Abstract
Regulation of phenotypic plasticity in smooth muscle requires an understanding of the mechanisms regulating phenotype-specific genes and the processes dysregulated during pathogenesis. Decades of study in airway smooth muscle has provided extensive knowledge of the gene expression patterns and signaling pathways necessary to maintain and alter smooth muscle cell phenotype. With this solid foundation, the importance and complexity of inheritable epigenetic modifications and mechanisms silencing gene expression have now emerged as fundamental components regulating aspects of inflammation, proliferation and remodeling.
Keywords: Airway smooth muscle, Asthma, Epigenetics, Inflammation, microRNA, Remodeling
1. Introduction
Epigenetics is often defined as the study of heritable changes of a phenotype, including altered patterns of gene expression within a specific cell type that result from changes in a chromosome without alterations in the DNA sequence [1]. In its natural state DNA is packaged into chromatin, an organized and dynamic protein-DNA complex consisting of DNA, histones and non-histone proteins. Epigenetic changes alter the accessibility of the proximal promoters and enhancer/silencer regions within the chromatin to transcription factors and may therefore regulate global gene activation or localized gene-specific transcription. In general, conformationally relaxed chromatin, known as euchromatin, is a hallmark of potentially active genes while compact chromatin, known as heterochromatin, is associated with transcriptionally silent genes. These epigenetic changes are mediated by a variety of molecular mechanisms, including post-transcriptional modification of the histone proteins, the involvement of small and other non-coding RNAs and DNA methylation. These changes can result in modifications to the structure of DNA itself upon DNA methylation or can alter the structure of chromatin through distinct modifications to scaffolding proteins, such as histones (See Figure 1).
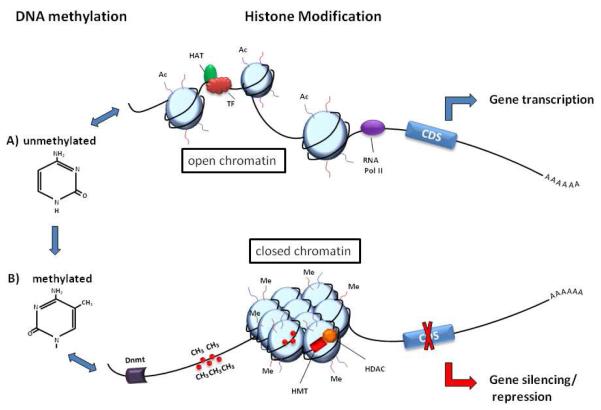
Fig 1. Epigenetic regulation of gene expression.
A) Histone modifications occur on the N-terminal tails of the four core histones. Histone acetylation (Ac) by histone acetyltransferases (HATs) stabilizes the unmethylated DNA in an open chromatin conformation to allow for transcription factor (TF) binding, recruitment of RNA polymerase II (RNAPol II), mRNA expression and the synthesis of proteins coded by the coding DNA sequence (CDS). B) DNA methylation occurs when DNA nucleotide methyltransferase (Dnmt) adds methyl groups (CH3) to cytosine residues at CpG sites. Association of methylated DNA with histone deacetylases (HDAC) and histones methylated (Me) by histone methyltransferases (HMT) closes the chromatin structure to inhibit TF binding and silence gene expression.
Although DNA methylation and histone modifications are regulated by different enzymes and chemical processes, there is evidence linking the two systems in co-operatively regulating patterns of gene expression. It appears that the relationship between these two epigenetic mechanisms can function in both directions, with histone methylation directing DNA methylation patterns and DNA methylation acting as a template to facilitate histone modifications following DNA replication [2]. microRNAs (miRNAs) most likely function independently of the other two epigenetic mechanisms. However, gene expression may be controlled by co-incident changes in DNA methylation and histone modification within a promoter region, accompanied by binding of miRNAs to mRNA transcripts targeted for gene-silencing. Individually, each of these regulatory mechanisms is complex, but when combined the complexity of the potential interactions provides unique challenges in dissecting and understanding the pathways involved. In recent years, understanding the mechanisms regulating the initiation, maintenance and heritability of epigenetic changes in cells that may undergo a phenotypic switch from a “normal” to “diseased” state has become a research focus for a variety of diseases.
In the lung, the role of epigenetic changes and miRNA-mediated gene-silencing has recently emerged as an important regulatory mechanism affecting smooth muscle cell phenotype and function. In this review, we will summarize general mechanisms of regulation by epigenetic factors and miRNA, followed by a discussion about what is known in smooth muscle cells using in vitro and in vivo models of asthma and the importance of these findings to future studies.
2. General mechanisms of epigenetic and miRNA-mediated regulation of gene expression
2.1. DNA methylation
The first epigenetic mechanism to be recognized was DNA methylation, a reversible modification of DNA structure in which a methyl group is added to cytosine residues. The majority of vertebrate genome methylation occurs at regions where a cytosine is followed directly by a guanine in the DNA sequence, also known as CpG sites. Methyl-C is prone to spontaneous deamination to thymine [3] and therefore CpG methylation frequently leads to mutation of these sites to TpG. As a result, the number of CpG occurrences in the genome is depleted from the value expected from random distribution [4]. Within the globally CpG poor genome are regions of, on average, 1000 base pairs with elevated C+G base composition, termed CpG islands (CGIs). In contrast to globally distributed CpGs, CpGs within CGIs are frequently unmethylated. Around 50% of all CGIs contain transcription start sites and coincide with gene promoters. Methylation of cytosines within these regions can lead to stable silencing of the associated promoter via direct inhibition of transcription factor binding and recruitment of methyl-binding domain containing chromatin remodeling complexes, including histone deacetylases. Histone deacteylases remove acetyl groups from histones, resulting in a closed chromatin structure. The remaining islands (“orphan CGIs”) are either intragenic or at the gene termini. These islands can be initiation sites for the transcription of non-coding RNAs (ncRNA) which, upon transcription, result in imprinting which is a silencing of a paternal/maternal allele such that only a single allele functions. For example, transcription of the ncRNA Air initiates from a CGI within exon 2 of the Igf2r gene and is only transcribed from the paternal chromosome. The resulting Air nc-RNA silences three protein-coding genes Igf2r, Slc22a2 and Slc22a3 only on the paternal allele. It is postulated that the Air promoter is silent on the maternal chromosome due to DNA methylation but this requires confirmation [5]. Furthermore, genome wide correspondence of “orphan CGIs” with RNA polymerase II binding and histone H3 lysine 4 trimethylation (H3K4me3) association suggests them to be novel promoters [4, 6, 7].
The enzymes responsible for DNA methylation are the DNA nucleotide methyltransferases (Dnmts). There are two families of Dnmts that are structurally and functionally distinct. The Dnmt3 family consists of Dnmt3a, 3b and 3L. Dnmt3a and Dnmt3b are active and establish the initial CpG pattern de novo, while Dnmt3L has no enzyme activity but immunoprecipitates with Dnmt3a/b and enhances their DNA methylation activity. The Dnmt1 family consists of Dnmt1 and Dnmt2. Dnmt1 maintains the de novo DNA methylation pattern during chromosome replication. Dnmt2, while having high sequence and structural similarity to DNA methyltransferases, does not methylate DNA[8]. Its exact role is still controversial but appears to involve methylation of tRNA[9, 10].
2.2. Histone modifications
The fundamental unit of chromatin is the nucleosome, which is composed of an octamer of four core histones (H3, H4, H2A and H2B) around which 147 base pairs of DNA are wrapped. The core histones are predominantly globular but also have unstructured N-terminal tails which contain multiple residues susceptible to modification. At least eight distinct types of post-translational histone modifications have been identified including acetylation, methylation, phosphorylation, ubiquitylation and sumoylation [11]. The combination of different N-terminal modifications, although generated by diverse molecular mechanisms, appear to interact to stabilize epigenetic changes and regulate gene expression [12]. Histone acetylation and histone methylation are the best characterized, with the modifications being regulated by histone acetyltransferases (HATs)/histone deacetylases (HDACs) and histone methyltransferases (HMTs)/histone demethylases respectively [11].
These enzymes are being increasingly implicated as both direct and indirect regulators of gene expression in a wide variety of cells types. In particular, hyperacetylation of histone lysine residues by HATs results in the open chromatin conformation which facilitates DNA interactions with the basal transcription machinery and increases gene transcription. The counterbalancing action of HATs/HDACs and HMTs/demethylases appears to fine tune gene expression to regulate important cellular process such as proliferation and differentiation, that when dysregulated can contribute to the pathogenesis of disease. As epigenetic changes are reversible, targeting and modifying the function of these enzymes has potential as a therapeutic target in a range of diseases.
