To the Editor:
Functional proteomics represents a powerful approach to understand the pathophysiology and therapy of cancer. However, comprehensive cancer proteomic data have been relatively limited. As a part of The Cancer Genome Atlas (TCGA) Project and other efforts, we have generated protein expression data over a large number of tumor and cell line samples using reverse-phase protein arrays (RPPAs). RPPA is a quantitative, antibody-based technology that can assess multiple protein markers in many samples in a cost-effective, sensitive and high-throughput manner1,2. This technology has been extensively validated for both cell line and patient samples3,4,5, and its applications range from building reproducible prognostic models6 to generating experimentally verified mechanistic insights7.
Our RPPA profiling platform includes extensively validated antibodies to nearly 200 proteins and phosphoproteins (Supplementary Methods and Supplementary Table 1). We are in the process of extending it to 500 independent proteins, covering all major signaling pathways, including PI3K, MAPK, mTOR, TGF-β, WNT, cell cycle, apoptosis, DNA damage, Hippo and Notch pathways. The current data release covers 4,379 tumor samples and consists of three parts (Supplementary Table 2). These are (i) TCGA tumor tissue sample sets: 3,467 samples from 11 cancer types, to be extended to 25 cancer types; (ii) independent tumor tissue sample sets: one endometrial tumor set (244 samples)7 and two ovarian tumor sets (99 and 130 samples, respectively)6, with other independent sets to be added soon; and (iii) tumor cell lines: 439 samples in four cell line sets, including both baseline and drug-treated cell lines. To our knowledge, this represents the largest publicly available collection of cancer functional proteomics data with parallel DNA and RNA data.
To facilitate broad access to these RPPA data sets, we developed a user-friendly data portal, The Cancer Proteome Atlas (TCPA; http://bioinformatics.mdanderson.org/main/TCPA:Overview). TCPA provides six modules: Summary, My Protein, Download, Visualization, Analysis and Cell Line (Fig. 1, i). The Summary module provides an overview of the RPPA data with detailed descriptions of each set (Fig. 1, ii). The Download module allows users to obtain any RPPA data set for analysis through a tree-view interface (Fig. 1, iii). The My Protein module provides detailed information about each RPPA protein: protein name, corresponding gene symbol, antibody status and source for the antibody. Users can examine the expression pattern of a protein of interest across different tumor types (for example, HER2 expression shown in Fig. 1, iv).
Figure 1. Overview of the TCPA data portal.
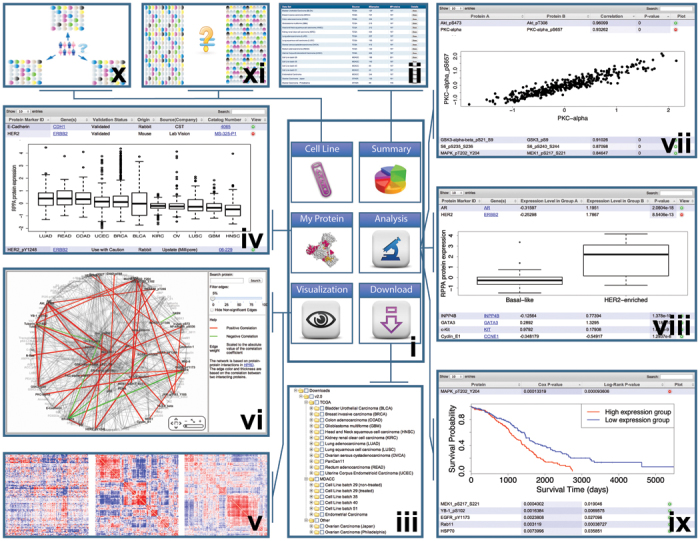
TCPA contains six modules (i): the Summary module (ii); the Download module (iii); the My Protein module, which has a table view (iv); the Visualization module, which has a “next-generation clustered heat map” view (v) and network view (vi); the Analysis module, which offers correlation analysis (vii), differential analysis (viii) and survival analysis (ix); and the Cell Line module, which offers cell line–patient BLAST analysis (x) and drug treatment effect analysis (xi).
The Visualization module provides two ways to examine global protein expression patterns in a specific RPPA data set. One is through a “next-generation clustered heat map” (Fig. 1, v), which allows users to zoom, navigate and scrutinize clustering patterns of samples or proteins and link those patterns to relevant biological information sources. The other is through a network view (Fig. 1, vi), which overlays the correlation between any two interacting partners in the protein interaction network (curated in the Human Protein Reference Database8).
