Abstract
Chemoenzymatic synthesis of SAM analogs
This study highlights a broadly applicable platform for the facile syntheses of SAM analogs that is directly compatible with downstream SAM utilizing enzymes. The ability to couple SAM synthesis and utilization in a single vessel circumvents issues associated with rapid SAM analog decomposition and thereby opens the door to the further interrogation of a wide range of SAM utilizing enzymes. As a proof of concept for the feasibility of natural product ‘alkylrandomization’, the coupled strategy was used to generate a small set of indolocarbazole analogs in conjunction with the rebeccamycin O-methyltransferase RebM.
Keywords: methionine adenosyltransferase, AdoMet synthetase, SAM synthetase, methyltransferase, natural product, alkylrandomization
Enzyme-catalyzed late stage group transfer-based tailoring reactions contribute to the structural and functional diversity of many complex natural products (NPs).[1] Representative examples of include amination, acylation, alkylation, glycosylation, halogenation, phosphorylation and sulfation and, in some cases, enzymes responsible for such transformations display notable flexibility with respect to their substrate scope.[2] Such promiscuity is an enabling feature of chemoenzymatic NP diversification platforms as exemplified by NP glycorandomization (a platform for differential glycosylation of natural products/drugs).[3] Among the examples above, enzyme-catalyzed alkylation is a highly prevalent occurrence that leads to NP N-, O-, S- and/or C-alkylation (Figure 1).[4] Thus, a platform to co-opt natural product methyltransferases (MTs) for broad natural product differential alkylation (alkylrandomization) is anticipated to dramatically expand the potential scope of NP chemical diversity.
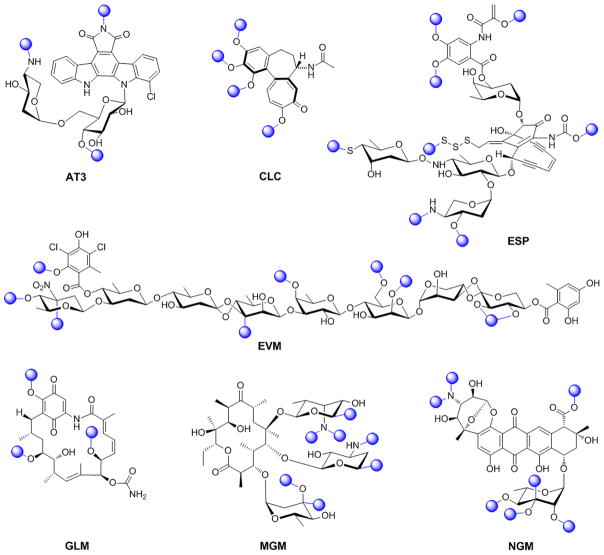
Figure 1.
Representative methylated natural products where methyl groups deriving from SAM via MT-catalyzed methylation are highlighted as spheres. AT3, AT2433; CLC, colchicine; ESP, esperamicin; EVM, evernimicin; GLM, geldamycin; MGM, megalomicin; NGM, nogalamycin.
The typical alkyl donor for MT-catalyzed alkylation is S-adenosyl-L-methionine (SAM) and importantly, we and others have demonstrated NP,[5] protein,[6] and nucleic acid[7] MTs to transfer alternative alkyl groups in the presence of suitably modified SAM analogs. However, reminiscent of the restriction imposed upon glycorandomization by sugar nucleotide availability, access to stable SAM analog arrays similarly restricts NP alkylrandomization development. Current state-of-art for SAM analog chemical synthesis relies upon multi-step syntheses of diastereomeric SAM analog mixtures and requires HPLC purification to remove starting material, S-adenosyl-homocysteine (SAH, a strong inhibitor of MTs).[8] The desired purified SAM analogs from this process are also markedly unstable, therby limiting their practical development as standalone synthetic reagents and/or biological probes. Within this context, a general platform to enable the generation and direct utilization of SAM analogs, beginning from stable precursors, would be considered advantageous.
To address this need, herein we describe the broad substrate specificity assessment of a representative set of five distinct methionine adenosyltransferases (MATs) from various sources. MAT (EC 2.5.1.6; also sometimes referred to as AdoMet synthetase or SAM synthetase) is the primary catalyst used for the biosynthesis of SAM from adenine triphosphate (ATP) and L-methionine (Met) (Scheme 2a) and there is preliminary evidence to suggest certain MATs to be capable of Met analog utilization.[6e,9] Consistent with this notion, the studies highlighted herein reveal human MAT II (hMAT2) to enable the cumulative synthesis of a broad panel of unnatural SAM analogs (29 analogs detected) starting from synthetic S/Se-alkylated Met analogs (44 analogs) or commercial sources (3 analogs). To demonstrate the feasibility of NP alkylrandomization, this study also highlights the subsequent generation of a small set of indolocarbazole analogs using a coupled hMAT2-RebM system, where RebM is the sugar C4′-O-MT involved in rebeccamycin biosynthesis.[5a,10] This study offers both a strategic advance in terms of NP diversification and reveals hMAT2 to display surprising permissivity.
