74), and notably cell cycle lesions were equally distributed across all grades (P = 0.9; Table 1, B). Hence, genomic lesions affecting the CDKN2A, RB, and CDK4 loci are significantly enriched among high-risk FL patients.
Table 1.
Cell cycle lesions are associated with patient risk by FLIPI
| FL classification | Diploid | Gain/Loss | Total (%) |
| A | |||
| FLIPI low | 20 | 6 | 26 (45) |
| FLIPI intermediate | 6 | 9 | 15 (26) |
| FLIPI high | 4 | 13 | 17 (29) |
| B | |||
| Grade 1 | 8 | 9 | 17 (29) |
| Grade 2 | 10 | 8 | 18 (31) |
| Grade 3A/B | 12 | 11 | 23 (39) |
(A) Cell cycle lesions are significantly associated with high-risk by FLIPI. P = 0.002. (B) Lesions affecting CDK4, RB1, and CDKN2a/b occur across all FL grades without predilection. P = 0.08.
Immunohistochemistry confirms frequent RB pathway lesions in FL
We retrieved an additional cohort of 105 paraffin-embedded and arrayed indolent FL samples (patient information for tissue microarrays [TMAs] overlap with aCGH cohorts summarized in Table S4). First, our genomic data implicate CDK4 as a target of gains in a large percentage of FL. We used CDK4 FISH to confirm the CDK4 gain in 24.5% of samples, and this includes an overlap group where both aCGH and FISH showed CDK4 gains (Table S4). This was also reflected at the protein level, and the CDK4 and CCND3 proteins were abundantly expressed in 67%, including 11 of the 13 CDK4 FISH-positive tumors (Fig. 3, A and B; and Table S4). CDK4/CCND3 for the active RB kinase and, accordingly, RB1 (Ser780) phosphorylation provide a summary readout of pathway activity. In an expanded series of 230 FLs, we observed RB phosphorylation in 46% (106/230) of the arrayed indolent FLs (Fig. 3 C), again including most cases with positive CDK4 FISH and elevated CDK4 and CCND3 protein levels (Fig. 3 D). Ki67 marks proliferating cells and is typically lower in FLs than in aggressive lymphomas (Llanos et al., 2001; Martinez et al., 2007). We confirm this in our TMA cohort and find that 21% of FLs had >50% Ki67-positive staining cells, indicating an extensive growth fraction (Fig. 3 E). These included most samples with increased CDK4, CCND3, and phosphorylated RB, indicating that loss of RB checkpoint drives proliferation but is not sufficient in all cases (Fig. 3 F). As expected, high Ki67 further corresponded to high risk disease by FLIPI (Fig. 3 G). Hence, the IHC data confirm functional RB pathway disruption in a large group of indolent FLs.
Figure 3.
TMA analysis for cell cycle control markers in FL identifies a group of P-RB–positive patients. (A–C) Representative TMA sections of 105 human FLs stained for CDK4 (A) and CCND3 (CYCLIN D3; B). An expanded series of 230 samples was stained for phosphorylated RB1 (C). (D) Heat map summary of the IHC stains on human FLs for the indicated proteins (negative stain: <20% of positive cells, positive: >20%, high: >50% of positive cells). (E) TMA stain for Ki67 in FL (low: <25% of positive cells, intermediate: 25–50% of positive cells, high: >50% of positive cells). (F) Association of high Ki67 stain (Ki67: >50% of cells) with expression status (positive/negative) of the indicated proteins (*, P < 0.05). (G) Association of high Ki67 stain (Ki67: >50% of cells) with copy number alterations affecting the CDKN2A/B, RB1, or CDK4 loci with high Ki67 stains (*, P < 0.05). P-value was calculated by unpaired Student’s t test.
Contribution of RB pathway lesions to BCL2-driven lymphomagenesis
We use a mouse model that recapitulates key aspects of the genetics and morphology of FL and is based on the adoptive transfer of vav-P-Bcl2 hematopoietic progenitor cells (HPCs; Egle et al., 2004; Oricchio et al., 2011; Fig. 4 A). We introduced MSCV vector constructs directing the expression of CDK4, a kinase-dead mutant form of CDK4 (CDK4 KD), mutant CCND3 (T283A), or c-MYC, and short-hairpin RNAs (shRNAs) targeting p16 or Rb1 into vavBcl2 transgenic hematopoietic precursors (Fig. 4, D–G). Mice receiving HPCs overexpressing CDK4 or mutant CCND3 develop tumors with high incidence and short latency compared with vector or CDK4 KD, although not as rapidly as c-MYC transduced HPCs (CDK4: n = 11, median latency 58 d [CDK4 vs. vector, P = 0.001]; CCND3 T283A: n = 10, median latency 87 d [CCND3 T283A vs. vector, P = 0.05]; vector controls: n = 24, 80% tumor free, CDK4 KD, n = 10, 100% tumor free [Cdk4 vs. vector, P = 0.001]; c-Myc: n = 7; Fig. 4 B). Similarly, knockdown of p16 (n = 11, median latency 70 d [shp16 vs. vector, P = 0.018]) and knockdown of Rb1 (n = 9, median latency 70 d [shRb vs. vector, P < 0.01]) strongly cooperate with Bcl2 in FL development (Fig. 4 B). Similar to the human tumors we analyzed by TMA and immunohistochemistry, tumors expressing CDK4, Cyclin D3, or shp16 showed high levels of phosphorylated Rb, were more proliferative by Ki67, and had individual mitotic figures (Fig. 4 C). Surface marker analysis confirms the B cell lymphoma pathology with varying degrees of T cell infiltration (unpublished data). c-MYC–expressing tumors resemble transformed FLs and are composed of large cells, with loss of peanut agglutinin (PNA) staining and a diffuse growth pattern. However, lymphomas harboring cell cycle lesions are composed of smaller B cells and maintain variable degrees of follicular organization that is best seen in the PNA stain (Fig. 4 C). Although transformed c-MYC–expressing vavBcl2 tumors were readily cultured and transplantable (5/5), FLs expressing CDK4 could not be cultured and did not engraft in syngeneic animals (0/5; unpublished data). Hence, the cell cycle activating lesions act as oncogenic drivers in the BCL2 context in vivo.
Figure 4.
Cell cycle lesions cooperate with Bcl2 in lymphomagenesis. (A) Diagram of the adoptive transfer model of FL. (B) Kaplan-Meier analysis showing time to tumor development after transplantation of vavP-Bcl2 transgenic HPC transduced with vector (black; n = 24) or CDK4 (blue; n = 11 [CDK4 vs. vector, P = 0.001]), kinase-dead CDK4 (CDK4-KD, gray; n = 6), c-MYC (n = 7) (red), CCND3 T283A (purple; n = 10 [CCND3 T283A vs. vector, P = 0.05]), and shRNAs against p16 (green; n = 11 [shp16 vs. vector, P = 0.018]) and Rb1 (pink; n = 9 [shRb vs. vector, P < 0.01]). P-value was derived using Log-Rank (Mantel-Cox) test. (C) Immunohistochemical analysis of murine lymphomas harboring the indicated genetic changes and high resolution inset. (D–G) Immunoblot on lysates of tumors expressing CDK4, CYCLIN D3 T283A, or shRNAs against p16 or Rb.