The Analysis module provides three analysis methods. (i) For correlation analysis, given a user-specified data set, correlations between any pair of proteins are presented in a table (Fig. 1, vii). Users can search the results by protein name, rank correlations or visualize the scatter plot of a correlation of interest (for example, there is a strong correlation between PKC-α and its phosphorylated form PKC-a_pS657 in endometrial cancer, as shown in Fig. 1, vii). (ii) For differential analysis, differentially expressed protein markers between two tumor types or subtypes can be identified. Given user-defined comparison groups, the results are displayed in a table view, and for a protein of interest, users can visualize the box plots for the comparison (for example, the much higher expression of HER2 in the HER2-enriched subtype of breast cancer than in the basal-like subtype shown in Fig. 1, viii). (iii) For survival analysis, protein markers or pathway events significantly correlated with patient survival can be identified. The table view shows the univariate Cox proportional hazards model, log rank–test P values and a Kaplan-Meier plot for each protein in the data set (for example, phosphorylated MAPK, MEK, EGFR and YB are the top predictors of patient survival in ovarian cancer, which suggests a strong prognostic value of the tyrosine kinase receptor–RAS–MAPK pathway in this disease, as shown in Fig. 1, ix).
The Cell Line module provides two analyses for RPPA data from tumor cell lines. (i) For cell line–patient BLAST, cell lines with RPPA profiles that are most similar to those of a patient sample of interest can be selected (Fig. 1, x). The returned cell lines are externally linked with Cancer Cell Line Encyclopedia (CCLE)9, from which selected mutations, transcriptomic profiles and sensitivity to specific drug treatments can be obtained. (ii) For drug treatment analysis, drug effects on RPPA profiles are provided (Fig. 1, xi).
Compared with other proteomic databases such as The Human Protein Atlas10, an advantage of TCPA is the availability of quantitative protein expression data over large cohorts of well-characterized TCGA patient tumors, with linked DNA and RNA analyses. TCPA allows the validation of findings from TCGA RPPA data through independent sample cohorts and will help users select model tumor cell lines for further functional investigation. TCPA complements nucleic acid–centric cancer genomic data resources such as the CCLE, the Memorial Sloan-Kettering Cancer Center's cBioPortal for Cancer Genomics, OncoMine and the UCSC Cancer Genomics Browser. TCPA is also complementary to other protein-driven resources such as the Human Protein Reference Database, search tool for the retrieval of interacting genes/proteins (STRING) and Human Interactome Project. We will include additional data sets from TCGA and other independent cancer studies as they become available, and we will also accept (and help curate as necessary) cancer proteomic data from other groups.
Author contributions
G.B.M. and H.L. conceived of and supervised the project. Y.L., R.A., Z.J., W.L., J.-Y.Y., R.G.W.V. and J.L. generated the data, and J.L., P.L.R., B.M.B., D.W.K., C.W., J.N.W., G.B.M. and H.L. developed the data portal. Y.L., G.B.M. and H.L. wrote the manuscript with input from all the other authors.
Supplementary information
Supplementary Table 2 and Supplementary Methods (PDF 53 kb)
The antibody list for TCGA RPPA datasets (XLSX 34 kb)
TCGA Consortium membership (XLSX 32 kb)
Acknowledgements
We gratefully acknowledge contributions from the TCGA Research Network and its TCGA Pan-Cancer Analysis Working Group (contributing consortium members are listed in Supplementary Note). The TCGA Pan-Cancer Analysis Working Group is coordinated by J.M. Stuart, C. Sander and I. Shmulevich. This study was supported by the US National Institutes of Health (U24CA143883 to J.N.W. and G.B.M and P30CA016672); UTMDACC–G.S. Hogan Gastrointestinal Research Fund and NCI/UTMDACC Uterine SPORE Career Development Award (to H.L.); Susan G. Komen Foundation (KG081694 to G.B.M.); and Lorraine Dell Program in Bioinformatics for Personalization of Cancer Medicine (to J.N.W.).
Competing interests
The authors declare no competing financial interests.
Footnotes
Jun Li and Yiling Lu: These authors contributed equally to this work
Contributor Information
Gordon B Mills, Email: gmills@mdanderson.org.
Han Liang, Email: hliang1@mdanderson.org.
References
- 1.Sheehan KM, et al. Mol. Cell. Proteomics. 2005;4:346–355. doi: 10.1074/mcp.T500003-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Spurrier B, Ramalingam S, Nishizuka S. Nat. Protoc. 2008;3:1796–1808. doi: 10.1038/nprot.2008.179. [DOI] [PubMed] [Google Scholar]
- 3.Tibes R, et al. Mol. Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 4.Hennessy BT, et al. Clin. Proteomics. 2010;6:129–151. doi: 10.1007/s12014-010-9055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishizuka S, et al. Proc. Natl. Acad. Sci. USA. 2003;100:14229–14234. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang, J.-Y. et al. J. Clin. Invest.10.1172/JCI68509 (15 August 2013).10.1172/JCI68509
- 7.Liang H, et al. Genome Res. 2012;22:2120–2129. doi: 10.1101/gr.137596.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prasad TSK, et al. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barretina J, et al. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uhlen M, et al. Nat. Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 2 and Supplementary Methods (PDF 53 kb)
The antibody list for TCGA RPPA datasets (XLSX 34 kb)
TCGA Consortium membership (XLSX 32 kb)