Scheme 2.
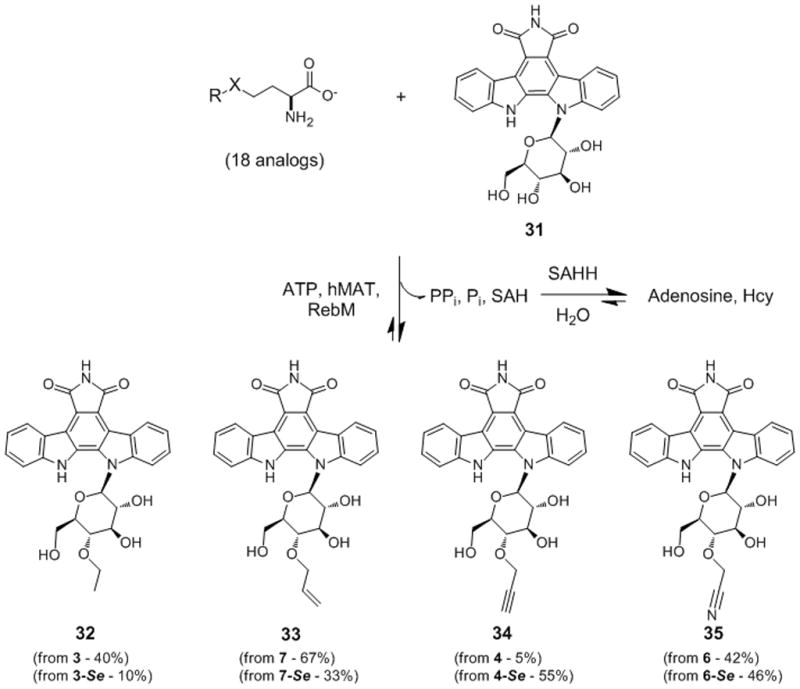
Products of hMAT2A-RebM coupled reaction in the presence of 3/3-Se, 7/7-Se, 4/4-Se, and 6/6-Se (see Figure 2). The percent product formation is noted in parentheses based upon the S/Se-Met analogs utilized (average percent error ≤5%, see Supporting Information).
Given the conflicting reports regarding the relative stability of S- versus Se-SAM[6c,6d,11] and the fact that Se-SAM is considered a better alkyl donor than S-SAM in MT-catalyzed reactions due to the inherently longer and weaker Se-C bond,[11b] both S- and Se-Met analogs were synthesized for this study. The general strategy for the synthesis of the S- and Se-Met analog panel employed direct or reductive alkylation of the corresponding L-homocysteine dimer (1 or 1-Se, Scheme 1b i and iii, respectively), L-Hcy (Scheme 1b, ii) or 2-Se (Scheme 1b, iv) following simple modifications of previously reported strategies.[12] The desired S- and Se-Met analogs were purified from crude reaction mixtures by reverse phase high pressure liquid chromatography (RP-HPLC), C18 RP flash chromatography or Dowex 50W8X-200 resin to afford isolated yields ranging from 20–90% (see Supporting Information Table S1).
Scheme 1.