The CDK4/6 kinase is a rational target in a subset of FLs
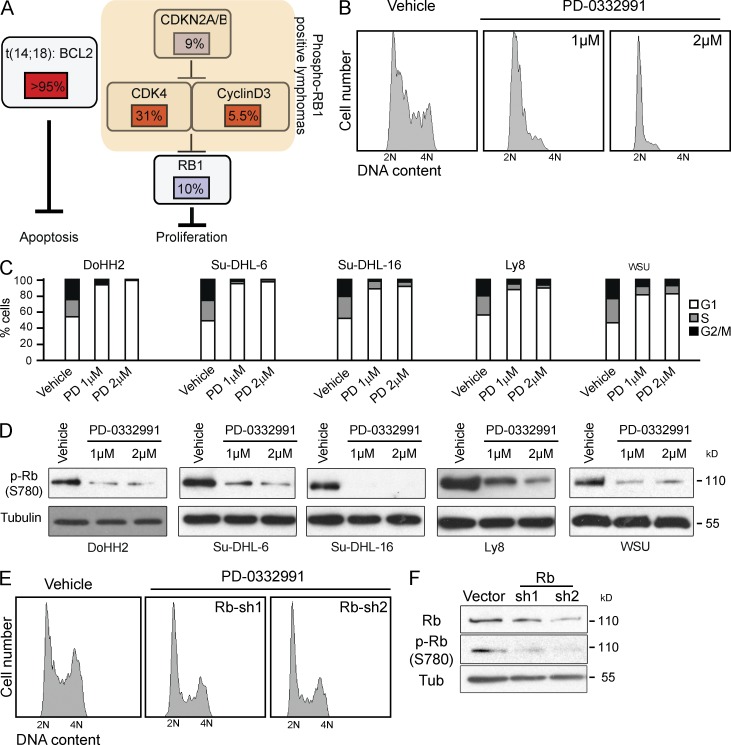
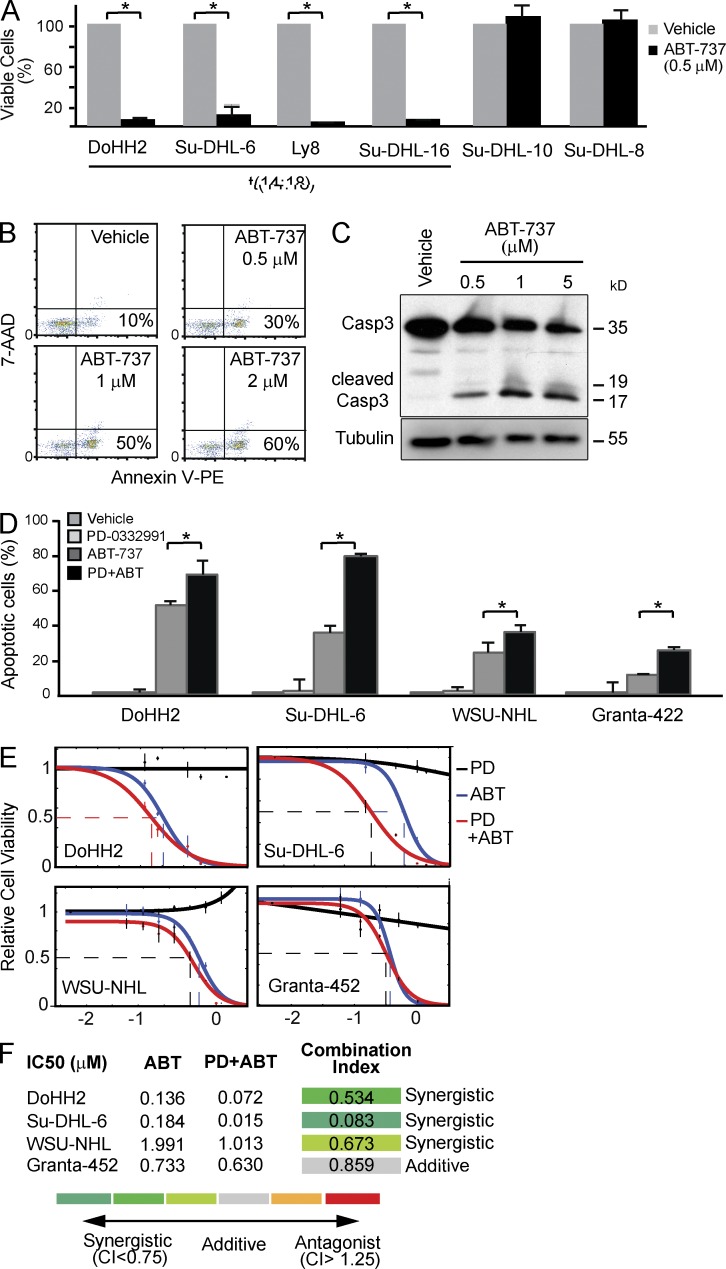
CDK4/6 acts as the RB kinase, and the frequent RB phosphorylation in indolent FL implies CDK4/6 as a therapeutic target in this disease (Fig. 5 A). We explored this hypothesis in all available t(14;18)-positive lines including DoHH2, Su-DHL-6, Su-DHL-16 and Ly 8, Granta-452, and WSU-NHL. All of these have elevated levels of CDK4, CDK6, and RB1 phosphorylation (S780, S795), and DoHH2 has a homozygous p16 deletion and a MYC gain/translocation, indicating that these changes are required for in vitro growth (Grønbaek et al., 2000). Growth or transplantation of primary human FLs is not possible (unpublished data). We first treated DoHH2, WSU-NHL, Su-DHL6, Su-DHL-16, and Ly8 cells with increasing doses of PD-0332991, which caused complete growth arrest, accumulation in G1 phase, and loss of RB1 phosphorylation (Fig. 5, B and D). The cytostatic effect of PD-0332991 was strictly dependent on RB1 and knockdown could bypass cell cycle arrest (Fig. 5, E and F). BH3 mimetic drugs like ABT-737 block BCL2 function by inserting into the BH3-binding groove of anti-apoptotic BCL2 proteins (Oltersdorf et al., 2005). Accordingly, ABT-737 has selective activity t(14;18)-positive lines in DoHH2, WSU-NHL, Su-DHL-6, SU-DHL-16, and LY8 cells (Fig. 6 A) and induces dose-dependent apoptosis by caspase cleavage and annexin induction (Fig. 6, B and C). ABT-737 and PD-0332991 show synergy against human FL-derived lines (Fig. 6 D). A detailed analysis of drug interaction based on cell viability (Fitzgerald et al., 2006) reveals combination indices < 0.75, indicating synergy for DoHH2, Su-DHL6, and WSU-NHL (P = 0.001), and an additive drug interaction in Granta 452 cells (combination index = 0.86; Fig. 6, E and F).
