Genome doubling and changes in genome size are fundamental evolutionary processes. Arabidopsis kamchatica has been reported to contain both diploid and tetraploid individuals (2 or 4 copies of every chromosome). We did find genome size differences among populations, and among populations genome size varied 7%. However, all sampled A. kamchatica plants from a wide geographic range were tetraploids. This level of intraspecific genome size variation in A. kamchatica is lower than in other Arabidopsis taxa. Due to its close relationship to A. thaliana, A. kamchatica has the potential to be very useful in the study of polyploidy and genome evolution.
Keywords: Allotetraploid, Arabidopsis halleri ssp. gemmifera, Arabidopsis kamchatica, Arabidopsis lyrata, C-value, 2C DNA content, flow cytometry, genome size, genome size variation
Abstract
Polyploidization and subsequent changes in genome size are fundamental processes in evolution and diversification. Little is currently known about the extent of genome size variation within taxa and the evolutionary forces acting on this variation. Arabidopsis kamchatica has been reported to contain both diploid and tetraploid individuals. The aim of this study was to determine the genome size of A. kamchatica, whether there is variation in ploidy and/or genome size in A. kamchatica and to study how genome size has evolved. We used propidium iodide flow cytometry to measure 2C DNA content of 73 plants from 25 geographically diverse populations of the putative allotetraploid A. kamchatica and its parents, Arabidopsis lyrata and Arabidopsis halleri. All A. kamchatica plants appear to be tetraploids. The mean 2C DNA content of A. kamchatica was 1.034 pg (1011 Mbp), which is slightly smaller than the sum of its diploid parents (A. lyrata: 0.502 pg; A. halleri: 0.571 pg). Arabidopsis kamchatica appears to have lost ∼37.594 Mbp (3.6 %) of DNA from its 2C genome. Tetraploid A. lyrata from Germany and Austria appears to have lost ∼70.366 Mbp (7.2 %) of DNA from the 2C genome, possibly due to hybridization with A. arenosa, which has a smaller genome than A. lyrata. We did find genome size differences among A. kamchatica populations, which varied up to 7 %. Arabidopsis kamchatica ssp. kawasakiana from Japan appears to have a slightly larger genome than A. kamchatica ssp. kamchatica from North America, perhaps due to multiple allopolyploid origins or hybridization with A. halleri. However, the among-population coefficient of variation in 2C DNA content is lower in A. kamchatica than in other Arabidopsis taxa. Due to its close relationship to A. thaliana, A. kamchatica has the potential to be very useful in the study of polyploidy and genome evolution.
Introduction
Polyploidy is one of the most important forces influencing plant diversification. Polyploidy was likely involved in 15 % of all recent angiosperm speciation events (Wood et al. 2009) and ancient polyploidy is apparent in all plant genomes sequenced to date (Jiao et al. 2011). Similarly, the majority of cultivated crops have undergone polyploidization during domestication (Otto and Whitton 2000). Polyploidy influences the ecology and physiology of plants by generating novel phenotypes that may influence mating system, habitat and geographical distribution (Levin 2002). It can have major genetic and genomic effects, such as altering chromosome segregation, masking deleterious mutations, influencing levels of genetic diversity, changing gene expression, causing rearrangements, gene loss and epigenetic changes, rewiring genetic networks, and altering rates of adaptation (Levin 2002; Adams and Wendel 2005; Chen 2007; De Smet and Van de Peer 2012; Madlung 2013). Ploidy variation has the potential to promote the origin of new species, but ploidy variation within species (or species complexes) may also be an important source of genetic and phenotypic variation (Thompson and Lumaret 1992). Thus, plant biodiversity cannot be understood without understanding the processes of polyploid evolution (Lutz 1907; Stebbins 1950; Grant 1981; Madlung 2013).
Polyploids are thought to experience high levels of genomic instability and undergo massive genetic and epigenetic changes within the first few generations after formation (Chen 2007). It is likely that a great deal of genomic and phenotypic diversity is generated and the majority of early generation polyploids are unable to survive in nature. However, if one or a few stable genotypes arise that happen to reconcile genomic incompatibilities, are vigorous and are well suited to survival in the prevailing habitat, polyploids can persist (Chen 2007; Madlung et al. 2012). After this rapid ‘genomic revolution’, it is likely that a slow process of diploidization begins, where gene duplicates may be silenced, lost or evolve new functions (Wolfe 2001). It is thought that nearly all angiosperms have experienced at least one polyploidy event in their evolutionary history (Wolfe 2001). However, due to extensive mutation, gene loss and rearrangements, these diploidized paleopolyploids, such as Arabidopsis thaliana, have only recently been recognized as whole-genome sequences became available for detailed analysis (Vision et al. 2000). Both the rapid genomic revolution and gradual process of diploidization are likely to result in variation and evolution in genome size as DNA is deleted, duplicated and rearranged, and variants are subject to genetic drift and selection.
Polyploidy can arise from the duplication of genomes within a single species (autopolyploidy) or through hybridization between two species, accompanied by chromosome doubling (allopolyploidy) (Levin 2002). Either allopolyploidy or autopolyploidy may arise via a single polyploidization event, like in Arabidopsis suecica (Säll et al. 2003; Jakobsson et al. 2006), or may have multiple origins (Soltis and Soltis 1999), as has been suggested for A. kamchatica (Shimizu-Inatsugi et al. 2009). Further, variation in ploidy level is frequently found within species both within and among populations (Schmuths et al. 2004; Marhold et al. 2010), and gene flow between ploidy levels is known to occur, either via a triploid bridge or through recurrent formation of unreduced gametes by diploids (Levin 2002; Husband 2004; Henry et al. 2005, 2009; Jørgensen et al. 2011). This gene flow from diploids to polyploids is likely an important source of genetic variation in polyploids (Jørgensen et al. 2011).
Arabidopsis kamchatica is an allotetraploid plant produced through hybridization through two closely related diploid taxa, Arabidopsis lyrata ssp. petraea and Arabidopsis halleri ssp. gemmifera (Shimizu et al. 2005; Shimizu-Inatsugi et al. 2009). Arabidopsis kamchatica has an amphi-Beringian distribution, and the pattern of genetic diversity suggests that it migrated northward out of Japan (or near Japan) to eastern Russia, across the Bering land bridge into Alaska, and down the west coast of Canada (Shimizu-Inatsugi et al. 2009). It has been suggested that A. kamchatica may have multiple origins through independent hybridization and polyploidization events (Shimizu-Inatsugi et al. 2009), and/or that it may hybridize with its diploid parental taxa (Shimizu-Inatsugi et al. 2009; Wang et al. 2010). Both of these processes have the potential to give rise to genome size variation. Further, A. kamchatica has been suggested to contain both diploid and tetraploid individuals (Dawe and Murray 1981; Wang et al. 2010). Because A. kamchatica is a close relative of the model plant, A. thaliana, a treasure trove of molecular research is easily applied to this organism, and development of A. kamchatica into a model system for the evolution of polyploidy has the potential to yield a great deal of insight into the evolution of polyploid genomes.
