Summary
We here show that PDCD4 inhibits NF-κB by a novel mechanism that is functionally significant. PDCD4 binds to p65, inhibiting its nuclear localization and inhibiting expression of NF-κB target mRNAs and proteins.
Abstract
PDCD4 is a tumor suppressor induced by apoptotic stimuli that regulates both translation and transcription. Previously, we showed that overexpression of PDCD4 leads to decreased anchorage-independent growth in glioblastoma (GBM)-derived cell lines and decreased tumor growth in a GBM xenograft model. In inflammatory cells, PDCD4 stimulates tumor necrosis factor-induced activation of the transcription factor NF-κB, an oncogenic driver in many cancer sites. However, the effect of PDCD4 on NF-κB transcriptional activity in most cancers including GBM is still unknown. We studied the effect of PDCD4 on NF-κB-dependent transcriptional activity in GBM by stably overexpressing PDCD4 in U251 and LN229 cells. Stable PDCD4 expression inhibits NF-κB transcriptional activation measured by a luciferase reporter. The molecular mechanism by which PDCD4 inhibits NF-κB transcriptional activation does not involve inhibited expression of NF-κB p65 or p50 proteins. PDCD4 does not inhibit pathways upstream of NF-κB including the activation of IKKα and IKKβ kinases or degradation of IκBα, events needed for nuclear transport of p65 and p50. PDCD4 overexpression does inhibit localization of p65 but not p50 in the nucleus. PDCD4 protein interacts preferentially with p65 protein as shown by co-immunoprecipitation and confocal imaging. PDCD4 overexpression inhibits the mRNA expression of two NF-κB target genes in a p65-dependent manner. These results suggest that PDCD4 can significantly inhibit NF-κB activity in GBM cells by a mechanism that involves direct or indirect protein–protein interaction independent of the expected mRNA-selective translational inhibition. These findings offer novel opportunities for NF-κB-targeted interventions to prevent or treat cancer.
Introduction
PDCD4 was first described as a protein induced by apoptotic stimuli (1) and subsequently shown to act as a tumor suppressor (2). Overexpressed PDCD4 inhibits and PDCD4 deficiency stimulates tumorigenesis and tumor progression in mouse models (3–5) and its loss is diagnostic for human cancer staging and prognostic for survival in colon, lung, liver, breast, glioma and esophageal cancers (6–11). Gene therapy with PDCD4 in an activated K-Ras model prevents lung carcinogenesis (12). We recently reported that PDCD4 leads to decreased anchorage-independent growth in glioblastoma (GBM)-derived cell lines as well as decreased tumor growth in a GBM xenograft model (13).
PDCD4 inhibits translation in an mRNA-selective way by interacting with translation initiation factor eIF4A and inhibiting its RNA helicase activity (14,15). The crystal structures of the functional MA3 domains of PDCD4 and of cocrystals with eIF4A have revealed details of how PDCD4 inhibits translation initiation (16–18). The RNA helicase activity of eIF4A in the eIF4F translation initiation complex is thought to be important for unwinding secondary structure in the 5′UTRs of certain oncogenic mRNAs (19) prior to arriving at the start codon. Translational targets of PDCD4 thus far reported include c-myb and p53 (20,21) as well as internal ribosome entry site-regulated apoptosis inhibitors (22). In addition to inhibiting mRNAs that are translational targets, PDCD4 inhibits, at least indirectly, mRNA expression of uPAR, lysyl oxidase and MAP4K (23–25). The responsible transcription factors have not been identified.
The transcription factor NF-κB acts as an oncogenic driver in many cancer sites. PDCD4 does not inhibit NF-κB in mouse JB6 cells where it inhibits AP-1 transactivation and transformation (26). The AP-1 inhibition by PDCD4 has been attributed to targeting Jun kinase (JNK) signaling (25,27). PDCD4 stimulates tumor necrosis factor-induced activation of NF-κB in inflammatory cells (28). PDCD4 inhibits cyclin D1 transcription in colon cancer cells by an IKK/NF-κB-dependent mechanism (29). Whether PDCD4 inhibits NF-κB transcriptional activity in other cancer sites and cells is still unknown. We asked whether and by what mechanism PDCD4 might regulate NF-κB-dependent transcriptional activity in malignant human GBM cell lines. Stable overexpression of PDCD4 in U251 and LN229 cells inhibits NF-κB transcriptional activation measured by a luciferase reporter. The mechanism of inhibition does not involve inhibited translation or transcription of NF-κB proteins p65 or p50 or of activating kinase IKKα/β but instead proceeds through interaction of PDCD4 protein with p65 to inhibit its nuclear localization. NF-κB target genes matrix metalloproteinase-9 (MMP-9) and vascular endothelial growth factor (VEGF) are identified as being PDCD4 regulated. This inhibition of p65-dependent transcription occurs independently of translational inhibition by Pdcd4. As these proteins are oncogenic mediators of invasion and angiogenesis, their suppressed expression may contribute to PDCD4 suppressed tumor growth and invasion.
Materials and methods
Cell culture and inhibitors
Human GBM cell lines U251 and LN229 cells (ATCC, Manassas, VA) were stably transfected with human PDCD4 in pFB-Blast (Invitrogen, Carlsbad, CA) expressing the human PDCD4 gene (24) to generate pooled clones (Supplementary Figure S1, available at Carcinogenesis Online). GBM cells stably expressing PDCD4 were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 2mM l-glutamine, 100 units/ml penicillin, 100 µg/ml streptomycin, 5 µg/ml Blasticidin S and 5% fetal bovine serum. Cell lines were routinely maintained at 37°C in a humidified 5% CO2 atmosphere. MG-132 proteasome inhibitor was purchased from Calbiochem (San Diego, CA).
Luciferase reporter assays
Cells (5 × 104) were transiently cotransfected with 1 µg of reporter plasmid, along with 0.05 µg of pRL-TK (Renilla luciferase) control plasmid using Fugene 6 reagent (Roche Applied Science, Indianapolis, IN). Cells were harvested 48 h after transfection and lysed in 100 µl of passive lysis buffer (Promega, Madison, WI). Aliquots (25 µl) of lysates were analyzed for luminescent signal with a FLUOstar Omega (BMG Labtech, Ortenberg, Germany).
Immunoblot assay and preparation of cytosolic and nuclear extracts
Cells were lysed by sonication in radioimmunoprecipitation assay (RIPA) buffer and protein was measured using Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford, IL). Equal amounts of protein were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred into nitrocellulose membranes (GE Healthcare, Uppsala, Sweden). The following antibodies were used: p65, phospho-p65 (Ser-536), p50, IκBα, IKKα, IKKβ, phospho-IκBα, phospho-IKKα/β, XIAP, BcLXL, CIAP-1, CIAP-2, cyclin D1, cyclin E, c-Rel, RelB, β-tubulin and eIF4A were obtained from Cell Signaling. VEGF and β-actin were purchased from Santa Cruz and p52, MMP-9, Lamin A from Abcam. Densities were measured using NIH ImageJ (Bethesda, MD). Subcellular fractionation was performed using NE-PER Nuclear and Cytoplasmic Extraction Reagents kit according to the manufacturer’s instructions (Thermo Scientific).
Immunoprecipitation assays and western blotting
GBM cells were seeded in 100mm plates and lysed in immunoprecipitation (IP) lysis buffer (Thermo Scientific). After removing insoluble materials by centrifugation, lysates were precleared using protein A sepharose. Ten percent of the supernatant was saved as input sample and the remainder was used for IP. Protein A sepharose beads were incubated with a monoclonal rabbit PDCD4 antibody (Novus Biological, Littleton, CO) or normal rabbit IgG (Cell Signaling Technology, Danvers, MA) at 4°C overnight. Following IP, the beads were washed three times with 250 µl of lysis buffer and boiled in sodium dodecyl sulfate sample buffer. The bound proteins were resolved on a 10% Tris-Bis NuPage gel (Invitrogen) and transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA). The membrane was blocked with 5% skim milk and incubated with appropriate dilutions of the primary antibody at room temperature for 3 h. Samples were analyzed by western blotting using the appropriate antibodies to detect protein expression. For reciprocal IP experiment, transient transfection with HA-tagged p65 into both GBM PDCD4 cells and immunoprecipitation with anti-HA antibody and whole cell lysate were followed by immunoblotting (IB) to detect PDCD4 and HA proteins.
