Abstract
The vascular system is organized throughout the plant body for transporting water, nutrients, and signaling molecules. During vascular development, xylem, phloem, and procambial/cambial cells are produced in a spatiotemporally organized manner. Several key regulators for xylem cell patterning and differentiation have been discovered, including auxin, cytokinin, CLE peptides, microRNAs, HD-ZIPIIIs, VNDs, and moving transcription factors SHR and AHLs. Recent studies are identifying functional interactions among these factors that ultimately determine xylem cell fate. This review focuses on regulatory networks underlying xylem cell fate determination in root vascular development.
Keywords: xylem, hormone, transcription factor, differentiation, patterning
INTRODUCTION
Cell fate determination is a fundamental mechanism underlying complex morphogenesis in multicellular organisms. Vascular tissues consist of phloem, xylem, and procambial cells. In Arabidopsis thaliana roots, the pattern of the central xylem axis, two phloem poles, and their intervening procambium is maintained during development, suggesting a robust mechanism for determining the spatial fate of each vascular cell (Cano-Delgado et al., 2010). Therefore, root vascular development is considered as an excellent system for studying cell fate determination (Bonke et al., 2003; Kubo et al., 2005). Recent studies on root vascular development have uncovered novel machineries regulating xylem cell fates in roots, such as cell-to-cell communication mediated by ligand–receptor interaction and intercellular movement of transcription factors (Hirakawa et al., 2011; Miyashima et al., 2012). We summarize recent advances on xylem cell fate determination in roots and discuss the regulatory networks controlling xylem cell fate determination.
CYTOKININ IS A CENTRAL REGULATOR OF PROTOXYLEM VESSEL CELL FATE IN ROOTS
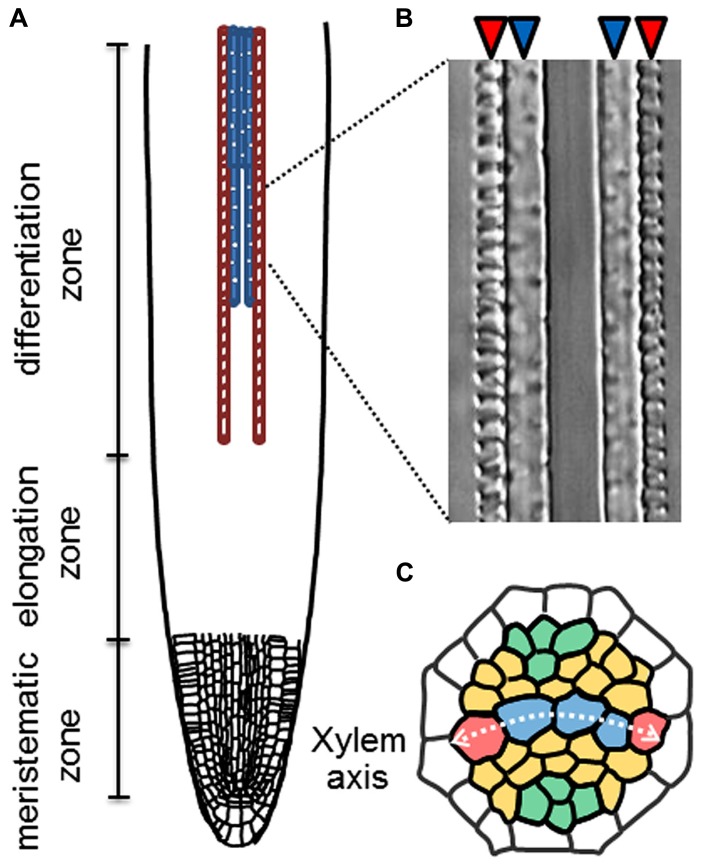
Root xylem vessels are classified into two types, protoxylem vessels and metaxylem vessels, which are equipped with a spiral-patterned and a pitted-patterned secondary cell-wall, respectively. The root vascular system is organized with precise cell patterning, in which five xylem vessel cells occupy the central xylem axis (Figure 1). In the xylem axis, two protoxylem vessels are always located on the outer side, and 2-4 metaxylem vessels are located on the inner side (Figure 1). It is widely recognized that root vascular cell identities are determined in the root apical meristem (RAM). The well-known vascular-specific marker genes ALTERED PHLOEM DEVELOPMENT (APL) and ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6 (AHP6) are expressed in phloem and protoxylem vessel cell files, respectively, in the RAM (Bonke et al., 2003; Mähönen et al., 2006).