The goal of this study was to investigate genome size variation in A. kamchatica using flow cytometry. We characterized the nuclear DNA content of A. kamchatica and its putative parental species, A. lyrata and A. halleri, in a total of 25 populations from North America, Europe and Japan. We used the results to determine whether there is variation in ploidy and/or genome size in A. kamchatica and its parents, and to determine how genome size has evolved in polyploids relative to their diploid parents.
Methods
Plant material
We estimated genome size from a total of 73 samples from A. kamchatica and its parental taxa A. lyrata (subspecies A. l. lyrata and A. l. petraea) and A. halleri ssp. gemmifera (Table 1, Fig. 1). All plants were germinated from seed and grown in the Institute for Arctic Biology Greenhouse at the University of Alaska Fairbanks. In populations with multiple samples, we sampled plants from different maternal families.
Table 1.
Collection locations, collectors and mean (±1 SE) genome size of each population. 1Assumes Glycine max ‘Polanka’ 2C DNA content of 2.5 pg (Doležel et al. 1994; Doležel and Greilhuber 2010). 2Populations with different letters have significantly different means (P < 0.05) in post hoc comparisons among A. kamchatica populations with >2 individuals. 3Conversion from pg to Mbp assuming Mbp = pg × 978 (Doležel et al. 2003).
| Taxon | Location | Latitude | Longitude | Collector/donor | Sample size | 2C DNA content (pg)1,2 | SE | Ploidy (2C) | 2C genome size (Mbp)3 |
|---|---|---|---|---|---|---|---|---|---|
| A. h. gemmifera | Japan | 34.93 | 133.63 | Fujita Corp. | 9 | 0.571 | 0.0127 | 2x | 558.35 |
| A. kamchatica | USA, Alaska | ||||||||
| Bear Creek | 65.41355 | −145.62545 | C. Parker | 1 | 1.013 | NA | 4x | 990.43 | |
| Chena River | 64.82 | −147.32 | N.T., D.E.W. | 4 | 1.023AB | 0.0035 | 4x | 1000.06 | |
| Fairbanks | 64.83333333 | −147.7 | C. Parker | 1 | 1.025 | NA | 4x | 1002.51 | |
| Goodnews Bay | 59.11666667 | −161.583333 | C. Parker | 5 | 1.016A | 0.0056 | 4x | 994.06 | |
| Grant Lagoon, Kodiak Island | 57.37 | −154.65 | C. Parker | 3 | 1.043B | 0.0054 | 4x | 1020.23 | |
| Liberty Falls | 61.62 | −144.55 | D.E.W. | 1 | 1.039 | NA | 4x | 1015.72 | |
| Portage Glacier | 60.79161667 | −148.9021333 | N.T., D.E.W. | 3 | 1.039AB | 0.0104 | 4x | 1016.02 | |
| Parks Highway | 63.25 | −149.25 | N.T., D.E.W. | 6 | 1.035AB | 0.0035 | 4x | 1012.30 | |
| Rainbow Ridge | 63.32 | −145.64 | N.T., D.E.W. | 3 | 1.032AB | 0.0053 | 4x | 1009.16 | |
| Shoup Bay | 61.13 | −146.59 | N.T., D.E.W. | 4 | 1.033AB | 0.0050 | 4x | 1010.30 | |
| Thompson Pass | 61.13 | −145.73 | N.T., D.E.W. | 1 | 1.033 | NA | 4x | 1010.20 | |
| Canada, Vancouver Island | |||||||||
| Strathcona Park | 49.82915 | −125.8728 | J.A.S., D.E.W. | 15 | 1.027AB | 0.0032 | 4x | 1004.24 | |
| Japan, Honshu Island | |||||||||
| Lake Biwa, Shinbo | 35.44444444 | 136.05 | H. Marui | 5 | 1.083C | 0.0027 | 4x | 1059.29 | |
| A. l. lyrata | USA, Michigan, Grand Mere | 42.01 | −86.54 | J.A.S. | 1 | 0.525 | NA | 2x | 513.27 |
| New York | 40 | −74 | T. Mitchell-Olds | 1 | 0.479 | NA | 2x | 468.72 | |
| Pennsylvania, Presque Isle | 42.14 | −80.11 | J.A.S. | 1 | 0.510 | NA | 2x | 499.19 | |
| Pennsylvania, Raccoon Creek | 40.51 | −80.34 | J.A.S. | 2 | 0.502 | 0.0097 | 2x | 491.38 | |
| Wisconsin | 44 | −89 | T. Mitchell-Olds | 1 | 0.498 | NA | 2x | 486.70 | |
| A. l. petraea | England, Exeter | 50.72 | −3.53 | T. Mitchell-Olds | 2 | 0.477 | 0.0082 | 2x | 466.68 |
| Germany, Plech | 49.65 | 11.47 | T. Mitchell-Olds | 2 | 0.494 | 0.0034 | 2x | 482.82 | |
| Iceland, Reykjavik, Esja Mountain | 64.2 | −21.7 | M. Schierup | 2 | 0.526 | 0.0010 | 2x | 513.96 | |
| Scotland, Braemer | 57.01 | −3.4 | R. Ennos | 1 | 0.514 | NA | 2x | 502.68 | |
| Austria, Mödling | 48.08 | 16.32 | S. Ansell | 1 | 0.922 | NA | 4x | 901.93 | |
| Germany, Dürn | 49.27 | 11.6 | T. Mitchell-Olds | 1 | 0.941 | NA | 4x | 920.06 |
Figure 1.
Map of collection localities of plants used for flow cytometry.
Ploidy determination
Chromosome counting is the traditional method for determining ploidy level of an organism; however, it is labour intensive and may be inaccurate in Arabidopsis species due to their very small chromosomes (ranging from 1.5 m to 2.8 µm in A. thaliana; Schweizer et al. 1987) and the high frequency of endopolyploidy (Galbraith et al. 1991; Melarango et al. 1993). Flow cytometry allows rapid analysis of thousands of nuclei per sample and high throughput of many samples (Kron et al. 2007). Therefore, we used flow cytometry to estimate genome size and infer DNA ploidy. Because flow cytometry reveals genome size rather than a count of chromosomes, ploidy must be verified by chromosome counts in at least a few samples. In our study, we included both diploid and tetraploid references from Arabidopsis locations where both flow cytometry and chromosome counts have previously been carried out (A. kamchatica from Japan, and A. l. petraea from Iceland and Austria; Table 1; Dart et al. 2004).
Flow cytometry
Each Arabidopsis sample was co-chopped and run with soybean leaf, Glycine max ‘Polanka’, as an internal reference standard. The standard was grown from the same seed stock previously quantified (Doležel et al. 1994). Young leaves were collected from each Arabidopsis plant and kept on ice until processing, which occurred within 3 h of leaf collection. For each plant, three fresh leaves were placed in a plastic Petri dish with approximately half as much fresh leaf tissue from G. max. Leaf tissue was chopped in the presence of 0.5 mL of cold chopping buffer using a fresh stainless-steel razor blade. The chopping buffer was modified from Otto (1990) Buffer I by adding 0.5 % v/v of Triton X-100 rather than Tween 20. When the leaves were well chopped, we added an additional 0.5 mL of cold chopping buffer. The sample was then filtered through a 30-µm Partec CellTrics® filter and centrifuged for 20 s at 3500 rpm. The supernatant was drawn off and 2 µL of RNase A was added to the pellet. The pellet was resuspended in 0.2 mL of propidium iodine staining buffer. The propidium iodine staining buffer (28.65 g of dibasic sodium phosphate, 190 mL of deionized water and 10 mL of propidium iodine stock, which consists of 5 mg of propidium iodine and 10 mL of deionized water) was modified from Otto (1990). Samples were stained in the dark for 40 min prior to performing flow cytometry.