Immunofluorescence
Stable PDCD4-expressing cells were grown on the two-well chamber slides in Dulbecco’s modified Eagle’s medium containing 5% fetal bovine serum, washed twice in phosphate-buffered saline, fixed in 4% paraformaldehyde and permeabilized in 0.5% Triton X-100. Cells were incubated with a mouse polyclonal anti-PDCD4 antibody (1:100 dilution; Abcam) and rabbit polyclonal anti-p65 antibody (1:100 dilution; Abcam). After being washed three times with phosphate-buffered saline, the slides were incubated with fluorescein isothiocyanate-conjugated goat anti-mouse RITC (PDCD4) and goat anti-rabbit FITC (p65) for 2 h (Molecular Probes). The cells were washed twice with phosphate-buffered saline and then mounted with antifading mounting fluid (Prolong; Molecular Probes). Fluorescence images were obtained with the 63× objective of a Zeiss LSM-5 confocal microscope at the National Cancer Institute Image Analysis Lab.
Quantitative real-time PCR
The mRNA was extracted from GBM cells with Trizol (Invitrogen). Reverse transcription reactions were performed using iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer’s instructions. Real-time PCR was performed on an iQ5 Real-Time PCR Detection System (Bio-Rad). To amplify mRNAs, we used iQ Sybr green supermix and the following primers: CIAP-1: forward, 5′-TCAGAAAGGAGT CTTGCTCGTGCT-3′, reverse, 5′-TATCCAGCATCAGGCCACAACAGA-3′; CIAP-2: forward, 5′-TCAGAAAGGAG TCTTGCTCGTGCT-3′, reverse, 5′-TATCCA GCATCAGGCCA CAACAGA-3′; XIAP: forward, 5′-TGTTTCAGCATCAA CACTGGCACG-3′, reverse, 5′-TGCATGACAACTAAAG CACCGCAC-3′; BcLXL: forward, 5′-TGGGCTCACTCTTCAGTCGGAAAT-3′, reverse, 5′-AT GTAGTGGTT CTGGTGGCAA-3′; cyclin D1: forward, 5′-TTGCAA GCAGGACTTTG AGGCAAG-3′, reverse, 5′-CAAACACCAGTTGGCACC AAAGGA-3′; cyclin E: forward, 5′-TGCAGAGCTGTTGGAT CTCTGT GT-3′, reverse, 5′-TGTCGCACCA CTGATACCCTGAAA-3′; MMP-9: forward, 5′-CTCAGGGAGTCTT CCATCACTTTC-3′, reverse, 5′-AGCATGAGAAAGGGCTTACACCAC-3′ and VEGF: forward, 5′-TGCCA GCA ACACTACCAC-3′, reverse, 5′-GAGTCATCTCCA GCATCC-3′. The GAPDH gene was used for normalization. The relative transcription level was calculated using the ΔΔC t method. Results were from three independent experiments and plotted as fold expression relative to control cell line arbitrarily set at 1.0. Statistical significance was determined by Student’s t-test analysis (*P < 0.05 and **P < 0.01).
Invasion assay
BD Biocoat Matrigel invasion chambers, control inserts and wells (BD Biosciences, Bedford, MA) were rehydrated in 500 µl serum-free medium at 37°C for 2 h. After removing rehydration medium, 750 µl medium plus 5% fetal bovine serum was added to each well of the 24-well plate followed immediately by the addition of 1.5×104 GBM cells in 500 µl serum-free medium plus each chamber and control insert. Plates were incubated for 22 h at 37°C. Non-invading cells were removed from the upper surface of the membrane by scrubbing the membrane with a cotton swab. Cells on the lower surface of the membrane were stained with the Diff Quik stain kit (IMEB, San Marcos, CA). Invading cells in five random fields were counted with a ×40 lens. Data are expressed as percent invasion through the Matrigel matrix membrane relative to migration through the control membrane.
Chromatin IP
A chromatin immunoprecipitation (ChIP) assay was conducted following the protocol provided by Millipore (Temecula, CA). DNA was purified with ‘QIAquick PCR purification kit’ (Qiagen) following the manufacturer’s recommendations and measured by iQ5 Real-Time PCR Detection System (Bio-Rad). The following primers were used: CIAP-1: forward, 5′-TCAGAAAG GAGTCTTGCTCGTGCT-3′, reverse, 5′-TATCCAGCATCAGGCCACAACAGA-3′; cyclin D1: forward, 5′- TTGCGGGTATTTTCTGAAGG -3′, reverse, 5′-CTCCCAC GAAACGCTA CTTC-3′; MMP-9: forward, 5′-GCCATGTC TGCTGTTTTCTAGAGG-3′, reverse, 5′-CACACTCCAGGCT CTGTCCTCTTT-3′ and VEGF: forward, 5′-AGACTCCACAGTGCATACGTG-3′, reverse, 5′- AGTGTG TCCCTCTGACAATG-3′.
Statistical analysis
Data are presented as mean ± SE. Statistical analysis was performed using Student’s t-test. All experiments represent at least there independent replications. The statistical significance of differences (*P < 0.05 and **P < 0.01) was indicated in figures by asterisks.
Results
PDCD4 inhibits NF-κB-dependent transcriptional activity in GBM cells
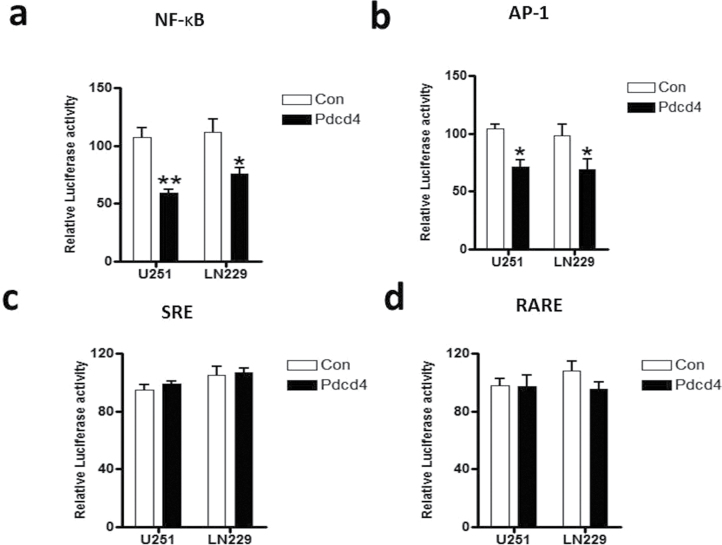
PDCD4 expression is lost in gliomas and in many other human cancers where PDCD4 loss is prognostic for poor survival (8). In gliomas, NF-κB is constitutively activated, the levels of NF-κB-regulated genes are elevated, and these circumstances are inversely correlated with patient prognosis. To investigate possible PDCD4 regulation of NF-κB, we generated cell lines stably overexpressing PDCD4 using U251 and LN229 human GBM cells. A PDCD4/pFB-blast retroviral vector was transfected into each of the two cell lines and stable lines were generated using Blasticidin S selection. Transfection with pFB-blast retroviral vector was used as a control. Blasticidin S-resistant colonies (pooled clones) were isolated and analyzed for PDCD4 expression using western blot analysis. Supplementary Figure S1, available at Carcinogenesis Online, shows substantially increased expression of PDCD4 following stable gene transfer into U251 and LN229 cells. Our previous studies with mouse preneoplastic JB6 cells showed that PDCD4 inhibits AP-1 but not NF-κB (26). To investigate the effect of PDCD4 on NF-κB transcriptional activity in GBM cells, we used an NF-κB luciferase reporter assay. As shown in Figure 1a, overexpression of PDCD4 significantly inhibited the transcriptional activity of NF-κB by 45 and 30% in U251 and LN229 cells, respectively. To ascertain specificity, we assayed three other transcriptional promoter reporters, AP-1, SRE (serum-response element) and RARE (retinoic acid receptor element) luciferase reporters for comparison with NF-κB. As in the case of mouse epidermal JB6 cells and mouse epidermis (4), PDCD4 also inhibited AP-1 activity in U251 and LN229 human glioma cells (Figure 1b). However, PDCD4 did not inhibit SRE or RARE dependent transcription (Figure 1c and d). These findings indicate relatively specific inhibition of NF-κB in addition to AP-1-dependent transcriptional activity by PDCD4 in GBM cells.
Fig. 1.
