Abstract
Pancreatic ductal adenocarcinomas (PDA) are extremely aggressive cancers and currently available therapies are only minimally effective in treating this disease. Tackling this devastating cancer has been a major challenge to the scientific and medical communities, in part due to its intense therapeutic resistance. One of the aspects of this tumor that contributes to its aggressive behavior is its altered cellular metabolism. Indeed, PDA cells seem to possess the ability to adapt their metabolism to the particular environment to which they are exposed, including utilizing diverse fuel sources depending on their availability. Moreover, PDA tumors are efficient at recycling various metabolic substrates through activation of different salvage pathways such as autophagy and macropinocytosis. Together, these diverse metabolic adaptations allow PDA cells to survive and thrive in harsh environments that may lack nutrients and oxygen. Not surprisingly, given its central role in the pathogenesis of this tumor, oncogenic Kras plays a critical role in much of the metabolic reprogramming seen in PDA. In this review, we discuss the metabolic landscape of PDA tumors, including the molecular underpinnings of the key regulatory nodes, and describe how such pathways can be exploited for future diagnostic and therapeutic approaches
Introduction
Pancreatic ductal adenocarcinomas epidemiology
With 45 220 new cases estimated for 2013 in the USA, pancreatic cancer is the 12th most common cancer, representing 2.7% of all the new cancer diagnoses in the USA (1,2). Despite not being one of the most prevalent cancers, it is by far one of the deadliest, with a 5 year survival of ~7% (2). It ranks fourth in cancer mortality and accounts for ~7% of all cancer-related deaths (1–3). With <10% of pancreatic cancers being diagnosed as localized disease (confined to primary site) (2), the majority of patients are not amenable to potentially curative surgical resection.
The normal pancreas is made up of two classes of cells: endocrine (hormone secreting) and exocrine (digestive enzyme producing). Depending on the cell of origin, pancreatic cancers can also be classified as endocrine or exocrine tumors. Roughly 90% of all pancreatic cancers are pancreatic ductal adenocarcinomas (PDA), an exocrine pancreatic tumor that resembles the cells lining the pancreatic duct (4,5). This tumor type will be the focus of this review.
Key characteristics of PDA
PDA have several defining features that influence its aggressive biology and resistance to multiple therapeutic modalities. These tumors are characterized by a distinct and exuberant stromal reaction (desmoplasia) (6,7), hypovascularization (8–11), genomic complexity (12–14) and an altered metabolism (15–17). This metabolic rewiring in PDA is critical to the growth of the tumor and is the subject of this review.
As mentioned above, PDA is characterized by a desmoplastic reaction, which often forms the bulk of the tumor mass (4,18). The PDA stroma is heterogeneous and is comprised of multiple cell types including pancreatic stellate cells, various leukocytes and endothelial cells, as well as a complex extracellular matrix (18–20). This dense fibrotic tissue, together with the poor vascularization limits access to the circulation, which has been shown to impair drug delivery (10,11,21). It also creates an hypoxic tumor microenvironment, known to negatively influence the response to radiotherapy in many cancer types (8,9,22). In addition, the PDA microenvironment is highly immunosuppressive (23–27), which has implications in immunotherapy for this disease (24).
The majority of PDA have Kras mutations, with 90% possessing activating mutations in this oncogene (28–31). In PDA, Kras is most commonly mutated at the G12 residue (G12D and G12V). This mutation affects the interaction site with GTPase activating proteins (GAPs), and therefore mutant proteins are guanosine triphosphate hydrolysis impaired, resulting in a constitutively active (guanosine triphosphate-bound) form of Kras. Like the other members of the RAS family of GTPases, Kras acts as a molecular switch, transducing signals from membrane-bound receptors to signaling pathways in the cell. By activating central signaling pathways in the cell such as mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) or PI3K/AKT pathways, Kras activity results in increased proliferation, cell growth, decreased apoptosis and increased invasiveness phenotypes (30,32,33). The Kras oncogene is considered a PDA driver mutation and, accordingly, it can be found mutated in early stages of tumor progression. Although rarely detectable in normal pancreas and chronic pancreatitis, the frequency of Kras mutation increases with the grade of the neoplasia to being nearly universal in advanced PDA (31). Consequently, its expression in the pancreas can drive the initiation of the disease in various mouse models (15,34–39).
In addition to Kras mutations, inactivating mutations or deletions are frequently seen in tumor suppressor genes, including p53, CDKN2A (INK4a/Arf) and Smad4 (21,40,41). Unfortunately, these recurrent genetic events have not provided any tractable therapeutic targets. Although there has been a significant effort to identify novel mutations in PDA tumors, such studies have not resulted in the identification of many recurrent driver events (42–44). However, it is now recognized that the PDA genomic landscape is highly complex with a particularly high rate of deletions and fold-back inversions (13), as well as frequent amplifications, deletions and complex rearrangements (12). In addition to a large amount of intertumoral heterogeneity, there is also significant intratumoral heterogeneity that may have implications on the intense therapeutic resistance (45).
Tumor metabolism
Normal, quiescent cells typically metabolize glucose to pyruvate, which can then enter the tricarboxylic acid (TCA) cycle where energy in the form of adenosine triphosphate (ATP) is efficiently produced by oxidative phosphorylation (46). Dividing cells, however, rely heavily on glycolysis, which allows for the production of energy along with building blocks required to generate a daughter cell (47). Similarly, metabolism in cancer cells is also altered to facilitate proliferation. An important distinction is that the metabolic networks in cancer cells are rewired to be independent of extracellular controls (46). In fact, the altered metabolism of tumor cells is now considered a hallmark of cancer (48). One of the main features of the metabolism of cancer cells is the emphasis on anabolic reactions required for de novo synthesis of proteins, nucleic acids and lipids (47,49,50). In order to fuel these processes, cancer cells may differ from normal cells in both the energy sources they utilize and how these fuel sources are metabolized (51–53). For example many cancer cells have developed a reliance on the amino acid glutamine in addition to glucose to help meet their biosynthetic needs (54,55). In this review, we will focus on the altered metabolism found in PDA, in particular those adaptations that are critical for the growth and maintenance of this aggressive tumor (Figure 1).
