Abstract
Proline racemase (PrdF) which is important for energy metabolism via the Stickland pathway and is unique to certain clostridia was investigated as a potential anti-C. difficile target, by examining its effects on the growth and virulence of Clostridium difficile. Inactivation of PrdF by insertional mutagenesis did not affect early logarithmic growth, but only attenuated growth in the mid- and late-logarithmic phases. There was no effect on virulence in vivo, suggesting that PrdF is also not required for C. difficile infection. These findings indicate that PrdF as well as other enzymes encoded by the proline-reductase operon are all non-essential and are unsuitable targets for anti-C. difficile drug discovery.
Keywords: Stickland metabolism, anti-C. difficile target discovery
Within the last decade Clostridium difficile, a spore-forming anaerobic bacterium, has emerged as the leading cause of healthcare-associated diarrhea, imposing considerable morbidity and mortality in hospitals (Rupnik et al. 2009). For example, in the United States alone an estimated >400,000 C. difficile infections (CDIs) occur annually, resulting in >14,000 deaths (McDonald et al. 2012; Rupnik et al. 2009). Due to the recent emergence of highly virulent epidemic strains (e.g. BI/NAP1/027), high rates of recurrence (20-25%) following vancomycin or metronidazole treatment and few therapeutic options, there is an urgent need for novel anti-C. difficile antimicrobials (Rupnik et al. 2009). Given that CDI arises from displacement of the microbiota, primarily by broad-spectrum antibiotics, it is required that new anti-C. difficile antibiotics exhibit narrow spectrum activity, preferably only targeting C. difficile to avoid further collateral damage to the microbiota (Rupnik et al. 2009). However, few studies have explored if clostridial specific proteins can be exploited to discover antibiotics that are selective for C. difficile. In this regard, we reasoned that one set of potential drug targets are the enzymes involved in the fermentation of amino acids via the Stickland pathway which is unique to Clostridia. It is thought that the Stickland pathway serves as the primary source of energy in C. difficile (Bouillaut et al. 2013; Jackson et al. 2006; Stickland 1935) and involves the oxidation of the Stickland donor amino acid (e.g. isoleucine) coupled to the reduction of the Stickland acceptor amino acids glycine and D-proline (Jackson et al. 2006). Whilst glycine reduction leads to substrate level phosphorylation, the reduction of D-proline is suggested to be coupled to generating the proton motive force (Jackson et al. 2006; Lovitt et al. 1987). L-proline undergoes racemization to D-proline, which is the substrate of the D-proline reductase in the Stickland reaction. Racemization is performed by the enzyme D-proline racemase (PrdF), which is located in the same operon (prd) as the D-proline reductase genes (Fig. 1A). Interestingly, this operon is induced in C. difficile in both the piglet and mouse models of CDI, suggesting a role for proline racemization in vivo (Janoir et al. 2013; Janvilisri et al. 2012). Inhibitors of energy metabolism have recently gained in interest as potential antimicrobials (Hurdle et al. 2011). Thus we sought to examine if PrdF could be a narrow spectrum anti-C. difficile drug target that could be important for the metabolism of C. difficile.
Fig.1.