Flow cytometry was performed on a BD Biosciences FACSAria flow cytometer (BD Biosciences, San Jose, CA, USA) equipped with FACSDiva Software (BD Biosciences), using a Coherent Sapphire Solid State laser (488 nm) as the excitation source. Noise signals derived from subcellular debris were eliminated by gating. Samples were run until 5000 Arabidopsis nuclei were scored. Since propidium iodide was used to stain the nuclei, fluorescence was measured using the R-phycoerythrin (PE) detector, which uses the 576/26 nm bandpass filter. 2C DNA content was estimated from gated fluorescence histograms of PE area (Fig. 2). Due to endopolyploidy, the populations of plant nuclei typically gave multiple peaks of fluorescence, representing 2C, 4C and 8C nuclei (and sometimes even higher endopolyploid levels) (Galbraith et al. 1991; Melarango et al. 1993). The 2C DNA content of each sample was calculated using the smallest of the peaks, and comparing it to the G. max standard ((sample fluorescence/soybean fluorescence) × 2.5 pg; Doležel et al. 1994). All samples had a coefficient of variance (CV) for relative fluorescence among nuclei that was <10 %; however, only 48 % of samples had a CV ≤5 %, as recommended (Doležel et al. 2007). We believe that this is due in part to the very small Arabidopsis genome (Doležel et al. 2007), as the larger soybean standard peak had a mean CV of 3.32 %, and only 6.1 % of the samples had a CV >5 %. All soybean samples had a CV ≤5.7 %. To ensure that genome size measurements were repeatable, eight samples were repeated on different days. Differences between repeat measurements never exceeded 1.1 %, indicating that genome size measurements were highly repeatable (Doležel et al. 2007).
Figure 2.
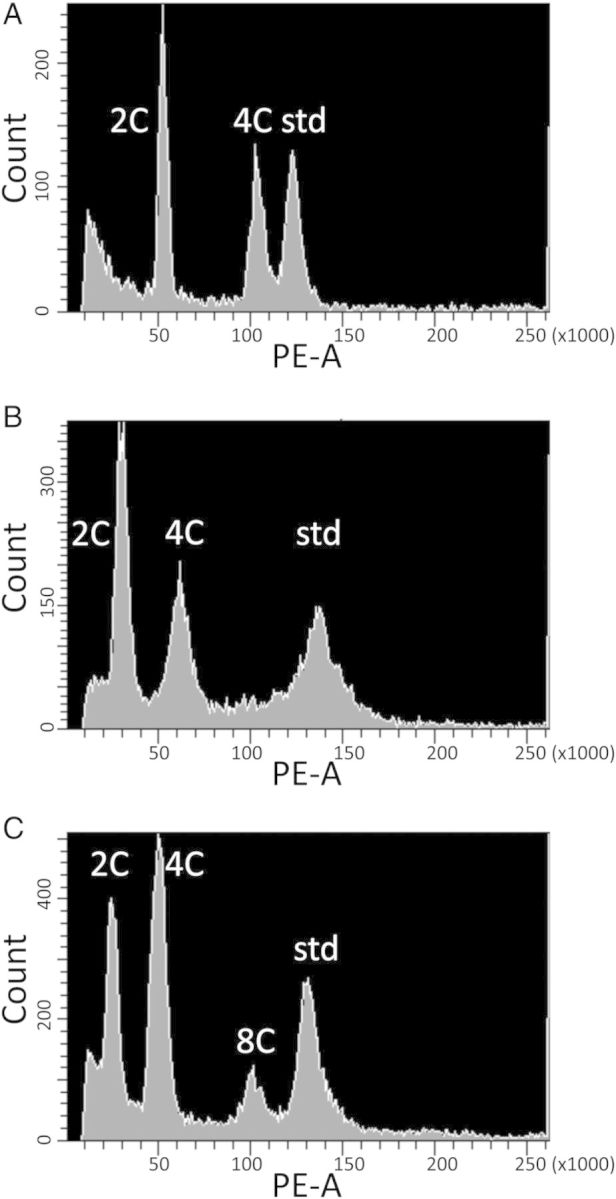
Fluorescence intensity histograms (PE-A) for (A) tetraploid A. kamchatica (2C = 4x = 32), (B) diploid A. h. gemmifera (2C = 2x = 16) and (C) diploid A. lyrata (2C = 2x = 16). Arabidopsis leaves show extensive endopolyploidy (Galbraith et al. 1991), and the 2C, 4C and 8C peaks are indicated, along with the soybean standard (std). The mean fluorescence of the smallest peak (2C) relative to the soybean peak was used to estimate 2C DNA content.
To determine whether the taxa differed in genome size, we used a linear mixed-effects model with species as the fixed effect, populations as the random effect and 2C DNA content (pg) as the dependent variable with the lme4 package (Bates 2005), implemented in R. The hypothesis test of the species effect was conducted with 5000 iterations of the parametric bootstrap approach based on the likelihood ratio statistics, D = −2 × (log-likelihood ratio), of Faraway (2006). To determine which species differed from one another in 2C DNA content, we performed Tukey's multiple comparison tests with an R package, multcomp (Hothorn et al. 2008). To determine whether populations of A. kamchatica differed in 2C DNA content, a second one-way ANOVA was performed with population as the fixed effect and 2C DNA content (pg) as the dependent variable. For this analysis we restricted our dataset to include only the nine A. kamchatica populations for which we had at least three samples. The mean number of samples per population was 5.3. Tukey's multiple comparison tests were performed to determine which A. kamchatica populations significantly differed from one another in genome size. In order to test the additivity of the tetraploid genome size, we examined a contrast null hypothesis, where the 1Cx genome size (i.e. the haploid genome size, sensu Greilhuber 2005) of A. kamchatica is the average of the two parental species, in the subset of data including A. kamchatica, A. h. gemifera and A. lyrata (two subspecies were combined). A linear mixed-effects model was fitted with 1Cx values as the dependent variable, population as a random effect and species as a fixed effect, and the linear contrast, (1Cx of A. kamchatica) = [(1Cx of A. h. gemifera) + (1Cx of A. lyrata)]/2, was tested with an R package, multcomp. For estimates of genome size diversity in each taxon, we used the CV among populations in 2C DNA content with the bias correction (Sokal and Rohlf 1995). To estimate genome size diversity in diploid A. thaliana, we used data from Schmuths et al. (2004) collected from 18 worldwide accessions using the same flow cytometry methods that we used.