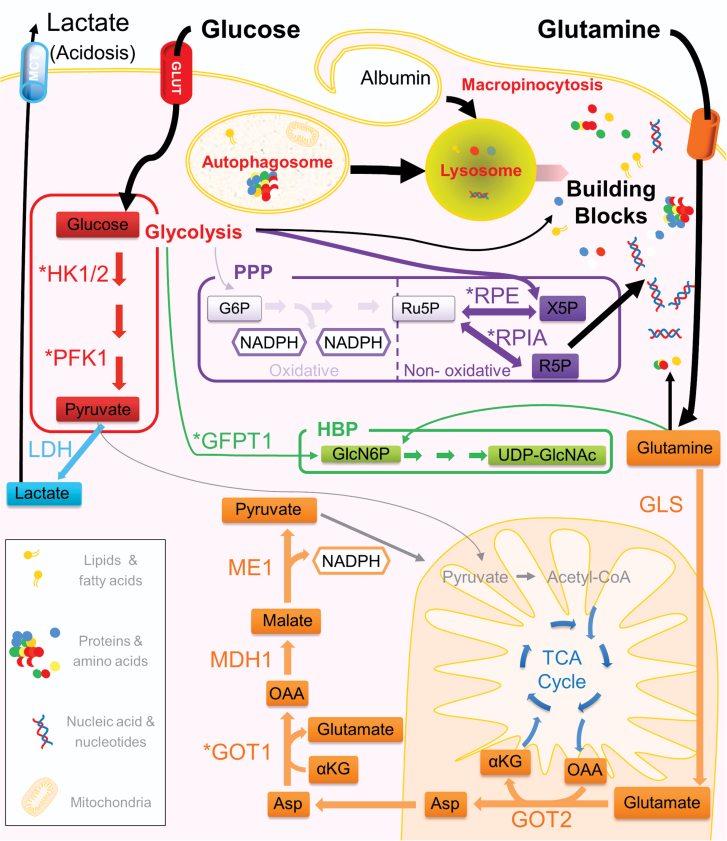
Fig. 1.
A typical PDA cell is depicted showing schematics of representative metabolic pathways that are altered in the disease in response to oncogenic Kras. Enzymes whose expression is increased by Kras are indicated with an asterisk. Increased glucose uptake fuels glycolysis (leading to increased lactate production), anabolic pathways such as the non-oxidative arm of the PPP (producing ribose for nucleotide biosynthesis) and the HBP (producing precursors for glycosylation). Glutamine is a key metabolite that is utilized to fuel the TCA cycle and maintains redox homeostasis in PDA through a novel pathway (shown in orange) that leads to NADPH production. PDA are efficient metabolic scavengers and use autophagy (intracellular) and macropinocytosis (extracellular) to provide metabolic substrates through cargo degradation via the lysosome. Lipids are also taken up extracellularly to provide fatty acids. HK1/2, hexokinase 1 and 2; PFK1, phosphofructokinase 1; ME1, malic enzyme; GOT1/GOT2, aspartate aminotransferase 1 and 2; MDH1, malate dehydrogenase; GLS, glutaminase; GFPT1, glutamine fructose-6-phosphate amidotransferase; RPIA, ribose 5-phosphate isomerase A; RPE, ribulose-5-phosphate-3-epimerase; Asp, aspartate; OAA, oxaloacetate; G6P, glucose 6-phosphate; Ru5P, ribulose 5-phosphate; R5P, ribose 5-phosphate; X5P, xylulose 5-phosphate. LDH, lactate dehydrogenase; αKG, alpha keto-glutarate; GlcN6P, glucosamine-6-phosphate; UPD-GlcNAc, uridine diphosphate N-acetylglucosamine; MCT, onocarboxylate transporter; GLUT, glucose transporter.
Kras-driven metabolic alterations in PDA
In addition to its well-studied roles in cancer cell proliferation, survival or metastasis, oncogenic Kras has been recently shown to have a key role in multiple aspects of PDA metabolism. In fact, it appears to have a prominent role in the metabolic rewiring of these tumors and this may be one of its critical roles in PDA pathogenesis.
Scavenging/recycling
Macroautophagy (hereafter referred to as autophagy) is a catabolic process that consists of the self-degradation of cellular organelles and molecular complexes (for review see ref. 56 ). In a cell, damaged or unnecessary organelles, proteins or protein aggregates are sequestered in a double membrane structure known as an autophagosome. The autophagosome eventually fuses with a lysosome creating an autolysosome, leading to the degradation and release of its contents. The degraded proteins (amino acids) and organelles (amino acids, lipids and nucleosides) are recycled back into the cytoplasm and used in the biosynthesis of proteins or nucleic acids, or for other anabolic or bioenergetic reactions. Because of its biological importance, the process is tightly controlled, with each step of autophagic progression being regulated by different complexes of proteins. Both the autophagic process and its regulation have been reviewed extensively elsewhere (57–62). There are three types of autophagy; macroautophagy, microautophagy and chaperone-mediated autophagy, which differ in terms of how cargo gets to the lysosome (63). Here, we will focus on macroautophagy.
Autophagy can act as a quality control mechanism in the cell by clearing damaged structures, including misfolded proteins, protein aggregates or dysfunctional organelles. Therefore, it is typically present at low levels in various tissues as a homeostatic mechanism. Different stimuli can trigger autophagy, thereby increasing it above baseline. One of the most well-studied and potent autophagy stimuli is starvation (lack of nutrients), which is regulated by the mammalian target of rapamycin (mTOR) complex (64). Other cellular stresses including protein damage, reactive oxygen species (ROS) or DNA damage are also known triggers of autophagy (58,65,66).
In cancer progression, autophagy has important but opposing roles (67,68). It has been demonstrated that autophagy can be both pro- and antitumorigenic. It is antitumorigenic due to the quality control function that it exerts: removing damaged organelles and protein aggregates, thereby mitigating oxidative stress, tissue damage and genomic instability; all protumorigenic factors that can promote tumor initiation. In established tumors, however, autophagy can fuel cellular proliferation in the nutrient-poor hypoxic regions in the tumor (68). Consistent with this, autophagy is found to be upregulated in tumors, particularly in nutrient-poor regions (69). It supports tumor cell survival via recycling cargo and generating substrates such as amino acids, fatty acids, nucleotides and ATP (70). Adding to this already complex role in cancer, autophagy is also linked with therapeutic resistance (71,72). In breast cancer for example autophagy upregulation increases resistance to hormonal therapy, playing a crucial role in the establishment of resistant tumors (73). Similarly, in lymphoma models, autophagy inhibition synergizes with cytotoxic chemotherapy (74). A recent study has shown that blocking autophagy in PDA cells via expression of a specific micro RNA enhances radiation response (75). However, it is important to note that there are some situations where inhibition of autophagy has been reported to mitigate the effect of a particular therapeutic agent in certain cancer types (71,76).
In Kras-driven tumors, autophagy has been shown to be required for tumor growth (77,78). Inhibiting autophagy also impairs Kras transformation of non-malignant breast cells (79). Similarly, transformation of mammary epithelial cells or immortalized mouse kidney cells by Hras also requires autophagy (80,81). Whereas in some cancer types, autophagy seems to be triggered as a reaction to various stressors (DNA damage, ROS, protein damage, lack of nutrients, etc.); in PDA, basal autophagy levels are unusually high (77). This appears to be cell autonomous, as high levels of autophagy are also observed in cell culture under nutrient-replete conditions. In PDA models, inhibition of autophagy both genetically (ATG5 depletion) or pharmacologically (chloroquine treatment) results in the inhibition of tumor growth in vitro and in vivo (77). The elevated basal autophagy appears to provide PDA cells with additional nutrients that can fuel the TCA cycle. Indeed, inhibition of autophagy resulted in decreased ATP production and impaired oxidative phosphorylation in PDA cells. Moreover, the impact of autophagy inhibition on PDA growth could be attenuated by adding back the metabolite pyruvate. Thus, autophagy appears to be a critical component of PDA metabolism. Similarly, autophagy inhibition in other tumor types has also been shown to impair mitochondrial metabolism (78) and in some contexts glycolysis (80,82).