2.3. miRNA-mediated gene silencing
miRNA are double-stranded RNA molecules found individually or as polycistronic clusters in introns, exons or in non-coding regions between protein-coding genes [13, 14]. They are members of a growing family of ncRNAs functioning throughout the genome to regulate epigenetic modifications, transcriptional and post-transcriptional events, maintain genomic integrity and to provide a defense from transpositional elements [15]. Indeed a large portion of non-coding sequences, once thought to be ‘junk’ DNA, are now known to contain ncRNA. miRNA themselves were first described in C.elegans, where lin-4 and let-7 were identified as repressors of regulatory proteins key to developmental timing [16, 17]. There are now more than 1900 mature human miRNA sequences [18], each predicted to target more than 100 mRNA for post-transcriptional regulation [13].
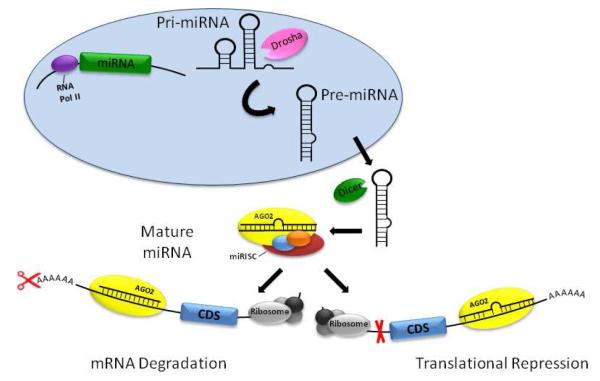
As shown in Figure 2, miRNA are initially transcribed by RNA polymerase II as a primary miRNA transcript and processed into a stem-loop structure of 70-100 nucleotides by the double-stranded-RNA specific ribonuclease (RNAse) III Drosha [19]. This precursor miRNA is exported into the cytoplasm via a GTP-dependent mechanism [20], where it is further cleaved by a second RNAse III Dicer [21]. This yields a mature miRNA duplex of 21-23 nucleotides bound by a miRNA-associated RNA-induced silencing complex (miRISC). A core catalytic component of this complex is Argonaute (AGO), a family of endonucleases with RNaseH-like activity [21] that ultimately use miRNA as a guide to bind complementary sequences in mRNA, leading to degradation of the mRNA transcript when binding in the seed sequence is complementary or translational repression when binding occurs with mismatches in seed sequence recognition [22]. In the canonical pathway, base-pairing is recognized between the 2-8 nucleotide seed sequence of the miRNA and the mRNA 3’untranslated region (UTR). In the non-canonical pathway, miRNA binding to its mRNA targets relies less on seed sequence homology and more on complementarity in coding regions or the 5′-UTR of the target transcript [23]. The importance of miRNA-mediated gene-silencing in smooth muscle cell phenotype is underscored in animals with smooth muscle targeted knock-out of Dicer, which impairs contractile differentiation of vascular smooth muscle cells [24] and inhibits smooth muscle cell development in the gastrointestinal tract [25].
Fig. 2. miRNA biogenesis and function.
miRNA are transcribed by RNA polymerase II (RNAPol II) as primary miRNA (pri-miRNA). The transcripts are cleaved by the ribonuclease Drosha to form precursor miRNA (pre-miRNA). Pre-miRNA are exported out of the nucleus and further cleaved by Dicer to yield a mature miRNA duplex bound by a miRNA-associated RNA-induced silencing complex (miRISC). Binding of complementary mRNA sequences by one strand of the miRNA leads to Argonaute (AGO2 in mammals) mediated cleavage, resulting in decreased mRNA stability and transcript cleavage or translational repression and reduced synthesis of proteins coded by the coding DNA sequence (CDS).
3. Phenotypic switching in smooth cells as a product of epigenetic dysregulation
Smooth muscle cells possess remarkable phenotypic plasticity in response to changes in the local environment and these phenotypic changes may be responsible for the development or progression of diseases including asthma [26], atherosclerosis [27] and pulmonary arterial hypertension (PAH) [28]. Although the mechanisms underlying these changes are still uncertain, evidence is emerging that epigenetic mechanisms may be at least partly responsible for the phenotypic changes seen in smooth muscle cells in these disease states including altered secretory potential, proliferation, extracellular matrix (ECM) production and vascular/airway remodeling.
3.1. Inflammatory mediators
Chronic inflammatory events are known to alter the phenotype of airway smooth muscle (ASM) cells [29, 30]. Differential expression of inflammatory mediators from diseased ASM cells has been primarily reported in the context of asthma using in vitro culture models. In the studies reported to date, ASM cells isolated from asthmatics secrete higher levels of inflammatory cytokines and chemokines than cells from non-asthmatic individuals, with no reports of repressed expression documented [31-33]. Descriptions of epigenetic mechanisms regulating inflammatory mediator expression in ASM cells from asthmatic patients are limited. However, these reports do suggest the importance of epigenetic dysregulation to smooth muscle cell phenotype in asthma.
CXCL8/IL8 is a chemokine involved in the initiation and maintenance of an inflammatory response via the recruitment of neutrophils. CXCL8 secretion from asthmatic ASM cells is increased compared to non-asthmatic cells. The mechanism involves increased binding of the transcriptional regulators NF-κB, C/EBPβ and RNA polymerase II to the unstimulated CXCL8 promoter [31]. Recent conference reports suggest that aberrant HAT binding and resultant histone H3 acetylation at the asthmatic CXCL8 promoter may be responsible for the increased binding of these transcriptional regulators [34]. In further support of histone acetylation dependent regulation of the CXCL8 promoter, Keslacy et al showed that the HDAC inhibitor trichostatin A (TSA) prevented the inhibitory effect of IFNγ on TNFα induced CXCL8 secretion [35].
The CC chemokine eotaxin is also hypersecreted from ASM cells derived from asthmatic donors compared to those from non-asthmatic donors, under unstimulated conditions [32]. Eotaxin is a potent eosinophil chemoattractant, critically involved in eosinophilic inflammatory diseases. The signaling responsible for asthmatic ASM cell hypersecretion is dependent on enhanced autocrine fibronectin secretion and subsequent signaling via α5β1 integrin. However, histone acetylation is also regulated at the eotaxin promoter in ASM cells. Specifically, TNFα induces histone H4 lysine 5 and lysine 12 acetylation (H4K5/14ac) at the eotaxin promoter [36]. It would be interesting to determine whether the enhanced autocrine fibronectin signaling of asthmatic ASM cells changes the acetylation status of histones at the eotaxin, and other cytokine promoters. Furthermore, the question remains whether changes in ECM composition alter epigenetic processes that contribute to altered inflammatory mediator secretion and disease pathogenesis? Interestingly expression of the eotaxin receptor CCR3 is also increased in asthmatic ASM cells [37]. The molecular mechanism in ASM cells is not reported, however CCR3 transcription in peripheral blood eosinophils is regulated by negative and positive GATA elements, with DNA methylation of CpG sites regulating occupancy of the sites [38]. Thus, speculatively, DNA methylation of the CCR3 gene may regulate its overexpression in ASM cells.
Finally, the CXC chemokine, CXCL10/IP10, is a potent chemoattractant for mast cells and T lymphocytes and is implicated in mast cell migration to ASM bundles [39]. A combined IFNγ/TNFα/IL-1β stimulus induces higher CXCL10 secretion from ASM cells isolated from asthmatic patients cells from non-asthmatic individuals. Differential basal expression was not reported [33]. Clarke et al have shown that both TNFα and IFNγ induce histone H4 (but not histone H3) acetylation at the CXCL10 promoter but whether this is exaggerated at the asthmatic CXCL10 promoter is unreported [39]. Future determination of how specific genes are epigenetically regulated may provide novel therapeutic targets.
3.2. Proliferation
ASM phenotype plasticity, characterized by reversible switching between contractile and proliferative phenotypes, contributes to increased ASM mass and airway hyperresponsiveness in asthma [40]. There is currently no direct evidence of a role for epigenetic regulation of ASM cell proliferation. However, vascular smooth muscle cells have been shown to undergo profound phenotypic changes during neointima formation within atherosclerotic plaques and in the development of PAH and there is some evidence suggesting that proliferation is under epigenetic control in these cells. Serum response factor (SRF) is an important mediator of transcriptional activation of genes that exhibit smooth muscle cell-restricted expression. Recent studies have determined that binding of SRF to DNA is associated with post-transcriptional histone modifications including di-methylation of lysine residues 4 and 79 on histone H3, acetylation of lysine 9 on histone 3 and acetylation of histone H4 [41]. As modulators of histone acetylation at the site of genes which control the cell-cycle, HDACs have also been implicated in regulating the proliferation of smooth muscle cells [42] with the HDAC inhibitor TSA being shown to inhibit proliferation [43].