Figure 5.
CDK4/6 inhibitors is effecting against human FL cells. (A) Summary of genetic data indicating the t(14;18) causing elevated BCL2 and genomic lesions activating the cell cycle as oncogenic drivers in a sizeable fraction of FLs. (B) Cell cycle profile of DoHH2 cells after 24 h of treatment with PD-0332991 or vehicle (DMSO). (C) Cell cycle profile of DoHH2, Su-DHL-6, Su-DHL-16, and Ly8 and WSU-NHL cell lines treated with PD-0332991 1 and 2 µM for 24 h. (D) Immunoblot for RB1 phosphorylation (Ser 780) in DoHH2, Su-DHL-6, Su-DHL-16, and Ly8 and WSU-NHL cells treated as indicated. (E) Cell cycle profile of DoHH2 cells expressing the RB1 sh treated with PD-0332991 (1 µM) or vehicle (DMSO). Cell cycle analysis has been repeated at least three times for each cell line. (F) Immunoblot for indicated proteins on lysates from DoHH2 cells expressing the RB1 shRNAs (sh1 and sh2).
Figure 6.
Synergy between CDK4/6 and BCL2 inhibition against human FL cells. (A) Viability in a panel of human lymphoma lines after 24 h of treatment with ABT-737; cell lines harboring the translocation t(14;18) are indicated (*, P = 0.001). Error bars represent the standard deviation of three independent experiments. (B) Apoptosis analysis of WSU-NHL treated with increasing ABT-737 doses measuring annexin-positive cells by flow cytometry. (C) Immunoblot for caspase 3 and cleaved caspase 3 in WSU-NHL cell line treated with increasing ABT-737 doses for 24 h. (D) DoHH2, Su-DHL-6, WSU-NHL, and Granta 452 cell apoptosis after in vitro treatment with PD-0332991 (1 µM), ABT-737 (100 nM), both drugs, or DMSO (*, P < 0.01). Error bars represent the standard deviation of three independent experiments. (E) IC50 curves for DoHH2, Su-DHL-6, WSU-NHL, and Granta-452 cell line treated with PD-0332991, ABT-737 alone, and with the drug combination. (F) Table summarizing the in vitro data. CI is the combination index, and values <0.75 indicate drug synergy (details in Materials and methods). Each experiment has been repeated at least three times, and the graphs represent an average of the experiments.
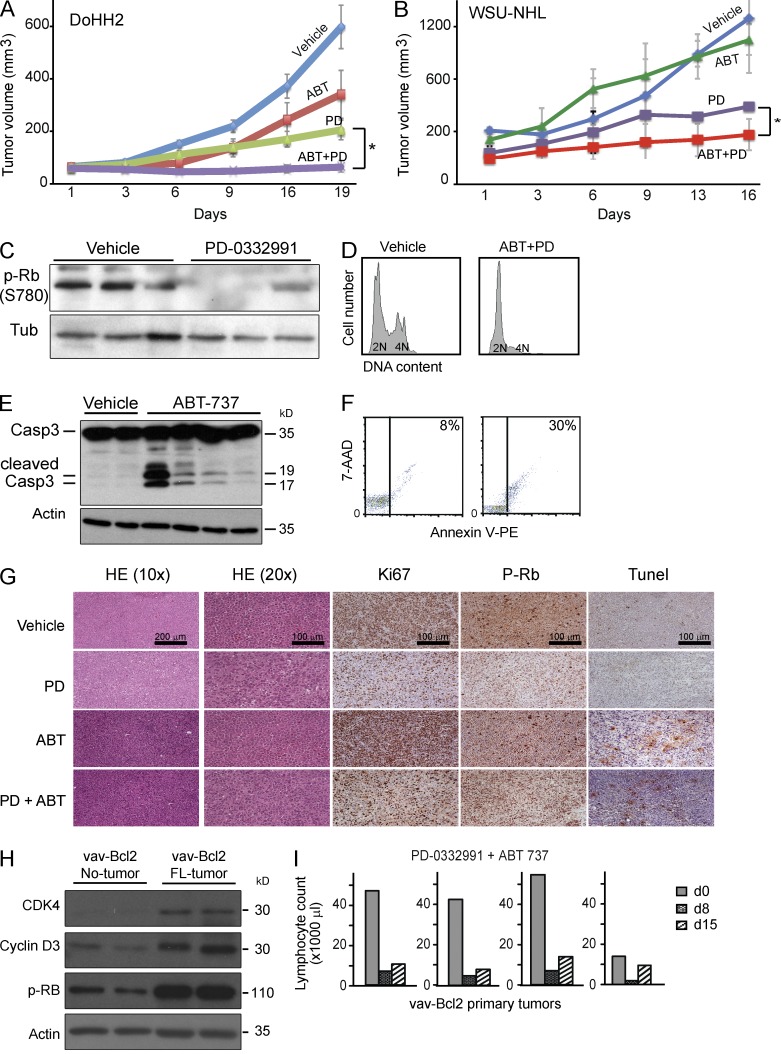
Next, we tested this synergistic drug interaction against xenografted DoHH2 and WSU-NHL lymphomas. In brief, xenografted tumors (>10 mm3) were treated with PD-0332991 (PD: 150 mg/kg, alternate days per os, days 1–16), ABT-737 (ABT: 50 mg/kg, alternate days, i.v., days 1–16), or both drugs. Individually, each drug somewhat restrained tumor growth during the treatment course, but residual DOHH2 and WSHU-NHL tumors rapidly expanded shortly thereafter. Strikingly, the combined treatment (PD: 150 mg/kg; ABT: 50 mg/kg, on alternate days 1–16) completely abrogated growth and caused lasting and complete responses (decrease by >95–100%; PD+ABT vs. PD or ABT, P < 0.004; Fig. 7, A and B). As expected, PD-0332991 caused complete loss of RB1 phosphorylation (Ser780), blocked cells in G1, and reduced Ki67 levels in vivo (Fig. 7, C and D). ABT-737 produced significant cell death by caspase cleavage, TUNEL stain, and annexin FACS (Fig. 7, E and G). We also tested PD-0332991 against primary mouse lymphomas because we noticed that these tumors spontaneously up-regulated CDK4 and CCND3 (Fig. 7 H). Consistent with the human data, we observed powerful therapeutic activity in these primary, indolent lymphomas (Fig. 7 I). At an effective combination dose (150 and 50 mg/kg) we did not observe toxicity, weight loss, organ damage, or changes in serum chemistry, and only temporary reductions in lymphocytes and platelet counts (unpublished data). Together, the results indicate a therapeutic window and significant activity of combined RB kinase and BCL2 inhibition in FL disease models.