FIGURE 1.
Xylem cell specification in roots. (A) Schematic illustration of longitudinal pattern of protoxylem (red) and metaxylem (blue) vessel cells in a root. (B) A Nomarski differential interference image of protoxylem (red arrowheads) and metaxylem vessels (blue arrowheads) in an Arabidopsis root. (C) Schematic illustration of radial cell patterning in the root stele. Red; protoxylem vessels (PX), blue; metaxylem vessels (MX), orange; procambial cells and green; phloem cells.
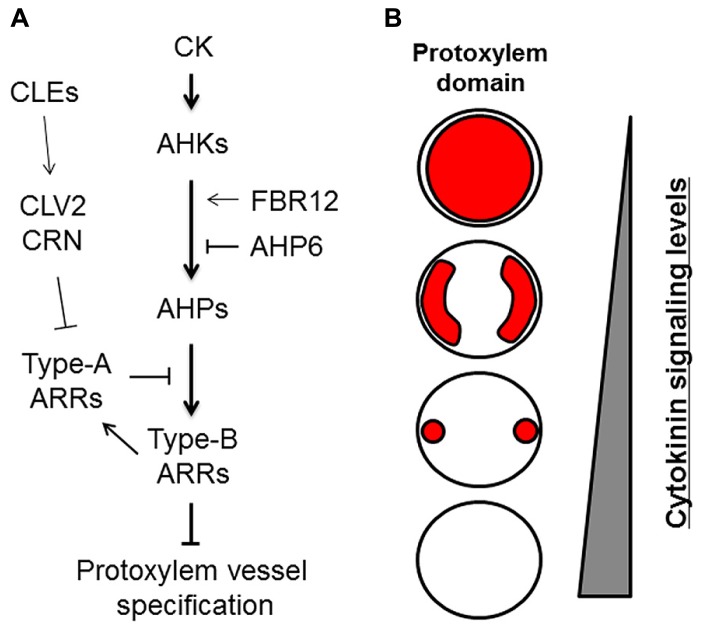
The plant hormone cytokinin (CK) has been implicated in the specification of protoxylem vessels in roots (Mähönen et al., 2000; Fukuda, 2004). Application of the synthetic CK benzyladenine causes the loss of root protoxylem vessels in a dose-dependent manner (Yokoyama et al., 2007; Kondo et al., 2011; Ren et al., 2013). Conversely, reduction of CK content by expressing CYTOKININ OXIDASE 2 (CKX2), which encodes a CK degradation enzyme, leads to the formation of extra protoxylem vessels (Mähönen et al., 2006). Thus, the number of protoxylem vessels depends on CK levels (Figure 2). CK signal transduction is initiated by the receptors ARABIDOPSIS HISTIDINE KINASE 2–4 (AHK2, AHK3, and CRE1/AHK4/WOL; Kieber and Schaller, 2014; Figure 2A). AHK mutants form extra protoxylem vessels adjacent to two original protoxylem vessels in the stele, due to the reduced CK sensitivity (Mähönen et al., 2006; Kondo et al., 2011; Figure 2B). The wol mutant displays a more severe phenotype, in which phloem cells are completely lost and only protoxylem vessels are formed in the stele (Scheres et al., 1995; Mähönen et al., 2006). The CK signal is transduced via phosphotransfer from AHKs to AHPs (Figure 2A). The AHP quintuple mutant (ahp1 ahp2 ahp3 ahp4 ahp5) exhibits the extra protoxylem phenotype similar to that of ahk mutants (Hutchison et al., 2006). These results indicate that CK negatively regulates protoxylem vessel formation via AHKs and AHPs. The atypical AHP, AHP6, lacks the histidine residue conserved among other AHPs. AHP6 is expressed in future protoxylem vessel cell files in RAM, and loss-of-function mutants often cause a partial loss of protoxylem vessels (Mähönen et al., 2006). These results strongly suggest that AHP6 functions as a pseudo-phosphotransfer protein that interferes with phosphorelay from AHKs to AHPs by competing with other AHPs, leading to inactivation of CK signaling in protoxylem vessel formation (Mähönen et al., 2006; Figure 2A).
FIGURE 2.
Roles of cytokinin signaling in protoxylem vessel specification. (A) Cytokinin (CK) signaling and its modulators involved in protoxylem vessel specification. (B) Schematic illustration of protoxylem domains (red) formed differentially depending on cytokinin signaling levels in the stele of roots.