PDCD4 specifically inhibits NF-κB transcriptional activity. PDCD4 suppresses transcriptional activity of NF-κB in glioma cells. Two glioma cell lines (U251 and LN229) stably expressing PDCD4 were cotransfected with pRL-TK Renilla plus (a) 5X NF-κB-luciferase, (b) AP-1 luciferase, (c) SRE (serum response element) luciferase and (d) RARE (retinoic acid receptor element) luciferase plasmids and analyzed by luciferase reporter activity assays. The mean activity ± SE are shown for three independent experiments, and statistical significance is represented by Student’s t-test (*significant difference P ≤ 0.05 and **significant difference P ≤ 0.01).
PDCD4 does not inhibit the expression or activation of kinases upstream of p65/p50 or phosphorylation of endogenous IKKα/β or IκBα
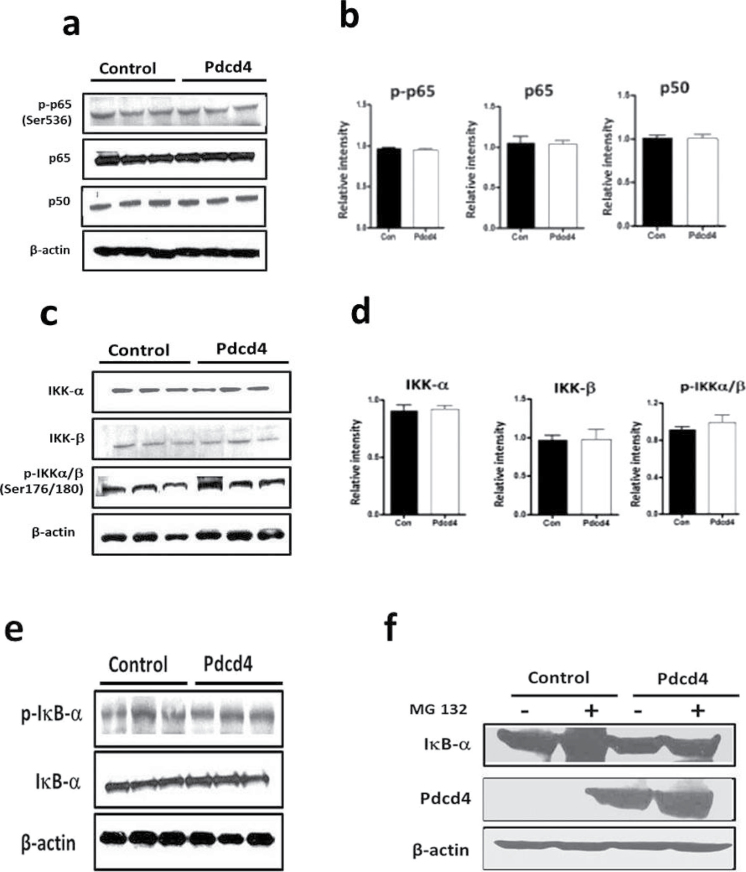
To investigate the mechanism by which PDCD4 inhibits NF-κB transcriptional activity, we examined the effect of PDCD4 on the expression of ‘canonical’ NF-κB proteins p65 (containing dimerization, DNA-binding and transactivation domains) and p50 (containing only dimerization and DNA-binding domains). PDCD4 overexpression does not change p65 protein expression (Figure 2a and b). The level of p50 protein, the p65 dimer partner often required for NF-κB transcription factor activity, was also unchanged by PDCD4. In addition, the levels of non-canonical NF-κB proteins Rel B and p52 were unchanged (data not shown). Optimal NF-κB activation is positively regulated by phosphorylation at multiple serine residues (e.g. Ser-276, Ser-311, Ser-468, Ser-529 and Ser-536) in functional domains of p65. Specifically, phosphorylation of p65 at the Ser-536 site within the transactivation domain is an event that facilitates nuclear import and transcriptional activity of p65 (30). Therefore, we examined the effect of PDCD4 on the phosphorylation of p65. Figure 2a and b shows that PDCD4 does not affect S-536 phosphorylation of p65. To identify the mechanism by which PDCD4 inhibits NF-κB transactivation, we determined whether PDCD4 affects upstream regulators of NF-κB. Because IKKα and IKKβ are major kinases for activating NF-κB proteins as well as phosphorylating IκBα (31), we investigated the effect of PDCD4 on IKKα and IKKβ phosphorylation and expression. As shown in Figure 2c and d, PDCD4 failed to inhibit phosphorylation of IKKα/β or total expression levels of IKKα and IKKβ. Phosphorylation of IκBα (Ser 32/36) is required for NF-κB activation and this phosphorylation results in the ubiquitination and subsequent proteasomal degradation of IκBα. The release of IκBα from the p65/p50 complex induces translocation of NF-κB into the nucleus and activates NF-κB-dependent gene transcription (32). PDCD4 does not increase IκBα protein expression or decrease phosphorylation of IκBα at Ser-32 or Ser-36 (Figure 2e). To confirm this result, we treated PDCD4-overexpressing cells with proteasome inhibitor MG-132, which inhibits the degradation of IκBα. Treatment of PDCD4-overexpressing cells with MG-132 produced accumulation of IκBα similar to that seen in vector control cells (Figure 2f). Overexpressing PDCD4 without MG-132 did not produce an increase in IκBα of a magnitude similar to that seen in vector control cells with proteasome inhibitor; this excludes the possibility that PDCD4 mimics the proteasome inhibitor by preventing degradation of IκBα. These results indicate that, in contrast to the observation in HT29 colon cancer cells (29), changes in upstream regulators of NF-κB do not contribute to the inhibition of NF-κB transactivation by PDCD4.
Fig. 2.
PDCD4 does not inhibit expression of NF-κB p65 or p50 proteins and activation of p65 or activation of kinases upstream of p65/p50. (a) Western blot analysis of PDCD4-overexpressing cells. The western blots were developed with phospho-p65 (Ser-536), total p65, p50 and β-actin antibodies. (b) Densitometry scans of western blot (n = 3). Error Bars represent mean ± SE from three independent experiments. (c) IKKα and IKKβ protein expression and IKKα/β (Ser-176/180) phosphorylation were determined by western blot analysis. (d) Densitometry scans of western blot (n = 3). Error bars represent mean ± SE from three independent experiments. (e) Total IκBα protein and phosphorylation of IκBα (Ser-32/34) were analyzed by western blotting. (f) PDCD4-overexpressing cells were treated with MG-132 for 12h. Total IκBα and PDCD4 protein expression were analyzed by western blotting.