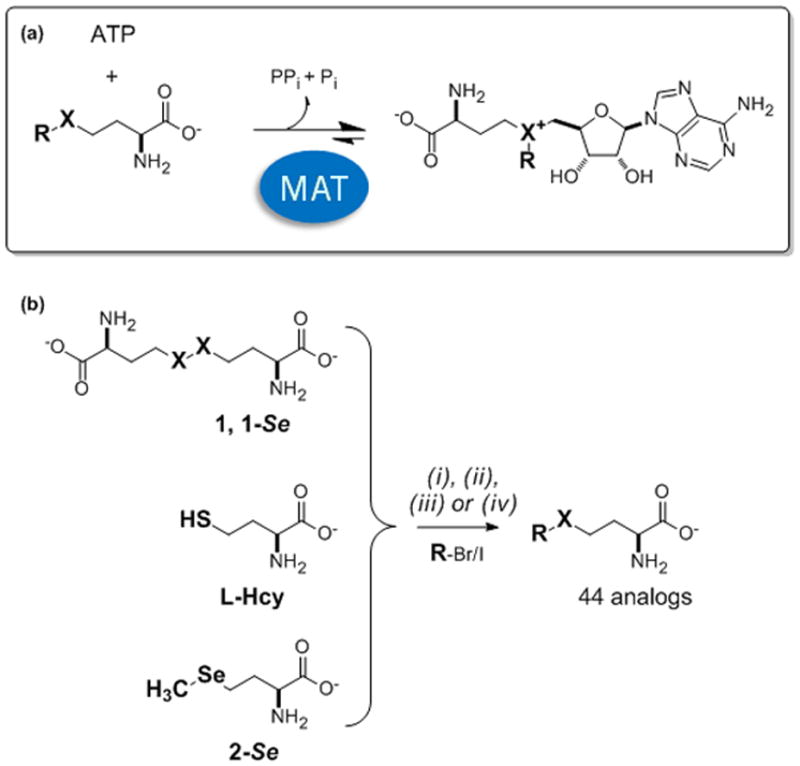
(a) Methionine adenosyltransferase (MAT)-catalyzed reaction, where L-Met (R=CH3, X=S) is the native substrate leading to native product S-adenosylmethionine (R=CH3, X=S). The variability of both R and X are assessed within the current study. (b) General methodology for the synthesis of S- and Se-L-Met analogs (X=S and Se, respectively) (i) Na, NH3, −78 °C with 1 or 1-Se; (ii) K2CO3, acetone with L-Hcy; (iii) NaBH4/NaOH, H2O, THF, 5 h with 1-Se; (iv) HCl, Reflux with 2-Se; see Supporting Information for additional information Table S1).
A representative set of MATs from variant sources, including bacterial, archaeal and mammalian orthologs, were selected for the initial assessment. Specifically, the exploratory set included human MAT II catalytic alpha and regulatory beta subunit (hMAT2),[13] human MAT II catalytic alpha subunit alone (hMAT2A),[13] human MAT I catalytic subunit alpha (hMAT1A),[13] Escherichia coli MAT (eMAT),[14] and the thermophilic Methanocaldococcus jannaschii MAT (mMAT).[15] The basis for inclusion of the human homologs derives from an interest to interrogate the substrate specificities of disease-relevant hMATs,[16] while the additional bacterial and archaeal homologs were selected to compare and contrast the current study with prior work relating to their substrate specificities.[9a, 17]
Standard uniform assay conditions (2 mM Met analog, 1 mM ATP, 5 μM MAT, 25 mM Tris, 5 mM MgCl2, 50 mM KCl, pH 8, 4 h) were adopted to facilitate this broad assessment. For the thermophilic counterpart (mMAT), reactions were conducted at 65 °C, while all other reactions were incubated at 37 °C. Production of SAM analogs was determined by a simple RP-HPLC end point assay to afford complete analysis of the desired SAM analog and/or any corresponding degradation products known to directly derive from the desired SAM analog intermediate. Putative SAM analog and SAM-derived degradative product formation was subsequently confirmed via HRMS analysis for all positive reactions (see Supporting Information Table S2).