In addition to utilizing intracellular substrates to recycle metabolites, PDA cells have the ability to take up and degrade extracellular macromolecules to fuel metabolism. Macropinocytosis is a form of endocytosis used by cells to engulf large portions of the extracellular space (83–85). Cells can extend their plasma membrane, folding it back onto the cell and creating a barrier around a portion of the extracellular fluid. This large, irregular, double membrane vesicle is known as a macropinosome. The macropinosome is internalized along with the extracellular fluid and associated molecules (proteins, bacteria, virus and even apoptotic bodies) (83–85) and then undergoes a step of maturation acquiring characteristics of an early endosome. It can then fuse with the lysosome, degrading its contents or, alternatively, it can be recycled back to the cell membrane, releasing the contents to the extracellular space.
It has been known for several decades that oncogenic Ras can promote macropinocytosis (86). Recently, PDA cell lines and tumors that possess activating Kras mutations were reported to show high levels of macropinocytosis (87). Macropinocytosis was shown to promote the uptake of extracellular albumin, which was then degraded in the lysosome. Interestingly, the amino acids that were produced by this degradation were shown to fuel cellular metabolism. In this elegant experiment, carbon-13 yeast protein was included in growth media. Kras-transformed cells consumed the protein and the liberated carbon-13 labeled amino acids were metabolized in the TCA cycle. Importantly, pharmacological inhibition of macropinocytosis showed significant antitumor responses in PDA xenografts (87).
In addition to recycling amino acids, Ras-transformed cells also appear to scavenge extracellular lipids as their primary source of fatty acids (88). Such lipids are hydrolyzed to form fatty acids and glycerol. The fatty acids can be used to fuel the TCA cycle, whereas the glycerol, the other product of lipid hydrolysis, can be converted into dihydroxyacetone, an intermediate of glycolysis (89). Alternatively, such fatty acids, which constitute a key component of the cellular membrane, can be used directly to make membranes in daughter cells (90).
Under normal conditions, the biosynthesis of fatty acids uses pyruvate, a product of glycolysis, which is converted to acetyl-CoA—a major precursor of fatty acids. These biosynthetic reactions require oxygen consumption and reduced nicotinamide adenine dinucleotide phosphate (NADPH). In cancer cells, scavenging substrates from the extracellular media to meet their fatty acid needs allows for the conservation of other rate-limiting molecules, such as NADPH, to be used for other functions such as redox balance (discussed below). Although this fatty acid scavenging appears to be a property of Ras-transformed cells, it has not yet been demonstrated specifically in PDA cells.
Together, the data from these studies illustrate that PDA cells are efficient in recycling and scavenging molecules to promote their own survival. Kras mutations appear to drive these mechanisms in the tumor, allowing tumor cells to adapt to environments where the access to nutrients can be diminished. Additionally, scavenging may allow for the conservation of energy and biomass so resources can be devoted to critical and rate-limiting processes such as NADPH biosynthesis, which will be discussed below. Importantly, these pathways have the potential for therapeutic targeting as chloroquine and its derivative hydroxychloroquine can inhibit autophagy through their interference with lysosomal acidification (71). Since autophagy and macropinocytosis converge at the level of the lysosome, these drugs would potentially attenuate both processes. Indeed, there are many clinical trials in various cancers, including PDA, incorporating hydroxychloroquine into the treatment regimen (http://clinicaltrials.gov/). Additionally, as macropinocytosis was shown to take up albumin in PDA, this could be used to help deliver drugs to the tumor. It is tempting to speculate that the recent success of the addition of nab-paclitaxel (an albumin bound paclitaxel) to gemcitabine in metastatic pancreatic cancer patients (91) was in part due to the increased delivery to the tumor by macropinocytosis. Lastly, as autophagy is often a reactive survival mechanism to metabolic stress, inhibiting this process using hydroxychloroquine, may increase the efficacy of inhibitors to other metabolic pathways.
Anabolic glucose metabolism
Like many other cancer types, PDA exhibit an elevated capacity for glucose uptake (15). Similar to the scavenging characteristics described previously, the metabolic changes involving glucose are also in part driven by oncogenic Kras. For example glycolysis is enhanced downstream of Kras in different and complementary ways. The glucose transporter GLUT1 is transcriptionally upregulated in response to Kras mutations in both PDA and other tumor types leading to increased glucose uptake (15,92). Additionally, key glycolysis enzymes such as HK1, HK2 and PFK1 are transcriptionally upregulated downstream of Kras activation (15,93,94). One of the fates of the glycolysis-derived pyruvate is its conversion to lactate by the enzyme lactate dehydrogenase. Oncogenic Kras has been shown to increase the expression of lactate dehydrogenase A (LDHA) through increasing its transcription (15,95). Additionally, PDA cells can upregulate LDHA levels and activity through posttranslational modification of the enzyme. Acetylation of LDHA at the lysine 5 residue reduces its activity and targets LDHA for lysosomal degradation. PDA cells and primary tumors were shown to have reduced levels of this particular acetylation, resulting in greater LDHA activity (96). Interestingly, a recent study suggested that lactate can be used as alternative fuel by certain PDA cells, thereby promoting proliferation (9). Given the elevated glycolysis in PDA, leading to increased lactate production, the ability to utilize lactate could provide an additional advantage to PDA cells. Indeed, the inhibition of glycolysis through suppressing LDHA expression by RNA interference decreased the growth of PDA cells (96).
Using a genetically engineered mouse model of PDA where KrasG12D expression is induced in the pancreas by feeding mice doxycycline, it was shown that oncogenic Kras is required for tumor maintenance, due in part to a Kras-specific rewiring of anabolic glucose metabolism (15). The metabolic rewiring by Kras is complex, and PDA metabolic pathways are divergent from normal cells in unique ways. In particular, the elevated glycolytic flux is shunted to various anabolic pathways. The pentose phosphate pathway (PPP) is a side branch of glycolysis that generates five carbon sugars (ribose-5-phosphate) from six carbon sugars, to be used in nucleotide biosynthesis. In parallel, it generates reducing equivalents in the form of NADPH for use in redox control and biosynthesis of different molecules such as fatty acids (47,97). The PPP can be visualized as having an oxidative and a non-oxidative arm. The oxidation steps use glucose-6-phosphate to generate two reducing equivalents in the form of NADPH. The non-oxidative reactions of the PPP are primarily used to produce ribose-5-phosphate. The non-oxidative reactions are also important to generate six carbon sugars from five carbon sugars, which can be used to support glycolysis. The oxidative and non-oxidative arm of PPP can be decoupled, and cells can regulate the relative contribution and output of the two arms of the PPP according to their needs (requiring more NADPH than ribose for example) (97).
In PDA, the PPP is important for tumor maintenance, again, as it generates the ribose moiety of DNA used to duplicate the genome during the generation of daughter cells. Unexpectedly, oncogenic Kras leads to an increase in flux specifically through the non-oxidative arm of PPP, resulting in the generation of more ribose 5-phosphate, which can be utilized for DNA/RNA biosynthesis. This occurs independently of the NADPH-generating oxidative arm. Kras activation leads to increased transcription of two non-oxidative PPP enzymes, ribose 5-phosphate isomerase A and ribulose-5-phosphate-3-epimerase, which lead to increased flux through the non-oxidative arm (15). Importantly, inhibition of this pathway through suppressing expression of either ribose 5-phosphate isomerase A or ribulose-5-phosphate-3-epimerase results in decreased PDA growth both in vitro and in vivo.