Typical AHPs activate transcription factors named type-B ARABIDOPSIS RESPONSE REGULATORs (ARRs), which are the final targets in CK signal transduction (Yokoyama et al., 2007; Argyros et al., 2008). Triple mutants of centrally acting type-B ARRs (arr1 arr10 arr12) develop ectopic protoxylem vessels similar to those of other CK-related mutants (Yokoyama et al., 2007; Ishida et al., 2008). Type-B ARRs directly up-regulate type-A ARRs, which negatively regulate CK signaling by interacting with AHPs and interfering with type-B ARR functions (To et al., 2007). Mutants for type-A ARRs exhibit elevated CK sensitivity (To et al., 2004) and have fewer protoxylem vessels in lateral roots but not in the primary root (Ren et al., 2009; Kondo et al., 2011). These studies indicate that the CK signaling cascade consisting of AHKs, AHPs, and ARRs has a central role in regulating protoxylem vessel cell specification (Figure 2).
MODULATORS OF CYTOKININ SIGNALING REGULATE PROTOXYLEM VESSEL FORMATION
Modulators of CK signaling are involved in the regulation of protoxylem vessel formation. There are 32 genes encoding CLAVATA3/EMBRYO SURROUNDING REGION-related (CLE) peptides in Arabidopsis (Ito et al., 2006; Jun et al., 2008). Many CLE peptides, including CLE10, inhibit protoxylem vessel formation in wild-type plants (Kondo et al., 2011). By contrast, CLE10 does not inhibit protoxylem vessel formation in the type-B arr10 arr12 double mutant (Kondo et al., 2011). Gene expression analysis shows that CLE10 down-regulates type-A ARRs. These results suggest that CLE10 activates CK signaling through the down-regulation of type-A ARRs, thereby suppressing protoxylem vessel formation (Kondo et al., 2011). Further genetic analysis suggests that CLAVATA2 (CLV2) may act as a receptor that mediates CLE10 signaling and regulates protoxylem vessel formation (Kondo et al., 2011).
A recent study reported that the loss-of-function mutant of FUMONISIN B1-RESISTANT 12 (FBR12) produced extra protoxylem vessels due to reduced CK sensitivity (Ren et al., 2013). FBR12 encodes a eukaryotic translation initiation factor (elF5A) that is believed to play various roles via interactions with different proteins and RNAs (Thompson et al., 2003; Jao and Chen, 2006; Feng et al., 2007). FBR12 physically and genetically interacts with CRE1/AHK4/WOL and AHPs, which results in enhanced CK signaling (Ren et al., 2013). The modulation of CK signaling by various factors at different signaling steps enables fine spatiotemporal regulation of the protoxylem vessel domain (Figure 2).
SPATIAL REGULATION OF THE CYTOKININ ACCUMULATION DOMAIN IN ROOTS
Precise protoxylem vessel patterning requires spatial control of CK accumulation. Reporter-GUS analysis using hormone-response markers shows that domains with high auxin and high CK levels are localized in xylem axis and procambium in the stele, respectively (Bishopp et al., 2011a; Figure 3). The auxin distribution pattern is formed by auxin lateral transport through auxin efflux carriers PIN-FORMED 3 (PIN3) and PIN7 (Bishopp et al., 2011a). High auxin level in the xylem axis directly up-regulates AHP6 expression in the protoxylem vessel position via auxin-responsive elements in its promoter (Bishopp et al., 2011a). NPA treatment, which inhibits polar auxin transport, blocks AHP6 expression in the protoxylem vessel position, resulting in the loss of protoxylem vessels (Bishopp et al., 2011a). AHP6 has a negative role in CK signaling (Mähönen et al., 2006). These results indicate that PIN-mediated polar auxin transport and auxin accumulation induces AHP6 expression, which in turn attenuates CK signaling at the protoxylem vessel position (Figure 3).
FIGURE 3.
A molecular mechanism determining xylem axis domain. Schematic illustration showing a machanism underlying the formation of xylem axis domain in the root stele. (1) Mutual inhibition of high auxin and cytokinin domain defines protoxylem vessel domain, in which auxin inhibits cytokinin signaling via AHP6 and cytokinin restricts the auxin accumulation via PIN3 and PIN7. (2) AHLs movement plays a role in the determination of not only protoxylem vessel but also metaxylem vessel domain, thereby establishing the boundary between xylem axis and procambium domain in root stele. (3) The combination of these regulations leads to the formation of spatially regulated xylem axis domain. The details are described in the main text.