Figure 7.
Dual RB kinase and BCL2 inhibition is safe and effective against xenografted FL cells. (A and B) Tumor volume curve for xenografted DoHH2 and WSU tumors treated with the indicated drugs for 20 d (five or six animals have been used in each treatment group). Error bars represent the standard deviations. (C) Immunoblot on lysates of tumors treated in vivo with PD-0332991 probed for RB1 phosphorylation (Ser780). (D) Cell cycle profile of tumors treated in vivo with vehicle or PD 0332991+ABT-737. (E) Immunoblot on lysates of tumors treated in vivo with ABT-737 cleaved caspase-3 (D). (F) Apoptosis analysis of tumors treated in vivo with vehicle or PD 0332991+ABT-737. (G) Immunohistochemical analysis of DoHH2 tumors harvested at the end of the treatment interval. (H) Western blot analysis of CDK4, CCND3, and phosphorylated RB expression in splenic B cells of vav-Bcl2 mice before and after spontaneous FL development. (I) Combined in vivo treatment of primary murine vavbcl2 lymphomas with combination of PD-0332991 and ABT-737 produces disease response quantified by blood counts.
DISCUSSION
RB pathway lesions in indolent FL
Our analysis identifies a pattern of mutually exclusive genomic alterations that impact the RB pathway in a sizeable fraction of indolent FLs. These include homozygous and heterozygous deletions of CDKN2A/p16 and RB, and a large region of chromosomal gains on Chromosome 12. The mutually exclusive relationship between losses of CDKN2A, RB, and the chromosome 12 amplifications implicate the RB kinase CDK4 as a relevant target at Chr. 12q13. Accordingly, CDK4 falls within the common minimal region of gain in all samples, and this does not rule out important roles for additional genes at this locus. Cell cycle lesions have been implicated in FL transformation, for example, most recently in Pinyol et al. (1998) and Pasqualucci et al. (2014). Individual lesions affecting CDKN2A or RB are relatively rare in the indolent disease (Lenz et al., 2008; Morin et al., 2011; Pasqualucci et al., 2011). This has led to the assumption that loss of cell cycle control mainly occurs upon transformation. However, the cumulative frequency of lesions affecting CDKN2A, RB1, and CDK4 indicates disruption of this cell cycle checkpoint in a sizeable fraction of indolent FLs. This view is further supported by abundant RB phosphorylation in up to 45% of FLs, increased levels of CDK4, and in vivo evidence that CDK4, CDKN2A, and RB impact BCL-2–driven FL development in vivo.
RB pathway lesions promote FL pathogenesis in vivo
In a murine FL model, all RB pathway lesions cooperate with BCL2 in lymphomagenesis. Most likely, RB inactivation overrides the growth inhibitory effects of both BCL2 and p21 in germinal center B cells (O’Reilly et al., 1996; Vairo et al., 1996, 2000; Phan et al., 2005). It is notable that RB inactivation does not lead to histologically transformed FLs in vivo, and this is in clear contradiction to enforced MYC expression. At least in the murine model, RB pathway lesions are not sufficient for disease transformation and this argues that additional changes contribute to lymphoma progression. It is also consistent with a view that RB pathway lesions facilitate FL evolution toward the more aggressive disease, and a serial analysis on a large cohort will be needed to address this (Bödör et al., 2013; Green et al., 2013; Okosun et al., 2014).
RB phosphorylation distinguishes two types of indolent FL with different clinical risk and therapeutic requirements
RB phosphorylation provides a facile summary readout of all upstream events that can influence the activity of the CDK4/CCND RB kinase including genetic and epigenetic changes (Sherr, 1996). We find increased RB phosphorylation in ∼45% of indolent FLs, and the number closely corresponds to the cumulative frequency of genomic lesions affecting CDK4 and CDKN2A. Although equally represented across histological grades (except for transformed FL which show the expected increase), the RB pathway lesions are strongly associated with elevated risk by FLIPI (Solal-Céligny et al., 2004). PD-0332991 is a selective inhibitor of the RB kinase CDK4/6 and is currently in clinical trials against several cancers, including mantle cell lymphoma, but patients with indolent FL have been specifically excluded (e.g., http://clinicaltrials.gov/show/NCT01111188; Fry et al., 2004; Flaherty et al., 2012). The cytostatic activity of PD-0332991 strictly depends on phosphorylated RB and the drug is inactive in its absence (Michaud et al., 2010). Therapeutic studies on indolent FLs are limited by lack of tractable models; notably, all FL cell lines represent the transformed state and carry cell cycle lesions similar to those seen in the human tumors. However, across the best available human models (DoHH2 and WSU-NHL), we find that PD-0332991 alone is mostly cytostatic, and that combinations with BCL2-inhibiting drugs (e.g., ABT-737 [Oltersdorf et al., 2005] or obatoclax) are synergistic in vitro and at least additive against xenografts in vivo and with acceptable toxicity. Together, we report the surprising observation of frequent and mutually exclusive RB pathway lesions (CDK4, CDKN2a/p16, RB, and CCND3) at the indolent stage of FL that is reflected in the RB phosphorylation state and identifies a high-risk group of FL patients who may benefit from CDK-directed therapies.
MATERIALS AND METHODS
Tumor ascertainment.
Newly diagnosed FLs collected at MSKCC since 1985 and evaluated by 3 hematopathologists were classified according to WHO criteria as nontransformed FL.
Array-CGH.
DNA from fresh frozen tissue or OCT-embedded was isolated and processed as previously described (Oricchio et al., 2011). In brief, labeling and hybridization was performed according to protocols provided by Agilent Technologies. Before submission to the Genomics core for labeling and hybridization to the 244K oligonucleotide array (Agilent Technologies), digestion efficiency of each DNA was checked by incubating 1 µg DNA with 1 µl HaeIII (10 U/µl) at 37°C for 2 h, and running in parallel the undigested and digested DNA (100 ng each) on a 1% agarose gel along with the 1 kb DNA ladder. Human male DNA (Promega) served as the reference DNA. Labeling and hybridization were performed according to protocols provided by Agilent Technologies. The slides were scanned at 5-µm resolution using the G2565 Microarray Scanner System (Agilent Technologies), and Feature Extraction software (v9.1; Agilent Technologies) was used to generate the data. Data are available on GEO under accession no. GSE40989. Copy Number Data from a second dataset (UNMC dataset) has been generated using GeneChip Human Mapping 250K Nsp SNP array (Affymetrix) as described in Bouska et al. (2014).
Copy number analysis.