PDCD4 inhibits nuclear localization of p65
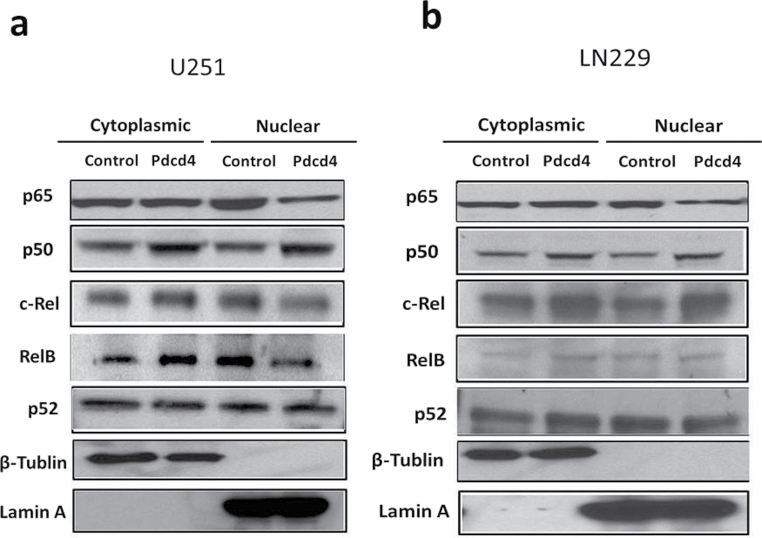
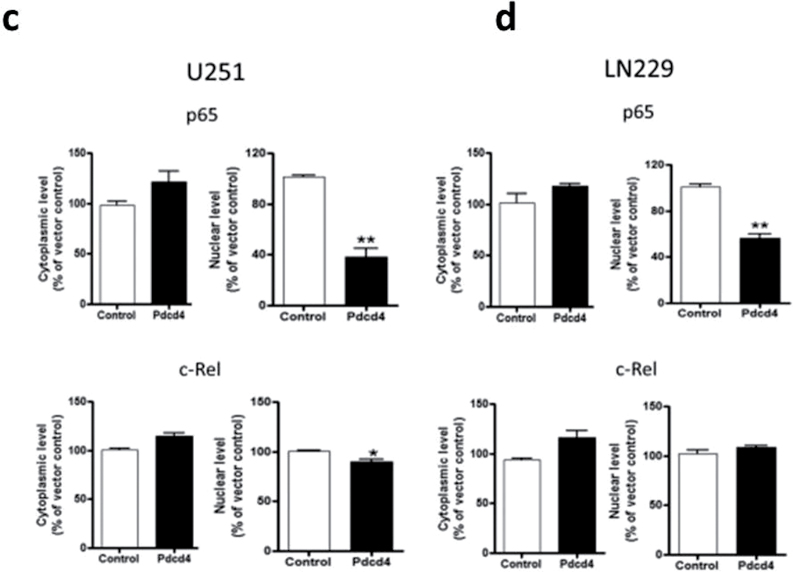
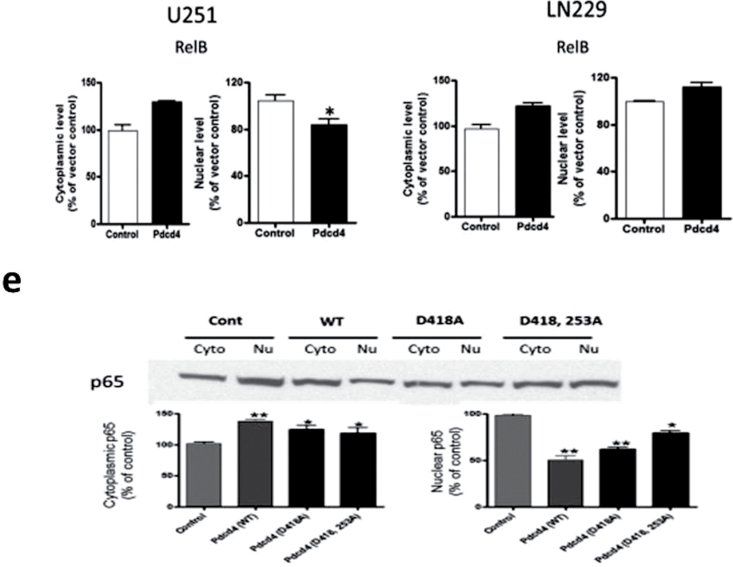
Because NF-κB transcriptional activation requires nuclear accumulation of NF-κB proteins, we asked whether PDCD4 overexpression alters the nuclear translocation of NF-κB. We fractionated cell lysates into cytoplasmic and nuclear fractions and determined the subcellular localization of NF-κB family proteins p65, p50, c-Rel, RelB and p52 by IB analysis. As shown in Figure 3a and b, PDCD4 overexpression substantially reduced the amount of p65 but produced no significant change in p50 or p52 protein levels in the nuclear fractions of the two GBM cell lines (Figure 3a–d). Nuclear c-Rel and RelB showed small but significant decreases in U251 but not in LN229. These results demonstrate that PDCD4 regulates NF-κB transcriptional activity by preferentially inhibiting p65 nuclear localization. To ascertain whether this inhibition of nuclear p65 by Pdcd4 occurs in a manner that is independent of the translational inhibition activity of Pdcd4, we cotransfected with mutants of Pdcd4 inactivated for interaction with translation factor eIF4A and for translational inhibition. Figure 3e shows that single and double translation mutants of PDCD4 significantly inhibited nuclear p65 levels. Thus, Pdcd4’s suppression of nuclear levels of p65 is not dependent on its translation-inhibiting activity.
Fig. 3.
PDCD4 inhibits nuclear localization of p65. (a and b) Subcellular localization of NF-κB family proteins in U251 and LN229 cells. Cytosolic and nuclear fractions of indicated cells were analyzed by IB analyses. Lamin A was used as a nuclear protein marker and β-tubulin was used as a loading control. (c and d) p65, c-Rel and RelB expression levels in the nucleus. Error bars represent mean ± SE from three independent experiments; *P < 0.05; **P < 0.01. (e) U251 cells were cotransiently transfected with HA-tagged p65 plasmid and PDCD4 mutant (PDCD4 D418 or PDCD4 D253A, D418A) plasmid for 24h. Nuclear extracts were prepared and analyzed for nuclear p65 expression by IB.
Interaction of PDCD4 with p65 in the cytoplasm of PDCD4-overexpressing cells
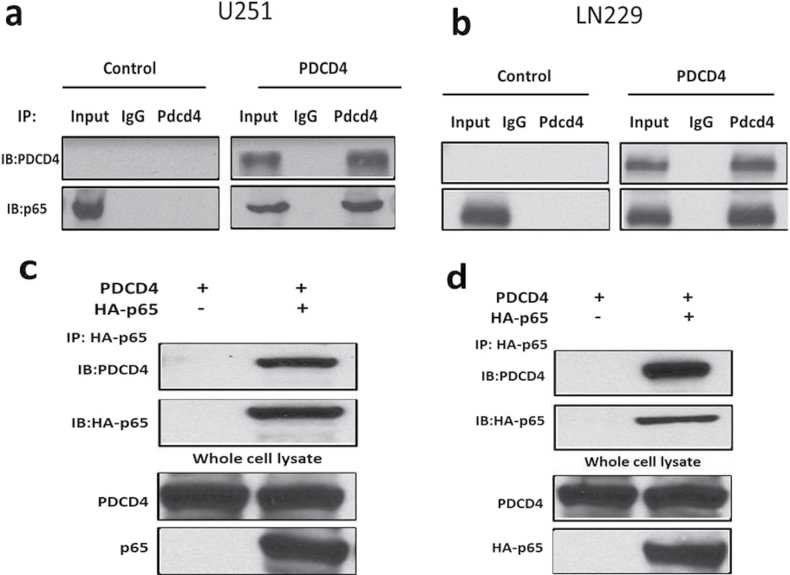
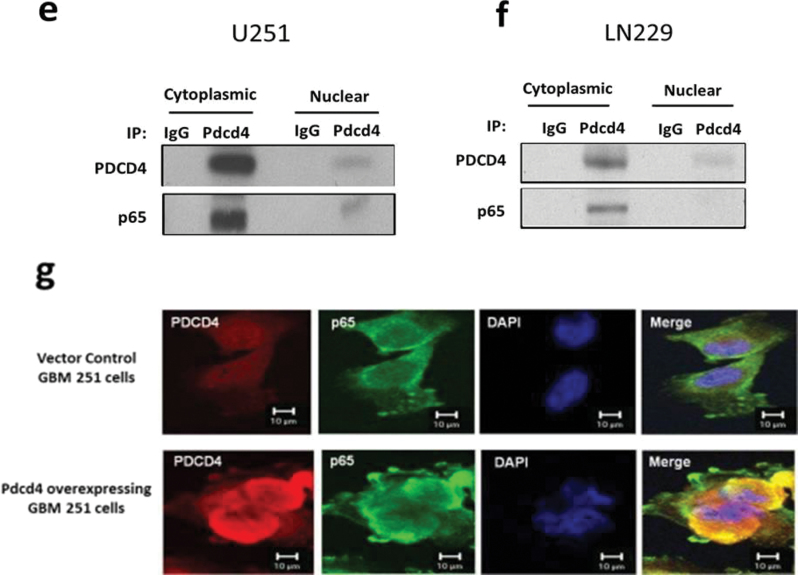
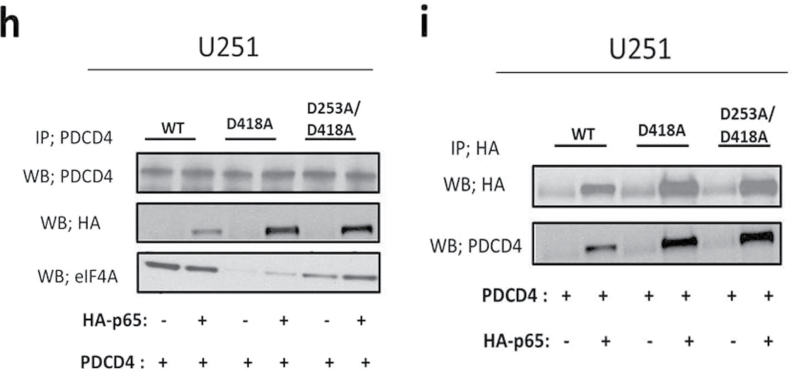
To investigate the mechanism by which PDCD4 inhibits p65 nuclear translocation, we tested the possibility that PDCD4 may directly interact with p65. The whole cell lysates of PDCD4-overexpressing and vector control GBM cells were immunoprecipitated with anti-PDCD4 antibody or with control rabbit sera, and immunoblotting was performed using anti-p65 and anti-PDCD4 antibodies. Figure 4a and b shows that endogenous p65 coprecipitates with PDCD4 in PDCD4-overexpressing cells, whereas IgG immunoprecipitations did not coprecipitate p65. A reciprocal experiment using lysates of PDCD4-overexpressing cells transfected with HA-p65 used anti-HA for immunoprecipitation. Figure 4c and d shows that HA-p65 physically interacts with PDCD4 protein. In contrast, when we performed IP with PDCD4 antibody and immunoblotted for IκBα, p50, c-Rel, RelB or p52, we found that PDCD4 did not interact with other NF-κB family proteins (Supplementary Figure S2, available at Carcinogenesis Online). Our results suggest that PDCD4 directly and specifically interacts with p65 in two GBM cell lines. To further investigate whether this interaction occurs in the cytoplasm, we performed immunoprecipitation with cytoplasmic and nuclear fractions using anti-PDCD4 antibody. Figure 4e and f shows that PDCD4 interaction with p65 was substantial in the cytoplasm upon PDCD4 overexpression. PDCD4 is known to localize both to the nucleus and the cytoplasm in cancer cells (10). Nuclear PDCD4 shows little or no binding to p65 in the nucleus of both lines overexpressing PDCD4 (Figure 