Conversely, PIN7 expression is regulated by CK (Bishopp et al., 2011a). The PIN7 expression domain overlaps the high CK-response domain, and PIN7 transcript levels are increased in response to CK treatment (Bishopp et al., 2011a). A recent study discusses a new technique for blocking symplastic connections by inducing the expression of mutated CALLOSE SYNTHASE 3 (CALS3), which substantially increases callose deposition at plasmodesmata (Vaten et al., 2011). Using this technique to inhibit symplastic transport revealed that basipetal transport of CK via the phloem is required for controlling the PIN7 expression domain (Bishopp et al., 2011b; Vaten et al., 2011). This result indicates that basipetal CK transport toward the RAM restricts the high auxin-response domain in the xylem axis by modulating auxin lateral transport (Bishopp et al., 2011b). Consequently, the fact that the ahk3 cre1 double mutant forms ectopic protoxylem vessels adjacent to the original protoxylem vessels (Mähönen et al., 2006; Kondo et al., 2011) can be explained because the high auxin-response domain in the xylem axis expands due to reduced CK signaling in that mutant (Mähönen et al., 2006; Bishopp et al., 2011a, b; Kondo et al., 2011; Figure 2B). Therefore, this mutually inhibitory feedback loop between auxin and CK allows precise establishment and maintenance of the protoxylem vessel position (Figure 3).
FACTORS THAT REGULATE METAXYLEM VESSEL FORMATION
The protoxylem domain is determined by the balance between auxin and CK; however, the molecular mechanisms that determine the metaxylem vessel domain remain unclear. Recently, Ursache et al. (2014) isolated mutants defective in TRP2, which is involved in tryptophan biosynthesis and tryptophan-mediated auxin biosynthesis. These mutants have a defect in metaxylem vessel formation but not in protoxylem vessel formation, suggesting an involvement of auxin biosynthesis in metaxylem vessel formation.
The conserved CLE–WOX signaling pathway is involved in metaxylem vessel development in rice (Oryza sativa; Chu et al., 2013). A rice CLE peptide named FON2-LIKE CLE PROTEIN2 (FCP2) negatively controls the expression of quiescent-center-specific-homeobox (QHB), which is an ortholog of AtWOX5 and is expressed in the QC and metaxylem precursor cells (Kamiya et al., 2003; Chu et al., 2013). Negative regulation of QHB by application of exogenous FCP2 causes the loss of metaxylem identity, leading to aberrant cell division in the metaxylem vessel position (Chu et al., 2013). As mentioned previously, CLE peptides can inhibit protoxylem vessel formation in Arabidopsis (Kondo et al., 2011). Therefore, the role of CLE signaling in the regulation of root xylem development is not conserved between Arabidopsis and rice.
BOUNDARY FORMATION BETWEEN XYLEM AND PROCAMBIUM DOMAINS
A recent study reported that the boundary between the procambium and xylem axis is determined by moving transcription factors named AT-HOOK MOTIF NUCLEAR LOCALIZED PROTEINs (AHLs; Zhou et al., 2013; Figure 3). In ahl3 and ahl4 single mutants, both ectopic protoxylem vessels and ectopic metaxylem vessels are formed in the procambial region adjacent to the xylem axis, suggesting an enlargement of the xylem axis domain (Zhou et al., 2013). This phenotype has some similarity with that of CK-defective mutants in terms of excess protoxylem vessel formation, but is distinctive in terms of extra metaxylem vessel formation adjacent to the original metaxylem. AHL4 is expressed in the procambium and its protein product can move into the xylem axis domain (Zhou et al., 2013). This intercellular movement is required for correct boundary formation between the procambium and the xylem (Zhou et al., 2013). The ahl3 ahl4 double mutant does not exhibit a more severe xylem phenotype compared with those of the single mutants, indicating that AHL3 and AHL4 function together (Zhou et al., 2013). AHL3 and AHL4 form a heterodimer, and have the potential to move from the procambium to the xylem (Zhou et al., 2013). High auxin and high CK-response domains are altered in ahl mutants (Zhou et al., 2013), but the relationship between AHLs and hormonal regulation of xylem axis formation is unknown. Further analysis of the function of AHLs may provide new insights into the mechanisms underlying boundary formation between xylem axis and the procambium.