To identify significantly amplified and deleted regions, we used the GISTIC algorithm (Beroukhim et al., 2010) through the GenePattern web service (http://genepattern.broadinstitute.org). GISTIC 2.0 (Beroukhim et al., 2010; Mermel et al., 2011) has been run on segmented copy number data generated for each dataset using the DNAcopy package from Bioconductor (Olshen et al., 2004). Recurrently amplified and deleted regions were selected if q < 0.05 or residual q < 0.1 (Beroukhim et al., 2010). Peaks identified by GISTIC span a variable number of genes (varying between 1 and ∼200). To focus on cancer-relevant genes in these regions, we intersected the set of genes from these regions with the Sanger Cancer Census gene list (Futreal et al., 2004). Both the full and cancer-specific list of genes contained in each region is provided in Table S2.
Next, we compared the two sets of GISTIC regions to identify which and how many regions determined on one dataset have at least one matching region in the other. Regions were considered matching if the corresponding peaks were overlapping or at maximum 5 MB apart. Overall, 67% of the regions from the MSKCC dataset have a match in the UNMC dataset and 46% of regions from the UNMC dataset have a match in the MSKCC dataset. Notably, all the most significant regions from the two datasets (residual q < 1−4) are matched.
Mutual exclusivity analysis.
To identify a significant mutually exclusive pattern of copy number alteration we used the algorithm MEMo (Ciriello et al., 2012). In brief, MEMo used as input the set of gene peaks of gain and loss we derived from the array-CGH analysis. Alterations targeting fully connected gene sets were tested for mutual exclusivity based on a random permutation of the observed genomic alterations. Genomic events were permuted using a Monte Carlo Markov Chain approach that preserves both the number of alteration per gene and the number of alterations per sample.
CCND3 sequencing and PCR amplification.
The exonic regions of interest (NCBI Human Genome Build 36.1) were broken into amplicons of 350 bp or less, and specific primers were designed using Primer 3 to cover the exonic regions plus at least 50 bp of intronic sequences on both sides of intron-exon junctions (Rozen and Skaletsky, 2000). M13 tails were added to facilitate Sanger sequencing. PCR reactions were performed in 384-well plates, in a Duncan DT-24 water bath thermal cycler, with 10 ng of template DNA (Repli-G Midi; QIAGEN) as template, using a touchdown PCR protocol with Kapa2G Fast HotStart Taq (Kapa Biosystems). The touchdown PCR method consisted of: 1 cycle of 95°C for 5 min; 3 cycles of 95°C for 30 s, 64°C for 15 s, and 72°C for 30 s; 3 cycles of 95°C for 30 s, 62°C for 15 s, and 72°C for 30 s; 3 cycles of 95°C for 30 s, 60°C for 15 s, and 72°C for 30 s; 37 cycles of 95°C for 30 s, 58°C for 15 s, and 72°C for 30 s; and 1 cycle of 70°C for 5 min. Templates were purified using AMPure (Beckman Coulter). The purified PCR reactions were split into two and sequenced bidirectionally with M13 forward and reverse primer and Big Dye Terminator kit (v.3.1; Applied Biosystems) at Beckman Coulter Genomics. Dye terminators were removed using the CleanSEQ kit (Beckman Coulter), and sequence reactions were run on an ABI PRISM 3730xl sequencing apparatus (Applied Biosystems).
CCND3 mutation detection.
Mutations were detected using an automated detection pipeline at the MSKCC Bioinformatics Core. Bi-directional reads and mapping tables (to link read names to sample identifiers, gene names, read direction, and amplicon) were subjected to a QC filter which excludes reads that have a mean phred score of <10 for bases 100–200. Passing reads were assembled against the reference sequences for each gene, containing all coding and UTR exons, including 5 Kb upstream and downstream of the gene, using command line Consed 16.0 (PMID: 9521923). Assemblies were passed on to Polyphred 6.02b (PMID: 9207020), which generated a list of putative candidate mutations, and to Polyscan 3.0 (PMID: 17416743), which generated a second list of putative mutations. The lists were merged together into a combined report, and the putative mutation calls were normalized to + genomic coordinates and annotated using the Genomic Mutation Consequence Calculator (PMID: 17599934). The resulting list of annotated putative mutations was loaded into a Postgres database along with select assembly details for each mutation call (assembly position, coverage, and methods supporting mutation call). To reduce the number of false positives generated by the mutation detection software packages, only point mutations which are supported by at least one bi-directional read pair and at least one sample mutation called by Polyphred were considered, and only the putative mutations, which are annotated as having nonsynonymous coding effects, occur within 11 bp of an exon boundary, or have a conservation score >0.699 were included in the final candidate list. Indels culled by any method were manually reviewed and included in the candidate list if found to hit an exon. All putative mutations were confirmed by a second PCR and sequencing reaction; matched normal tissue DNA was not available but the reported mutations have never been seen as normal variation (1000 Genomes Project Consortium et al., 2012). All traces for mutation calls were manually reviewed.
Gene expression array and analysis.
Affymetrix U133A gene chips were used for gene expression profiling of 72 FL samples. The transcriptome analyses were performed using RMA system software packages (version 2.8.0) including Bioconductor (version 2.0). Raw gene expression data from Affymetrix CEL files were analyzed using the affy R library from Bioconductor to generate probe level intensity. Gene-centric expression values were generated using a CDF file based on remapping of probes to the human genome 36.1. The raw data for 72 FL patient samples (CEL file) and the normalized matrix have been deposited in GEO (GSE37088).
Clinical data analysis.
Collection of clinical information of all FL cases is under MSKCC IRB waiver approval. Clinical data for UNMC are available in (Bouska et al., 2014). Statistical methods: the association between the alteration in cell cycle genes (binary yes/no) and GRADE and FLIPI were compared using Fisher’s exact test. FLIPI model 2004 has been used to classified FL patients (Solal-Céligny et al., 2004). Overall survival is summarized using Kaplan-Meier curves and tests of association between the categorical variables (alteration, GRADE, and FLIPI).
Immunohistochemical and TMA methods.
TMAs were constructed as previously published (Schatz et al., 2011), but using a fully automated Beecher Instrument, ATA-27. The study cohort was comprised of FL consecutively ascertained at MSKCC between 1985 and 2000. Additional clinical details are listed in Table S1. Use of tissue samples was approved with an Institutional Review Board Waiver and approval of the Human Biospecimen Utilization Committee. All cancer biopsies were evaluated at MSKCC, and the histological diagnosis was based on hematoxylin and eosin (H&E) staining. TMAs were stained with the P-Rb (Ser780) and Cyclin D3 antibody (1:500; Cell Signaling Technology), with CDK4 (DCS-31; 1:500; Invitrogen), and with Ki67 and Ventana Discovery XT, using protocol 67: CC1, standard, at 1:100 dilution overnight at 4°C, and then with a secondary anti–rabbit antibody (Vector Laboratories) at 1:200 dilution for 60 min. The negative controls were performed on the same slides as the FL-TMAs. Each slide with TMA spots also had spleen, kidney, and/or lymph node controls. Breast cancer and sarcoma samples have been used as positive staining control. Positive score is defined with >20% of cells expressing CDK4, Cyclin D3, and P-RB; negative score for samples with 0–20% of signal and high levels in samples with >50% of positive cells.