3.3. Remodeling
Inappropriate remodeling of airways or blood vessel walls in asthma or vascular disease may result from an imbalance in the production of ECM proteins and their degradation by matrix metalloproteinases. In asthmatic ASM cells, altered patterns of ECM production including increased collagen I deposition, appear to contribute to increased proliferative responses [29, 44, 45] but there is as yet no evidence for an epigenetic mechanism regulating ECM production. Histone acetylation modifications have been implicated in the regulation and/or functional expression of a number of matrix metalloproteinases including MMP1, MMP2, MMP3, MMP9 and MMP13 suggesting that epigenetic changes may be intrinsically involved in airway or blood vessel remodeling. However, these observations have primarily been made in tumor cell lines and evidence is not yet available from primary smooth muscle cells [46].
Dysregulation of genes involved in smooth muscle cell proliferation and remodeling in asthma has received little attention, however we have recently demonstrated a role for epigenetics in the regulation of VEGF expression in this disease[47]. VEGF is a key angiogenic molecule that is overexpressed in asthmatic sputum and tissue and contributes to the increased number and size of airway blood vessels in asthma [48]. VEGF secretion in ASM cells from asthmatic patients is greater than from non-asthmatic controls [49] due to differential histone H3 lysine 9 methylation (H3K9me3) modulating Sp1 and RNA polymerase II binding to the VEGF promoter[47].
There is much more information known about epigenetic dysregulation of vascular remodeling. Endothelin-1 (ET-1) is a potent vasoconstrictor and co-mitogen for vascular smooth muscle (VSM), where it plays a well-known role in vascular remodeling in PAH. ET-1 is overexpressed in the lungs of patients with PAH [50, 51] but this has not been pinpointed to the smooth muscle cells and little attention has been paid to the molecular mechanism regulating the increased expression. However, cooperative acetylation of NF-κB and histone H4 at the preproET-1 promoter are responsible for the synergistic expression of ET-1 in response to TNFα and IFNγ, therefore it could be speculated that aberrant histone acetylation mediates ET-1 overexpression in PAH.
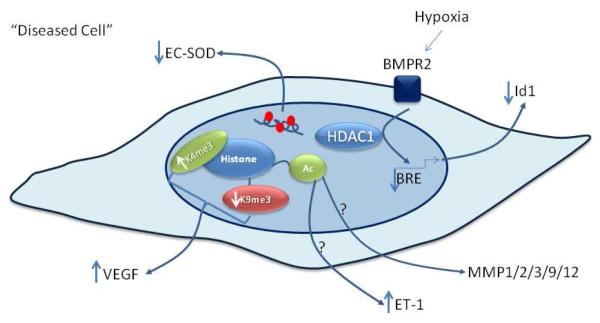
Bone morphogenetic protein (BMP) signaling represents another example of epigenetic dysregulation in PAH. BMP receptor 2 (BMPR2) mutations are causal of familial PAH and an important target of BMP signaling are the Id1 genes. Hypoxia is a potential “second hit” in familial PAH and is implicated in the pathogenesis of non-familial PAH. In both familial and non-familial PAH, hypoxia can contribute to the observed vascular remodeling [52]. Hypoxia suppresses BMP signaling and Id1 expression in pulmonary artery smooth muscle (PASM) cells, via increased HDAC1 recruitment to the BMP response element (BRE) region of the Id1 promoter. Furthermore, hypoxia induced repression of Id1 expression is abrogated by the HDAC inhibitor TSA [52]. Whether hypoxia regulates transcription of other remodeling genes in PAH or in other lung diseases via regulation of HDACs is unclear.
Extracellular superoxide dismutase (EC-SOD) appears critically involved in the development of PAH in vivo. Its overexpression attenuates the development of PAH secondary to pulmonary fibrosis in a bleomycin model via protection against medial, adventitial and intimal layer remodeling of the pulmonary vessels wall [53]. EC-SOD overexpression also attenuates chronic hypoxia induced PAH [54] and EC-SOD gene transfer ameliorates monocrotaline induced PAH, reducing medial wall thickening and pulmonary microvessel muscularization [55]. EC-SOD is more highly expressed in human PASM cells than pulmonary artery endothelial (PAEC) cells, due to increased methylation of the 5′-flanking region of the EC-SOD gene in PAECs causing repressed transcription [56]. It is possible that repression of EC-SOD synthesis by differential CpG methylation of the ECSOD gene in the PASM cells may contribute to the development of PAH.
The epigenetic mechanisms contributing to the regulation of genes involved in remodeling of smooth cells are summarized in Fig 3.
Fig. 3. Epigenetic regulation of genes associated with remodeling.
Histone lysine methylation regulates differential VEGF expression from ASM cells isolated from non-asthmatic donors compared to non-asthmatic donors. Histone acetylation is involved in the transcriptional regulation of Endothelin-1 (ET-1) and MMPs but its involvement in dysregulation of these genes expression in disease has yet to be investigated. Increased CpG methylation of the 5′ flanking region of the extracellular superoxide dismutase (EC-SOD) gene represses EC-SOD expression and may be implicated in pulmonary arterial hypertension pathogenesis.
4. miRNA mediating ASM cell functions
The role of miRNA-mediated gene silencing mechanisms in regulating smooth muscle cell functions was first described in vascular smooth muscle cells. These studies described upregulation of miR-21 following balloon injury, established that miR-21 stimulates vascular smooth muscle cell proliferation and determined that knockdown of miR-21 expression negatively affects neointimal formation [57]. From these results, it quickly became apparent that miRNA represented a key mechanism regulating smooth muscle cell phenotypic plasticity. This was solidified by the identification of miR-143 and miR-145 as modulators of vascular smooth muscle cell plasticity [58] and response to injury [59] that directly target the network of transcription factors important for smooth muscle-specific gene expression including SRF, myocardin and Krüppel-like factor (KLF) 4 [58]. Subsequent studies have now described miRNA expression and it role in phenotypic regulation of smooth muscle cell fate in airway smooth muscle [60, 61], smooth muscle from the gastrointestinal tract [25] and in uterine smooth muscle [55]. The implications of miRNA functions to pulmonary vascular disease have recently been thoroughly reviewed [28]. The purpose here will be to highlight the studies in ASM cells, summarized in Table I, and how the miRNA identified to date can provide insight into normal and pathogenic ASM functions.
Table I.
Functions of miRNA Reported in Airway Smooth Muscle
| Inflammatory Response |
Proliferation | Remodeling | Innervation |
|---|---|---|---|
| let-7 | miR-25 | miR-16 | miR-206 |
| miR-25 | miR-25 | ||
| miR-133a | miR-26a | ||
| miR-143/miR-145 | miR-133a | ||
| miR-146a | miR-146a |
4.1. miR-25 in ASM cell phenotype
Work by Kuhn et al was the first to identify a set of miRNA regulated by inflammatory mediators in human ASM cells [60]. Using the combined pro-inflammatory stimulus consisting of IFNγ/TNFα/IL-1β, miRNA array profiling demonstrated global downregulation of miRNA expression. This suggests that, at least in vitro, the initial inflammatory response of ASM is a widespread increase in gene expression, as a result of repressed gene-silencing and perhaps epigenetic modifications. Further analysis of specific miRNA in this study identified miR-25 as having a broad role in regulating ASM cell phenotype. Inhibition of miR-25 reduced expression of myosin heavy chain, decreased eotaxin and RANTES release, and had a broad impact on expression of genes involved in ECM turnover and remodeling. One target of miR-25’s actions is KLF-4, which is also targeted by miR-145 [62], miR-146a [63] and miR-1 [64]. KLF-4 acts as a transcriptional repressor of smooth-muscle specific gene expression by recruiting histone H4 deacetylase activity to smooth muscle cell genes, thereby blocking SRF association with methylated histones and CArG box chromatin [41]. Recent conference reports further established the role of miR-25 in phenotypic plasticity by demonstrating that miR-25 expression reduces proliferation of ASM cells, possibly through inhibition of cyclin D1 [65]. The role of miR-25 in vivo has yet to be reported. However, recent profiling studies of miRNA expression have shown that, as expected, miR-25 expression is significantly downregulated with prolonged OVA-sensitization and challenge [66].