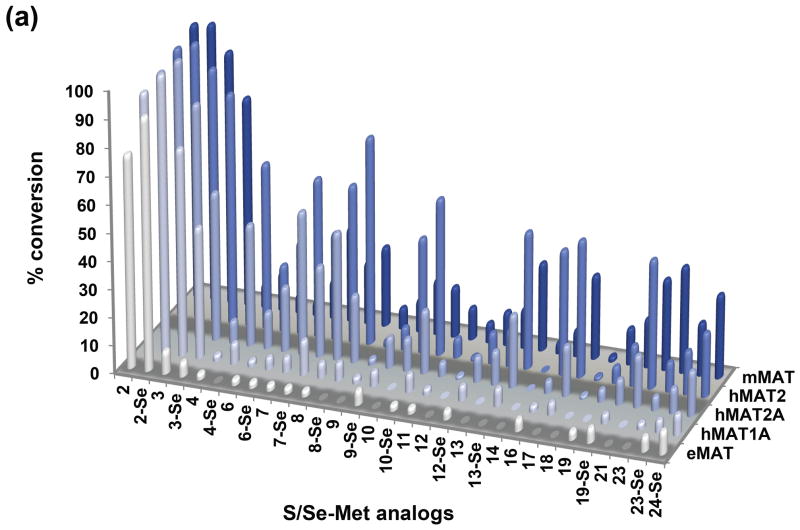
A cumulative comparison of the substrate specificity for all five MAT model systems based upon RP-HPLC is illustrated in Figure 2 wherein observed 5′-methyl-thio(seleno)-5′-deoxyadenosine (MTA) production (via RP-HPLC) was interpreted as product based upon the well-established SAM decay pathways indicating MTA to directly derive from SAM (not ATP).[6d,11a,18] This cumulative analysis revealed hMAT2 to be the most permissive of the MATs tested at 37 °C, while the thermophilic mMAT also displayed notable permissivity at 65 °C. Of the 47 putative substrates tested with hMAT2, 10 led to appreciable (>50%) SAM analog production, an additional 8 led to moderate (25–50%) conversion, while 11 offered detectable product (<25%) under the conditions described. In general, smaller alkyl substitutions were better tolerated, suggesting steric infringement to possibly prohibit larger substitutions. Interestingly, in the case where direct comparisons could be made, the degree of unsaturation correlated with a reduction in turnover (e.g., propyl > allyl > propargyl). Also, in most cases, Se- analogs were preferred over their S- comparators, most notably for hMAT2. Consistent with previous studies,[19] the regulatory beta subunit of MAT2A improved the overall proficiency of hMAT2 and, in some cases, this increase in proficiency translated to a slight increase in permissivity in the current study. Finally, the addition of inorganic pyrophosphatase in an attempt to drive the reaction via degradation of the pyrophosphate co-product did not appreciably increase turnover.
Figure 2.
(a) Turnover of S/Se-Met analogs to the corresponding S(Se)AM analogs catalyzed by selected MATs based upon RP-HPLC (average percent error ≤5%, see Supporting information). No product formation was observed in the absence of MAT, S/Se-Met analogs or ATP. (b) Structures of the S/Se-Met analogs listed in Figure 2a (X=S, Se). (A color rendering of Figure 2a is provided in Supporting Information).
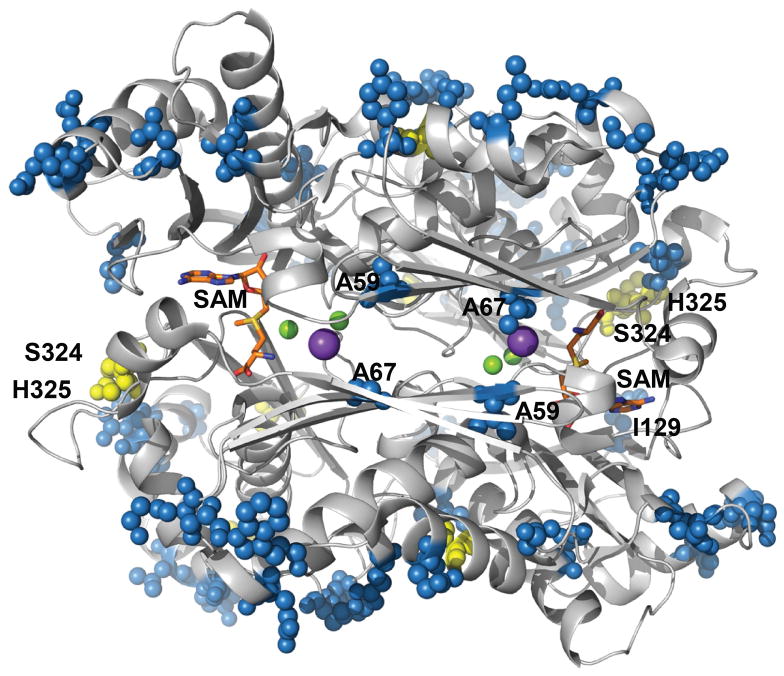
Based on these results, the corresponding MATs can be generally classified as having broad (hMAT2/hMAT2A), medium (mMAT) or stringent (eMAT and hMAT1A) substrate specificities. Superimposition of available X-ray structures (hMAT1A, hMAT2A and eMAT) reveal a root mean square deviation (rmsd) of 0.268 Å between hMAT2A and hMAT1A (sequence identity of ~85%) and ~0.56 Å between eMAT and hMAT1A or hMAT2A (sequence identity of ~58%) (Supporting Information Figure S3). Structural comparison of the permissive (hMAT2A) and stringent (hMAT1A and eMAT) MATs highlights key distinctions among residues (Figure 3). Interestingly, while hMAT2A active site mutagenesis has led to a slight improvement in turnover with an unnatural analog,[6e] the current structural comparison suggests the broad substrate specificity of hMAT2A to be predominately mediated by residues within the secondary shell and/or on the solvent-exposed surface distal to active-site loops that likely contribute to interdomain movement/dynamics. Whether or not these residues contribute to the hMAT2 alpha/beta interface is currently unknown.