4e and f). These findings suggest that PDCD4 interacts with p65, either directly or through a third partner, in the cytoplasm thus inhibiting net nuclear translocation (involving import or export) of p65. To further confirm the PDCD4–p65 interaction in the cytoplasm, we used a multicolor confocal immunofluorescence microscope to ascertain the subcellular localizations of PDCD4 and p65 in U251 PDCD4-overexpressing cells. Figure 4g depicts the immunofluorescence signal for PDCD4 (red), which was distributed in both the cytoplasm and nuclei in overexpressing PDCD4 cells, whereas the signal for p65 (green) was mainly localized to the cytoplasm. The colocalized signal (yellow) for PDCD4 and p65 was found almost exclusively in the cytoplasm, with little or no colocalization signal observed in the nucleus (blue). These results demonstrate that cytoplasmic sequestration of NF-κB p65 by PDCD4 interaction contributes to inhibition of NF-κB-dependent gene transcription. Based on the results shown above (Figure 3e), we would expect translational mutants of Pdcd4 to associate with p65 in the cytoplasm and thereby inhibit p65 nuclear translocation. To ascertain whether Pdcd4 mutants can bind to p65 in U251 cells, we cotransfected HA-p65 with Pdcd4 single and double MA3 mutants. The cell lysates were immunoprecipitated with PDCD4 antibody and immunoblotted for HA-p65 or eIF4A. Figure 4h shows that Pdcd4 single and double mutants bind to HA-p65 but show relatively little association with eIF4A. A reciprocal experiment using HA immunoprecipitation confirmed that Pdcd4 single and double mutants interact with HA-p65 protein (Figure 4i). These results indicate that PDCD4 interaction resulting in cytoplasmic sequestration of p65 occurs by a mechanism that is independent of PDCD4’s translation-inhibiting activity.
Fig. 4.
PDCD4 interacts with p65 in the cytoplasm. (a and b) Total protein extracts from U251 and LN229 vector control and PDCD4-overexpressing cells were immunoprecipitated with anti-PDCD4 (C-terminal) or anti-IgG antibodies. Precipitated PDCD4 and p65 proteins were detected by western blot after IP with anti-PDCD4. (c and d) Transfection with HA-tagged p65 plasmid into U251 and LN229 PDCD4-overexpressing cells and immunoprecipitation with anti-HA antibody (upper panel) and whole cell lysate (bottom panel) were followed by western blotting to detect PDCD4 and HA proteins. (e and f) Cytoplasmic and nuclear extracts were immunoprecipitated with anti-PDCD4 (C-terminal) or anti-IgG antibodies and were analyzed by western blotting using antibodies against PDCD4 and p65. (g) U251 PDCD4-overexpressing cells were immunostained with anti-PDCD4, anti-p65 antibodies and 4′,6-diamidino-2-phenylindole (red for PDCD4, green for p65 and blue for 4′,6-diamidino-2-phenylindole). Stained cells were visualized using an LSM-5 fluorescence 9 microscope. (h) U251 cells were cotransiently transfected with HA-tagged p65 and WT PDCD4 or mutant (PDCD4 D418, PDCD4 D253A, D418A) plasmid for 24h, immunoprecipitated with anti-PDCD4 antibody and analyzed by western blotting using antibodies against HA (upper panel) or endogenous eIF4A protein (bottom panel). (i) For the reciprocal IP experiment, we followed the cotransfection of HA-p65 and WT or mutant PDCD4 with immunoprecipitation using anti-HA antibody and western blotting to detect HA and PDCD4 protein.
PDCD4 inhibits mRNA expression of invasion-related but not apoptosis- or cell proliferation-related NF-κB target genes
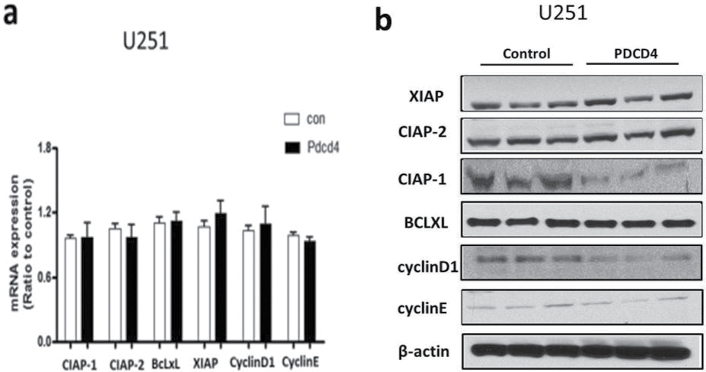
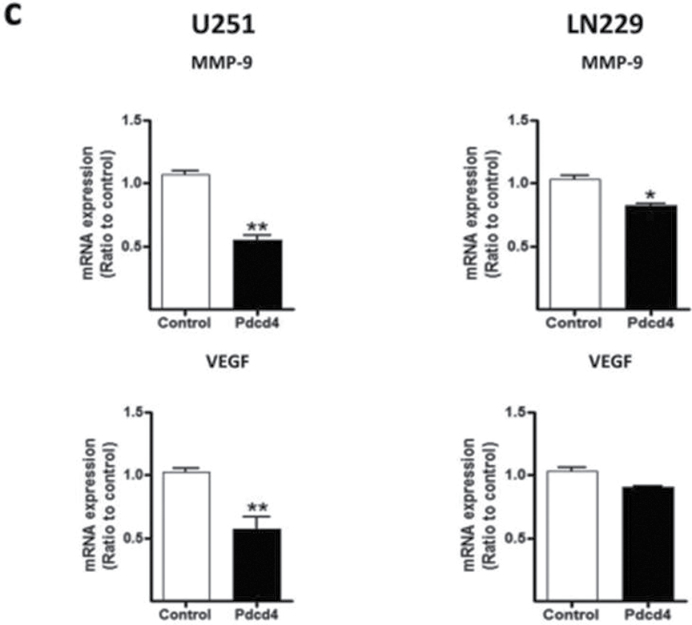
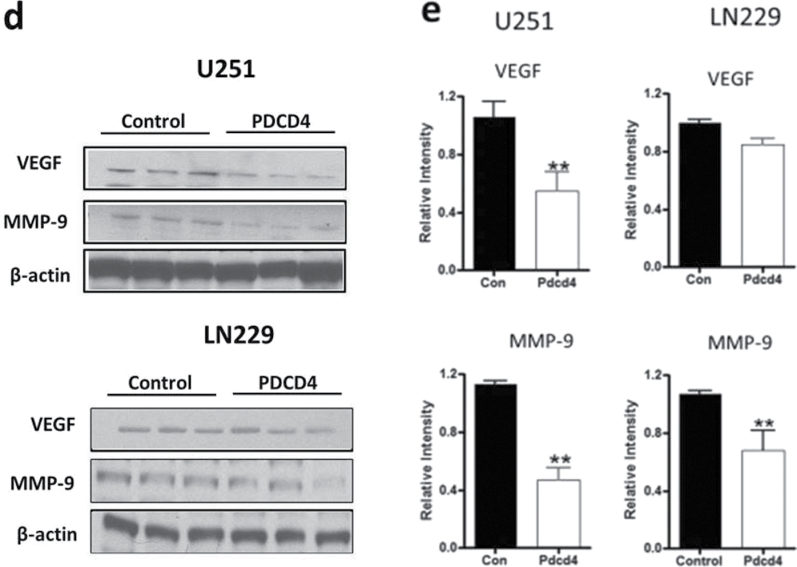
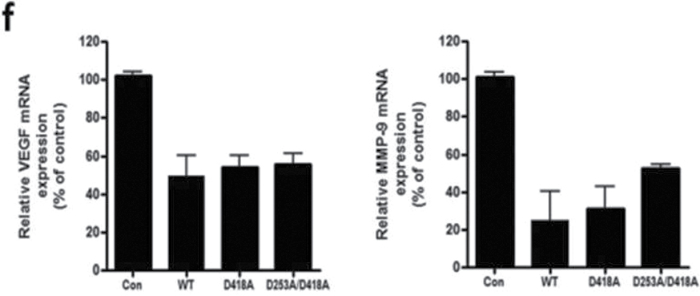
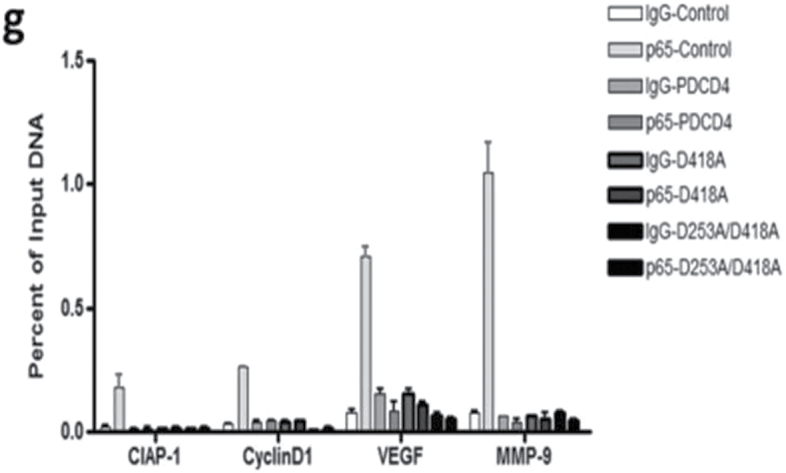
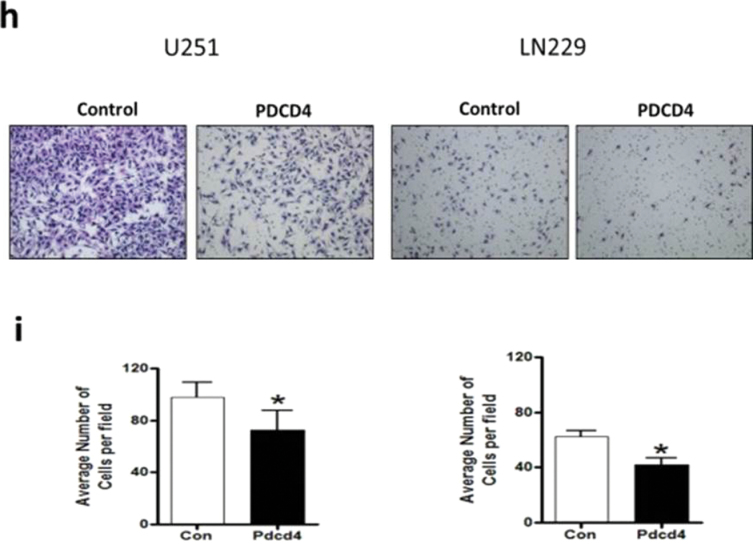