Results
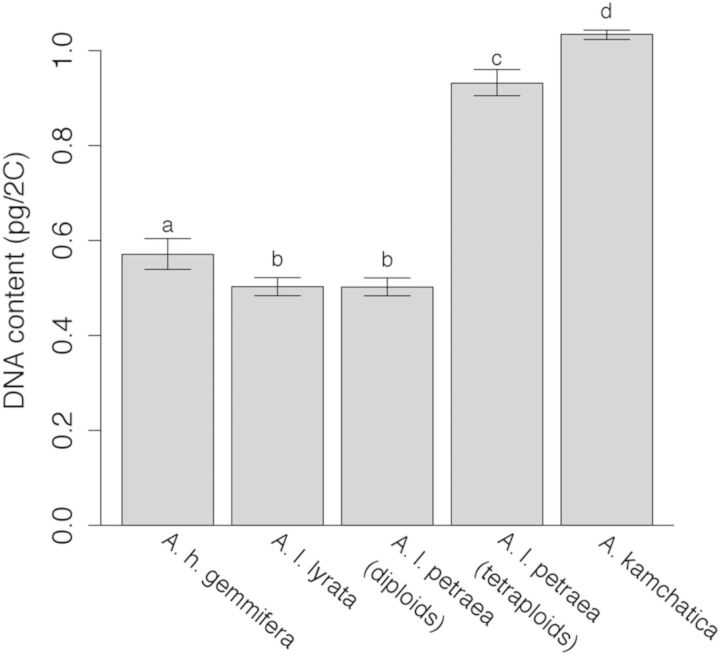
We found that 2C DNA content in A. kamchatica populations varied from 1.013 to 1.083 pg/2C, with a mean 2C DNA content of 1.034 ± 0.005 pg/2C (mean ± SE). Arabidopsis kamchatica and two of the A. lyrata ssp. petraea samples (Austria and Dürn, Germany) had approximately double the genome size of the other A. lyrata (ssp. lyrata and ssp. petraea) and A. halleri ssp. gemmifera samples (Fig. 3, Table 1). These taxa significantly differed in nuclear DNA content (D = 136.18, df = 1, P < 0.0002). These results, when taken together with chromosome counts and flow cytometry results conducted by Dart et al. (2004) in some of the same collections we used, suggest that the majority of A. l. lyrata, A. l. petraea and A. h. gemmifera are diploids, while A. kamchatica and two A. l. petraea samples are tetraploids (Fig. 3; Table 1).
Figure 3.
Estimates of 2C DNA content (pg) of each taxon and the 95 % confidence intervals of the estimates. Letters indicate significant differences (P< 0.05) based on Tukey's post hoc comparisons.
There was significant variation in genome size among A. kamchatica populations (F8,39 = 15.7, P < 10−9). Post hoc tests indicate that the genome size of the Japanese A. kamchatica population (Shinbo) was significantly larger than the North American populations. The Canadian A. kamchatica population did not differ in genome size from the Alaskan populations. However, two of the six Alaskan populations differed in genome size; the nuclear DNA content of the Goodnews Bay population was 3 % smaller than that of the Grant Lagoon population. Despite the minor amounts of variation among populations, none of the A. kamchatica plants sampled appear to be diploid.
The 2C DNA content of A. l. lyrata (0.503 pg/2C, 95 % CI [0.484, 0.522]) and diploid A. l. petraea (0.502 pg/2C, 95 % CI [0.484, 0.521]) did not significantly differ from one another (Fig. 3). The A. h. gemmifera genome (0.571 pg/2C, 95 % CI [0.539, 0.604]) was 14 % larger than A. l. petraea and A. l. lyrata (Fig. 3). We did not have enough samples/population of these taxa to analyse differences among populations.
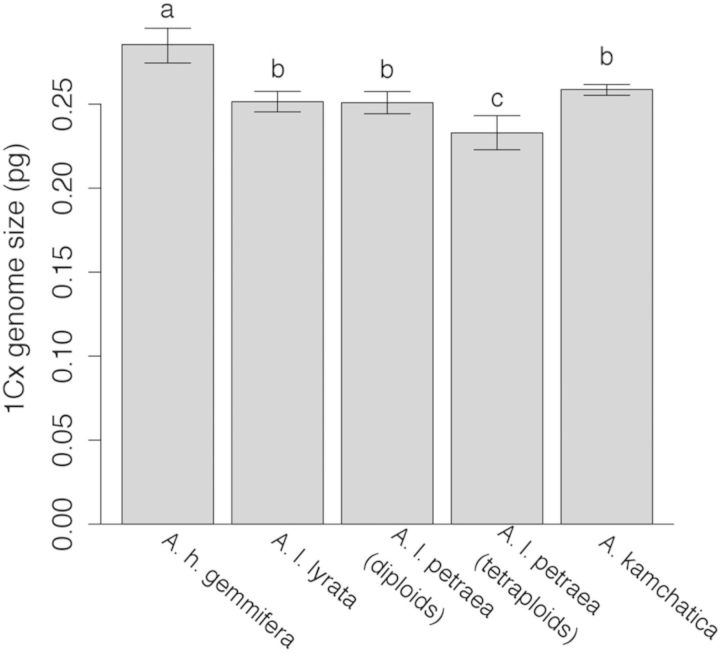
Arabidopsis kamchatica appears to have been derived through allopolyploidy from A. lyrata and A. h. gemmifera (Shimizu-Inatsugi et al. 2009). Thus, if polyploidization was recent, and there were no subsequent changes in genome size, we would predict that the genome size of the allotetraploid should be equal to the sum of the two parental taxa. Further, the 1Cx genome size (i.e. the haploid genome size, sensu Greilhuber 2005) should be an average of its parents. However, A. kamchatica, on average, is slightly smaller than expected. Comparing the 1Cx genome sizes of A. kamchatica to its parents (Fig. 4), we can see that the A. kamchatica 1Cx genome size is intermediate to its parents, but less than the average of its parents (A. kamchatica: 0.259 pg; mean of parents: 0.268 pg, z = −2.81, P = 0.0049). Further, it is not significantly different from the smaller parent, A. lyrata (Fig. 4), suggesting that A. kamchatica may have lost DNA. Arabidopsis kamchatica appears to have lost ∼37.594 Mbp/2C of DNA or 3.6 % of its genome. Autotetraploid A. l. petraea also appears to have lost DNA. The mean 1Cx genome size of tetraploid A. l. petraea (0.233 pg, 95 % CI [0.223, 0.243]) is less than the 1Cx content of diploid A. l. petraea (0.251 pg, 95 % CI [0.244, 0.258]), a loss of ∼70.366 Mbp/2C, or 7.2 % of the genome.
Figure 4.
Estimates of 1Cx (haploid) genome size (pg) of each taxon. The error bars are 95 % confidence intervals of the estimates. Letters indicate significant differences (P < 0.05) based on Tukey's post hoc comparisons.