4.2. miRNA targeting RhoA
Up-regulation of RhoA is associated with enhanced ASM contractility in animal models of asthma [67] and has gained momentum as a potential therapeutic target [68]. In cardiac myocytes, upregulation of miR-133 during hypertrophy corresponds to a decrease in RhoA expression [69] and binding sites for miR-133 in the RhoA 3’UTR have been predicted. In ASM, miR-133a targeting RhoA expression was demonstrated both in vitro in ASM cells and in OVA-sensitized animals [70]. Antagomirs, which are modified oligonucleotides designed to target specific miRNA sequences, are commonly used as molecular inhibitors of miRNA expression and functions. Expression of a miR-133a specific antogomir upregulated RhoA in ASM cells and provided evidence of direct targeting of RhoA by miR-133a. However, in OVA-sensitized mice both RhoA and miR-133a expression were increased. This may be due to regulation of miR-133a expression by IL-13, which mediates allergic inflammation in the OVA model. The future of this approach to asthma treatment relies on additional data exploring the efficacy of miR-133a inhibition in vivo and in other animal models of asthma.
4.3 miR-206 in ASM innervation
Innervation of the respiratory tract regulates smooth muscle tone by controlling reflex responses necessary for coughing and sneezing [19]. Changes in innervation are associated with allergic airway disease [71] and in a mouse asthma model, expression of the p75 neurotrophin receptor contributes to neuronal hyperreactivity in the airway [72]. Radzikinas et al (2011) recently determined that ASM innervation and branch formation is dependent on brain-derived neurotrophic factor (BDNF) [73]. This work identified targeting of the BDNF transcript by miR-206, which had previously been identified as one of the miRNA induced during skeletal muscle differentiation [74]. miR-206 expression in ASM is blocked by sonic hedgehog signaling, which is activated during innervation to increase BDNF. While many studies to date have focused on role of miRNA expression on ASM remodeling events associated with the inflammatory response, the results from this study underscore the need to not overlook interactions of ASM with surrounding tissues and cell types critical to lung function.
4.3. miR-26a in hypertrophy
In asthmatics, inflammatory events increase epithelial mucosal secretions, induce airway constriction leading to hyperreactivity, and result in clinically relevant airway remodeling evidenced by hyperplasia and hypertrophy of ASM [75]. Mohamed et al (2010) used mechanical stretch as an in vitro model to identify miRNA candidates involved in ASM cell hypertrophy [76]. Microarray analysis identified miR-16, miR-26a and miR-140 as potential candidates. However, only inhibition of miR-26a affected cell size and protein synthesis, while forced expression of miR-26a promoted hypertrophy. Glycogen synthase kinase 3β, a recognized mediator of ASM hypertrophy in animal models [77], was consequently identified as a target of miR-26a-mediated silencing. Further studies have verified the role of miR-26a in hypertrophy using ASM cells isolated from desmin null mice, which exhibit hypertrophy and express upregulated miR-26a [78] and have established that the early growth responsive protein-1 activates the miR-26a promoter via ERK 1/2. The notion that mitogenic or inflammatory signaling pathways may transcriptionally or post-transcriptionally regulate miRNA expression in ASM cells is intriguing and is supported by recent studies of miR-146a regulation in ASM cells. Work by Larner-Svensson et al (2010) found that IL-1β stimulated miR-146a expression was regulated transcriptionally by the NF-κB pathway through IκB kinase, while the JNK 1/2 and MEK 1/2 signaling pathways regulated miR-126a post-transcriptionally via its primary miRNA sequence [79].
4.4. Interactions with the miR-143/miR-145 cluster in ASM cells
In light of the established roles of the miR-143/miR-145 cluster in phenotypic plasticity and response of vascular smooth muscle cells to injury [58, 59], it is worth noting that a detailed analysis of airway architecture and phenotype in miR-143/miR-145 knockout animals [80] has not been reported. Since these animals are viable and do not display any obvious abnormalities, one can conclude that the miR-143/miR-145 cluster is not essential for development. Upregulation of miR-143 and miR-145 was reported with long term OVA-sensitization [66] and miR-145 was increased 5-fold in profiling studies using the house dust mite (HDM) model [81], but data in asthmatic patients is lacking. That is not to say that miR-143/miR-145 expression is not an important mediator of ASM phenotype. Both miR-143 and miR-145 are expressed in human asthmatic biopsies containing a heterogeneous cell population, although their expression levels were not significantly different compared to control patients or with corticosteroid therapy [61]. miR-143 and miR-145 were also down-regulated with pro-inflammatory stimulation in human ASM, although again not significantly when compared to non-treated cultures [60].
5. Epigenetics and asthma : potential for therapeutic targeting
Epigenetic regulation affects the transcriptional program of an individual cell without altering the underlying genetic code and importantly, these changes are reversible. A number of studies have suggested that epigenetic changes may be one of the factors responsible for the increasing prevalence of asthma in developed countries [82]. Many of these changes aresuggested to be influenced by prenatal exposure to environmental factors including diet, exposure to cigarette smoke and air pollution and have been recently reviewed in detail by Durham et al (2011) and Lovinsky-Desir and Miller (2012) [83, 84].
Although the data relating to epigenetic regulation of gene expression in individual cell types in asthma, such as ASM cells, is limited and is often restricted to the analysis of a single gene in a single cell type, the more global epigenetic changes induced by environmental exposure have the potential to simultaneously regulate expression of a variety of genes across multiple cell types.
Epidemiological studies have identified a role for maternal diet in the development of asthma [85, 86] with in utero exposure to folate being associated with increased risk of developing the disease. In vivo studies in ovalbumin challenged mice showed increased airway inflammation, Th2 cytokines, serum IgE and hyperresponsiveness in pups of mothers fed a high methylation diet [87]. The dietry change appears to promote an altered pattern of methylation at specific CpG motifs resulting in differential expression of genes which regulate the Th2 immune response and airway remodeling.
There is also continuing interest in the association of prenatal exposure to cigarette smoke with development of childhood asthma [88]. Although not all studies have reported a direct association between smoking and global DNA methylation [89], epigenetic mechanisms have been implicated in global and gene-specific changes in DNA methylation in children exposed to tobacco smoking in utero [90]. Smoking is believed to interfere with DNA methyltransferase binding resulting in hypomethylation of DNA [91]. As DNA methylation is often a mechanism for repressing or silencing gene expression, the loss of repression may result in an increase in expression of pro-inflammatory mediators and genes which regulate airway remodeling. In addition to altering DNA methylation patterns, cigarette smoke also downregulates expression of the histone deacetylase, HDAC2. The loss of this deacetylase activity may contribute to the altered HAT/HDAC ratio seen in both children [92] and adults [93] with asthma and, as described in a recent conference report, the elevated global HAT activity may subsequently lead to increased expression of pro-inflammatory genes including CXCL8 [34].
By assessing the links between environmental exposure and the onset of asthma, DNA and histone methylation as well as histone acetylation have been identified as potential targets for therapeutic intervention in asthma. From a therapeutic standpoint, steroids and β2 agonists are currently the mainstay of asthma therapy. Although these drugs were not developed with epigenetic targeting in mind, they have been shown to reduce global HAT activity and increase global HDAC activity [94]. In addition, dexamethasone, fluticasone, salbutamol and salmeterol have all been previously shown to inhibit TNFα induced H4K5/12Ac at the eotaxin promoter in ASM cells [36].
There is now clear evidence that histone acetylation plays a role in the transcriptional regulation of cytokine and chemokine expression, and in a recent conference report it is emerging that histone acetylation is altered at the promoters of cells isolated from asthmatic patients[34]. We know that global HAT activity is increased, and global HDAC activity decreased in biopsy tissue from asthmatic patients [94], but it is still unclear whether this results in changes in a single cell type e.g. ASM cells alone or alters gene regulation patterns in numerous cells types in the airways. As discussed above, the epigenetic regulation of a small number of genes with a potential role in asthma has been investigated in ASM cells and all have been found to be hypersecreted in asthmatic cells. If the global HAT/HDAC balance is altered to favor acetylation in asthma, a global increase in transcription would be expected, including increased transcription of anti-inflammatory and anti-proliferative genes. It will therefore be interesting to determine whether the epigenetic mechanisms which promote the asthmatic phenotype result from changes at promoters which regulate pro-inflammatory and pro-remodeling genes while those with anti-inflammatory and anti-proliferative roles are protected from the effects of increased global HAT activity. Although the anti-inflammatory effects of steroids are undoubtedly beneficial in asthma, their inhibitory effects on global histone acetylation, which may have widespread effects on numerous gene promoters including those that are potentially protective in asthma, suggest that in future it may be beneficial to attempt to design drugs capable of targeting therapy to specific promoters.