NF-κB functions as an oncogenic driver by activating the expression of genes that are antiapoptotic or proproliferative or genes that stimulate invasion or angiogenesis (33). We performed quantitative reverse transcription–polymerase chain reaction (qRT–PCR) analyses to examine the expression levels of four apoptosis-associated NF-κB target genes including CIAP-1, CIAP-2, XIAP and BcLXL in PDCD4-overexpressing and vector control U251 cells. As shown in Figure 5a, when compared with vector control cells, PDCD4 overexpression did not produce a change in CIAP-1, CIAP-2, XIAP or BcLXL mRNA levels (Figure 5a). Protein expression was also unchanged except for that of CIAP-1, which decreased with PDCD4 overexpression (Figure 5b). Assessment of proliferation-associated NF-κB target genes cyclin D1 and cyclin E also showed no change at the mRNA level with cyclin D1 decreasing at the protein level (Figure 5a and 5b). These results indicate that PDCD4 does not appear to regulate mRNA expression of potential NF-κB-dependent antiapoptotic- or cell proliferation-related genes in GBM cells. Among other NF-κB target genes, MMP-9 is a major regulator of invasive malignant potential in several cancer sites (34), and angiogenesis factor VEGF promotes invasion and metastasis of cancer cells (35). To determine whether PDCD4 inhibits NF-κB-regulated genes known to be involved in invasion such as MMP-9 and VEGF, we measured endogenous levels of MMP-9 and VEGF mRNA by qRT–PCR in U251 and LN229 cells. PDCD4 significantly inhibited the MMP-9 mRNA expression in U251 cells by >40% compared with vector control cells. VEGF mRNA level was also decreased >40% by PDCD4 in U251 cells. In LN229 cells, PDCD4 produced a statistically significant decrease only in MMP-9 mRNA expression (Figure 5c). PDCD4 significantly inhibited MMP-9 protein levels in both U251 and LN229 cells and VEGF protein in U251 cells (Figure 5d and e). Having observed that translation mutants of PDCD4 bind p65 and reduce its nuclear level (Figures 3 and 4), we would expect Pdcd4 mutants to inhibit transcription of NF-κB target genes such as MMP-9 and VEGF in a translation-independent manner. Figure 5f shows that transfection of single or double mutants of Pdcd4 into U251 cells also inhibits MMP-9 and VEGF mRNA expression compared with vector control. This result suggests that PDCD4’s suppression of NF-κB target genes such as MMP-9 and VEGF can be attributed to a translation-independent mechanism. To investigate whether PDCD4 inhibits transcriptional activity of MMP-9 and VEGF by suppressing the NF-κB p65 binding activity of NF-kB on the endogenous target gene promoters, we conducted a ChIP with p65 antibody in U251 vector control and PDCD4-overexpressing cells. As seen in Figure 5g, p65 binds to endogenous MMP-9, VEGF, cyclin D1 and CIAP-1 gene promoters with substantially greater binding seen for MMP-9 and VEGF promoters. In wild-type (WT) PDCD4-overexpressing cells, p65 binding was reduced to background levels for all four gene promoters. Like WT, translation mutants of PDCD4 reduced the binding of p65 to background levels for all four genes (Figure 5g). The ChIP assay thus supports a functional role for PDCD4 in inhibiting NF-κB/p65-dependent transcription of the target genes MMP-9 and VEGF independently of PDCD4-regulated translation. To ask whether the decreased expression of these invasion-inducing proteins is biologically significant, we performed a Matrigel invasion assay in GBM cells. The results showed that PDCD4 significantly suppressed cell invasion in both GBM lines (Figure 5h and i). These observations suggest that PDCD4 inhibits GBM cell invasion at least in part by suppressing expression of NF-κB target genes such as MMP-9 and VEGF. The lack of PDCD4 regulation of CIAP-1 and cyclin D1 mRNA levels despite moderate p65 promoter binding and PDCD4 inhibition (Figure 5a and f) may suggest that these mRNA levels are controlled primarily by other Rel family proteins, by transcription factors other than NF-κB, or posttranscriptionally.
Fig. 5.