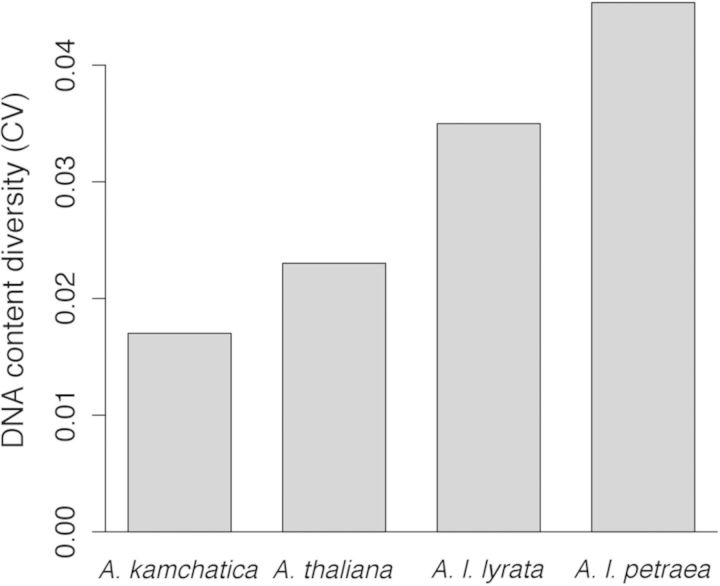
We were able to estimate genome size diversity (i.e. the CV in 2C DNA content) in A. kamchatica, A. l. petraea, A l. lyrata and A. thaliana, which were all sampled from multiple populations (A. thaliana data were from Schmuths et al. 2004). Arabidopsis kamchatica has the lowest diversity of all the Arabidopsis taxa studied (Fig. 5), including A. thaliana (Schmuths et al. 2004).
Figure 5.
Genome size diversity in Arabidopsis taxa, measured as CV in 2C DNA content. Only diploid A. thaliana and A. l. petraea are included because there were too few tetraploids to estimate CV (two from each taxon).
Discussion
Reliability of ploidy estimates
Our genome size estimates are very similar to those of Dart et al. (2004) for diploid and tetraploid collections in common (Table 1), suggesting that our results are reliable. Using both chromosome counting and flow cytometry, Dart et al. (2004) found that plants from Japan (Shinbo) and Austria are tetraploid (2n = 4x = 32) with genome sizes of 1.1 pg/2C (Japan) and 0.9 pg/2C (Austria), while plants from Iceland are diploid (2n = 2x = 16) with a genome size of 0.52 pg/2C. The small differences between our data and those of Dart et al. (2004) are likely due to the fact that Dart et al. (2004) used fluorescent beads as an internal size standard, whereas we used leaf tissue from G. max. While beads are sufficient for ploidy determination, leaf tissue is the preferred internal size standard for absolute genome size estimation because staining variation can be taken into account (Doležel et al. 2007).
No ploidy variation within A. kamchatica
Several previous reports have suggested that A. kamchatica contains both diploid and tetraploid individuals (Dawe and Murray 1981; Wang et al. 2010). While many species show a mix of ploidy levels, even within a population, these are likely autopolyploids (Schmuths et al. 2004; Jørgensen et al. 2011). Given that A. kamchatica is an allopolyploid, diploids spontaneously produced from tetraploids would likely have low vigour and fertility (Kerber 1964; Ladizinsky and Fainstein 1978), as allopolyploidization appears to rapidly result in gene silencing and gene loss for numerous loci (Kashkush et al. 2002; Adams and Wendel 2005). Our data from 52 A. kamchatica specimens representing most of the species' range found no evidence of diploid A. kamchatica, and we suggest that the species is likely to be entirely tetraploid. If diploids are present, they are likely to be in very low frequencies, and not maintained by selection.
Deeper investigation into previous reports also suggests that there is no good evidence for the presence of diploid A. kamchatica. Dawe and Murray (1981) report chromosome counts from three diploid (2n = 2x = 16) and two tetraploid A. kamchatica samples (2n = 4x = 32). Arabidopsis kamchatica is very difficult to morphologically distinguish from mostly diploid A. lyrata; however, molecular data suggest that the two species have distinct geographical ranges (Schmickl et al. 2010). The tetraploid counts reported by Dawe and Murray (1981) are within the species range of A. kamchatica suggested by Schmickl et al. (2010), whereas two of the three diploid counts are from plants growing north of the Brooks Range in Alaska and are probably A. l. petraea (Schmickl et al. 2010) or A. media (Mulligan 1995). One of the diploid counts (originally reported in Dawe and Murray 1979) comes from well within A. kamchatica's range in interior Alaska, near several of our collections (63°02′N, 145°29′W), and was likely taken from A. kamchatica. However, Mulligan (1995) claims that the diploid report is an error, and that the voucher in ALA indicates that 2n = 32 (tetraploid), not 2n = 16 (diploid). Other chromosome counts reported for A. kamchatica by Mulligan (1995) are all tetraploid, and he suggests that the species is entirely tetraploid.
Wang et al. (2010) claim to have detected both diploid and tetraploid A. kamchatica in Taiwan using flow cytometry and sequencing of nuclear DNA from 98 genes. They suggest that diploids have a ‘mosaic genome’ of the two parental species. Although this would be very interesting if confirmed, more complete evidence is desirable. First, their flow cytometry runs seem to lack an internal standard. The absolute value of nucleus fluorescence cannot reliably be used to estimate genome size as this value shifts due to variation in sample preparation, staining and analysis (Doležel et al. 2007). This shift can be seen by comparing Fig. S1A and S1B in Wang et al (2010), which were presented as evidence of diploid and tetraploid A. kamchatica. Further, their DNA sequence data do not provide any evidence of ploidy since only a single clone per PCR reaction was sequenced, ensuring that only a single homeologue (randomly chosen from one of the two parental genomes of tetraploids) could be obtained from each individual (Wang et al. 2010). Although we have not sampled A. kamchatica from Taiwan for our study, the ‘mosaic genome’ of purported diploid A. kamchatica can possibly be explained by misinterpretation of flow cytometry data and randomly sequencing only one of the two homeologues from each gene.
DNA content variation within A. kamchatica
We appear to have identified variation in the 2C DNA content among A. kamchatica populations. Greilhuber (2005) suggested that a great deal of apparent within-species, within-ploidy variation in genome DNA content estimated by flow cytometry is due to methodological artefacts. For instance, different levels of anthocyanins, tannic acid and other secondary metabolites in leaves can influence fluorescence and apparent DNA content (Loureiro et al. 2006; Bennett et al. 2008). Following best-practice recommended protocols (Doležel et al. 2007), we used an internal size standard co-chopped with each sample, we used Otto's buffer, which reduces the effects of tannic acid (Loureiro et al. 2006), and leaves were not pigmented. Further, repeated measurements of the same plant on different days produced very similar DNA content estimates (<1.1 % variation). Thus the variation we observed should be biologically real (Schmuths et al. 2004). However, co-chopping two putatively different samples from different populations would further increase certainty that differences among populations are not artefactual (Greilhuber 2005).