Fluorescence in situ hybridization.
FISH was performed using BAC clones spanning CDK4 in 12q13: RP11-571M6 (part of a 1Mb clone set supplied by the Wellcome Trust Sanger Institute) and RP11-970A5 (from BACPAC resources, Children’s Hospital at Oakland Research Center). The BACs were labeled by nick translation with Red-dUTP (Enzo Life Sciences, supplied by Abbott Molecular, Inc.), and hybridized with chromosome 12 centromeric repeat pα12H8, labeled with Orange-dUTP, as reference. The baseline autofluorescence of many tumor cores was quite high, and the orange dye was used instead of the more usual Green-dUTP in an attempt to improve probe visibility. The TMA slides were hybridized according to standard procedures. In brief, the slides were dewaxed, microwaved in 10 mM sodium citrate buffer, pH 6.0, and treated with pepsin-HCl. Slides with probe mix were co-denatured on a HYBrite automated hybridizing station (Abbott Molecular Inc.) at 80°C for 8 min, and then incubated for 2–3 d at 37°C. After coverslip removal, the slides were washed using standard nonformamide stringency washes, stained with DAPI, and then mounted in antifade solution (VECTASHIELD; Vector Laboratories). The slides were scanned at low resolution using a Metafer MetaCyte automated scanning system (Metasystems Group Inc.), comprised of an epifluorescence microscope (Axioplan 2i; Carl Zeiss) equipped with a megapixel charge-coupled device camera (CV-M4+CL, JAI) and motorized stage (Martzhauser). The scanned image was then segmented into individual cores using the MetaCyte TMA tool, and high-resolution compressed image z-stacks (9 × 0.5 µm) were captured using FISH imaging software (5.2; Isis). Orange signals were pseudocolored in green for clarity, and CDK4 and reference signals were counted for each image and the ratio calculated for each core.
Xenografts and treatment studies.
Xenografts and treatment studies were generated as previously described (Schatz et al., 2011). In brief, s.c. injection of 5 Mio DoHH2 and WSU-NHL human lymphoma cells mixed with Matrigel (BD) into the flanks of mice NOD/SCID (NOD.CB17-Prkdcscid/J). Upon tumor size, 100-mm3 mice were treated daily by gavage with 150 mg/kg PD-0332991 (Chemitek) diluted in 50 mmol/liter sodium lactate, pH 4, for 2 wk and with ABT-737 50 mg/kg dilute in PTD solution (30% propylene glycol, 5% Tween 80, and 65% D5W). The control animals were treated with 50 mmol/liter sodium lactate, pH 4, PTD solution, and 5% Dextrose in water solution, respectively. Tumors were weighed and volumes were measured twice weekly.
Cell culture, cell cycle, and viability assays.
Lymphoma cell lines DoHH2, Ly-8, Su-DHL-6, Su-DHL-16, Su-DHL-8, and Su-DHL-10 and WSU-NHL were maintained in RPMI 1640 with 10% fetal bovine serum, 1% l-Glutamine, and 1% penicillin/streptomycin. Human lymphoma cell lines treated with PD-0332991 and ABT-737 were analyzed for cell viability and cell cycle assays. In brief, cells were seed at 5 × 105/ml and they were treated with PD-0332991 and/or ABT-737. After 24 h of treatment, cell viability was assessed using CellTiterGlo reagent (Promega). The data were analyzed for drug interactions and the Combination Index (CI) was calculated as reported in Fitzgerald et al., (2006).
Western blot and flow cytometry analysis.
Immunoblots were performed from whole cell lysates or tissue samples as previously described (Wendel et al., 2004). In brief, 30 µg protein/sample was resolved on SDS-PAGE gels and transferred to Immobilon-P membranes (Millipore). Antibodies were against Rb (Cell Signaling Technology), Phospho-Rb S780 (Cell Signaling Technology), Phospho-Rb S795 (Cell Signaling Technology), CDK4 (Cell Signaling Technology), Cyclin D3 (Cell Signaling Technology), Cyclin D1 (Cell Signaling Technology), Cyclin D2 (Cell Signaling Technology), p16 (Santa Cruz Biotechnology, Inc.), Caspase 3 (Cell Signaling Technology), and Tubulin (1:5,000; Sigma-Aldrich). Enhanced chemiluminescence was used for detection (ECL; GE Healthcare). Cell cycle profile and apoptosis analyses has been performed using Guava Cell Cycle reagent (Millipore) Guava Nexin reagent (Millipore) and expression was assessed through FACS analysis (Guava EasyCyte; Millipore). Experiments have been performed three times or more.
Generation of mice.
The vavP-Bcl2 mouse model of FL (Egle et al., 2004) was adapted to the adoptive transfer approach using retrovirally transduced HPCs (Wendel et al., 2004). In brief, we isolated vavPBcl2 transgenic fetal liver cells from vavPBcl2 heterozygous animals at embryonic day 13.5 (E13.5). The HPCs are grown for 4 d in a specially adapted growth medium containing IL3, stem cell factor, and IL6 and are retrovirally transduced with MSCV vectors directing the expression of genes or shRNAs of interest. We transplant genetically modified HPCs into lethally irradiated, syngeneic wild-type recipients and monitor disease onset weekly by palpation. Data were analyzed in Kaplan-Meier format using the log-rank (Mantel-Cox) test for statistical significance. All experiments were approved by the MSKCC IACUC committee (protocol #07–01-002). Plasmids expressing human short hairpins for RB1 and overexpressing CDK4 were provided by A. Chicas, S. Lowe, and D. Ciznadid (MSKCC, New York, NY).
Online supplemental material.
Table S1 shows array-CGH analysis of 64 (MSKCC dataset) and 198 (UNMC dataset) FLs. Table S2 shows a summary of genome-wide mutually exclusive analysis in 292 FLs. Table S3 shows a summary of reported Cyclin D3 mutations in lymphoma. Table S4 shows a summary of aCGH, FISH, and IHC analyses for CDK4, Cyclin D3, and RB in our FL cohorts. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20132120/DC1.
Supplementary Material
Acknowledgments
We thank A. Chicas, S. Lowe, and D. Ciznadid for reagents, J. Maragulia for assembling clinical data, and A. Koff for editorial advice. We also thank the MSK research animal facility and, in particular, HuiYong Zhao of the Antitumor Assessment Core facility, Katy Huberman of the Geoffrey Beene Translational Oncology Core Facility, Jiao Maria of the pathology core facility, and Margaret Leversha Molecular Cytogenetics Core Facility.