MOLECULAR SWITCHES FOR PROTOXYLEM AND METAXYLEM VESSEL CELL FATE
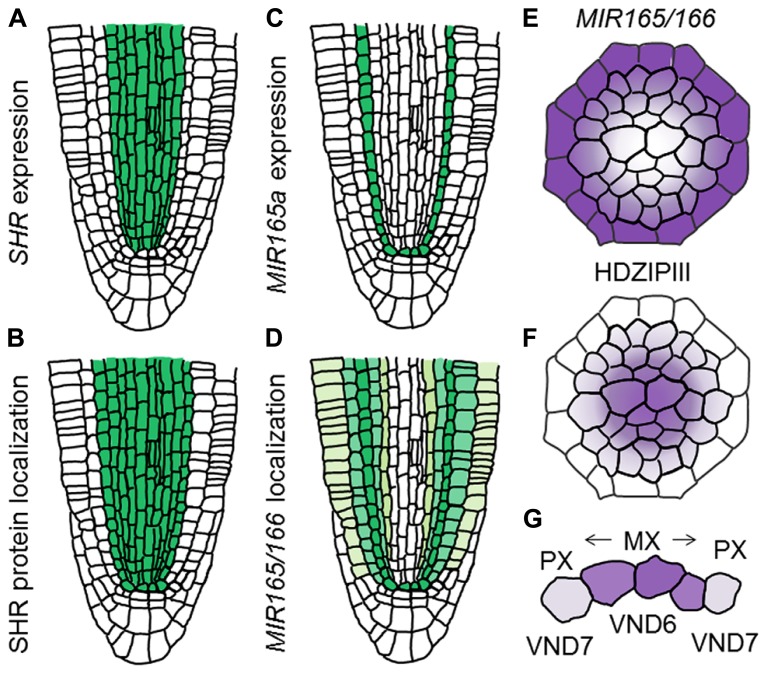
The GRAS-family transcription factor SHORT-ROOT (SHR), which is known to establish the identity of endodermis and cortex (Helariutta et al., 2000; Nakajima et al., 2001; Gallagher et al., 2004; Cui et al., 2007), also functions in the regulation of protoxylem and metaxylem specification (Carlsbecker et al., 2010). The shr mutant forms metaxylem vessels at the protoxylem position, indicating a switch of vessel types from protoxylem to metaxylem (Carlsbecker et al., 2010). SHR moves from the stele to the endodermis and induces expression of miR165 and miR166, in co-operation with SCARECROW (SCR; Carlsbecker et al., 2010; Figures 4A–C). Then, miR165 and miR166 move from the endodermis to the stele, leading to their higher accumulation in the outer region than in the inner domain of the stele (Carlsbecker et al., 2010; Figures 4D,E). miR165 and miR166 destabilize the mRNAs of class III homeodomain-leucine zipper (HD-ZIPIII) family genes, which include AtHB8, PHABULOSA (PHB), PHAVOLUTA, REVOLUTA, and CORONA/AtHB15 (Prigge et al., 2005; Carlsbecker et al., 2010). This results in higher expression of these genes in the inner domain of the stele (Carlsbecker et al., 2010; Figure 4F). The miR165/miR166-insensitive mutant phb-7d exhibits ectopic metaxylem vessel formation at the protoxylem vessel position, similarly to that of the shr mutant (Carlsbecker et al., 2010). Conversely, quadruple mutants for HD-ZIPIIIs only produce protoxylem vessels in the xylem axis, which is confirmed by the loss of metaxylem vessel marker ACAULIS 5 (ACL5) and ectopic expression of protoxylem vessel marker AHP6 (Mähönen et al., 2006; Muniz et al., 2008; Carlsbecker et al., 2010). These results indicate that HD-ZIPIII genes ultimately determine the xylem vessel types; high expression induces metaxylem vessels, whereas low expression induces protoxylem vessels (Miyashima et al., 2011).
FIGURE 4.
Determination of xylem vessel types in the xylem axis. (A–D) Schematic illustration showing difference between gene expression and localization of gene products for SHR and MIR165/166 in the RAM (based on the data reported in Carlsbecker et al., 2010). (A) Expression pattern of SHR. (B) Localization of SHR protein. (C) Expression pattern of MIR165a. (D) Localization of MIR165/166. (E,F) Schematic illustration showing localization patterns of MIR165/166 (E) and HD-ZIPIII transcripts (F). (G) Schematic illustration of xylem vessel type determination. Vessel types are determined by VND6 and VND7 as well as HD-ZIPIII gradients.