The 2C DNA content of Japanese A. kamchatica appears to be slightly larger than North American A. kamchatica. This observed genome size difference may differentiate the two A. kamchatica subspecies: A. kamchatica ssp. kamchatica and A. kamchatica ssp. kawasakiana. Our Japanese A. kamchatica samples are from subspecies A. k. kawasakiana, whereas the rest of our samples represent subspecies A. k. kamchatica from North America. These two subspecies differ in habitat, morphology and nucleotide allele frequencies (Shimizu-Inatsugi et al. 2009; Higashi et al. 2012), and Shimizu-Inatsugi et al. (2009) suggested that A. k. kawasakiana may represent a distinct origin of A. kamchatica. The difference in genome size between the Japanese A. k. kawasakiana and North American A. k. kamchatica potentially supports that hypothesis. Alternatively, ongoing hybridization between A. kamchatica and its diploid parent, A. h. gemmifera, in Asia (Wang et al. 2010) could increase the genome size in Asia by reintroducing homeologues that may have been deleted in the allotetraploid.
Other possible explanations for the genome size differences between Japan and North America include biogeographic history and selection. It has been suggested that time-limited environments may select for a smaller genome with more rapid cell division (reviewed in Šmarda and Bureš 2010). As A. kamchatica expanded north out of Japan and across the cold Bering land bridge into North America (Shimizu-Inatsugi et al. 2009), a smaller genome may have been favoured due to the short growing season. Interestingly, despite the difference in genome size, Japanese and North American samples appear to have lost similar numbers of genes (P. L. Chang, unpubl. res.). Our sampling from Japan was very limited. A thorough investigation of genome size variation from throughout Japan, accompanied by an investigation of introgression and deletions, is needed for a thorough understanding of genome size evolution in this species.
Within-species variation in nuclear genome size may be an important source of genetic diversity, especially if it is associated with phenotypic and ecological variation (Levin 2002; Matsushita et al. 2012). Although we did find significant levels of genome size diversity in the allotetraploid A. kamchatica, levels of genome size diversity were much lower than in the diploid Arabidopsis taxa studied (Fig. 5). This is consistent with the low levels of nucleotide diversity in A. kamchatica relative to the other taxa studied (Shimizu-Inatsugi et al. 2009). Although nucleotide diversity is generated by point mutations, while genome size variation is generated by indels, changes in repetitive DNA and transposon activity (Šmarda and Bureš 2010; Long et al. 2013), the two forms of genetic diversity are likely to be governed by many of the same population genetic processes such as mating system, biogeography and demographic history (Loveless and Hamrick 1984; Ingvarsson 2002; Glémin et al. 2006; Duchoslav et al. 2013).
Loss of DNA in tetraploids
The DNA content of tetraploid A. kamchatica was slightly less than expected based on the sum of the two parental taxa. It is possible that this apparent loss in DNA content could be artefactual, due to differences between species in plant secondary compounds (Greilhuber 2005). However, rapid loss of DNA after polyploidization appears to be common in polyploids, as the 1Cx genome size has been shown to decrease as the ploidy level increases (Bennett and Thomas 1991; Raina et al. 1994; Ozkan et al. 2001; Leitch and Bennett 2004; Angulo and Dematteis 2013; Duchoslav et al. 2013). Bennett and Thomas (1991) suggest that these changes in DNA content may have adaptive significance, perhaps because the rate of cell division is slowed considerably as genome size increases (Bennett 1972) and it may be beneficial to remove unnecessary DNA when ploidy level is high.
The majority of genome size variation within plant species at a single ploidy level is due to variation in amounts of repetitive DNA such as transposable elements, ribosomal genes and centromeric repeats (Levin 1993; Davison et al. 2007; Šmarda and Bureš 2010; Long et al. 2013). However, polyploids may also lose considerable amounts of functional DNA either because it is not necessary to have two copies or because it may allow the two parental genomes to resolve incompatibilities (Kashkush et al. 2002; Adams and Wendel 2005; Buggs et al. 2012). Whole-genome sequencing of A. kamchatica, and comparison to its parental taxa, suggests that each of three accessions from different geographic regions lost ∼463 of more than 60 000 total genes (∼2 % of assembled genes; P. L. Chang, unpubl. res.). Considering that our flow cytometry estimate of the A. kamchatica genome size was 3.6 % smaller than expected based on the sum of the parental genomes, the total amount of DNA lost is comparable to the percent of genes lost. This suggests that DNA was lost from both genic regions and non-functional regions in A. kamchatica.
Arabidopsis l. petraea tetraploids appear to have lost considerably more DNA than A. kamchatica. Although these plants are thought to be A. l. petraea autotetraploids, they may have experienced hybridization and introgression of DNA from A. arenosa (Jørgensen et al. 2011; Schmickl and Koch 2011), which has a genome size that is 13 % smaller than A. l. petraea (Jørgensen et al. 2011). Using DNA content numbers from Jørgensen et al. (2011), A. l. petraea tetraploid genomes are just slightly smaller than expected from the sum of diploid A. l. petraea and diploid A. arenosa genomes: observed tetraploid A. l. petraea relative genome size 0.44; vs expected diploid A. l. petraea 0.23 + diploid A. arenosa 0.20 = 0.43 (data are presented as a ratio of the sample peak over the internal standard peak, and cannot be converted to picograms since the 2C DNA content of the standard, Ilex crenata, is unknown; Jørgensen et al. 2011). The apparent loss of DNA in tetraploid A. l. petraea may thus be largely due to hybridization rather than gradual DNA loss through diploidization.
Conclusions
Contrary to some prior reports, all A. kamchatica plants in our samples appear to be tetraploid. We found that the allotetraploid, A. kamchatica, has a genome size that is just slightly less than the sum of its diploid parental taxa, A. l. petraea and A. h. gemmifera. Genome size diversity was lower in A. kamchatica than in other Arabidopsis taxa. However, there was some variation in genome size, where North American populations of A. k. kamchatica seem to have lost slightly more DNA than the Japanese population of subspecies A. k. kawasakiana. The development of A. kamchatica into a model system for the study of polyploidy has the potential to yield a great deal of insight, as its parental taxa have been well studied at both the ecological and genetic levels, and myriad molecular tools from A. thaliana are available.
Sources of Funding
This research was supported by the National Center for Research Resources (NCRR) at the National Institutes of Health (NIH) (Alaska INBRE) [RR016466 and 5P20RR016466] and the National Science Foundation (NSF) Experimental Program to Stimulate Competitive Research (Alaska EPSCoR) [0346770].
Contributions by the Authors
Plants were collected by J.A.S., D.E.W. and N.T. Methods were developed and debugged by J.A.S., G.J.H. and N.T. J.A.S. collected the data. J.A.S. and N.T. analysed the data and produced figures. D.E.W. conceived and wrote the manuscript, which was edited by J.A.S., G.J.H. and N.T.
Conflicts of Interest Statement
None declared.
Acknowledgements
We thank Beth Dunkel for helping with collection of the flow cytometry data. We thank J. Doležel for generously providing seeds of the genome size standard, Glycine max ‘Polanka’. We thank Mark Wright at the UAF IAB Greenhouse for growing and maintaining plants.