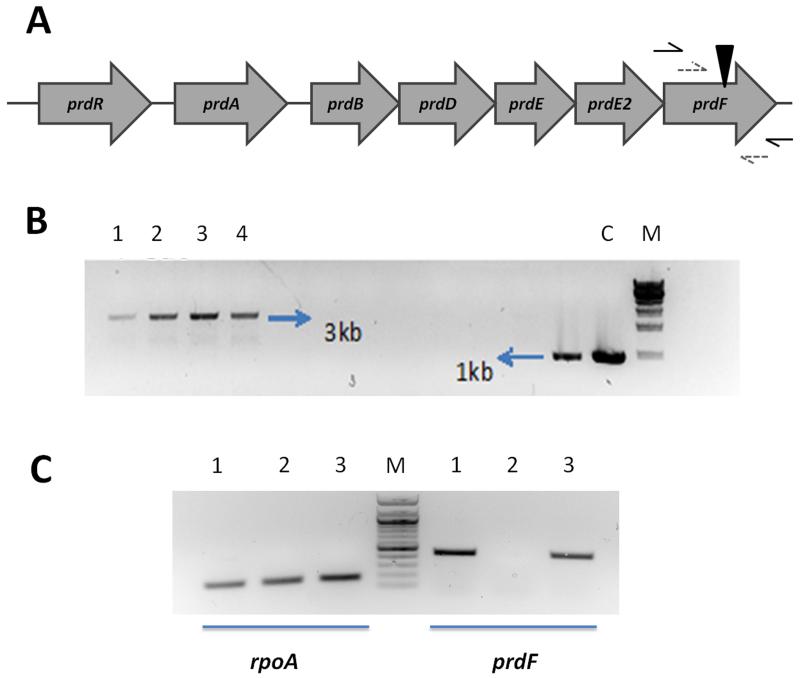
(A) The location of prdF in the prd operon. Black triangle indicates the insertion site of the group II intron. (B) PCR verification of intron insertion into prdF. Primer sites are indicated as black arrows in Fig. 1A. Lanes: 1–4, 4 independent clones of JIR8094ΔprdF; C, wild-type JIR8094; and M, 1 kb DNA marker. (C) Verification of transcript production by reverse transcription PCR. Primer sites are indicated as dashed gray arrows in Fig. 1A. Lanes: 1, RNA from wild-type JIR8094; 2, RNA from JIR8094ΔprdF; and 3, RNA from prdF complemented strain JIR8094ΔprdF_prdF; M, 100 bp DNA marker. The housekeeping gene rpoA was used as control.
Strains of C. difficile were routinely grown in pre-reduced Brain Heart Infusion (BHI) broth or agar, or a previously reported defined medium (Karasawa et al. 1995) at 37°C in a Whitley A35 anaerobic workstation (Don Whitley Scientific), whilst Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) broth or agar. All strains and plasmids used in this study are listed in Table 1. Insertional mutagenesis of prdF in C. difficile JIR8094 was achieved using the ClosTron system as previously described (Heap et al. 2010). Firstly, the intron retargeting region was designed by Perutka algorithm implemented at ClosTron.com and the resulting 344 bp target region was synthesized and cloned into pMTL007C-E2 by DNA2.0 Inc (Heap et al. 2010). The resulting plasmid pMTL007C-E2ΔprdF was conjugated into C. difficile JIR8094 as previously described (Heap et al. 2010). Intron integration mutants were selected on BHI agar containing erythromycin (2.5 μg/ml). Disruption of prdF was confirmed by PCR and sequencing of the amplicon using primer pairs flanking either side of the integration site (Fig. 1B). The ΔprdF mutant was then complemented with a wild type copy of prdF joined to the prdA promoter carried on the shuttle plasmid pMTL84151 (Heap et al. 2009). RNA of wild type, PrdF mutant and complementary cells were extracted and cDNA produced using random hexamers from Promega. The restoration of functional PrdF transcript was confirmed by PCR, with an elongation time of 30 sec, using primer pairs flanking either side of the integration site and both located within the prdF gene. The housekeeping gene rpoA served as a control. The effects of these genetic manipulations on the growth of strains were then analyzed. Overnight C. difficile cultures were washed once and resuspended in defined medium at 10-fold of start culture volume; 1 mL aliquots were then added to 24-well plates; 3 μg/mL of thiamphenicol was added to the media for plasmid maintenance. The absorbance at 600 nm was recorded automatically every 30 min over a 12 h period, with shaking before each read, in a Biotek Synery 2 plate reader. Doubling times were calculated using GraphPad prism 5 from values in the linear region of the curve for the respective strains. Since it has been proposed that proline reduction is coupled to the generation of a proton motive force, we determined minimum inhibitory concentrations in defined medium as previously described (Wu et al. 2013) for molecules known to affect bioenergetic properties of the membrane. To determine whether mutants were virulent, their pathogenicity were examined in the hamster model of CDI as previously described (Wu et al. 2013). Animal studies were approved by The Institutional Animal Care and Use Committee of The University of Texas at Arlington. Statistical analyses of data from all experiments were perfromed in GraphPad prism 5.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α |
fhuA2 Δ(argF-lacZ)U169 phoA glnV44 φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 |
New England Biolabs, USA |