VASCULAR-RELATED NAC-DOMAIN 6 (VND6) and VND7, which belong to the NAM, ATAF1/2, and CUC2 (NAC) transcription factor family, are master regulators for xylem cell differentiation and determine the cell fate of the metaxylem and protoxylem vessels, respectively (Kubo et al., 2005). Both VND6 and VND7 directly up-regulate genes involved in programmed cell death and secondary cell-wall thickening (Ohashi-Ito et al., 2010; Yamaguchi et al., 2011). VND6 is expressed in the central metaxylem vessels, whereas VND7 is expressed primarily in the protoxylem vessel cell files (Kubo et al., 2005). Overexpression of VND6 and VND7 leads to ectopic formation of metaxylem and protoxylem vessel elements, respectively (Kubo et al., 2005; Yamaguchi et al., 2010a). Conversely, expression of VND6 and VND7 fused with the chimera repression domain SRDX under control of their own promoters causes a failure of central metaxylem and protoxylem vessel development, respectively (Kubo et al., 2005). However, loss-of-function mutants for VND6 and VND7 do not show any defect in root xylem development (Kubo et al., 2005), indicating that seven VND family members function differently but in some cases redundantly in the regulation of xylem cell differentiation. VND-interacting 2 (VNI2) was identified as an interacting protein with VND7 by a yeast two-hybrid screen (Yamaguchi et al., 2010b). VNI2 negatively regulates xylem vessel differentiation in opposition to VND7 (Yamaguchi et al., 2010b). Further analyses are required to elaborate the relationship between VNDs, VNIs, and HD-ZIPIIIs in the context of switching xylem vessel types (Figure 4G).
CONCLUDING REMARKS
Xylem cell fate is regulated by spatiotemporal actions of various signaling factors. Mutual inhibition between CK and auxin determines the precise xylem vessel domains, in particular protoxylem vessels. Some CLE peptides play a role in fine-tuning the CK signal. The movement of AHLs defines the boundary between the procambial domain and the xylem domain, thereby establishing the xylem axis in root stele. The opposite movement of SHR and miR165/miR166 between outer endodermis and inner stele ultimately regulates the level of HD-ZIPIII proteins, resulting in the fate determination of different xylem vessel types. Finally, the master transcription factors VND6 and/or VND7 execute the program of metaxylem and protoxylem vessel differentiation, respectively. Collectively, in roots, xylem cell fates are controlled precisely by a regulatory network consisting of hormone signaling pathways and transcription factors in a hierarchical organization.
However, the basic root vascular pattern is determined during embryogenesis. Therefore, to understand the regulation of xylem cell fate, we should elucidate the mechanism underlying the onset of vascular cells in early embryos. Recent studies demonstrated that two bHLH transcription factors, LONESOME HIGHWAY (LHW) and TARGET OF MONOPTEROS 5 (TMO5), play crucial roles in the initiation of vascular cells (De Rybel et al., 2013; Ohashi-Ito et al., 2013). Further functional analysis of downstream targets of these transcription factors may provide novel insights into understanding the determination of xylem cell fates.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported partly by Grants-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan (NC-CARP project) to Hiroo Fukuda, from the Japan Society for the Promotion of Science (23227001 to Hiroo Fukuda, and JSPS Research Fellowships for Young Scientists to Takayuki Tamaki).