Another glucose-dependent pathway that is upregulated by oncogenic Kras in PDA is the hexosamine biosynthesis pathway (HBP). This pathway leads to production of uridine diphosphate–N-acetylglucosamine and other nucleotide hexosamines, which are the major substrates for glycosylation of proteins and lipids, including many cytoplasmic and nuclear proteins on their serine or threonine residues (98). The HBP uses glucose and glutamine to generate uridine diphosphate–N-acetylglucosamine, acting as a bridge between glycolysis and glutaminolysis. Glucose entry into the HBP is regulated by its first and rate-limiting enzyme, glutamine fructose-6-phosphate amidotransferase (GFPT1). This enzyme catalyzes the conversion of fructose-6-phosphate and glutamine to glucosamine-6-phosphate and glutamate, respectively (99). In a negative-feedback loop, the HBP final product, uridine diphosphate–N-acetylglucosamine, inhibits GFPT1 activity self-regulating this pathway (100).The HBP coordinates nutrient uptake, partially through modulating the glycosylation and membrane localization of growth factor receptors (101). Additionally, the HBP and protein glycosylation have been found to be increased in different cancers (98,102). In general, glycosylation is a protumorigenic modification that triggers cancer-related phenotypes such as motility, proliferation and angiogenesis (103,104). In PDA, oncogenic Kras increases glucose flux through the HBP by upregulating GFPT1 expression (15). Consistent with this, suppression of Kras resulted in a decrease in total cellular O-linked glycosylation in PDA cells. Inhibition of the HBP in PDA cells by RNA interference-mediated suppression of GFPT1 resulted in a decrease in clonogenic growth and xenograft growth in mice (15).
One of the interesting aspects of many of the glucose metabolic changes seen in PDA is that they appear to be driven by the activation of the Raf/MEK/extracellular signal-regulated kinase pathway by oncogenic Kras (15). Indeed, pharmacological inhibition of MEK leads to a decrease in many of the rate-limiting enzymes that are upregulated by Kras and decreases in the respective downstream metabolites. This effect on transcription is mediated by the c-Myc oncogene as a significant fraction of the metabolic gene transcripts that were upregulated by oncogenic Kras have Myc binding elements in their promoters. Functionally, RNA interference-mediated suppression of c-Myc expression resulted in decreased expression of the same metabolic enzymes regulated by Kras and the corresponding downstream metabolites (15). Although there are no clinical grade inhibitors to most of the metabolic enzymes discussed above (GFPT1, ribose 5-phosphate isomerase A, ribulose-5-phosphate-3-epimerase), MEK inhibitors are being used in clinical trials for multiple tumor types (105). Therefore, utilizing MEK inhibitors based on their ability to inhibit anabolic glucose metabolism in PDA in combination with other therapeutic agents may be a useful strategy. In fact, the available data suggest potentially attractive therapeutic combinations. For example as MEK signaling pathways were shown to control expression of key enzymes of the non-oxidative arm of the PPP (15), a significant source of ribonucleotides for de novo DNA synthesis in the cell, combining DNA damaging therapies such as radiation with MEK inhibitors, could potentially achieve a synergetic effect.
Glutamine metabolism
Glutamine is the most abundant free amino acid in the blood (106). It has been studied for its role in cancer due to the fact that it appears to be required for the growth of many tumor types (107). It is a significant source of carbon in cancer cells, supporting anabolic processes through glutaminolysis (glutamine metabolism generating α-ketoglutarate) (54). Glutamine is also a precursor of glutathione (through its conversion to glutamate by glutaminase), a major cellular antioxidant. It is also the source of amino groups for non-essential amino acids such as alanine, aspartate, serine and glycine. The nitrogen group, released in the conversion of glutamine to glutamate also feeds the synthesis of nucleotides and the HBP. For a comprehensive review on glutamine metabolism see (54,108).
As discussed above, oncogenic Kras in PDA does not impact flux through the oxidative and NADPH-generating arm of PPP (15). Additionally, glucose deprivation, while impairing PDA growth, has minimal effects on cellular redox state in PDA cells (16). Together, this suggests that PDA utilize alternative mechanisms to maintain redox balance. Indeed, glutamine withdrawal results in a significant increase in ROS in PDA cells and it was shown that glutamine metabolism is critical for redox balance in PDA (16).
Canonical metabolism of the glutamine carbon skeleton generates α-ketoglutarate in the mitochondria to fuel the TCA cycle, and this relies on glutamate dehydrogenase (GLUD1) for the conversion of glutamate to α-ketoglutarate (54). The α-ketoglutarate can be used for anapleurosis, ultimately leading to the generation of intermediates used for biomass such as nucleic acids, proteins and lipids (109–111). In contrast, PDA cell lines do not rely on GLUD1. Instead they use the aspartate transaminase (GOT2) to generate anaplerotic α-ketoglutarate in the mitochondria. This reaction simultaneously creates aspartate from oxaloacetate, which is released into the cytoplasm. Cytosolic aspartate is then acted on by the cytosolic aspartate aminotransferase (GOT1) to convert glutamine-derived aspartate back into oxaloacetate. This oxaloacetate is subsequently converted into malate by malate dehydrogenase (MDH1) and then pyruvate and NADPH by malic enzyme. Indeed, this pathway appears to be a critical source of cytosolic NADPH in PDA cells (16). Importantly, this alternative branch of glutamine metabolism is required for PDA growth as depletion of any of the key enzymes of this pathway (GOT1, MDH1 or malic enzyme) suppressed in vitro growth and xenograft growth (16). Moreover, the NADPH produced by this pathway was critical to support PDA growth via its role in redox balance, as restoration of redox state by adding N-acetylcysteine or reduced glutathione could rescue growth upon suppression of any of these key enzymes. Moreover, this alternative glutamine metabolism was shown to be Kras dependent, as oncogenic Kras drives the expression of GOT1 while repressing GLUD1 in PDA cells (16). Interestingly, GOT1 was dispensable for the proliferation of normal cells, indicating that this pathway may be therapeutically tractable. Although no bona fide GOT1 inhibitors are currently available, other key enzymes in the glutamine metabolism have known inhibitors. One example is glutaminase and its inhibitor bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES), that decreases PDA cell viability in vitro (16). As seen in this same study, a major outcome of the unique glutamine metabolism in PDA is the production of NADPH, which results in an increased ability of PDA cells to cope with oxidative damage. One could think of a combined therapy targeting glutamine metabolism (such as glutaminase inhibition) along with classical ROS generating therapies such as radiation or chemotherapy. These therapies should synergize and effectively kill PDA cells.