Literature Cited
- Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Current Opinion in Plant Biology. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Angulo MB, Dematteis M. Nuclear DNA content in some species of Lessingianthus (Vernonieae, Asteraceae) by flow cytometry. Journal of Plant Research. 2013;126:461–468. doi: 10.1007/s10265-012-0539-x. [DOI] [PubMed] [Google Scholar]
- Bates D. Fitting linear mixed models in R. R News. 2005;5:27–30. [Google Scholar]
- Bennett MD. Nuclear DNA content and minimum generation time in herbaceous plants. Proceedings of the Royal Society B Biological Sciences. 1972;181:109–135. doi: 10.1098/rspb.1972.0042. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Price HJ, Johnston JS. Anthocyanin inhibits propidium iodide DNA fluorescence in Euphorbia pulcherrima: implications for genome size variation and flow cytometry. Annals of Botany. 2008;101:777–790. doi: 10.1093/aob/mcm303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett ST, Thomas SM. Karyological analysis and genome size in Milium (Gramineae) with special reference to polyploidy and chromosomal evolution. Genome. 1991;34:868–878. [Google Scholar]
- Buggs RJA, Chamala S, Wu W, Tate JA, Schnable PS, Soltis DE, Soltis PS, Barbazuk WB. Rapid, repeated, and clustered loss of duplicate genes in allopolyploid plant populations of independent origin. Current Biology. 2012;22:248–252. doi: 10.1016/j.cub.2011.12.027. [DOI] [PubMed] [Google Scholar]
- Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annual Review of Plant Biology. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dart S, Kron P, Mable BK. Characterizing polyploidy in Arabidopsis lyrata using chromosome counts and flow cytometry. Canadian Journal of Botany. 2004;82:185–197. [Google Scholar]
- Davison J, Tyagi A, Comai L. Large-scale polymorphism of heterochromatic repeats in the DNA of Arabidopsis thaliana. BMC Plant Biology. 2007;7:44. doi: 10.1186/1471-2229-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe JC, Murray DF. In IOPB chromosome number reports LXIII. Taxon. 1979;28:265–279. [Google Scholar]
- Dawe JC, Murray DF. 1981. Atlas of chromosome numbers document for the Alaskan flora http://www.uaf.edu/museum/collections/herb/links-and-references/chromatl.html .
- De Smet R, Van De Peer Y. Redundancy and rewiring of genetic networks following genome-wide duplication events. Current Opinion in Plant Biology. 2012;15:168–176. doi: 10.1016/j.pbi.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Doležel J, Greilhuber J. Nuclear genome size: are we getting closer? Cytometry Part A. 2010;77A:635–642. doi: 10.1002/cyto.a.20915. [DOI] [PubMed] [Google Scholar]
- Doležel J, Doleželová M, Novák FJ. Flow cytometric estimation of nuclear DNA amount in diploid bananas (Musa acuminata and M. balbisiana) Biologia Plantarum. 1994;36:351–357. [Google Scholar]
- Doležel J, Bartos J, Voglmayr H, Greilhuber J. Nuclear DNA content and genome size of trout and human. Cytometry Part A. 2003;51A:127–128. doi: 10.1002/cyto.a.10013. [DOI] [PubMed] [Google Scholar]
- Doležel J, Greilhuber J, Suda J. Estimation of nuclear DNA content in plants using flow cytometry. Nature Protocols. 2007;2:2233–2244. doi: 10.1038/nprot.2007.310. [DOI] [PubMed] [Google Scholar]
- Duchoslav M, Šafářová L, Jandová M. Role of adaptive and non-adaptive mechanisms forming complex patterns of genome size variation in six cytotypes of polyploid Allium oleraceum (Amaryllidaceae) on a continental scale. Annals of Botany. 2013;111:419–431. doi: 10.1093/aob/mcs297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraway JJ. Extending the linear model with R. New York: Taylor and Francis; 2006. [Google Scholar]
- Galbraith DW, Harkins KR, Knapp S. Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiology. 1991;96:985–989. doi: 10.1104/pp.96.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S, Bazin E, Charlesworth D. Impact of mating systems on patterns of sequence polymorphism in flowering plants. Proceedings of the Royal Society B Biological Sciences. 2006;273:3011–3019. doi: 10.1098/rspb.2006.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant V. Plant speciation. New York: Columbia University Press; 1981. [Google Scholar]
- Greilhuber J. Intraspecific variation in genome size in angiosperms: identifying its existence. Annals of Botany. 2005;95:91–98. doi: 10.1093/aob/mci004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry IM, Dilkes BP, Young K, Watson B, Wu H, Comai L. Aneuploidy and genetic variation in the Arabidopsis thaliana triploid response. Genetics. 2005;170:1979–1988. doi: 10.1534/genetics.104.037788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry IM, Dilkes BP, Tyagi AP, Lin HY, Comai L. Dosage and parent-of-origin effects shaping aneuploid swarms in A. thaliana. Heredity. 2009;103:458–468. doi: 10.1038/hdy.2009.81. [DOI] [PubMed] [Google Scholar]
- Higashi H, Ikeda H, Setoguchi H. Population fragmentation causes randomly fixed genotypes in populations of Arabidopsis kamchatica in the Japanese Archipelago. Journal of Plant Research. 2012;125:223–233. doi: 10.1007/s10265-011-0436-8. [DOI] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models. Biometrical Journal. 2008;50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- Husband BC. The role of triploid hybrids in the evolutionary dynamics of mixed-ploidy populations. Biological Journal of the Linnean Society. 2004;82:537–546. [Google Scholar]
- Ingvarsson PK. A metapopulation perspective on genetic diversity and differentiation in partially self-fertilizing plants. Evolution. 2002;56:2368–2373. doi: 10.1111/j.0014-3820.2002.tb00162.x. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Hagenblad J, Tavaré S, Säll T, Halldén C, Lind-Halldén C, Nordborg M. A unique recent origin of the allotetraploid species Arabidopsis suecica: evidence from nuclear DNA markers. Molecular Biology and Evolution. 2006;23:1217–1231. doi: 10.1093/molbev/msk006. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Jørgensen MH, Ehrich D, Schmickl R, Koch MA, Brysting AK. Interspecific and interploidal gene flow in central european Arabidopsis (Brassicaceae) BMC Evolutionary Biology. 2011;11:346. doi: 10.1186/1471-2148-11-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics. 2002;160:1651–1659. doi: 10.1093/genetics/160.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerber ER. Wheat: reconstitution of the tetraploid component (AABB) of hexaploids. Science. 1964;143:253–255. doi: 10.1126/science.143.3603.253. [DOI] [PubMed] [Google Scholar]
- Kron P, Suda J, Husband BC. Applications of flow cytometry to evolutionary and population biology. Annual Review of Ecology, Evolution and Systematics. 2007;38:847–876. [Google Scholar]
- Ladizinsky G, Fainstein R. A case of genome partition in polyploid oats. Theoretical and Applied Genetics. 1978;51:159–160. doi: 10.1007/BF00273140. [DOI] [PubMed] [Google Scholar]