Although most observations of inflammatory mediator epigenetic modulation involve changes in histone acetylation, the importance of other epigenetic modifications should not be overlooked. As an example, VEGF hypersecretion in asthmatic smooth muscle cells is regulated differently and appears to be dependent on histone lysine methylation and a loss of repression of VEGF [47]. Thus, drugs which modulate histone methylation may also prove to be an important therapeutic target for treating asthma.
6. miRNA in asthma
Expression profiling has been used as an approach to identify differential miRNA expression in disease and has proved to be particularly useful in generating data from mouse models of asthma. A summary of the key miRNA identified in these studies can be found in Table II. Garbacki and colleagues(2011) performed one of the most thorough studies to date profiling miRNA expression in the lung following short, intermediate and long-term exposure to OVA [66]. Both induction and repression of miRNA expression was noted at each time point with miR-146b the only miRNA remaining upregulated throughout the entire time course. In contrast, miR29c was progressively downregulated with prolonged OVA exposure. miR-29 could be an intriguing candidate for novel epigenetic therapies since the miR-29 family (miR-29a, b, c) targets the DNA methyltransferases Dnmt3a and Dnmt3b and forced expression of miR-29s restores normal DNA methylation patterns in lung cancer cell lines [95]. miRNA profiling studies are most useful when changes in expression can be functionally verified with identification of specific mRNA targets. Garbacki et al (2011) experimentally verified targeting of miRNA to the 3’UTR of potential mRNA candidates and used bioinformatics analysis to categorize the types of regulatory pathways affected during the progression of disease in this mouse model [66]. Their analysis highlighted the differences in cell cycle signaling, inflammatory response and ECM regulation at each time point with changes in matrix metalloproteinases, IL-13 and TGFβ signaling common to all stages of the disease. miRNA expression has also been profiled using house dust mite (HDM) as a model of human disease. Studies by Mattes et al (2009) observed a significant increase in miR-16, miR-21 and miR-126 in the airway wall 24 hours after HDM challenge [96]. Silencing miR-126 with intranasal administration of antagomirs suppressed that asthmatic phenotype by diminishing IL-5 and IL-13 secretion, reducing airway hyperreactivity and eosinophilia. This was one of the first reports to suggest that an inhaled targeted delivery of miRNA inhibitors could affect lung function. This effect, however, appears to be specific to short term HDM exposure since inhibition of miR-126 in a chronic model of OVA sensitization suppressed eosinophil recruitment but did not affect airway remodeling [81]. Longer term exposure to HDM in a subsequent study identified upregulation of let-7b, miR-21 and miR-145 [81]. However, only inhibition of miR-145 with intranasal antagomirs suppressed allergic inflammation and reduced airway remodeling. These effects were comparable to dexamethasone treatment. Thus, it remains to be seen whether targeting miRNA expression can improve existing treatment paradigms or stand-alone as alternatives.
Table II.
miRNA identified in Animal Models of Asthma
| IL-13 | HDM | OVA | |||
|---|---|---|---|---|---|
| ST | LT | ST | LT | ||
| let-7 | ↓ | ↑ | |||
| miR-16 | ↑ | ||||
| miR-21 | ↑ | ↑ | ↑ | ↑ | |
| miR-29c | ↓ | ↓ | |||
| miR-126 | ↑ | ||||
| miR-145 | ↑ | ↑ | |||
| miR-146a | ↑ | ↑ | |||
ST, short-term; LT, long-term; ↓ downregulated; ↑upregulated
Targeting IL-13 expression in mouse models of asthma has also proven successful using miRNA-based mimics. Kumar et al (2011) reported that increased IL-13 production in human T cells was inversely correlated with expression of the let-7 family of miRNA and identified targeting of let-7 to the IL-13 3’UTR [97]. Intranasal administration of chemically modified oligonucleotides designed to mimic let-7 reduced IL-13 levels in mice acutely sensitized to OVA and reduced hallmarks of allergic inflammation, including mucus secretion, fibrosis, airway inflammation and hyperreactivity. Thus, let-7 represents a promising target for miRNA-based interventions, although the importance of let-7 expression to other animal models of asthma or to human disease remains to be determined. In a transgenic model of IL-13-induced allergic asthma, overexpression of miR-21 was again identified and found to target IL-12p35 [98]. Targeted ablation of miR-21 in OVA-sensitized mice leads to increased Th1 responses with increased IFNγ and reduced eosinophilia consistent with suppressed IL-12p35 expression [98]. Regulation of Th1 and Th2 responses by miR-21 in the lung represents a paradigm shift in what is known about polarization of T cells during the allergic immune response.
In human asthmatic airways, miRNA expression has been measured in lung biopsies from mild asthmatics but no significant changes in miRNA expression were noted in expression of 227 profiled miRNA [61]. This same study analyzed the effect of inhaled budesonide treatment and after 1 month of treatment did not find any significant difference in miRNA expression despite improved lung function in the patients. This result was surprising but perhaps not all that unexpected since asthma is a complex, multi-cellular, progressive disease and miRNA expression data was obtained from airway biopsies containing a mixture of cell types at only one time point. Indeed, the authors went on to analyze miRNA expression in individual cell types from the lung, including epithelial cells, ASM cells, alveolar macrophages and lung fibroblasts and reported distinct miRNA expression profiles in each cell type. This suggests that cell-specific gene silencing may be a significant mechanism used to regulate gene expression in a heterogeneous population. Analysis of miRNA expression in severe asthmatics or in vitro in individual cell-types, such as ASM cells, from asthmatic patients will provide much needed data on the importance of cell-type specific miRNA expression to the pathogenesis of asthma. As an example, studies in alveolar epithelial cells and macrophages have shown that miR-146a expression negatively regulates IL-8 and RANTES release [99], while evidence in ASM cells does not supports a role for miR-146a in IL-6 or IL-8 release [79].
Another approach to understanding the role of miRNA expression in asthma is to identify miRNA binding sites in known or potential susceptibility genes. This can be done in silico using miRNA target site prediction algorithms such as TargetScan [100], MiRanda [101] and PicTar [102]. In this way, miRNA binding sites have been identified on the class 1 histocompatibility antigen G (HLA-G). HLA-G was described as an asthma susceptibility gene in families with a history of asthma where the child’s HLA-G genotype correlated with hyperresponsiveness of the mother [103]. Prediction algorithms identified binding sites for miR-148a, miR-148b and miR-152 in the HLA-G 3’UTR and this result was verified experimentally to demonstrate that these miRNA target HLA-G for gene-silencing [55]. Subsequent experiments identified miRNA binding sites in single nucleotide polymorphisms of HLA-G and suggest that miRNA binding at these sites contributes to HLA-G expression [104].
7. Future directions
Epigenetic and miRNA research is already contributing to an improved understanding of the mechanisms that regulate the development of many diseases including asthma, cancer and vascular disease [105]. In many disciplines, these studies are still in their infancy, focusing on the changes which regulate expression of a single gene within a single diseased cell type. Future studies will need to build on these early observations to assess epigenetic and miRNA-mediated changes across a range of genes and cells types within diseased tissues as well as gaining clearer insights into the more complex interactions which occur when multiple mechanisms contribute to altered patterns of gene expression within diseased cells.
A number of research tools have been developed to characterize and functionally analyze the wide variety of epigenetic changes which occur throughout the entire genome. This research, known as epigenomics, should provide insights into the importance of epigenetic modifications in both normal development and the transition to diseases states and has the potential to detect quantitative changes, multiplex modifications and also identify regulatory sequences that all combine to control gene expression [106]. Investigating epigenetic changes throughout the genome is likely to be a complex process given the large number of potential modifications and the dynamic nature of the epigenomic landscape, as this may result in the epigenomic profile changing during disease progression. The same can be said for miRNA-mediated changes in gene expression since one miRNA is predicted to target hundreds of mRNA transcripts [13]. There is also evidence that the enzymes responsible for epigenetic modifications are themselves targets of miRNA-mediated gene silencing [95, 107], and interactions between these processes are an opportunity for new avenues of investigation in the lung. The few examples of cell-type specificity that have been highlighted here suggest that global miRNA expression profiling in heterogeneous tissues may not prove useful in delineating individual effects of miRNA on specific cell types.
The currently available epigenetic therapies target one specific type of epigenetic abnormality, but it is becoming clear that many complex diseases may result from a combination of diverse epigenetic modifications. DNA methyltransferase inhibitors or HDAC inhibitors may be effective therapies in diseases such as cancer which result from abnormal silencing of gene expression but these drugs would be ineffective in diseases resulting from an increase in gene expression. Although data on disease-associated epigenetic changes in smooth muscle cells is limited, most of the functional and phenotypic changes in these cells have been associated with gene activation rather than gene repression, and therefore drugs which inhibit histone acetylation or promote histone methylation to silence hyperactive genes may be more effective.