Although the NADPH produced by this novel pathway is critical for PDA via its role in maintaining reduced glutathione pools, NADPH has many key roles in normal and particularly in tumor cells. NADPH is required for many aspects of biosynthesis and is thought to be limiting for tumor growth. A dividing cell for example requires significant pools of NADPH for the synthesis of fatty acids when compared with a normal cell in quiescence (47,97). Additionally, DNA synthesis, and particularly the conversion of ribonucleotides to deoxyribonucleotides (via ribonucleotide reductase), requires NADPH as the electron donor, explaining in part the large quantities of NADPH required by proliferating cells (97). Although the data suggest a critical role of NADPH production in PDA redox balance, NADPH is probably needed for all of these biosynthetic processes in PDA and detailed studies assessing the utilization of NADPH under different conditions during PDA pathogenesis are needed to define its precise roles.
ROS balance
ROS are produced during cellular metabolism. They can cause oxidative damage in cells (lipid, protein and DNA oxidation—the latter is a major cause of mutations), which in normal cells may lead to cell death through apoptosis when in excess (112–114). It is important to note that ROS are not simply toxic byproducts of cellular metabolism or damaged mitochondria. In fact, ROS are also important signaling molecules that have roles in such diverse processes as inflammation, immune response, adhesion or cellular migration (115–117).
Cancer cells often have higher levels of ROS than normal counter parts, as a result of either mitochondrial damage, increased metabolic rates or elevated expression of oxidizing enzymes (118–120). Indeed, ROS have been shown to act as signaling molecules in different cancer-related cellular behaviors. ROS generated by tumor cells can promote cell motility (and ultimately metastasis) in a cell autonomous or non-autonomous way by acting in the stroma and impairing stromal mobility-inhibitory activity (115,121,122). In fact, ROS have been shown to be critical for Kras transformation (123) and growth of Kras-transformed PDA (124). For example superoxide (O2−), a byproduct of mitochondrial respiration, is a prosurvival factor in PDA as scavenging O2− inhibits cell growth (125,126). Moreover, oncogenic Kras promotes the production of superoxide by increasing levels of NADPH oxidase 2 (NOX2), an enzyme that transfers electrons from NADPH, coupling them to molecular oxygen. This activity appears to be important for PDA growth as inhibition of NOX2 in PDA cell lines impairs clonogenic growth (124). Similarly, the ROS produced by NOX4 appears to promote PDA survival as demonstrated by suppression of NOX4 expression (127).
Given the aforementioned roles of ROS in various cellular processes, it is not surprising that ROS would have both pro- and antitumorigenic effects. Indeed, this delicate balance is probably why PDA and other tumors have developed multiple mechanisms to balance the production and scavenging of these molecules. The redox state in a cell is determined by multiple factors including the balance of NADP+/NADPH to maintain pools of reduced glutathione (128). The reduced species (NADPH) allow for the maintenance of pools of reduced glutathione, which is critical for glutathione oxidation, a key pathway to reduce the levels of peroxide in the cell. As discussed above, one of the main roles of glutamine in PDA is to produce NADPH through a novel metabolic pathway to allow proper redox balance (16). Another way by which PDA can regulate the redox state is through NRF2, a transcription factor targeting genes in drug metabolism and ROS response (129,130). In the cell, NRF2 is stabilized upon different stresses. It then accumulates in the cell nucleus to promote transcription of multiple target genes involved in the ROS response including glutathione reductase, superoxide dismutase 3, NQO1 and thioredoxin (130). Recently, it was shown that the Kras oncogene, expressed at physiological levels, can upregulate NRF2, leading to decreased cellular ROS levels (131). Consistent with this, NRF2 is found overexpressed in PDA (132), and the higher availability of NRF2 in PDA could represent an increased potential of these cells to respond to ROS, thereby protecting PDA cells against their potentially detrimental effects. In addition to Kras, other oncogenes such as B-Raf and c-Myc can induce NRF2 expression to promote ROS detoxification. Importantly, NRF2 was shown to be critical for Kras-driven tumorigenisis in PDA models (131). Together, these data show that PDA have developed multiple mechanisms to control ROS levels and this has significant implications on tumor progression.
Hypoxia
As described above, areas within PDA tumors are hypoxic (8,9), which is probably a result of the hypovascular nature of the tissue. This hypoxic microenvironment has implications on its complex biology and in particular its cellular metabolism. Indeed, a recent study characterizing the hypoxic regions in PDA tumors suggested that hypoxia enhanced activity of the HBP (9).
The hypoxia inducible factor 1 alpha (HIF1α) is a central player in hypoxia response. At low oxygen concentrations in the cell, HIF1α is stabilized and translocates into the nucleus where it acts as a transcription factor triggering many hypoxic responses. The regulation of HIF1α stabilization is complex and very tightly regulated by ubiquitin-mediated proteosomal degradation (for review please see ref. 133). One of the consequences of HIF1α activation is the metabolic rewiring allowing a cell to subsist in low-oxygen environment. Namely, it leads to an increase in glycolysis by upregulating key genes such as HK, pyruvate kinase M2 and LDHA among others (133), as well as a shift to the non-oxidative arm of PPP by upregulation of transkelotases (TKT and TKTL2) expression (134). HIF1α also coordinates a decrease in the entry of glucose carbon into the TCA cycle, a beneficial adaptation under hypoxia due to the lack of oxygen, which acts as an electron acceptor during oxidative phosphorylation. One of the mechanisms by which HIF1α accomplishes this decrease in glucose flux through the TCA cycle is by increasing transcription of pyruvate dehydrogenase kinase (135). Pyruvate dehydrogenase kinase phosphorylates and inhibits pyruvate dehydrogenase, the enzyme that converts pyruvate to acetyl-CoA.
In colon cancers with Kras mutations, target genes of both HIF1α and HIF2α expression are induced downstream of oncogenic Kras (95), creating a state of ‘aerobic-hypoxia’. This is consistent with the findings that Ras-transformed cells have similar metabolic profiles as hypoxic cells (55). In PDA, HIF1α has been shown to mediate metabolic changes downstream of MUC1 (136) and interference with HIF1α function has antitumor effects on PDA cells in vitro and in vivo (137,138). Another hypoxia response induced by HIF1α activity is the activation of autophagy (139,140). As autophagy is a critical mechanism for PDA cell metabolism under normoxic conditions, it may have additional roles in the setting of hypoxia. Indeed, autophagy appears to promote tumor-initiating cells in PDA cultures under hypoxic conditions (141).
PDA tumor suppressor genes are linked to metabolism
Oncogenic Kras, a key driver of PDA pathogenesis and a regulator of PDA metabolism, exists in the context of additional genetic alterations in tumor suppressor genes that in many cases have been shown to constrain malignant progression (142). Importantly, several of these genes have links to cellular metabolism (143). Thus, the metabolic consequences of Kras mutations in PDA or even Kras-independent metabolic changes are probably modulated by the constellation of tumor suppressor gene alterations. Additionally, these tumor suppressor mutations may alter the dependency of a tumor on particular metabolic pathways.