- Leitch IJ, Bennett MD. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society. 2004;82:651–663. [Google Scholar]
- Levin DA. S-gene polymorphism in Phlox drummondii. Heredity. 1993;71:193–198. [Google Scholar]
- Levin DA. The role of chromosomal change in plant evolution. Oxford: Oxford University Press; 2002. [Google Scholar]
- Long Q, Rabanal FA, Meng DZ, Huber CD, Farlow A, Platzer A, Zhang QR, Vilhjálmsson BJ, Korte A, Nizhynska V, Voronin V, Korte P, Sedman L, Mandáková T, Lysak MA, Seren Ü, Hellmann I, Nordborg M. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nature Genetics. 2013;45:884–890. doi: 10.1038/ng.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro J, Rodriguez E, Doležel J, Santos C. Flow cytometric and microscopic analysis of the effect of tannic acid on plant nuclei and estimation of DNA content. Annals of Botany. 2006;98:515–527. doi: 10.1093/aob/mcl140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveless MD, Hamrick JL. Ecological determinants of genetic structure in plant populations. Annual Review of Ecology and Systematics. 1984;15:65–95. [Google Scholar]
- Lutz AM. A preliminary note on the chromosomes of Oenothera lamarckiana and one of its mutants, O. gigas. Science. 1907;26:151–152. doi: 10.1126/science.26.657.151. [DOI] [PubMed] [Google Scholar]
- Madlung A. Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity. 2013;110:99–104. doi: 10.1038/hdy.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A, Henkhaus N, Jurevic L, Kahsai EA, Bernhard J. Natural variation and persistent developmental instabilities in geographically diverse accessions of the allopolyploid Arabidopsis suecica. Physiologia Plantarum. 2012;144:123–133. doi: 10.1111/j.1399-3054.2011.01526.x. [DOI] [PubMed] [Google Scholar]
- Marhold K, Kudoh H, Pak JH, Watanabe K, Španiel S, Lihová J. Cytotype diversity and genome size variation in eastern Asian polyploid Cardamine (Brassicaceae) species. Annals of Botany. 2010;105:249–264. doi: 10.1093/aob/mcp282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita SC, Tyagi AP, Thornton GM, Pires JC, Madlung A. Allopolyploidization lays the foundation for evolution of distinct populations: evidence from analysis of synthetic Arabidopsis allohexaploids. Genetics. 2012;191:535–547. doi: 10.1534/genetics.112.139295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melarango JE, Mehrotra B, Coleman A. Relationship between endopolyploidy and cell cize in epidermal tissue of Arabidopsis. The Plant Cell. 1993;5:1661–1668. doi: 10.1105/tpc.5.11.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan GA. Synopsis of the genus Arabis (Brassicaceae) in Canada, Alaska and Greenland. Rhodora. 1995;97:109–163. [Google Scholar]
- Otto FJ. DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. In: Darzynkiewickz Z, Crissman H, editors. Methods in cell biology. San Diego: Academic Press; 1990. pp. 105–110. [DOI] [PubMed] [Google Scholar]
- Otto S, Whitton J. Polyploid incidence and evolution. Annual Reviews of Genetics. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Ozkan H, Levy AA, Feldman M. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops–Triticum) group. The Plant Cell. 2001;13:1735–1747. doi: 10.1105/TPC.010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina SN, Parida A, Koul KK, Salimath SS, Bisht MS, Raja V, Khoshoo TN. Associated chromosomal DNA changes in polyploids. Genome. 1994;37:560–564. doi: 10.1139/g94-080. [DOI] [PubMed] [Google Scholar]
- Säll T, Jakobsson M, Lind-Halldén C, Halldén C. Chloroplast DNA indicates a single origin of the allotetraploid Arabidopsis suecica. Journal of Evolutionary Biology. 2003;16:1019–1029. doi: 10.1046/j.1420-9101.2003.00554.x. [DOI] [PubMed] [Google Scholar]
- Schmickl R, Koch MA. Arabidopsis hybrid speciation processes. Proceedings of the National Academy of Sciences of the USA. 2011;108:14192–14197. doi: 10.1073/pnas.1104212108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmickl R, Jørgensen M, Brysting A, Koch M. The evolutionary history of the Arabidopsis lyrata complex: a hybrid in the amphi-Beringian area closes a large distribution gap and builds up a genetic barrier. BMC Evolutionary Biology. 2010;10:98. doi: 10.1186/1471-2148-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmuths H, Meister A, Horres R, Bachmann K. Genome size variation among accessions of Arabidopsis thaliana. Annals of Botany. 2004;93:317–321. doi: 10.1093/aob/mch037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer D, Ambros P, Gründler P, Varga F. Attempts to relate cytological and molecular chromosome data of Arabidopsis thaliana to its genetic linkage map. Arabidopsis Information Service. 1987;25:27–34. [Google Scholar]
- Shimizu KK, Fujii S, Marhold K, Watanabe K, Kudoh H. Arabidopsis kamchatica (fisch. Ex dc.) K. Shimizu & Kudoh and A. kamchatica subsp. kawasakiana (Makino) K. Shimizu & Kudoh, new combinations. Acta Phytotaxonomica et Geobotanica. 2005;56:163–172. [Google Scholar]
- Shimizu-Inatsugi R, Lihova J, Iwanaga H, Kudoh H, Marhold K, Savolainen O, Watanabe K, Yakubov VV, Shimizu KK. The allopolyploid Arabidopsis kamchatica originated from multiple individuals of Arabidopsis lyrata and Arabidopsis halleri. Molecular Ecology. 2009;18:4024–4048. doi: 10.1111/j.1365-294X.2009.04329.x. [DOI] [PubMed] [Google Scholar]
- Šmarda P, Bureš P. Understanding intraspecific variation in genome size in plants. Preslia. 2010;82:41–61. [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry. 3rd edn. New York: W. H. Freeman; 1995. p. 58. [Google Scholar]
- Soltis DE, Soltis PS. Polyploidy: recurrent formation and genome evolution. Trends in Ecology and Evolution. 1999;14:348–352. doi: 10.1016/s0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. Variation and evolution in plants. New York: Columbia University Press; 1950. [Google Scholar]
- Thompson JD, Lumaret R. The evolutionary dynamics of polyploid plants: origins, establishment and persistence. Trends in Ecology and Evolution. 1992;7:302–307. doi: 10.1016/0169-5347(92)90228-4. [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Wang W-K, Ho C-W, Hung K-H, Wang K-H, Huang C-C, Araki H, Hwang C-C, Hsu T-W, Osada N, Chiang T-Y. Multilocus analysis of genetic divergence between outcrossing Arabidopsis species: evidence of genome-wide admixture. New Phytologist. 2010;188:488–500. doi: 10.1111/j.1469-8137.2010.03383.x. [DOI] [PubMed] [Google Scholar]
- Wolfe KH. Yesterday's polyploids and the mystery of diploidization. Nature Reviews Genetics. 2001;2:333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- Wood TE, Takebayashi N, Mayrose I, Barker MS, Greenspoon PB, Rieseberg LH. The frequency of polyploid speciation in vascular plants. Proceedings of National Academy of Sciences of the USA. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]