In contrast, the promise of developing viable therapeutic interventions based miRNA-mediated gene silencing is quickly becoming a reality and the lung represents an ideal target for localized delivery. One of the first examples of RNA interference therapy delivered antisense oligonucleotides intranasally to inhibit respiratory syncytial virus replication in mice [88]. Indeed, miRNA represent a versatile target for therapeutic intervention with means that either inhibit or mimic miRNA function. Pre-clinical studies targeting inhibition of miR-208 and miR-15 in heart failure[108] and ischemic injury [109] are currently underway, while miRNA replacement therapy for either let-7 or miR-34a is a strategy being explored in non-small cell lung cancer. The identification of small-molecule inhibitors is as yet an unexplored option that may yield compounds effective in modifying epigenetic changes and/or miRNA expression. Continued study of these processes in ASM using both in vitro and in vivo methods, coupled with future additional drug discovery and development may yield exciting new therapeutic approaches to lung disease.
Acknowledgements
R.L. Clifford is supported by The Wellcome Trust, C.A. Singer is supported by the National Institutes of Health HL080960 and grants from the National Center for Research Resources RR016464and the National Institute of General Medical Sciences GM103440. A.E. John. is supported by the Medical Research Council.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–3. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- [3].Walser JC, Ponger L, Furano AV. CpG dinucleotides and the mutation rate of non-CpG DNA. Genome Res. 2008;18:1403–14. doi: 10.1101/gr.076455.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Medvedeva YA, Fridman MV, Oparina NJ, Malko DB, Ermakova EO, Kulakovskiy IV, et al. Intergenic, gene terminal, and intragenic CpG islands in the human genome. BMC Genomics. 2010;11:48. doi: 10.1186/1471-2164-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Seidl CI, Stricker SH, Barlow DP. The imprinted Air ncRNA is an atypical RNAPII transcript that evades splicing and escapes nuclear export. EMBO J. 2006;25:3565–75. doi: 10.1038/sj.emboj.7601245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–22. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11:1068–75. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- [8].Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure. 2008;16:341–50. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–8. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- [10].Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, et al. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 2007;21:261–6. doi: 10.1101/gad.1472907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- [12].Nightingale KP, O’Neill LP, Turner BM. Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Curr Opin Genet Dev. 2006;16:125–36. doi: 10.1016/j.gde.2006.02.015. [DOI] [PubMed] [Google Scholar]
- [13].Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- [14].Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- [15].Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–39. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- [16].Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- [17].Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- [18].Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Canning BJ. Reflex regulation of airway smooth muscle tone. J Appl Physiol. 2006;101:971–85. doi: 10.1152/japplphysiol.00313.2006. [DOI] [PubMed] [Google Scholar]
- [20].Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. Rna. 2004;10:185–91. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- [23].Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879–84. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Albinsson S, Skoura A, Yu J, DiLorenzo A, Fernandez-Hernando C, Offermanns S, et al. Smooth muscle miRNAs are critical for post-natal regulation of blood pressure and vascular function. PLoS One. 2011;6:e18869. doi: 10.1371/journal.pone.0018869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Park C, Yan W, Ward SM, Hwang SJ, Wu Q, Hatton WJ, et al. MicroRNAs dynamically remodel gastrointestinal smooth muscle cells. PLoS One. 2011;6:e18628. doi: 10.1371/journal.pone.0018628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dekkers BG, Bos IS, Zaagsma J, Meurs H. Functional consequences of human airway smooth muscle phenotype plasticity. Br J Pharmacol. 2011 doi: 10.1111/j.1476-5381.2011.01773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Alexander MR, Owens GK. Epigenetic control of smooth muscle cell differentiation and phenotypic switching in vascular development and disease. Annu Rev Physiol. 2012;74:13–40. doi: 10.1146/annurev-physiol-012110-142315. [DOI] [PubMed] [Google Scholar]
- [28].Joshi SR, McLendon JM, Comer BS, Gerthoffer WT. MicroRNAs-control of essential genes: Implications for pulmonary vascular disease. Pulm Circ. 2011;1:357–64. doi: 10.4103/2045-8932.87301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hirst SJ, Twort CH, Lee TH. Differential effects of extracellular matrix proteins on human airway smooth muscle cell proliferation and phenotype. Am J Respir Cell Mol Biol. 2000;23:335–44. doi: 10.1165/ajrcmb.23.3.3990. [DOI] [PubMed] [Google Scholar]
- [30].Halayko AJ, Amrani Y. Mechanisms of inflammation-mediated airway smooth muscle plasticity and airways remodeling in asthma. RespirPhysiol Neurobiol. 2003;137:209–22. doi: 10.1016/s1569-9048(03)00148-4. [DOI] [PubMed] [Google Scholar]
- [31].John AE, Zhu YM, Brightling CE, Pang L, Knox AJ. Human airway smooth muscle cells from asthmatic individuals have CXCL8 hypersecretion due to increased NF-kappa B p65, C/EBP beta, and RNA polymerase II binding to the CXCL8 promoter. J Immunol. 2009;183:4682–92. doi: 10.4049/jimmunol.0803832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chan V, Burgess JK, Ratoff JC, O’Connor BJ, Greenough A, Lee TH, et al. Extracellular matrix regulates enhanced eotaxin expression in asthmatic airway smooth muscle cells. Am J Respir Crit Care Med. 2006;174:379–85. doi: 10.1164/rccm.200509-1420OC. [DOI] [PubMed] [Google Scholar]
- [33].Brightling CE, Ammit AJ, Kaur D, Black JL, Wardlaw AJ, Hughes JM, et al. The CXCL10/CXCR3 axis mediates human lung mast cell migration to asthmatic airway smooth muscle. Am J Respir Crit Care Med. 2005;171:1103–8. doi: 10.1164/rccm.200409-1220OC. [DOI] [PubMed] [Google Scholar]
- [34].John AE, Clifford RL, Brightling CE, Knox AJ. American Thoracic Society. Am J Respir Crit Care Med; New Orleans: 2010. Basal Hypersecretion of CXCL8 in Asthmatic Human Airway Smooth Muscle Cells is Associated with Increased Binding of Histone Acetyltransferases to the CXCL8 Promoter and Acetylation of Histone H3; p. A2145. [Google Scholar]
- [35].Keslacy S, Tliba O, Baidouri H, Amrani Y. Inhibition of tumor necrosis factor-alpha-inducible inflammatory genes by interferon-gamma is associated with altered nuclear factor-kappaB transactivation and enhanced histone deacetylase activity. Mol Pharmacol. 2007;71:609–18. doi: 10.1124/mol.106.030171. [DOI] [PubMed] [Google Scholar]
- [36].Nie M, Knox AJ, Pang L. beta2-Adrenoceptor agonists, like glucocorticoids, repress eotaxin gene transcription by selective inhibition of histone H4 acetylation. J Immunol. 2005;175:478–86. doi: 10.4049/jimmunol.175.1.478. [DOI] [PubMed] [Google Scholar]
- [37].Joubert P, Lajoie-Kadoch S, Labonte I, Gounni AS, Maghni K, Wellemans V, et al. CCR3 expression and function in asthmatic airway smooth muscle cells. J Immunol. 2005;175:2702–8. doi: 10.4049/jimmunol.175.4.2702. [DOI] [PubMed] [Google Scholar]