Figure 3.
Mapping of residue positions that differ among eMAT (PDB ID:1RG9), hMAT2A (PDB ID:2PO2) and hMAT1A (PDB ID:2OBV) on the hMAT2A homodimeric structure. Conserved residues among all three MATs are indicated by grey ribbons; residues that differ among all three MATs are highlighted as ball and stick models; and conserved residues among ‘stringent‘ MATs (hMAT1A and eMAT) which differ from the more permissive hMAT2A are highlighted by by yellow ball and stick models. (A color rendering of Figure 3 is provided in the Supporting Information)
To subsequently assess the feasibility of ‘alkylrandomization’ via a single vessel coupled MAT/MT reaction, the rebeccamycin MT RebM was selected as the representative MT. RebM is the sugar C4′-O-MT involved in rebeccamycin biosynthesis and has previously been demonstrated to tolerate both alternative acceptors and alkyl donors.[5a,10] The coupled system also employed standard assay conditions (2 mM Met analog, 1 mM ATP, 50 μM rebeccamycin congener 31, 5 μM hMAT2, 10 μM RebM, 5 μM SAH-hydrolase, 25 mM Tris, 5 mM MgCl2, 50 mM KCl, pH 8, 24 hat 37 °C) and included only S/Se-Met analogs that afforded ≥20% turnover with hMAT2 under standard conditions (Figure 2). From this pilot study, 8 out of 18 of the selected S/Se-Met series led to the production of the corresponding differentially alkylated indolocarbazoles in appreciable yields (≥40%, Scheme 2 and Supporting Information Figure S4b). Importantly, inclusion of SAH-hydrolase (SAHH) in the coupled reaction, to prevent product inhibition associated with most if not all MTs,[20] improved overall product yields by 15–40%. Notably, this is the first report of MT-catalyzed acetonitrilylation (35 deriving from L-Met analog 6 or 6-Se).
In conclusion, the broad substrate specificity analysis presented revealed hMAT2/2A and mMAT to display notably broad substrate tolerance. Of these, hMAT2A is over-expressed in a number of tumor types,[21] where inhibition of hMAT2A by small molecule inhibitors or siRNA affords dramatic tumor reduction.[22] The discovered substrate malleability of hMAT2A in conjunction with aberrant cancer-specific hMAT2A overexpression therefore may offer the potential to employ suitable S/Se-Met analogs as cell-based metabolic probes of the role of methylation in cancer. In addition, the work highlighted herein enables one of the first the facile syntheses of SAM analogs directly compatible with downstream SAM utilizing enzymes including MTs and is advantageous over other recently reported chemoenzymatic strategies that depend upon synthetic non-native alkylated amino acid alkyl donors and nucleoside acceptors.[6f] As such, this platform circumvents a major liability of prior SAM analog/MT strategies, namely SAM analog decomposition, and thereby opens the door to the further interrogation of a host of MTs which operate upon macromolecular (protein/nucleic acid) and small molecule (natural products) substrates. Finally, the strategy presented unveils a single vessel proof of concept for natural product ‘alkylrandomization’ which, while currently somewhat limited via enzyme specificity, is expected to be further advanced via MT/MAT directed evolution and/or structure-based engineering in a manner reminiscent to that used for advancing glycorandomization.[3,23]
Supplementary Material
Acknowledgments
This work was supported by NIH RO1 CA84374 (JST) and the National Center for Advancing Translational Sciences (UL1TR000117).
Contributor Information
Dr. Shanteri Singh, Center for Pharmaceutical Research and Innovation, College of Pharmacy, University of Kentucky, Lexington, KY 40536 (USA). Department of Pharmaceutical Sciences, College of Pharmacy, University of Kentucky, 789 South Limestone Street, Lexington, KY 40536 (USA).
Dr. Jianjun Zhang, Center for Pharmaceutical Research and Innovation, College of Pharmacy, University of Kentucky, Lexington, KY 40536 (USA). Department of Pharmaceutical Sciences, College of Pharmacy, University of Kentucky, 789 South Limestone Street, Lexington, KY 40536 (USA)
Tyler D. Huber, Center for Pharmaceutical Research and Innovation, College of Pharmacy, University of Kentucky, Lexington, KY 40536 (USA). Department of Pharmaceutical Sciences, College of Pharmacy, University of Kentucky, 789 South Limestone Street, Lexington, KY 40536 (USA)
Manjula Sunkara, Division of Cardiovascular Medicine, Gill Heart Institute, University of Kentucky, Lexington, KY 40536 (USA).