One such example is the tumor suppressor p53, which is well known for its roles in apoptosis and growth arrest. As a transcription factor, it can promote the transcription of genes that lead to growth arrest at G1 phase, having a cell cycle ‘check-point’ function that guards cells against genotoxic insult coming from various sources such as irradiation, hypoxia or drug-induced genotoxic damage. Alternatively, it can promote apoptosis in response to certain stimuli (144–147). p53 is the most commonly mutated gene in cancer, with over half of all human tumors possessing mutations altering p53 function (144). In PDA, p53 is mutated in the majority of tumors (41), and its frequency increases with tumor progression (40,148,149). As further proof of its important role in PDA progression, deletion or mutation of p53 accelerates the development of PDA tumors in Kras-driven genetic mouse models of the disease (35,150).
p53 has been shown to regulate multiple aspects of cellular metabolism. Overall, the net effect of p53 expression appears to promote oxidative phosphorylation and attenuate glycolysis although there are certainly particular tissues and cellular contexts where this may differ. The reader is directed to several excellent reviews on this topic (151–155). Therefore, in cancers where p53 would be absent or mutated (such as the majority of PDA), one would expect to see increases in glycolysis and decreases in oxidative phosphorylation. For example p53 promotes expression of SCO2, a member of the COX-2 assembly involved in the electron transport chain that increases levels of mitochondrial respiration while decreasing glycolysis (156). Furthermore, glutamine is a major fuel source of the TCA cycle in many cancers as it can supply TCA cycle intermediates(105). p53 can upregulate glutaminase 2, which increases the conversion of glutamine to glutamate that can be ultimately used to make αKG (157,158). The net effect of this can lead to increased flux through the TCA cycle and increased oxidative phosphorylation.
There are multiple ways by which p53 can attenuate glycolysis, including promoting the inactivation of phosphoglycerate mutase (159) and repressing the glucose transporters GLUT1 and GLUT4 (160). p53 also upregulates TP53-induced glycolysis and apoptosis regulator, a fructose-2,6-bisphosphatase that lowers the intracellular concentration of fructose-2,6-bisphosphate resulting in a decrease in glycolysis and an increase in oxidative phosphorylation (161). Furthermore, p53 also influences the PPP, by binding to and inhibiting the activation of glucose-6-phosphate dehydrogenase, the first rate-limiting step enzyme of the PPP (162).
Additionally, recent data have suggested that p53 status may influence the role of autophagy in PDA progression using a PDA genetically engineered mouse model where Kras is activated concurrently with ATG5 or ATG7 loss and both copies of p53 are embryonically deleted. In the setting of combined loss of p53 and autophagy impairment, tumor formation may actually be accelerated. This is in contrast to tumors where Kras is activated and autophagy abrogated in the setting of an intact p53 locus. Here, tumor progression is completely inhibited (163). One issue not addressed in this study concerns the way p53 is mutated/lost in a physiological cancer setting. In this study, the homozygous deletion of p53 embryonically creates a situation where p53 is never present in the tissue prior to tumor development. Although these data are certainly of great interest, it would be valuable to study the role of autophagy in PDA progression in a more physiological setting, with p53 being lost as the tumor progresses. Indeed data from our group have shown that ATG5 loss in a PDA genetically engineered mouse model where p53 is lost stochastically by loss of heterozygosity significantly impairs PDA progression (unpublished data).
Other tumor suppressor genes involved in PDA also have roles in cellular metabolism. This includes the liver kinase B1 (LKB1), a serine threonine kinase that phosphorylates adenosine monophosphate-activated protein kinase (AMPK) (164). Patients with germline mutations of LKB1 (Peutz-Jeghers syndrome) have a significantly increased risk of developing PDA (165) and LKB1 haploinsufficiency cooperates with oncogenic Kras in a PDA mouse model (166). Although the metabolic consequences of LKB1 loss in PDA have not been well studied, downstream effectors of LKB1 such as AMPK have critical roles in metabolism, suggesting that PDA metabolism may be altered upon LKB1 loss. For example low energy levels result in stable AMPK phosphorylation and activation by LKB1 (164). In turn, AMPK stimulates ATP-producing catabolic pathways (glycolysis and fatty acid oxidation) and attenuates ATP-consuming anabolic pathways (lipogenesis, protein synthesis) (167,168). In fact, this switch to fatty acid oxidation has the net benefit of increasing NADPH levels in cells (169). Lastly, LKB1–AMPK signaling also positively regulates autophagy through activation of ULK1 and through inhibition of the mTOR pathway (168,170), suggesting that the LKB1 mutant tumors may have reduced autophagy, compared with tumors with LKB1 intact. Therefore, it would be interesting to understand how LKB1 mutant PDA tumors deal with energy or other metabolic stressors.
Other metabolic pathways
Lipids and fatty acids
Lipids and fatty acids are important for tumor cell growth and as mentioned previously, Ras-transformed cells have developed mechanisms to scavenge free fatty acids from the extracellular environment (85). The role of individual fatty acids in PDA metabolism has recently begun to be explored. Interestingly, there are both pro- and antitumorigenic properties of different fatty acid species described in various cancers making the biology complex (171,172).
Several recent studies have shown that PDA tumors have lower levels of fatty acids than corresponding normal tissues. For example in a proton magnetic resonance study (HNMR), lipids, choline-containing compounds and fatty acids were found decreased in pancreatic cancer compared with normal pancreatic tissue (173). Palmitoleic acid was found to be decreased in rat models of PDA (bearing oncogenic Kras), both in the tumor tissue and in the serum, compared with control animals (174). In another study that combined transcriptomic and proteomic approaches to compare PDA with matched non-tumor tissue from the same patients, fatty acids were shown to be consistently decreased in PDA (175). The proposed mechanism was that specific lipases (PNLOP, CLPS, PNLIPRP1 and PNLIPRP2) are downregulated in PDA, thereby resulting in a decrease in the content of free fatty acids. These include palmitic and stearic acids, two saturated fatty acids shown to induce apoptosis and inhibit proliferation of PDA cell lines (175). Along these lines, other fatty acids with antitumorigenic properties have been described such as N-3 polyunsaturated acids, which can impair PDA progression (176).
Lipids, however, can also be protumorigenic, with a high-fat diet promoting tumor growth in murine models of pancreatic cancer (177). Although this may not be due to direct effects of the lipids on the tumor cells themselves, there is evidence for direct protumorigenic effects of fatty acids in PDA. Indeed, lipid metabolism is likely to be an important source of energy in PDA and consistent with this, PDA cell lines treated with oleic and linoleic acid display increased proliferative rates (178). The metabolism of fatty acids can also generate ROS as it is the case with linoleic acid (179), a potentially pro- or antitumorigenic factor in PDA (depending on the levels of ROS), which could also explain why in some contexts fatty acids are growth suppressive and in others growth promoting.
Even if reducing the levels of certain fatty acids is apparently important for PDA, it remains to be determined which fatty acids are cytotoxic for tumor cells and which fatty acids provide the tumor with metabolic substrates. One possible explanation for the fatty acid level reduction in PDA could be that they are being rapidly metabolized by the tumor cells. It would therefore be of great interest to understand the complex role of lipids in PDA through detailed metabolic studies.