- [38].Uhm TG, Lee SK, Kim BS, Kang JH, Park CS, Rhim TY, et al. CpG Methylation at GATA Elements in the Regulatory Region of CCR3 Positively Correlates with CCR3 Transcription. Exp Mol Med. 2012 doi: 10.3858/emm.2012.44.4.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Clarke DL, Clifford RL, Jindarat S, Proud D, Pang L, Belvisi M, et al. TNFalpha and IFNgamma synergistically enhance transcriptional activation of CXCL10 in human airway smooth muscle cells via STAT-1, NF-kappaB, and the transcriptional coactivator CREB-binding protein. J Biol Chem. 2010;285:29101–10. doi: 10.1074/jbc.M109.0999952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bousquet J, Jeffery PK, Busse WW, Johnson M, Vignola AM. Asthma. From bronchoconstriction to airways inflammation and remodeling. Am J Respir Crit Care Med. 2000;161:1720–45. doi: 10.1164/ajrccm.161.5.9903102. [DOI] [PubMed] [Google Scholar]
- [41].McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest. 2006;116:36–48. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sambucetti LC, Fischer DD, Zabludoff S, Kwon PO, Chamberlin H, Trogani N, et al. Histone deacetylase inhibition selectively alters the activity and expression of cell cycle proteins leading to specific chromatin acetylation and antiproliferative effects. J Biol Chem. 1999;274:34940–7. doi: 10.1074/jbc.274.49.34940. [DOI] [PubMed] [Google Scholar]
- [43].Okamoto H, Fujioka Y, Takahashi A, Takahashi T, Taniguchi T, Ishikawa Y, et al. Trichostatin A, an inhibitor of histone deacetylase, inhibits smooth muscle cell proliferation via induction of p21(WAF1) J Atheroscler Thromb. 2006;13:183–91. doi: 10.5551/jat.13.183. [DOI] [PubMed] [Google Scholar]
- [44].Araujo BB, Dolhnikoff M, Silva LF, Elliot J, Lindeman JH, Ferreira DS, et al. Extracellular matrix components and regulators in the airway smooth muscle in asthma. Eur Respir J. 2008;32:61–9. doi: 10.1183/09031936.00147807. [DOI] [PubMed] [Google Scholar]
- [45].Johnson PR, Burgess JK, Underwood PA, Au W, Poniris MH, Tamm M, et al. Extracellular matrix proteins modulate asthmatic airway smooth muscle cell proliferation via an autocrine mechanism. J Allergy Clin Immunol. 2004;113:690–6. doi: 10.1016/j.jaci.2003.12.312. [DOI] [PubMed] [Google Scholar]
- [46].Pons D, de Vries FR, van den Elsen PJ, Heijmans BT, Quax PH, Jukema JW. Epigenetic histone acetylation modifiers in vascular remodelling: new targets for therapy in cardiovascular disease. Eur Heart J. 2009;30:266–77. doi: 10.1093/eurheartj/ehn603. [DOI] [PubMed] [Google Scholar]
- [47].Clifford RL, John AE, Brightling CE, Knox AJ. Abnormal Histone Methylation Is Responsible for Increased Vascular Endothelial Growth Factor 165a Secretion from Airway Smooth Muscle Cells in Asthma. J Immunol. 2012 doi: 10.4049/jimmunol.1103641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].McDonald DM. Angiogenesis and remodeling of airway vasculature in chronic inflammation. Am J Respir Crit Care Med. 2001;164:S39–45. doi: 10.1164/ajrccm.164.supplement_2.2106065. [DOI] [PubMed] [Google Scholar]
- [49].Simcock DE, Kanabar V, Clarke GW, Mahn K, Karner C, O’Connor BJ, et al. Induction of angiogenesis by airway smooth muscle from patients with asthma. Am J Respir Crit Care Med. 2008;178:460–8. doi: 10.1164/rccm.200707-1046OC. [DOI] [PubMed] [Google Scholar]
- [50].Cacoub P, Dorent R, Nataf P, Carayon A, Riquet M, Noe E, et al. Endothelin-1 in the lungs of patients with pulmonary hypertension. Cardiovasc Res. 1997;33:196–200. doi: 10.1016/s0008-6363(96)00189-7. [DOI] [PubMed] [Google Scholar]
- [51].Giaid A, Yanagisawa M, Langleben D, Michel RP, Levy R, Shennib H, et al. Expression of endothelin-1 in the lungs of patients with pulmonary hypertension. N Engl J Med. 1993;328:1732–9. doi: 10.1056/NEJM199306173282402. [DOI] [PubMed] [Google Scholar]
- [52].Stenmark KR, Fagan KA, Frid MG. Hypoxia-induced pulmonary vascular remodeling: cellular and molecular mechanisms. Circ Res. 2006;99:675–91. doi: 10.1161/01.RES.0000243584.45145.3f. [DOI] [PubMed] [Google Scholar]
- [53].Van Rheen Z, Fattman C, Domarski S, Majka S, Klemm D, Stenmark KR, et al. Lung extracellular superoxide dismutase overexpression lessens bleomycin-induced pulmonary hypertension and vascular remodeling. Am J Respir Cell Mol Biol. 2011;44:500–8. doi: 10.1165/rcmb.2010-0065OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Nozik-Grayck E, Suliman HB, Majka S, Albietz J, Van Rheen Z, Roush K, et al. Lung EC-SOD overexpression attenuates hypoxic induction of Egr-1 and chronic hypoxic pulmonary vascular remodeling. Am J Physiol Lung Cell Mol Physiol. 2008;295:L422–30. doi: 10.1152/ajplung.90293.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kamezaki F, Tasaki H, Yamashita K, Tsutsui M, Koide S, Nakata S, et al. Gene transfer of extracellular superoxide dismutase ameliorates pulmonary hypertension in rats. Am J Respir Crit Care Med. 2008;177:219–26. doi: 10.1164/rccm.200702-264OC. [DOI] [PubMed] [Google Scholar]
- [56].Zelko IN, Stepp MW, Vorst AL, Folz RJ. Histone acetylation regulates the cell-specific and interferon-gamma-inducible expression of extracellular superoxide dismutase in human pulmonary arteries. Am J Respir Cell Mol Biol. 2011;45:953–61. doi: 10.1165/rcmb.2011-0012OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–88. doi: 10.1161/CIRCRESAHA.106.141986. Epub 2007 May 3. [DOI] [PubMed] [Google Scholar]
- [58].Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–10. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–78. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kuhn AR, Schlauch K, Lao R, Halayko AJ, Gerthoffer WT, Singer CA. MicroRNA expression in human airway smooth muscle cells: Role of miR-25 in regulation of airway smooth muscle mhenotype. Am J Respir Cell Mol Biol. 2010;42:506–13. doi: 10.1165/rcmb.2009-0123OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Williams AE, Larner-Svensson H, Perry MM, Campbell GA, Herrick SE, Adcock IM, et al. MicroRNA expression profiling in mild asthmatic human airways and effect of corticosteroid therapy. PLoS ONE. 2009;4:e5889. doi: 10.1371/journal.pone.0005889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–58. doi: 10.1016/j.cell.2009.02.038. Epub 2009 Apr 30. [DOI] [PubMed] [Google Scholar]
- [63].Sun SG, Zheng B, Han M, Fang XM, Li HX, Miao SB, et al. miR-146a and Kruppel-like factor 4 form a feedback loop to participate in vascular smooth muscle cell proliferation. EMBO Rep. 2011;12:56–62. doi: 10.1038/embor.2010.172. Epub 2010 Nov 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Xie C, Huang H, Sun X, Guo Y, Hamblin M, Ritchie RP, et al. MicroRNA-1 regulates smooth muscle cell differentiation by repressing Kruppel-like factor 4. Stem Cells Dev. 2011;20:205–10. doi: 10.1089/scd.2010.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Munguia R, Kuhn AR, Singer CA. American Thoracic Society. Am J Respir Crit Car Med; New Orleans, LA: 2010. Expression of microRNA-25 inhibits airway smooth muscle cell proliferation; p. A2011. [Google Scholar]
- [66].Garbacki N, Di Valentin E, Huynh-Thu VA, Geurts P, Irrthum A, Crahay C, et al. MicroRNAs profiling in murine models of acute and chronic asthma: a relationship with mRNAs targets. PLoS ONE. 2011;6:e16509. doi: 10.1371/journal.pone.0016509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Chiba Y, Ueno A, Shinozaki K, Takeyama H, Nakazawa S, Sakai H, et al. Involvement of RhoA-mediated Ca2+ sensitization in antigen-induced bronchial smooth muscle hyperresponsiveness in mice. Respir Res. 2005;6:4. doi: 10.1186/1465-9921-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Schaafsma D, Gosens R, Zaagsma J, Halayko AJ, Meurs H. Rho kinase inhibitors: a novel therapeutical intervention in asthma? Eur J Pharmacol. 2008;585:398–406. doi: 10.1016/j.ejphar.2008.01.056. Epub 2008 Mar 18. [DOI] [PubMed] [Google Scholar]
- [69].Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–8. doi: 10.1038/nm1582. Epub 2007 Apr 29. [DOI] [PubMed] [Google Scholar]
- [70].Chiba Y, Tanabe M, Goto K, Sakai H, Misawa M. Down-regulation of miR-133a contributes to up-regulation of Rhoa in bronchial smooth muscle cells. Am J Respir Crit Care Med. 2009;180:713–9. doi: 10.1164/rccm.200903-0325OC. [DOI] [PubMed] [Google Scholar]
- [71].Undem BJ, Kajekar R, Hunter DD, Myers AC. Neural integration and allergic disease. J Allergy Clin Immunol. 2000;106:S213–20. doi: 10.1067/mai.2000.110153. [DOI] [PubMed] [Google Scholar]