Katherine Hurley, Department of Biochemistry, University of Wisconsin-Madison, Madison, WI 53705 (USA).
Dr. Randal D. Goff, Western Wyoming Community College, 2500 College Dr. Rock Springs, WY 82902-0428
Dr. Guojun Wang, Department of Pharmaceutical Sciences, College of Pharmacy, University of Kentucky, 789 South Limestone Street, Lexington, KY 40536 (USA)
Wen Zhang, Molecular and Cellular Biochemistry, University of Kentucky, College of Medicine, University of Kentucky, Lexington, KY 40536 (USA).
Prof. Chunming Liu, Molecular and Cellular Biochemistry, University of Kentucky, College of Medicine, University of Kentucky, Lexington, KY 40536 (USA)
Prof. Jürgen Rohr, Department of Pharmaceutical Sciences, College of Pharmacy, University of Kentucky, 789 South Limestone Street, Lexington, KY 40536 (USA)
Prof. Steven G. Van Lanen, Department of Pharmaceutical Sciences, College of Pharmacy, University of Kentucky, 789 South Limestone Street, Lexington, KY 40536 (USA)
Prof. Andrew J. Morris, Division of Cardiovascular Medicine, Gill Heart Institute, University of Kentucky, Lexington, KY 40536 (USA)
Prof. Jon S. Thorson, Center for Pharmaceutical Research and Innovation, College of Pharmacy, University of Kentucky, Lexington, KY 40536 (USA). Department of Pharmaceutical Sciences, College of Pharmacy, University of Kentucky, 789 South Limestone Street, Lexington, KY 40536 (USA).
References
- 1.a) Lamb SS, Wright GD. Proc Natl Acad Sci USA. 2005;102:519–520. doi: 10.1073/pnas.0408858102. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Olano C, Mendez C, Salas JA. Nat Prod Rep. 2010;27:571–616. doi: 10.1039/b911956f. [DOI] [PubMed] [Google Scholar]
- 2.a) Banik JJ, Craig JW, Calle PY, Brady SF. J Am Chem Soc. 2010;132:15661–15670. doi: 10.1021/ja105825a. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kwon SJ, Mora-Pale M, Lee MY, Dordick JS. Curr Opin Chem Biol. 2012;16:186–195. doi: 10.1016/j.cbpa.2012.02.001. [DOI] [PubMed] [Google Scholar]; c) Li TL, Liu YC, Lyu SY. Curr Opin Chem Biol. 2012;16:170–178. doi: 10.1016/j.cbpa.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 3.a) Gantt RW, Peltier-Pain P, Thorson JS. Nat Prod Rep. 2011;28:1811–1853. doi: 10.1039/c1np00045d. [DOI] [PubMed] [Google Scholar]; b) Gantt RW, Peltier-Pain P, Cournoyer WJ, Thorson JS. Nat Chem Biol. 2011;7:685–691. doi: 10.1038/nchembio.638. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Gantt RW, Peltier-Pain P, Singh S, Zhou M, Thorson JS. Proc Natl Acad Sci USA. 2013;110:7648–7653. doi: 10.1073/pnas.1220220110. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zhou M, Hamza A, Zhan CG, Thorson JS. J Nat Prod. 2013;76:279–286. doi: 10.1021/np300890h. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Peltier-Pain P, Marchillo K, Zhou M, Andes DR, Thorson JS. Org Lett. 2012;14:5086–5089. doi: 10.1021/ol3023374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liscombe DK, Louie GV, Noel JP. Nat Prod Rep. 2012;29:1238–1250. doi: 10.1039/c2np20029e. [DOI] [PubMed] [Google Scholar]