REFERENCES
- Argyros R. D., Mathews D. E., Chiang Y. H., Palmer C. M., Thibault D. M., Etheridge N., et al. (2008). Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20 2102–2116 10.1105/tpc.108.059584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopp A., Help H., El-Showk S., Weijers D., Scheres B., Friml J., et al. (2011a). A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr. Biol. 21 917–926 10.1016/j.cub.2011.04.017 [DOI] [PubMed] [Google Scholar]
- Bishopp A., Lehesranta S., Vaten A., Help H., El-Showk S., Scheres B., et al. (2011b). Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr. Biol. 21 927–932 10.1016/j.cub.2011.04.049 [DOI] [PubMed] [Google Scholar]
- Bonke M., Thitamadee S., Mähönen A. P., Hauser M. T., Helariutta Y. (2003). APL regulates vascular tissue identity in Arabidopsis. Nature 426 181–186 10.1038/nature02100 [DOI] [PubMed] [Google Scholar]
- Cano-Delgado A., Lee J. Y., Demura T. (2010). Regulatory mechanisms for specification and patterning of plant vascular tissues. Annu. Rev. Cell Dev. Biol. 26 605–637 10.1146/annurev-cellbio-100109-104107 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A., Lee J. Y., Roberts C. J., Dettmer J., Lehesranta S., Zhou J., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465 316–321 10.1038/nature08977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Liang W., Li J., Hong F., Wu Y., Wang L., et al. (2013). A CLE-WOX signalling module regulates root meristem maintenance and vascular tissue development in rice. J. Exp. Bot. 64 5359–5369 10.1093/jxb/ert301 [DOI] [PubMed] [Google Scholar]
- Cui H., Levesque M. P., Vernoux T., Jung J. W., Paquette A. J., Gallagher K. L., et al. (2007). An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316 421–425 10.1126/science.1139531 [DOI] [PubMed] [Google Scholar]
- De Rybel B., Moller B., Yoshida S., Grabowicz I., Barbier De Reuille P., Boeren S., et al. (2013). A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev. Cell 24 426–437 10.1016/j.devcel.2012.12.013 [DOI] [PubMed] [Google Scholar]
- Feng H., Chen Q., Feng J., Zhang J., Yang X., Zuo J. (2007). Functional characterization of the Arabidopsis eukaryotic translation initiation factor 5A-2 that plays a crucial role in plant growth and development by regulating cell division, cell growth, and cell death. Plant Physiol. 144 1531–1545 10.1104/pp.107.098079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H. (2004). Signals that control plant vascular cell differentiation. Nat. Rev. Mol. Cell Biol. 5 379–391 10.1038/nrm1364 [DOI] [PubMed] [Google Scholar]
- Gallagher K. L., Paquette A. J., Nakajima K., Benfey P. N. (2004). Mechanisms regulating SHORT-ROOT intercellular movement. Curr. Biol. 14 1847–1851 10.1016/j.cub.2004.09.081 [DOI] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., et al. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101 555–567 10.1016/S0092-8674(00)80865-X [DOI] [PubMed] [Google Scholar]
- Hirakawa Y., Kondo Y., Fukuda H. (2011). Establishment and maintenance of vascular cell communities through local signaling. Curr. Opin. Plant Biol. 14 17–23 10.1016/j.pbi.2010.09.011 [DOI] [PubMed] [Google Scholar]
- Hutchison C. E., Li J., Argueso C., Gonzalez M., Lee E., Lewis M. W., et al. (2006). The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18 3073–3087 10.1105/tpc.106.045674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida K., Yamashino T., Yokoyama A., Mizuno T. (2008). Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 49 47–57 10.1093/pcp/pcm165 [DOI] [PubMed] [Google Scholar]
- Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N., et al. (2006). Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313 842–845 10.1126/science.1128436 [DOI] [PubMed] [Google Scholar]
- Jao D. L., Chen K. Y. (2006). Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J. Cell. Biochem. 97 583–598 10.1002/jcb.20658 [DOI] [PubMed] [Google Scholar]
- Jun J. H., Fiume E., Fletcher J. C. (2008). The CLE family of plant polypeptide signaling molecules. Cell. Mol. Life Sci. 65 743–755 10.1007/s00018-007-7411-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya N., Nagasaki H., Morikami A., Sato Y., Matsuoka M. (2003). Isolation and characterization of a rice WUSCHEL-type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. Plant J. 35 429–441 10.1046/j.1365-313X.2003.01816.x [DOI] [PubMed] [Google Scholar]
- Kieber J. J., Schaller G. E. (2014). Cytokinins. Arabidopsis Book 12:e0168 10.1199/tab.0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y., Hirakawa Y., Kieber J. J., Fukuda H. (2011). CLE peptides can negatively regulate protoxylem vessel formation via cytokinin signaling. Plant Cell Physiol. 52 37–48 10.1093/pcp/pcq129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., et al. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19 1855–1860 10.1101/gad.1331305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen A. P., Bishopp A., Higuchi M., Nieminen K. M., Kinoshita K., Tormakangas K., et al. (2006). Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311 94–98 10.1126/science.1118875 [DOI] [PubMed] [Google Scholar]