- [72].Kerzel S, Path G, Nockher WA, Quarcoo D, Raap U, Groneberg DA, et al. Pan neurotrophin receptor p75 contributes to neuronal hyperreactivity and airway inflammation in a murine model of experimental asthma. Am J Respir Cell Mol Biol. 2003;28:170–8. doi: 10.1165/rcmb.4811. [DOI] [PubMed] [Google Scholar]
- [73].Radzikinas K, Aven L, Jiang Z, Tran T, Paez-Cortez J, Boppidi K, et al. A Shh/miR-206/BDNF cascade coordinates innervation and formation of airway smooth muscle. J Neurosci. 2011;31:15407–15. doi: 10.1523/JNEUROSCI.2745-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–87. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ebina M, Takahashi T, Chiba T, Motomiya M. Cellular hypertrophy and hyperplasia of airway smooth muscles underlying bronchial asthma. A 3-D morphometric study. Am Rev Respir Dis. 1993;148:720–6. doi: 10.1164/ajrccm/148.3.720. [DOI] [PubMed] [Google Scholar]
- [76].Mohamed JS, Lopez MA, Boriek AM. Mechanical stretch up-regulates microRNA-26a and induces human airway smooth muscle hypertrophy by suppressing glycogen synthase kinase-3beta. J Biol Chem. 2010;285:29336–47. doi: 10.1074/jbc.M110.101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Bentley JK, Deng H, Linn MJ, Lei J, Dokshin GA, Fingar DC, et al. Airway smooth muscle hyperplasia and hypertrophy correlate with glycogen synthase kinase-3(beta) phosphorylation in a mouse model of asthma. Am J Physiol Lung Cell Mol Physiol. 2009;296:L176–84. doi: 10.1152/ajplung.90376.2008. Epub 2008 Nov 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Mohamed JS, Hajira A, Li Z, Paulin D, Boriek AM. Desmin regulates airway smooth muscle hypertrophy through early growth-responsive protein-1 and microRNA-26a. J Biol Chem. 2011;286:43394–404. doi: 10.1074/jbc.M111.235127. Epub 2011 Sep 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Larner-Svensson HM, Williams AE, Tsitsiou E, Perry MM, Jiang X, Chung KF, et al. Pharmacological studies of the mechanism and function of interleukin-1beta-induced miRNA-146a expression in primary human airway smooth muscle. Respir Res. 2010;11:68. doi: 10.1186/1465-9921-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–8. doi: 10.1038/cdd.2009.153. Epub 2009 Oct 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Collison A, Herbert C, Siegle JS, Mattes J, Foster PS, Kumar RK. Altered expression of microRNA in the airway wall in chronic asthma: miR-126 as a potential therapeutic target. BMC Pulm Med. 2011;11:29. doi: 10.1186/1471-2466-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Eder W, Ege MJ, von Mutius E. The asthma epidemic. N Engl J Med. 2006;355:2226–35. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- [83].Durham AL, Wiegman C, Adcock IM. Epigenetics of asthma. Biochim Biophys Acta. 2011;1810:1103–9. doi: 10.1016/j.bbagen.2011.03.006. [DOI] [PubMed] [Google Scholar]
- [84].Lovinsky-Desir S, Miller RL. Epigenetics, asthma, and allergic diseases: a review of the latest advancements. Curr Allergy Asthma Rep. 2012;12:211–20. doi: 10.1007/s11882-012-0257-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Whitrow MJ, Moore VM, Rumbold AR, Davies MJ. Effect of supplemental folic acid in pregnancy on childhood asthma: a prospective birth cohort study. Am J Epidemiol. 2009;170:1486–93. doi: 10.1093/aje/kwp315. [DOI] [PubMed] [Google Scholar]
- [86].Haberg SE, London SJ, Stigum H, Nafstad P, Nystad W. Folic acid supplements in pregnancy and early childhood respiratory health. Arch Dis Child. 2009;94:180–4. doi: 10.1136/adc.2008.142448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Hollingsworth JW, Maruoka S, Boon K, Garantziotis S, Li Z, Tomfohr J, et al. In utero supplementation with methyl donors enhances allergic airway disease in mice. J Clin Invest. 2008;118:3462–9. doi: 10.1172/JCI34378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [88].Bitko V, Musiyenko A, Shulyayeva O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2005;11:50–5. doi: 10.1038/nm1164. Epub 2004 Dec 26. [DOI] [PubMed] [Google Scholar]
- [89].Hillemacher T, Frieling H, Moskau S, Muschler MA, Semmler A, Kornhuber J, et al. Global DNA methylation is influenced by smoking behaviour. Eur Neuropsychopharmacol. 2008;18:295–8. doi: 10.1016/j.euroneuro.2007.12.005. [DOI] [PubMed] [Google Scholar]
- [90].Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180:462–7. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Franco R, Schoneveld O, Georgakilas AG. Panayiotidis MI. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. 2008;266:6–11. doi: 10.1016/j.canlet.2008.02.026. [DOI] [PubMed] [Google Scholar]
- [92].Su RC, Becker AB, Kozyrskyj AL, Hayglass KT. Altered epigenetic regulation and increasing severity of bronchial hyperresponsiveness in atopic asthmatic children. J Allergy Clin Immunol. 2009;124:1116–8. doi: 10.1016/j.jaci.2009.08.033. [DOI] [PubMed] [Google Scholar]
- [93].Ito K, Lim S, Caramori G, Chung KF, Barnes PJ, Adcock IM. Cigarette smoking reduces histone deacetylase 2 expression, enhances cytokine expression, and inhibits glucocorticoid actions in alveolar macrophages. FASEB J. 2001;15:1110–2. [PubMed] [Google Scholar]
- [94].Ito K, Caramori G, Lim S, Oates T, Chung KF, Barnes PJ, et al. Expression and activity of histone deacetylases in human asthmatic airways. Am J Respir Crit Care Med. 2002;166:392–6. doi: 10.1164/rccm.2110060. [DOI] [PubMed] [Google Scholar]
- [95].Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. Epub 2007 Sep 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci U S A. 2009;20:20. doi: 10.1073/pnas.0905063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Kumar M, Ahmad T, Sharma A, Mabalirajan U, Kulshreshtha A, Agrawal A, et al. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J Allergy Clin Immunol. 2011;128:1077–85.e1-10. doi: 10.1016/j.jaci.2011.04.034. Epub 2011 May 25. [DOI] [PubMed] [Google Scholar]
- [98].Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Perry MM, Moschos SA, Williams AE, Shepherd NJ, Larner-Svensson HM, Lindsay MA. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol. 2008;180:5689–98. doi: 10.4049/jimmunol.180.8.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- [101].John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. Epub 2004 Oct 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. Epub 2005 Apr 3. [DOI] [PubMed] [Google Scholar]
- [103].Nicolae D, Cox NJ, Lester LA, Schneider D, Tan Z, Billstrand C, et al. Fine mapping and positional candidate studies identify HLA-G as an asthma susceptibility gene on chromosome 6p21. Am J Hum Genet. 2005;76:349–57. doi: 10.1086/427763. Epub 2004 Dec 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Castelli EC, Moreau P, Oya e Chiromatzo A, Mendes-Junior CT, Veiga-Castelli LC, Yaghi L, et al. In silico analysis of microRNAS targeting the HLA-G 3′ untranslated region alleles and haplotypes. Hum Immunol. 2009;70:1020–5. doi: 10.1016/j.humimm.2009.07.028. [DOI] [PubMed] [Google Scholar]
- [105].Hamm CA, Costa FF. The impact of epigenomics on future drug design and new therapies. Drug Discov Today. 2011;16:626–35. doi: 10.1016/j.drudis.2011.04.007. [DOI] [PubMed] [Google Scholar]
- [106].Callinan PA, Feinberg AP. The emerging science of epigenomics. Hum Mol Genet. 2006;15 doi: 10.1093/hmg/ddl095. Spec No 1:R95-101. [DOI] [PubMed] [Google Scholar]
- [107].Roccaro AM, Sacco A, Jia X, Azab AK, Maiso P, Ngo HT, et al. microRNA-dependent modulation of histone acetylation in Waldenstrom macroglobulinemia. Blood. 2010;116:1506–14. doi: 10.1182/blood-2010-01-265686. Epub 2010 Jun 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Montgomery RL, Hullinger TG, Semus HM, Dickinson BA, Seto AG, Lynch JM, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–47. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Hullinger TG, Montgomery RL, Seto AG, Dickinson BA, Semus HM, Lynch JM, et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res. 2012;110:71–81. doi: 10.1161/CIRCRESAHA.111.244442. Epub 2011 Nov 3. [DOI] [PMC free article] [PubMed] [Google Scholar]