- 5.a) Zhang C, Weller RL, Thorson JS, Rajski SR. J Am Chem Soc. 2006;128:2760–2761. doi: 10.1021/ja056231t. [DOI] [PubMed] [Google Scholar]; b) Stecher H, Tengg M, Ueberbacher BJ, Remler P, Schwab H, Griengl H, Gruber-Khadjawi M. Angew Chem Int Ed Engl. 2009;48:9546–9548. doi: 10.1002/anie.200905095. [DOI] [PubMed] [Google Scholar]; c) Winter JM, Chiou G, Bothwell IR, Xu W, Garg NK, Luo M, Tang Y. Org Lett. 2013;15:3774–3777. doi: 10.1021/ol401723h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Islam K, Zheng W, Yu H, Deng H, Luo M. ACS Chem Biol. 2011;6:679–684. doi: 10.1021/cb2000567. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang R, Zheng W, Yu H, Deng H, Luo M. J Am Chem Soc. 2011;133:7648–7651. doi: 10.1021/ja2006719. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Bothwell IR, Islam K, Chen Y, Zheng W, Blum G, Deng H, Luo M. J Am Chem Soc. 2012;134:14905–14912. doi: 10.1021/ja304782r. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Willnow S, Martin M, Luscher B, Weinhold E. Chembiochem. 2012;13:1167–1173. doi: 10.1002/cbic.201100781. [DOI] [PubMed] [Google Scholar]; e) Wang R, Islam K, Liu Y, Zheng W, Tang H, Lailler N, Blum G, Deng H, Luo M. J Am Chem Soc. 2013;135:1048–1056. doi: 10.1021/ja309412s. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Thomsen M, Vogensen SB, Buchardt J, Burkart MD, Clausen RP. Org Biomol Chem. 2013;11:7606–7610. doi: 10.1039/c3ob41702f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Dalhoff C, Lukinavicius G, Klimasauskas S, Weinhold E. Nat Prot. 2006;1:1879–1886. doi: 10.1038/nprot.2006.253. [DOI] [PubMed] [Google Scholar]; b) Dalhoff C, Lukinavicius G, Klimasauskas S, Weinhold E. Nat Chem Biol. 2006;2:31–32. doi: 10.1038/nchembio754. [DOI] [PubMed] [Google Scholar]; c) Klimasauskas S, Weinhold E. Trends Biotechnol. 2007;25:99–104. doi: 10.1016/j.tibtech.2007.01.006. [DOI] [PubMed] [Google Scholar]; d) Lukinavicius G, Lapiene V, Stasevskij Z, Dalhoff C, Weinhold E, Klimasauskas S. J Am Chem Soc. 2007;129:2758–2759. doi: 10.1021/ja0691876. [DOI] [PubMed] [Google Scholar]; e) Dalhoff C, Huben M, Lenz T, Poot P, Nordhoff E, Koster H, Weinhold E. Chembiochem. 2010;11:256–265. doi: 10.1002/cbic.200900349. [DOI] [PubMed] [Google Scholar]; f) Motorin Y, Burhenne J, Teimer R, Koynov K, Willnow S, Weinhold E, Helm M. Nucleic Acids Res. 2011;39:1943–1952. doi: 10.1093/nar/gkq825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ueland PM. Pharmacol Rev. 1982;34:223–285. [PubMed] [Google Scholar]
- 9.a) Lu ZJ, Markham GD. J Biol Chem. 2002;277:16624–16631. doi: 10.1074/jbc.M110456200. [DOI] [PubMed] [Google Scholar]; b) Ottink OM, Nelissen FH, Derks Y, Wijmenga SS, Heus HA. Anal Biochem. 2010;396:280–283. doi: 10.1016/j.ab.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 10.a) Sanchez C, Butovich IA, Brana AF, Rohr J, Mendez C, Salas JA. Chem Biol. 2002;9:519–53. doi: 10.1016/s1074-5521(02)00126-6. [DOI] [PubMed] [Google Scholar]; b) Hyun CG, Bililign T, Liao JC, Thorson JS. Chembiochem. 2003;4:114–117. doi: 10.1002/cbic.200390004. [DOI] [PubMed] [Google Scholar]; c) Zhang C, Albermann C, Fu X, Peters NR, Chisholm JD, Zhang G, Gilbert EJ, Wang PG, Van Vranken DL, Thorson JS. Chembiochem. 2006;7:795–804. doi: 10.1002/cbic.200500504. [DOI] [PubMed] [Google Scholar]; d) Singh S, McCoy JG, Zhang C, Bingman CA, Phillips GN, Jr, Thorson JS. J Biol Chem. 2008;283:22628–22636. doi: 10.1074/jbc.M800503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Iwig DF, Booker SJ. Biochemistry. 2004;43:13496–13509. doi: 10.1021/bi048693+. [DOI] [PubMed] [Google Scholar]; b) Iwig DF, Grippe AT, McIntyre TA, Booker SJ. Biochemistry. 2004;43:13510–13524. doi: 10.1021/bi048692h. [DOI] [PubMed] [Google Scholar]