| CA434 | HB101 containing R207 | (Heap et al. 2010) |
| C. difficile strains | ||
| JIR8094 | Derived from CD630; erythromycin susceptible | (O’Connor et al. 2006) |
| JIR8094ΔprdF | JIR8094 prdF::erm | This study |
| JIR8094_pMTL | JIR8094 containing plasmid pMTL84151 | This study |
| JIR8094ΔprdF_pMTL | JIR8094 prdF::erm containing plasmid pMTL84151 | This study |
| JIR8094ΔprdF_prdF | Complemented JIR8094ΔprdF mutant | This study |
| Plasmids | ||
| pMTL007C-E2ΔprdF | ClosTron vector containing group II intron targeted to prdF | This study |
| pMTL84151 | E. coli-C. difficile shuttle plasmid with MCS | (Heap et al. 2009) |
| pMTL84151_prdF | prdF expressed by prdA promoter | This study |
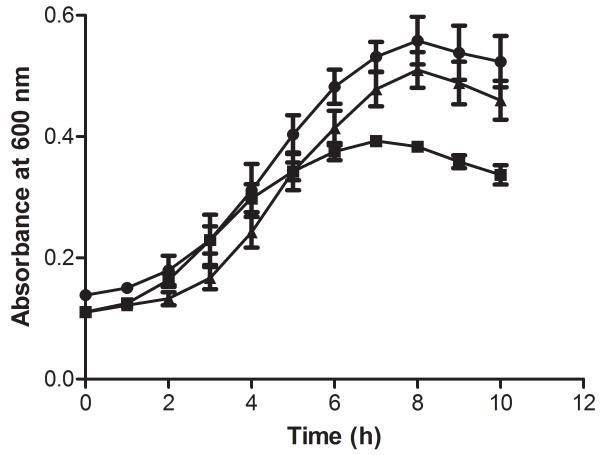
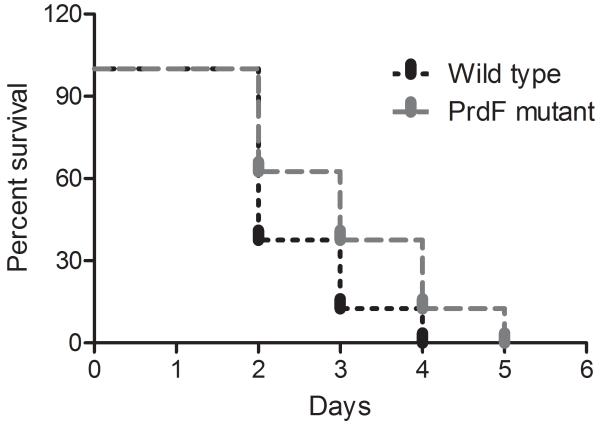
Bioinformatic analysis of the prdF of C. difficile revealed it possesses a homolog in humans (NP_653182) which is only 36% identical to C. difficile PrdF, has no racemase activity and key genetic differences in their active sites (Visser et al. 2012), which may enable the rationale design of inhibitors of PrdF. Ideally, a good anti-C. difficile drug target should be absent in key intestinal organisms. Blasting of the C. difficile prdF against genomes of key gut anaerobes revealed that they lacked a prdF homolog. These species include Lactobacillus johnsonii, Bacteroides ovatus and B. fragilis, Bifidobacterium breve, Actinomycetes viscosus, Porphyromonas uenonis and Fusobacterium nucleatum. C. perfringens that does not perform the Stickland reaction also lacked a homolog of prdF, whereas C. sporogenes which uses the Stickland reaction carried a homolog that was 60% identical. This would suggest that the PrdF enzyme could be a desirable target for a narrow-spectrum anti-C. difficile agent. However, as we were able to genetically inactivate this gene by insertional mutagenesis, this indicates that PrdF is not essential for the survival of C. difficile i.e. if it was essential inactivated mutants would not grow on BHI agar. Insertional inactivation of prdF was confirmed as an increase in the size of PCR amplicons generated with prdF specific primers (i.e. 1 kb in wild type and approximately 3 kb in insertion mutants in Fig. 1B). In defined medium, the growth of prdF insertion mutants were only decreased in the late logarithmic phase, compared to wild type JIR8094 (Fig. 2), suggesting that PrdF is partially involved in mediating optimal growth of C. difficile. To confirm that the growth defect was due to racemase inactivation, the wild type copy of prdF was ectopically expressed and driven by the prdA promoter in the complementary plasmid, designated pMTL84151_prdF. Restoration of functional prdF RNA was confirmed by reverse transcription PCR with a pair of RT-PCR primers, with results showing ~500 bp for wild type and complementary strains, but no band for the insertion mutant (Fig. 1C). The empty pMTL84151 vector was also conjugated into the wild type and prdF mutant. The resulting strains JIR8094_pMTL and JIR8094ΔprdF_pMTL, together with complementary strain JIR8094ΔprdF_prdF, were tested for growth in defined medium. Under the identical test conditions, in the same plate, there was no significant difference in growth rates in the early logarithmic phases as the doubling times for the wild-type and prdF mutant were 145.5 ± 7.4 min and 142.1 ± 4.4 min respectively. However, after 4 hours, the growth of the prdF mutant decelerated (Fig. 2), implying a requirement for prdF in mid- and late logarithmic metabolism. Indeed, at 8 hours the wild-type reached its highest absorbance at 600 nm of 0.56 ± 0.022, whereas the mutant was 0.38 ± 0.003; these differences were found to be statistically significant with P value <0.0001, from t-tests. Growth was restored to wild type levels by introducing the native prdF gene into the mutant, since the complemented strain had a doubling time of 135.4 ± 3.5 min and final absorbance of 0.51 ± 0.017 at 8 h. Use of the prdA promoter to drive the ectopic expression of prdF, and observation of growth restoration in the complemented strain, also suggests that other genes in the prd operon were expressed under the test conditions, as the D-proline product is utilized by D-proline reductase. Therefore, our findings largely confirm those recently reported by Bouillaut et al. (2013), suggesting that the Stickland metabolism is required for optimal growth of C. difficile, and herein we show that this defect appears in the mid- and late logarithmic stages of growth at least when prdF is inactivated (Fig. 2). However, toxin production appeared unaffected compared to wild type at 48 h (data not shown). Further study in hamsters also showed that there was no effect on virulence as no significant difference (P=0.19) was observed between the survival of hamsters infected with prdF mutant and wild type C. difficile (Fig. 3); thus PrdF also appears unsuitable for anti-virulence strategies although it is induced in vivo in two other animal models (Janoir et al. 2013; Janvilisri et al. 2012). If D-proline reduction is coupled to the generation of the proton motive force, as suggested (Jackson et al. 2006; Lovitt et al. 1987), it is plausible that inactivation of this pathway may alter the susceptibility of cells to membrane-active molecules. However, the prdF mutant did not show any significant changes in their susceptibility to several membrane-active molecules and ionophores, which might imply that the reduction of D-proline does not substantially contribute to the proton motive force in C. difficile, at least under the test conditions in defined medium. The MICs of known membrane-active molecules against wild type JIR8094 and JIR8094ΔprdF were as follows: nisin (0.4 and 0.4 μg/ml), monensin (0.12 and 0.12 μg/ml), nigericin (0.06 and 0.08 μg/ml) and valinomycin (0.75 and 0.5 μg/ml).
Fig.2.
Growth curve of JIR8094_pMTL (•), JIR8094ΔprdF_pMTL (■), and JIR8094ΔprdF_prdF (▲). Values shown are means ± standard error of the mean of 3 biological repeats; no significant difference exists between the doubling time means (P = 0.45) for all strains, as determined by one-way ANOVA.
Fig.3.
Survival of hamsters infected with wild-type Clostridium difficile JIR8094 and PrdF mutant. Each group consisted of 8 animals; no significant difference (P = 0.19) was noted as determined by Mantel–Cox test.
Very recently, the deletion of Stickland pathway enzymes D-proline reductase and glycine reductases by Bouillaut et al. (2013) provided the first indication that this pathway is not essential for the survival of C. difficile. Since the essentiality of PrdF was not analyzed in their work, our studies provide further evidence that all key enzymes in this pathway are not essential to C. difficile in vitro. We also now show that there is no effect on the pathogenicity of C. difficile. Our work also raises the question of whether D-proline reduction is coupled to the generation of the proton motive force in C. difficile, as previously suggested (Jackson et al. 2006; Lovitt et al. 1987), or implies it only makes a marginal contribution. Thus, it appears that further studies will be needed to elucidate the impact of the Stickland pathway on the metabolism of C. difficile, particularly metabolic changes in the mid and late logarithmic stages. Albeit, the present study concludes that these enzymes do not represent attractive targets for discovering novel drugs that could be selective treatments for CDI.