- Mähönen A. P., Bonke M., Kauppinen L., Riikonen M., Benfey P. N., Helariutta Y. (2000). A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 14 2938–2943 10.1101/gad.189200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashima S., Koi S., Hashimoto T., Nakajima K. (2011). Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138 2303–2313 10.1242/dev.060491 [DOI] [PubMed] [Google Scholar]
- Miyashima S., Sebastian J., Lee J. Y., Helariutta Y. (2012). Stem cell function during plant vascular development. EMBO J. 32 178–193 10.1038/emboj.2012.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz L., Minguet E. G., Singh S. K., Pesquet E., Vera-Sirera F., Moreau-Courtois C. L., et al. (2008). ACAULIS5 controls Arabidopsis xylem specification through the prevention of premature cell death. Development 135 2573–2582 10.1242/dev.019349 [DOI] [PubMed] [Google Scholar]
- Nakajima K., Sena G., Nawy T., Benfey P. N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413 307–311 10.1038/35095061 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K., Oda Y., Fukuda H. (2010). Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22 3461–3473 10.1105/tpc.110.075036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K., Oguchi M., Kojima M., Sakakibara H., Fukuda H. (2013). Auxin-associated initiation of vascular cell differentiation by LONESOME HIGHWAY. Development 140 765–769 10.1242/dev.087924 [DOI] [PubMed] [Google Scholar]
- Prigge M. J., Otsuga D., Alonso J. M., Ecker J. R., Drews G. N., Clark S. E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17 61–76 10.1105/tpc.104.026161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B., Chen Q., Hong S., Zhao W., Feng J., Feng H., et al. (2013). The Arabidopsis eukaryotic translation initiation factor eIF5A-2 regulates root protoxylem development by modulating cytokinin signaling. Plant Cell 25 3841–3857 10.1105/tpc.113.116236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B., Liang Y., Deng Y., Chen Q. G., Zhang J., Yang X. H., et al. (2009). Genome-wide comparative analysis of type-A Arabidopsis response regulator genes by overexpression studies reveals their diverse roles and regulatory mechanisms in cytokinin signaling. Cell Res. 19 1178–1190 10.1038/Cr.2009.88 [DOI] [PubMed] [Google Scholar]
- Scheres B., Dilaurenzio L., Willemsen V., Hauser M. T., Janmaat K., Weisbeek P., et al. (1995). Mutations affecting the radial organization of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121 53–62 [Google Scholar]
- Thompson G. M., Cano V. S., Valentini S. R. (2003). Mapping eIF5A binding sites for Dys1 and Lia1: in vivo evidence for regulation of eIF5A hypusination. FEBS Lett. 555 464–468 10.1016/S0014-5793(03)01305-X [DOI] [PubMed] [Google Scholar]
- To J. P., Deruere J., Maxwell B. B., Morris V. F., Hutchison C. E., Ferreira F. J., et al. (2007). Cytokinin regulates type-A Arabidopsis Response Regulator activity and protein stability via two-component phosphorelay. Plant Cell 19 3901–3914 10.1105/tpc.107.052662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To J. P., Haberer G., Ferreira F. J., Deruere J., Mason M. G., Schaller G. E., et al. (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16 658–671 10.1105/tpc.018978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursache R., Miyashima S., Chen Q., Vaten A., Nakajima K., Carlsbecker A., et al. (2014). Tryptophan-dependent auxin biosynthesis is required for HD-ZIP III-mediated xylem patterning. Development 141 1250–1259 10.1242/dev.103473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaten A., Dettmer J., Wu S., Stierhof Y. D., Miyashima S., Yadav S. R., et al. (2011). Callose biosynthesis regulates symplastic trafficking during root development. Dev. Cell 21 1144–1155 10.1016/j.devcel.2011.10.006 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Goue N., Igarashi H., Ohtani M., Nakano Y., Mortimer J. C., et al. (2010a). VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 153 906–914 10.1104/pp.110.154013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Ohtani M., Mitsuda N., Kubo M., Ohme-Takagi M., Fukuda H., et al. (2010b). VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22 1249–1263 10.1105/tpc.108.064048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Mitsuda N., Ohtani M., Ohme-Takagi M., Kato K., Demura T. (2011). VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 66 579–590 10.1111/j.1365-313X.2011.04514.x [DOI] [PubMed] [Google Scholar]
- Yokoyama A., Yamashino T., Amano Y., Tajima Y., Imamura A., Sakakibara H., et al. (2007). Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol. 48 84–96 10.1093/pcp/pcl040 [DOI] [PubMed] [Google Scholar]
- Zhou J., Wang X., Lee J. Y., Lee J. Y. (2013). Cell-to-cell movement of two interacting AT-Hook factors in Arabidopsis root vascular tissue patterning. Plant Cell 25 187–201 10.1105/tpc.112.102210 [DOI] [PMC free article] [PubMed] [Google Scholar]