- 12.a) Jiracek J, Collinsova M, Rosenberg I, Budesinsky M, Protivinska E, Netusilova H, Garrow TA. J Med Chem. 2006;49:3982–3989. doi: 10.1021/jm050885v. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yi Y, Fa SX, Cao W, Zeng LW, Wang MX, Xu HP, Zhang X. Chem Commun. 2012;48:7495–7497. doi: 10.1039/c2cc33760f. [DOI] [PubMed] [Google Scholar]
- 13.Strausberg RL, et al. Proc Natl Acad Sci USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markham GD, Deparasis J, Gatmaitan J. J Biol Chem. 1984;259:4505–4507. [PubMed] [Google Scholar]
- 15.Bult CJ, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 16.Lu SC, Mato JM. Physio Rev. 2012;92:1515–1542. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yarlett N, Garofalo J, Goldberg B, Ciminelli MA, Ruggiero V, Sufrin JR, Bacchi CJ. Biochim Biophys Acta. 1993;1181:68–76. doi: 10.1016/0925-4439(93)90092-f. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman JL. Biochemistry. 1986;25:4444–4449. doi: 10.1021/bi00363a041. [DOI] [PubMed] [Google Scholar]
- 19.LeGros HL, Jr, Halim AB, Geller AM, Kotb M. J Biol Chem. 2000;275:2359–2366. doi: 10.1074/jbc.275.4.2359. [DOI] [PubMed] [Google Scholar]
- 20.Coward JK, Slisz EP. J Med Chem. 1973;16:460–463. doi: 10.1021/jm00263a008. [DOI] [PubMed] [Google Scholar]
- 21.a) Yang H, Huang ZZ, Wang J, Lu SC. FASEB. 2001;15:1507–1516. doi: 10.1096/fj.01-0040com. [DOI] [PubMed] [Google Scholar]; b) Chen H, Xia M, Lin M, Yang HP, Kuhlenkamp J, Li T, Sodir NM, Chen YH, Josef-Lenz H, Laird PW, Clarke S, Mato JM, Lu SC. Gastroenterology. 2007;133:207–218. doi: 10.1053/j.gastro.2007.03.114. [DOI] [PubMed] [Google Scholar]; c) Zhang T, Zheng ZC, Liu YQ, Zhang JJ, Zhao Y, Liu Y, Zhu HT, Zhao GH, Liang JW, Li Q, Xu HM. Acta Histochem. 2013;115:48–55. doi: 10.1016/j.acthis.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 22.a) Liu Q, Wu K, Zhu Y, He Y, Wu J, Liu Z. Hepatology Res. 2007;37:376–388. doi: 10.1111/j.1872-034X.2007.00041.x. [DOI] [PubMed] [Google Scholar]; b) Zhang W, Sviripa V, Chen X, Shi J, Yu T, Hamza A, Ward ND, Kril LM, Vander Kooi CW, Zhan CG, Evers BM, Watt DS, Liu C. ACS Chem Biol. 2013;8:796–803. doi: 10.1021/cb3005353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.a) Barton WA, Lesniak J, Biggins JB, Jeffrey PD, Jiang J, Rajashankar KR, Thorson JS, Nikolov DB. Nat Struct Biol. 2001;8:545–551. doi: 10.1038/88618. [DOI] [PubMed] [Google Scholar]; b) Barton WA, Biggins JB, Jiang J, Thorson JS, Nikolov DB. Proc Natl Acad Sci USA. 2002;99:13397–13402. doi: 10.1073/pnas.192468299. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hoffmeister D, Yang J, Liu L, Thorson JS. Proc Natl Acad Sci USA. 2003;100:13184–13189. doi: 10.1073/pnas.2235011100. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Yang J, Liu L, Thorson JS. Chembiochem. 2004;5:992–996. doi: 10.1002/cbic.200400041. [DOI] [PubMed] [Google Scholar]; e) Yang J, Fu X, Liao J, Liu L, Thorson JS. Chem & Biol. 2005;12:657–664. doi: 10.1016/j.chembiol.2005.04.009. [DOI] [PubMed] [Google Scholar]; f) Moretti R, Thorson JS. J Biol Chem. 2007;282:16942–16947. doi: 10.1074/jbc.M701951200. [DOI] [PubMed] [Google Scholar]; g) Williams GJ, Zhang C, Thorson JS. Nat Chem Biol. 2007;3:657–662. doi: 10.1038/nchembio.2007.28. [DOI] [PubMed] [Google Scholar]; h) Moretti R, Chang A, Peltier-Pain P, Bingman CA, Phillips GN, Jr, Thorson JS. J Biol Chem. 2011;286:13235–13243. doi: 10.1074/jbc.M110.206433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.