This work is supported by grants from the NCI (R01-CA142798-01), a P30 supplemental award (H.-G. Wendel), the American Cancer Society grant (RSG-13-048-01-LIB to H.-G. Wendel), the Leukemia Research Foundation (H.-G. Wendel), the Louis V. Gerstner Foundation (H.-G. Wendel), the Geoffrey Beene Cancer Center, the Society of MSKCC (H.-G. Wendel), the Starr Cancer Consortium (grant I4-A410 to H.-G. Wendel and J.H. Schatz), a grant from the American cancer Society (H.-G. Wendel), the Lymphoma Research Foundation (W.-C. Chan), the NCI SPECS grant (UO1 CA 84967 to W.-C. Chan), a SCOR grant from The Leukemia and Lymphoma Society (R.S.K. Chaganti), the Lymphoma Research Foundation (J.H. Schatz), and the MSKCC Translational-Integrative Research Fund (J.H. Schatz). H.-G. Wendel and E. Oricchio are Scholar and Special Fellow of the Leukemia Lymphoma Society. This work is supported by an NCI careeer developmental grant (1k99CA175179-01A1 to E. Oricchio).
The authors declare no competing financial interests.
Author contributions: E. Oricchio, G. Ciriello, and J.H. Schatz conducted experiments and were involved in the experimental design and data analysis; M. Jiang and M.H. Boice assisted with experiments; A. Heguy directed sequence analyses; A. Viale directed gene expression and array CGH generation; E.de Stanchina directed in vivo drug studies; A. Bouska, T. McKeithan; and W.-C. Chan provided array CGH data for the UNMC cohort; J. Teruya-Feldstein performed and analyzed TMA studies; C. Sander directed computational analyses; V.E. Seshan performed clinical data analysis; W. Tam provided pathological analyses and samples; R.S.K. Chaganti directed array-CGH and expression studies; and H.-G. Wendel and E. Oricchio designed the study and wrote the paper.
Footnotes
Abbreviations used:
- FL
- follicular lymphoma
- FLIPI
- FL international prognostic index
- HPC
- hematopoietic progenitor cell
- MEMo
- mutual exclusivity module
- RB
- retinoblastoma
- TMA
- tissue microarray
References
- 1000 Genomes Project Consortium, Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature. 491:56–65 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R., Mermel C.H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J.S., Dobson J., Urashima M., et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature. 463:899–905 10.1038/nature08822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bödör C., Grossmann V., Popov N., Okosun J., O’Riain C., Tan K., Marzec J., Araf S., Wang J., Lee A.M., et al. 2013. EZH2 mutations are frequent and represent an early event in follicular lymphoma. Blood. 122:3165–3168 10.1182/blood-2013-04-496893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouska A., McKeithan T.W., Deffenbacher K.E., Lachel C., Wright G.W., Iqbal J., Smith L.M., Zhang W., Kucuk C., Rinaldi A., et al. 2014. Genome-wide copy-number analyses reveal genomic abnormalities involved in transformation of follicular lymphoma. Blood. 123:1681–1690 10.1182/blood-2013-05-500595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G., Cerami E., Sander C., Schultz N. 2012. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 22:398–406 10.1101/gr.125567.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egle A., Harris A.W., Bath M.L., O’Reilly L., Cory S. 2004. VavP-Bcl2 transgenic mice develop follicular lymphoma preceded by germinal center hyperplasia. Blood. 103:2276–2283 10.1182/blood-2003-07-2469 [DOI] [PubMed] [Google Scholar]
- Elenitoba-Johnson K.S., Gascoyne R.D., Lim M.S., Chhanabai M., Jaffe E.S., Raffeld M. 1998. Homozygous deletions at chromosome 9p21 involving p16 and p15 are associated with histologic progression in follicle center lymphoma. Blood. 91:4677–4685 [PubMed] [Google Scholar]
- Fitzgerald J.B., Schoeberl B., Nielsen U.B., Sorger P.K. 2006. Systems biology and combination therapy in the quest for clinical efficacy. Nat. Chem. Biol. 2:458–466 10.1038/nchembio817 [DOI] [PubMed] [Google Scholar]
- Flaherty K.T., Lorusso P.M., Demichele A., Abramson V.G., Courtney R., Randolph S.S., Shaik M.N., Wilner K.D., O’Dwyer P.J., Schwartz G.K. 2012. Phase I, dose-escalation trial of the oral cyclin-dependent kinase 4/6 inhibitor PD 0332991, administered using a 21-day schedule in patients with advanced cancer. Clin. Cancer Res. 18:568–576 10.1158/1078-0432.CCR-11-0509 [DOI] [PubMed] [Google Scholar]
- Fry D.W., Harvey P.J., Keller P.R., Elliott W.L., Meade M., Trachet E., Albassam M., Zheng X., Leopold W.R., Pryer N.K., Toogood P.L. 2004. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol. Cancer Ther. 3:1427–1438 [PubMed] [Google Scholar]
- Futreal P.A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M.R. 2004. A census of human cancer genes. Nat. Rev. Cancer. 4:177–183 10.1038/nrc1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M.R., Gentles A.J., Nair R.V., Irish J.M., Kihira S., Liu C.L., Kela I., Hopmans E.S., Myklebust J.H., Ji H., et al. 2013. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 121:1604–1611 10.1182/blood-2012-09-457283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grønbaek K., de Nully Brown P., Møller M.B., Nedergaard T., Ralfkiaer E., Møller P., Zeuthen J., Guldberg P. 2000. Concurrent disruption of p16INK4a and the ARF-p53 pathway predicts poor prognosis in aggressive non-Hodgkin’s lymphoma. Leukemia. 14:1727–1735 10.1038/sj.leu.2401901 [DOI] [PubMed] [Google Scholar]
- Hans C.P., Weisenburger D.D., Vose J.M., Hock L.M., Lynch J.C., Aoun P., Greiner T.C., Chan W.C., Bociek R.G., Bierman P.J., Armitage J.O. 2003. A significant diffuse component predicts for inferior survival in grade 3 follicular lymphoma, but cytologic subtypes do not predict survival. Blood. 101:2363–2367 10.1182/blood-2002-07-2298 [DOI] [PubMed] [Google Scholar]
- Hsi E.D., Mirza I., Lozanski G., Hill J., Pohlman B., Karafa M.T., Coupland R. 2004. A clinicopathologic evaluation of follicular lymphoma grade 3A versus grade 3B reveals no survival differences. Arch. Pathol. Lab. Med. 128:863–868 [DOI] [PubMed] [Google Scholar]
- Lenz G., Wright G.W., Emre N.C., Kohlhammer H., Dave S.S., Davis R.E., Carty S., Lam L.T., Shaffer A.L., Xiao W., et al. 2008. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc. Natl. Acad. Sci. USA. 105:13520–13525 10.1073/pnas.0804295105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llanos M., Alvarez-Argüelles H., Alemán R., Oramas J., Diaz-Flores L., Batista N. 2001. Prognostic significance of Ki-67 nuclear proliferative antigen, bcl-2 protein, and p53 expression in follicular and diffuse large B-cell lymphoma. Med. Oncol. 18:15–22 10.1385/MO:18:1:15 [DOI] [PubMed] [Google Scholar]