Nicotinamide adenine dinucleotide biosynthesis
Nicotinamide adenine dinucleotide (NAD) is a crucial cofactor in redox reactions in many metabolic pathways (180). NAD can be found in oxidized (NAD+) or reduced (NADH) forms. It is a crucial factor for both the TCA cycle and glycolysis, with NAD reduction being required for the conversion of glyceraldehyde-3-phosphate into 1,3-biphosphoglycerate. It also is used as an electron donor to make ATP via oxidative phosphorylation. NAD has other roles in the cell other than its oxidative potential. NAD regulates transcription factors involved in pathways linked to inflammation, cell cycle progression, apoptosis, metabolism or DNA repair. For a review see ref. 181.
There are two main pathways for NAD synthesis in a cell: de novo synthesis, from the amino acids tryptophan or aspartic acid or the salvage pathway, where NAD is recycled from compounds containing nicotinamide (180,182). The salvage synthesis pathway is essential to maintain NAD levels in mammals and is dependent on nicotinamide phosphoribosyltransferase, the rate-limiting enzyme that converts nicotinamide to nicotinamide mononucleotide. Blocking this enzyme with a specific chemical inhibitor (FK866) will lower NAD levels in cell, which can lead to cell death in many cancer types (183). As cancer cells rely on NAD availability, targeting NAD indirectly through nicotinamide phosphoribosyltransferase can be seen as a possible therapeutic target (184,185). A recent study demonstrated that PDA cells depend on NAD metabolism for their survival, with promising preclinical results when targeting nicotinamide phosphoribosyltransferase both genetically and with FK866 (184).
Conclusions
There has been much work done recently to define the spectrum of metabolic changes in PDA. Importantly, many of these metabolic changes appear to facilitate or be required for growth. Because tumor specific events, such as Kras mutations, play a critical role in driving much of the metabolic alterations in PDA and moreover because some of these alterations have been shown to be dispensable in normal cells, there is an exciting opportunity for therapeutic intervention. Indeed, the unique metabolism of PDA cells that allows their survival and proliferation could also be seen as their Achilles heel. As discussed in this review, our understanding of the complex metabolism of PDA suggests a number of rationally designed combination therapies that could be utilized in this deadly disease. Because metabolic enzymes are catalytic, they are potentially amenable to inhibition using small molecules and therefore we will probably see an increasing amount of metabolic-targeted therapies in the coming years. As metabolism is a complex and interconnected network, many unanswered questions still remain. However, as our disease models are improving and our capacity to measure complex metabolic reactions in aggregate are becoming more accessible and more robust, the future holds great promise.
Funding
National Institutes of Health/National Cancer Institute (1R01CA157490); American Cancer Society (RSG-13-298-01-TBG); Department of Defense Discovery Award (W81XWH-12-1-0459); the Lustgarten Foundation for Pancreatic Cancer Research to A.C.K.
Acknowledgements
We apologize for the omission of any primary references. We thank Costas Lyssiotis and Haoqiang Ying for critical reading of this manuscript and Celana Higginbottom for administrative support.
Conflict of Interest Statement: Alec C. Kimmelman is a consultant for Forma Therapeutics.
Glossary
Abbreviations:
- AMPK
adenosine monophosphate-activated protein kinase
- ATP
adenosine triphosphate
- HBP
hexosamine biosynthesis pathway
- HIF
hypoxia inducible factor
- LDHA
lactate dehydrogenase A
- LKB1
liver kinase B1
- MEK
mitogen-activated protein kinase kinase
- NAD
nicotinamide adenine dinucleotide
- NADPH
reduced nicotinamide adenine dinucleotide phosphate
- NOX
NADPH oxidase
- PDA
pancreatic ductal adenocarcinoma
- PPP
pentose phosphate pathway
- ROS
reactive oxygen species
- TCA
tricarboxylic acid.
References
- 1. Siegel R., et al. (2013). Cancer statistics, 2013. CA. Cancer J. Clin., 63, 11–30 [DOI] [PubMed] [Google Scholar]
- 2. National Cancer Institute (2012). SEER Cancer Statistics Factsheets: Pancreas Cancer. National Cancer Institute, Bethesda, MD: http://seer.cancer.gov/statfacts/html/pancreas.html (6 May 2014, date last accessed). [Google Scholar]
- 3. Hidalgo M. (2010). Pancreatic cancer. N. Engl. J. Med., 362, 1605–1617 [DOI] [PubMed] [Google Scholar]
- 4. Vincent A., et al. (2011). Pancreatic cancer. Lancet, 378, 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li D., et al. (2004). Pancreatic cancer. Lancet, 363, 1049–1057 [DOI] [PubMed] [Google Scholar]
- 6. Erkan M., et al. (2012). StellaTUM: current consensus and discussion on pancreatic stellate cell research. Gut, 61, 172–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chu G.C., et al. (2007). Stromal biology of pancreatic cancer. J. Cell. Biochem., 101, 887–907 [DOI] [PubMed] [Google Scholar]
- 8. Koong A.C., et al. (2000). Pancreatic tumors show high levels of hypoxia. Int. J. Radiat. Oncol. Biol. Phys., 48, 919–922 [DOI] [PubMed] [Google Scholar]
- 9. Guillaumond F., et al. (2013). Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc. Natl. Acad. Sci. U. S. A., 110, 3919–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Olive K.P., et al. (2009). Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science, 324, 1457–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Provenzano P.P., et al. (2012). Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell, 21, 418–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kimmelman A.C., et al. (2008). Genomic alterations link Rho family of GTPases to the highly invasive phenotype of pancreas cancer. Proc. Natl. Acad. Sci. U. S. A., 105, 19372–19377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Campbell P.J., et al. (2010). The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature, 467, 1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Y.H., et al. (2012). Inhibition of non-homologous end joining repair impairs pancreatic cancer growth and enhances radiation response. PLoS One, 7, e39588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ying H., et al. (2012). Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell, 149, 656–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Son J., et al. (2013). Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature, 496, 101–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lyssiotis C.A., et al. (2013). Pancreatic cancers rely on a novel glutamine metabolism pathway to maintain redox balance. Cell Cycle, 12, 1987–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feig C., et al. (2012). The pancreas cancer microenvironment. Clin. Cancer Res., 18, 4266–4276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Neesse A., et al. (2011). Stromal biology and therapy in pancreatic cancer. Gut, 60, 861–868 [DOI] [PubMed] [Google Scholar]
- 20. Apte M.V., et al. (2012). Pancreatic stellate cells: a starring role in normal and diseased pancreas. Front. Physiol., 3, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oberstein P.E., et al. (2013). Pancreatic cancer: why is it so hard to treat? Therap. Adv. Gastroenterol., 6, 321–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Milosevic M., et al. (2012). Tumor hypoxia predicts biochemical failure following radiotherapy for clinically localized prostate cancer. Clin. Cancer Res., 18, 2108–2114 [DOI] [PubMed] [Google Scholar]
- 23. Heller A., et al. (2010). Immunogenicity of SEREX-identified antigens and disease outcome in pancreatic cancer. Cancer Immunol. Immunother., 59, 1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bazhin A.V., et al. (2013). Overcoming immunosuppression as a new immunotherapeutic approach against pancreatic cancer. Oncoimmunology, 2, e25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bayne L.J., et al. (2012). Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell, 21, 822–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pylayeva-Gupta Y., et al. (2012). Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell, 21, 836–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clark C.E., et al. (2007). Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res., 67, 9518–9527 [DOI] [PubMed] [Google Scholar]
- 28. Almoguera C., et al. (1988). Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell, 53, 549–554 [DOI] [PubMed] [Google Scholar]
- 29. Uemura T., et al. (2004). Detection of K-ras mutations in the plasma DNA of pancreatic cancer patients. J. Gastroenterol., 39, 56–60 [DOI] [PubMed] [Google Scholar]