PDCD4 inhibits the mRNA expression of putative NF-κB target genes involved in invasion but not those involved in apoptosis or cell proliferation. (a) Antiapoptotic genes (CIAP-1, CIAP-2, XIAP and BcLXL) and cell proliferation-associated genes (cyclin D1 and cyclin E) mRNA levels were analyzed by qRT–PCR. Expression levels were normalized to GAPDH RNA expression. Results were from three independent experiments plotted as fold expression relative to vector control arbitrarily set at 1.0. (b) Protein expression of antiapoptotic genes and proliferation-associated genes. Expression level of antiapoptotic genes (CIAP-1, CIAP-2, XIAP and BcLXL) and cell proliferation genes (cyclin D1 and cyclin E) were determined by western blot analysis. (c) Total RNA extract from U251 and LN229 cells and MMP-9 and VEGF mRNA expression levels were determined by qRT–PCR analysis. Error bars represent mean ± SE from three independent experiments; *P < 0.05; **P < 0.01. (d) Expression levels of MMP-9 and VEGF were analyzed by western blotting. Expression level of β-actin served as loading control. (e) Densitometry of VEGF and MMP-9 protein level in U251 (left panel) and LN229 (right panel) cells. (f) U251 cells (1×107) were seeded in 100mm plates and cotransiently transfected with PDCD4 WT or mutant (PDCD4 D418, PDCD4 D253A, D418A) and HA-p65 plasmid using FuGene HD transfection reagent for 24h. Cells were collected to detect MMP-9 and VEGF mRNA expression by qRT–PCR. (g) U251 vector control and WT or translation mutant PDCD4-overexpressing (1×107) cells were seeded in 100mm dishes, allowed to grow for 24h and processed for ChIP assay using anti-p65 antibody. Immunoprecipitated chromatin was analyzed by quantitative PCR using specific primers for CIAP-1, Cyclin D1, VEGF and MMP-9 promoters. All values are shown as the mean ± SEM of p65 bound as percent of input DNA for three independent experiments. (h) U251 and LN229 cells were allowed to invade in a collagen-coated transwell migration chamber; invasive cells were fixed, stained with crystal violet and optical density was measured. (i) Average number of invaded cells as percent in U251 (left panel) and LN229 (right panel) cells. Error bars represent mean ± SE from three independent experiments; *P < 0.05.
Discussion
This inquiry into the regulation of NF-κB-dependent transcription by the tumor suppressor PDCD4 has revealed an unexpected activity of PDCD4 and an unexpected mechanism of action. Unlike the finding for immune cells, PDCD4 inhibits NF-κB-dependent transcription in two GBM cell lines. Instead of regulating translation of NF-κB proteins, PDCD4 interacts directly or indirectly with one of them. As observed for HT29 colon cancer cells (29), PDCD4 inhibits NF-κB transactivation, but unlike in HT29 cells, PDCD4 does not inhibit the expression or activation of IKKα/β kinase. Instead, PDCD4 substantially suppresses the localization of p65 but not p50, c-Rel, RelB or p52 in the nucleus. This inhibition of nuclear transport occurs through interaction of PDCD4 with p65 but minimally with other NF-κB proteins as seen through co-immunoprecipitation using antibody to either PDCD4 or tagged p65. Confocal microscopy places the merged PDCD4-bound p65 outside the nucleus in the perinuclear region. Two invasion-inducing genes often regulated by NF-κB namely MMP-9 and VEGF show PDCD4-reduced mRNA expression. This activity of PDCD4 to inhibit p65-dependent transcription occurs independently of translation inhibition as translationally inactivated PDCD4 mutants bind to p65 and suppress p65 nuclear levels and target gene expression. PDCD4 also inhibits invasion in GBM cell lines as it does in other human cancer cell lines (24). The PDCD4-regulated availability of nuclear p65 appears to be limiting for mRNA expression of invasion-inducing NF-κB target genes MMP-9 and VEGF but not for cyclin D1 and CIAP-1. The latter two, however, are targeted at the protein expression level and their downregulation might therefore contribute, along with that of MMP-9 and VEGF, to the tumor suppressing activity of PDCD4.
PDCD4 regulates mRNA expression of several reported target genes
Among the genes whose mRNA expression is regulated by PDCD4 are p53-regulated cell cycle inhibitor p21 that is upregulated by PDCD4 knockdown (36), and several PDCD4-downregulated mRNAs including invasion-associated plasminogen activator receptor uPAR (23), a protein kinase upstream of c-Jun N-terminal kinase MAP4K/HPK1 (25), endothelial cell carbonic anhydrase (37), and hypoxia and invasion-associated lysyl oxidase (24). With the exception of PDCD4-altered p21 transcription, the responsible transcription factors have not been identified although Sp1 and Sp3 appear to be candidates for regulating uPAR (23). Although translational regulation by PDCD4 of transcription factors or their activators has been proposed to account for altered mRNA expression, such a mechanism has not been demonstrated for most of the above PDCD4 targets. PDCD4 regulates translation of two transcription factors, namely p53 and c-myb (20,21). Wedeken and coworkers have demonstrated that PDCD4 knockdown stimulated translation of p53 (21) and consequently stimulated transcription of p53 target genes such as p21 in HeLa cells (36).
There is a precedent for PDCD4 working through direct binding to a transcription factor
Although PDCD4 is thought to work primarily by inhibiting translation initiation in an mRNA-selective manner, there is a precedent for direct binding to a transcription factor. PDCD4 directly interacts with the helix-loop-helix DNA-binding domain of Twist1 to inhibit its binding to the promoter of Y-box-binding protein (YB-1) to consequently downregulate YB-1 mRNA expression in prostate cancer cells (38). Twist1 and target YB-1 appear to have oncogenic activity in prostate cancer cells and tissues. In the case of Twist1, unlike p65, the functionally significant binding to PDCD4 occurs in the nucleus. Mapping the binding domain on p65 as well as the binding domain on PDCD4 will be of interest for future studies. Of note is the observation that the PDCD4-binding domain on p65 appears not to be shared by c-Rel or RelB. Identification of the p65-binding domain associated with cytoplasmic retention or sequestration may provide insight into the regulation of NF-κB nuclear translocation, whether import or export. The failure of PDCD4 to inhibit translocation of p50 to the nucleus suggests that dimerization of p50 with p65 is not necessary for p50 translocation. Moreover, the lack of PDCD4 binding to p50 is consistent with the uninhibited nuclear localization of p50 if such interaction with NF-κB proteins produces sequestration in the cytoplasm.
The inhibition by PDCD4 of NF-κB-dependent gene regulation may contribute to the mechanism by which PDCD4 inhibits invasion
A number of genes that regulate apoptosis, cell proliferation, invasion or angiogenesis are known to be NF-κB target genes in one or more models or cancer sites (32). Of those we analyzed in PDCD4-overexpressing cells, only MMP-9 and VEGF showed inhibited mRNA expression (although PDCD4 inhibited the binding of p65 to endogenous promoters for CIAP-1 and cyclin D1 as well as MMP-9 and VEGF). VEGF and MMP-9 are known to be important in invasion in the case of many human cancer cells (39). Both MMP-9 and VEGF are overexpressed in GBM tissues that also show overactivated NF-κB (40). mRNA and protein levels of both are downregulated by PDCD4 in GBM cells. Transcription of genes such as cyclin D1 having NF-κB-binding sites, but not attenuated by PDCD4, may be predominantly regulated by other transcription factors as described by others (41) or by other NF-κB proteins. Both cyclin D1 and CIAP-1 show downregulation at the protein but not mRNA level when PDCD4 is overexpressed, thus might function in PDCD4-mediated tumor suppression. Apoptosis inhibitor XIAP, but not CIAP-1, has been found by Liwak et al. (22) to show PDCD4-regulated translation via internal ribosome entry sites in HEK 293 cells. Cyclin D1 is translationally regulated in some models (42). It is noteworthy that other tumor suppressors among them Fbw7 (43) and p53 have also been reported to target NF-κB (44).
Therapeutic implications for targeting NF-κB
Because NF-κB functions as an oncogenic driver in many human cancer sites, it is an attractive target for cancer prevention or treatment. However, as NF-κB also regulates normal immune response, targeting it can produce immunotoxicity. A number of NF-κB-targeting drugs have been used in clinical trials, with sometimes disappointing results (45). Thus, novel ways to target NF-κB are sought. Stabilizing or mimicking PDCD4 or its binding to p65 to sequester p65 in the cytoplasm may present a useful alternative.
Supplementary material
Supplementary Figures S1 and S2 can be found at http://carcin.oxfordjournals.org/
Funding
National Cancer Institute , National Institute of Health Intramural Funding (ZIA BC 010026).
Supplementary Material
Acknowledgements
We thank Ms Kim Peifley for technical assistance with confocal microscopy and Drs Peter Johnson and Yinling Hu for comments on the manuscript.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- ChIP
chromatin immunoprecipitation
- GBM
glioblastoma
- IB
immunoblotting
- IP
immunoprecipitation
- MMP
matrix metalloproteinase
- qRT–PCR
quantitative reverse transcription–polymerase chain reaction
- VEGF
vascular endothelial growth factor
- WT
wild-type.