- Martin A.R., Weisenburger D.D., Chan W.C., Ruby E.I., Anderson J.R., Vose J.M., Bierman P.J., Bast M.A., Daley D.T., Armitage J.O. 1995. Prognostic value of cellular proliferation and histologic grade in follicular lymphoma. Blood. 85:3671–3678 [PubMed] [Google Scholar]
- Martinez A.E., Lin L., Dunphy C.H. 2007. Grading of follicular lymphoma: comparison of routine histology with immunohistochemistry. Arch. Pathol. Lab. Med. 131:1084–1088 [DOI] [PubMed] [Google Scholar]
- Matsushime H., Ewen M.E., Strom D.K., Kato J.Y., Hanks S.K., Roussel M.F., Sherr C.J. 1992. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell. 71:323–334 10.1016/0092-8674(92)90360-O [DOI] [PubMed] [Google Scholar]
- Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. 2011. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12:R41 10.1186/gb-2011-12-4-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud K., Solomon D.A., Oermann E., Kim J.S., Zhong W.Z., Prados M.D., Ozawa T., James C.D., Waldman T. 2010. Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 70:3228–3238 10.1158/0008-5472.CAN-09-4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin R.D., Johnson N.A., Severson T.M., Mungall A.J., An J., Goya R., Paul J.E., Boyle M., Woolcock B.W., Kuchenbauer F., et al. 2010. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 42:181–185 10.1038/ng.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin R.D., Mendez-Lago M., Mungall A.J., Goya R., Mungall K.L., Corbett R.D., Johnson N.A., Severson T.M., Chiu R., Field M., et al. 2011. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 476:298–303 10.1038/nature10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrove E.A., Caldon C.E., Barraclough J., Stone A., Sutherland R.L. 2011. Cyclin D as a therapeutic target in cancer. Nat. Rev. Cancer. 11:558–572 10.1038/nrc3090 [DOI] [PubMed] [Google Scholar]
- Naderi S., Gutzkow K.B., Låhne H.U., Lefdal S., Ryves W.J., Harwood A.J., Blomhoff H.K. 2004. cAMP-induced degradation of cyclin D3 through association with GSK-3β. J. Cell Sci. 117:3769–3783 10.1242/jcs.01210 [DOI] [PubMed] [Google Scholar]
- O’Reilly L.A., Huang D.C., Strasser A. 1996. The cell death inhibitor Bcl-2 and its homologues influence control of cell cycle entry. EMBO J. 15:6979–6990 [PMC free article] [PubMed] [Google Scholar]
- Okosun J., Bödör C., Wang J., Araf S., Yang C.Y., Pan C., Boller S., Cittaro D., Bozek M., Iqbal S., et al. 2014. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 46:176–181 10.1038/ng.2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen A.B., Venkatraman E.S., Lucito R., Wigler M. 2004. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 5:557–572 10.1093/biostatistics/kxh008 [DOI] [PubMed] [Google Scholar]
- Oltersdorf T., Elmore S.W., Shoemaker A.R., Armstrong R.C., Augeri D.J., Belli B.A., Bruncko M., Deckwerth T.L., Dinges J., Hajduk P.J., et al. 2005. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 435:677–681 10.1038/nature03579 [DOI] [PubMed] [Google Scholar]
- Oricchio E., Nanjangud G., Wolfe A.L., Schatz J.H., Mavrakis K.J., Jiang M., Liu X., Bruno J., Heguy A., Olshen A.B., et al. 2011. The Eph-receptor A7 is a soluble tumor suppressor for follicular lymphoma. Cell. 147:554–564 10.1016/j.cell.2011.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L., Trifonov V., Fabbri G., Ma J., Rossi D., Chiarenza A., Wells V.A., Grunn A., Messina M., Elliot O., et al. 2011. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 43:830–837 10.1038/ng.892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L., Khiabanian H., Fangazio M., Vasishtha M., Messina M., Holmes A.B., Ouillette P., Trifonov V., Rossi D., Tabbò F., et al. 2014. Genetics of follicular lymphoma transformation. Cell Rep. 6:130–140 10.1016/j.celrep.2013.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan R.T., Saito M., Basso K., Niu H., Dalla-Favera R. 2005. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat. Immunol. 6:1054–1060 10.1038/ni1245 [DOI] [PubMed] [Google Scholar]
- Pinyol M., Cobo F., Bea S., Jares P., Nayach I., Fernandez P.L., Montserrat E., Cardesa A., Campo E. 1998. p16(INK4a) gene inactivation by deletions, mutations, and hypermethylation is associated with transformed and aggressive variants of non-Hodgkin’s lymphomas. Blood. 91:2977–2984 [PubMed] [Google Scholar]
- Relander T., Johnson N.A., Farinha P., Connors J.M., Sehn L.H., Gascoyne R.D. 2010. Prognostic factors in follicular lymphoma. J. Clin. Oncol. 28:2902–2913 10.1200/JCO.2009.26.1693 [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H.J. 2000. Primer3 on the WWW for general users and for biologist programmers. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Krawetz S., Misener S., Humana Press, Totowa, NJ: 365–386 [DOI] [PubMed] [Google Scholar]
- Schatz J.H., Oricchio E., Wolfe A.L., Jiang M., Linkov I., Maragulia J., Shi W., Zhang Z., Rajasekhar V.K., Pagano N.C., et al. 2011. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J. Exp. Med. 208:1799–1807 10.1084/jem.20110846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C.J. 1996. Cancer cell cycles. Science. 274:1672–1677 10.1126/science.274.5293.1672 [DOI] [PubMed] [Google Scholar]
- Solal-Céligny P., Roy P., Colombat P., White J., Armitage J.O., Arranz-Saez R., Au W.Y., Bellei M., Brice P., Caballero D., et al. 2004. Follicular lymphoma international prognostic index. Blood. 104:1258–1265 10.1182/blood-2003-12-4434 [DOI] [PubMed] [Google Scholar]
- Staudt L.M. 2007. A closer look at follicular lymphoma. N. Engl. J. Med. 356:741–742 10.1056/NEJMcibr067155 [DOI] [PubMed] [Google Scholar]
- Vairo G., Innes K.M., Adams J.M. 1996. Bcl-2 has a cell cycle inhibitory function separable from its enhancement of cell survival. Oncogene. 13:1511–1519 [PubMed] [Google Scholar]
- Vairo G., Soos T.J., Upton T.M., Zalvide J., DeCaprio J.A., Ewen M.E., Koff A., Adams J.M. 2000. Bcl-2 retards cell cycle entry through p27(Kip1), pRB relative p130, and altered E2F regulation. Mol. Cell. Biol. 20:4745–4753 10.1128/MCB.20.13.4745-4753.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel H.G., De Stanchina E., Fridman J.S., Malina A., Ray S., Kogan S., Cordon-Cardo C., Pelletier J., Lowe S.W. 2004. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 428:332–337 10.1038/nature02369 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.