Acknowledgements
Funding was provided by the National Center for Complementary and Alternative Medicine/National Institutes of Health Grant 5R01AT006732 and the University of Texas at Arlington.
Footnotes
Disclaimer
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest
None to declare.
References
- Bouillaut L, Self WT, Sonenshein AL. Proline-dependent regulation of Clostridium difficile Stickland metabolism. J. Bacteriol. 2013;195(4):844–854. doi: 10.1128/JB.01492-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, Scott JC, Minton NP. The ClosTron: Mutagenesis in Clostridium refined and streamlined. J. Microbiol. Methods. 2010;80(1):49–55. doi: 10.1016/j.mimet.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Heap JT, Pennington OJ, Cartman ST, Minton NP. A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods. 2009;78(1):79–85. doi: 10.1016/j.mimet.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Hurdle JG, O’Neill AJ, Chopra I, Lee RE. Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 2011;9(1):62–75. doi: 10.1038/nrmicro2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S, Calos M, Myers A, Self WT. Analysis of proline reduction in the nosocomial pathogen Clostridium difficile. J. Bacteriol. 2006;188(24):8487–8495. doi: 10.1128/JB.01370-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoir C, Deneve C, Bouttier S, Barbut F, Hoys S, Caleechum L, Chapeton-Montes D, Pereira FC, Henriques AO, Collignon A, Monot M, Dupuy B. Adaptive strategies and pathogenesis of Clostridium difficile from in vivo transcriptomics. Infect. Immun. 2013;81(10):3757–3769. doi: 10.1128/IAI.00515-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janvilisri T, Scaria J, Teng CH, McDonough SP, Gleed RD, Fubini SL, Zhang S, Akey B, Chang YF. Temporal differential proteomes of Clostridium difficile in the pig ileal-ligated loop model. PLoS One. 2012;7(9):e45608. doi: 10.1371/journal.pone.0045608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasawa T, Ikoma S, Yamakawa K, Nakamura S. A defined growth medium for Clostridium difficile. Microbiology. 1995;141(2):371–375. doi: 10.1099/13500872-141-2-371. [DOI] [PubMed] [Google Scholar]
- Lovitt RW, Kell DB, Morris JG. The physiology of Clostridium sporogenes NCIB 8053 growing in defined media. J Appl Bacteriol. 1987;62(1):81–92. doi: 10.1111/j.1365-2672.1987.tb02383.x. [DOI] [PubMed] [Google Scholar]
- McDonald LC, Lessa F, Sievert D, Wise M, Herrera R, Gould C, Malpiedi P, Dudeck M, Srinivasan A, Fridkin S, Cardo D. Vital signs: preventing Clostridium difficile infections. MMWR. 2012;61(9):157–162. [PubMed] [Google Scholar]
- O’Connor JR, Lyras D, Farrow KA, Adams V, Powell DR, Hinds J, Cheung JK, Rood JI. Construction and analysis of chromosomal Clostridium difficile mutants. Mol. Microbiol. 2006;61(5):1335–1351. doi: 10.1111/j.1365-2958.2006.05315.x. [DOI] [PubMed] [Google Scholar]
- Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 2009;7(7):526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- Stickland LH. Studies in the metabolism of the strict anaerobes (genus Clostridium): The oxidation of alanine by Cl. sporogenes. IV. The reduction of glycine by Cl. sporogenes. Biochem. J. 1935;29(4):889–898. doi: 10.1042/bj0290889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser WF, Verhoeven-Duif NM, de Koning TJ. Identification of a human trans-3-hydroxy-L-proline dehydratase, the first characterized member of a novel family of proline racemase-like enzymes. J. Biol. Chem. 2012;287(26):21654–21662. doi: 10.1074/jbc.M112.363218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Cherian PT, Lee RE, Hurdle JG. The membrane as a target for controlling hypervirulent Clostridium difficile infections. J. Antimicrob. Chemother. 2013;68(4):806–815. doi: 10.1093/jac/dks493. [DOI] [PMC free article] [PubMed] [Google Scholar]