References
- 1. Shibahara K., et al. (1995). Isolation of a novel mouse gene MA-3 that is induced upon programmed cell death. Gene, 166, 297–301 [DOI] [PubMed] [Google Scholar]
- 2. Cmarik J.L., et al. (1999). Differentially expressed protein Pdcd4 inhibits tumor promoter-induced neoplastic transformation. Proc. Natl Acad. Sci. USA, 96, 14037–14042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hilliard A., et al. (2006). Translational regulation of autoimmune inflammation and lymphoma genesis by programmed cell death 4. J. Immunol., 177, 8095–8102 [DOI] [PubMed] [Google Scholar]
- 4. Jansen A.P., et al. (2005). Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res., 65, 6034–6041 [DOI] [PubMed] [Google Scholar]
- 5. Schmid T., et al. (2008). Translation inhibitor Pdcd4 is targeted for degradation during tumor promotion. Cancer Res., 68, 1254–1260 [DOI] [PubMed] [Google Scholar]
- 6. Afonja O., et al. (2004). Induction of PDCD4 tumor suppressor gene expression by RAR agonists, antiestrogen and HER-2/neu antagonist in breast cancer cells. Evidence for a role in apoptosis. Oncogene, 23, 8135–8145 [DOI] [PubMed] [Google Scholar]
- 7. Chen Y., et al. (2003). Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J. Pathol., 200, 640–646 [DOI] [PubMed] [Google Scholar]
- 8. Gao F., et al. (2007). Frequent loss of PDCD4 expression in human glioma: possible role in the tumorigenesis of glioma. Oncol. Rep., 17, 123–128 [PubMed] [Google Scholar]
- 9. Hiyoshi Y., et al. (2009). MicroRNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clin. Cancer Res., 15, 1915–1922 [DOI] [PubMed] [Google Scholar]
- 10. Mudduluru G., et al. (2007). Loss of programmed cell death 4 expression marks adenoma-carcinoma transition, correlates inversely with phosphorylated protein kinase B, and is an independent prognostic factor in resected colorectal cancer. Cancer, 110, 1697–1707 [DOI] [PubMed] [Google Scholar]
- 11. Zhang H., et al. (2006). Involvement of programmed cell death 4 in transforming growth factor-beta1-induced apoptosis in human hepatocellular carcinoma. Oncogene, 25, 6101–6112 [DOI] [PubMed] [Google Scholar]
- 12. Jin H., et al. (2006). Aerosol delivery of urocanic acid-modified chitosan/programmed cell death 4 complex regulated apoptosis, cell cycle, and angiogenesis in lungs of K-ras null mice. Mol. Cancer Ther., 5, 1041–1049 [DOI] [PubMed] [Google Scholar]
- 13. Gaur A.B., et al. (2011). Downregulation of Pdcd4 by mir-21 facilitates glioblastoma proliferation in vivo . Neuro. Oncol., 13, 580–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang H.S., et al. (2004). A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Mol. Cell. Biol., 24, 3894–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang H.S., et al. (2003). The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol. Cell. Biol., 23, 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. LaRonde-LeBlanc N., et al. (2007). Structural basis for inhibition of translation by the tumor suppressor Pdcd4. Mol. Cell. Biol., 27, 147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chang J.H., et al. (2009). Crystal structure of the eIF4A-PDCD4 complex. Proc. Natl Acad. Sci. USA, 106, 3148–3153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Loh P.G., et al. (2009). Structural basis for translational inhibition by the tumour suppressor Pdcd4. EMBO J., 28, 274–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sonenberg N., et al. (1998). The mRNA 5’ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol., 10, 268–275 [DOI] [PubMed] [Google Scholar]
- 20. Singh P., et al. (2011). Pdcd4 directly binds the coding region of c-myb mRNA and suppresses its translation. Oncogene, 30, 4864–4873 [DOI] [PubMed] [Google Scholar]
- 21. Wedeken L., et al. (2011). Tumor suppressor protein Pdcd4 inhibits translation of p53 mRNA. J. Biol. Chem., 286, 42855–42862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liwak U., et al. (2012). Tumor suppressor PDCD4 represses internal ribosome entry site-mediated translation of antiapoptotic proteins and is regulated by S6 kinase 2. Mol. Cell. Biol., 32, 1818–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leupold J.H., et al. (2007). Tumor suppressor Pdcd4 inhibits invasion/intravasation and regulates urokinase receptor (u-PAR) gene expression via Sp-transcription factors. Oncogene, 26, 4550–4562 [DOI] [PubMed] [Google Scholar]
- 24. Santhanam A.N., et al. (2010). Pdcd4 repression of lysyl oxidase inhibits hypoxia-induced breast cancer cell invasion. Oncogene, 29, 3921–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang H.S., et al. (2006). Tumorigenesis suppressor Pdcd4 down-regulates mitogen-activated protein kinase kinase kinase kinase 1 expression to suppress colon carcinoma cell invasion. Mol. Cell. Biol., 26, 1297–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang H.S., et al. (2001). A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation. Oncogene, 20, 669–676 [DOI] [PubMed] [Google Scholar]
- 27. Bitomsky N., et al. (2004). Transformation suppressor protein Pdcd4 interferes with JNK-mediated phosphorylation of c-Jun and recruitment of the coactivator p300 by c-Jun. Oncogene, 23, 7484–7493 [DOI] [PubMed] [Google Scholar]
- 28. Sheedy F.J., et al. (2010). Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol., 11, 141–147 [DOI] [PubMed] [Google Scholar]
- 29. Guo X., et al. (2011). AKT Activation by Pdcd4 Knockdown Up-Regulates Cyclin D1 Expression and Promotes Cell Proliferation. Genes Cancer, 2, 818–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dai C., et al. (2011). Inhibition of proinflammatory RANTES expression by TGF-beta1 is mediated by glycogen synthase kinase-3beta-dependent beta-catenin signaling. J. Biol. Chem., 286, 7052–7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Adli M., et al. (2010). IKKalpha and IKKbeta each function to regulate NF-kappaB activation in the TNF-induced/canonical pathway. PLoS One, 5, e9428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hayden M.S., et al. (2004). Signaling to NF-kappaB. Genes Dev., 18, 2195–2224 [DOI] [PubMed] [Google Scholar]
- 33. Ghosh S., et al. (2002). Missing pieces in the NF-kappaB puzzle. Cell, 109 (suppl.), S81–S96 [DOI] [PubMed] [Google Scholar]
- 34. Nyberg P., et al. (2002). MMP-9 activation by tumor trypsin-2 enhances in vivo invasion of human tongue carcinoma cells. J. Dent. Res., 81, 831–835 [DOI] [PubMed] [Google Scholar]
- 35. Su J.L., et al. (2006). The VEGF-C/Flt-4 axis promotes invasion and metastasis of cancer cells. Cancer Cell, 9, 209–223 [DOI] [PubMed] [Google Scholar]
- 36. Bitomsky N., et al. (2008). siRNA-mediated knockdown of Pdcd4 expression causes upregulation of p21(Waf1/Cip1) expression. Oncogene, 27, 4820–4829 [DOI] [PubMed] [Google Scholar]
- 37. Lankat-Buttgereit B., et al. (2004). Pdcd4 inhibits growth of tumor cells by suppression of carbonic anhydrase type II. Mol. Cell. Endocrinol., 214, 149–153 [DOI] [PubMed] [Google Scholar]
- 38. Shiota M., et al. (2009). Programmed cell death protein 4 down-regulates Y-box binding protein-1 expression via a direct interaction with Twist1 to suppress cancer cell growth. Cancer Res., 69, 3148–3156 [DOI] [PubMed] [Google Scholar]
- 39. Liu Y., et al. (2011). Expression of VEGF and MMP-9 and MRI imaging changes in cerebral glioma. Oncol. Lett., 2, 1171–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zanotto-Filho A., et al. (2011). NFκB inhibitors induce cell death in glioblastomas. Biochem. Pharmacol., 81, 412–424 [DOI] [PubMed] [Google Scholar]
- 41. Zhang H.S., et al. (2012). PAX2 protein induces expression of cyclin D1 through activating AP-1 protein and promotes proliferation of colon cancer cells. J. Biol. Chem., 287, 44164–44172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rosenwald I.B., et al. (1993). Elevated levels of cyclin D1 protein in response to increased expression of eukaryotic initiation factor 4E. Mol. Cell. Biol., 13, 7358–7363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arabi A., et al. (2012). Proteomic screen reveals Fbw7 as a modulator of the NF-κB pathway. Nat. Commun., 3, 976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ghose J., et al. (2011). Regulation of miR-146a by RelA/NFkB and p53 in STHdh(Q111)/Hdh(Q111) cells, a cell model of Huntington’s disease. PLoS One, 6, e23837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Staudt L.M. (2010). Oncogenic activation of NF-kappaB. Cold Spring Harb. Perspect. Biol., 2, a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.