- 30. Pylayeva-Gupta Y., et al. (2011). RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer, 11, 761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Löhr M., et al. (2005). Frequency of K-ras mutations in pancreatic intraductal neoplasias associated with pancreatic ductal adenocarcinoma and chronic pancreatitis: a meta-analysis. Neoplasia, 7, 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Campbell P.M., et al. (2004). Oncogenic Ras and its role in tumor cell invasion and metastasis. Semin. Cancer Biol., 14, 105–114 [DOI] [PubMed] [Google Scholar]
- 33. Cox A.D., et al. (2010). Ras history: The saga continues. Small GTPases, 1, 2–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aguirre A.J., et al. (2003). Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev., 17, 3112–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hingorani S.R., et al. (2005). Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell, 7, 469–483 [DOI] [PubMed] [Google Scholar]
- 36. Ijichi H., et al. (2006). Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev., 20, 3147–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guerra C., et al. (2007). Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell, 11, 291–302 [DOI] [PubMed] [Google Scholar]
- 38. Gidekel Friedlander S.Y., et al. (2009). Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell, 16, 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hingorani S.R., et al. (2003). Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell, 4, 437–450 [DOI] [PubMed] [Google Scholar]
- 40. Pellegata N.S., et al. (1994). K-ras and p53 gene mutations in pancreatic cancer: ductal and nonductal tumors progress through different genetic lesions. Cancer Res., 54, 1556–1560 [PubMed] [Google Scholar]
- 41. Hezel A.F., et al. (2006). Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev., 20, 1218–1249 [DOI] [PubMed] [Google Scholar]
- 42. Hudson T.J., et al. (2010). International network of cancer genome projects. Nature, 464, 993–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cowley M.J., et al. (2013). Understanding pancreatic cancer genomes. J. Hepatobil. Pancreat. Sci, 20, 549–556 [DOI] [PubMed] [Google Scholar]
- 44. Biankin A.V., et al. ; Australian Pancreatic Cancer Genome Initiative. (2012). Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature, 491, 399–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yachida S., et al. (2010). Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature, 467, 1114–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vander Heiden M.G., et al. (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science, 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lunt S.Y., et al. (2011). Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol., 27, 441–464 [DOI] [PubMed] [Google Scholar]
- 48. Hanahan D., et al. (2011). Hallmarks of cancer: the next generation. Cell, 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 49. Vander Heiden M.G. (2011). Targeting cancer metabolism: a therapeutic window opens. Nat. Rev. Drug Discov., 10, 671–684 [DOI] [PubMed] [Google Scholar]
- 50. Ward P.S., et al. (2012). Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell, 21, 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Le A., et al. (2012). Conceptual framework for cutting the pancreatic cancer fuel supply. Clin. Cancer Res., 18, 4285–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. White E. (2013). Exploiting the bad eating habits of Ras-driven cancers. Genes Dev., 27, 2065–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. DeBerardinis R.J., et al. (2008). The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab., 7, 11–20 [DOI] [PubMed] [Google Scholar]
- 54. Hensley C.T., et al. (2013). Glutamine and cancer: cell biology, physiology, and clinical opportunities. J. Clin. Invest., 123, 3678–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fan J., et al. (2013). Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol. Syst. Biol., 9, 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang Z., et al. (2010). Eaten alive: a history of macroautophagy. Nat. Cell Biol., 12, 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Levine B., et al. (2008). Autophagy in the pathogenesis of disease. Cell, 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kroemer G., et al. (2010). Autophagy and the integrated stress response. Mol. Cell, 40, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mizushima N., et al. (2011). The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol., 27, 107–132 [DOI] [PubMed] [Google Scholar]
- 60. Boya P., et al. (2013). Emerging regulation and functions of autophagy. Nat. Cell Biol., 15, 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Das G., et al. (2012). Regulation and function of autophagy during cell survival and cell death. Cold Spring Harb. Perspect. Biol., 4,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Klionsky D.J., et al. (2011). A comprehensive glossary of autophagy-related molecules and processes (2nd edition). Autophagy, 7, 1273–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang Z., et al. (2010). Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol., 22, 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Neufeld T.P. (2010). TOR-dependent control of autophagy: biting the hand that feeds. Curr. Opin. Cell Biol., 22, 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dodson M., et al. (2013). Cellular metabolic and autophagic pathways: traffic control by redox signaling. Free Radic. Biol. Med., 63, 207–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rodriguez-Rocha H., et al. (2011). DNA damage and autophagy. Mutat. Res., 711, 158–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kimmelman A.C. (2011). The dynamic nature of autophagy in cancer. Genes Dev., 25, 1999–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. White E. (2012). Deconvoluting the context-dependent role for autophagy in cancer. Nat. Rev. Cancer, 12, 401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Degenhardt K., et al. (2006). Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell, 10, 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rabinowitz J.D., et al. (2010). Autophagy and metabolism. Science, 330, 1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Amaravadi R.K., et al. (2011). Principles and current strategies for targeting autophagy for cancer treatment. Clin. Cancer Res., 17, 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sui X., et al. (2013). Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis., 4, e838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Samaddar J.S., et al. (2008). A role for macroautophagy in protection against 4-hydroxytamoxifen-induced cell death and the development of antiestrogen resistance. Mol. Cancer Ther., 7, 2977–2987 [DOI] [PubMed] [Google Scholar]
- 74. Amaravadi R.K., et al. (2007). Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J. Clin. Invest., 117, 326–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang P., et al. (2013). MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology, 145, 1133–1143.e12 [DOI] [PubMed] [Google Scholar]
- 76. Liu E.Y., et al. (2012). Autophagy and cancer–issues we need to digest. J. Cell Sci., 125(Pt 10), 2349–2358 [DOI] [PubMed] [Google Scholar]
- 77. Yang S., et al. (2011). Pancreatic cancers require autophagy for tumor growth. Genes Dev., 25, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Guo J.Y., et al. (2013). Autophagy is required for mitochondrial function, lipid metabolism, growth, and fate of KRAS(G12D)-driven lung tumors. Autophagy, 9, 1636–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kim M.J., et al. (2011). Involvement of autophagy in oncogenic K-Ras-induced malignant cell transformation. J. Biol. Chem., 286, 12924–12932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lock R., et al. (2011). Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol. Biol. Cell, 22, 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Guo J.Y., et al. (2011). Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev., 25, 460–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wei H., et al. (2011). Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev., 25, 1510–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. McMahon H.T., et al. (2011). Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol., 12, 517–533 [DOI] [PubMed] [Google Scholar]
- 84. Swanson J.A., et al. (1995). Macropinocytosis. Trends Cell Biol., 5, 424–428 [DOI] [PubMed] [Google Scholar]
- 85. Lim J.P., et al. (2011). Macropinocytosis: an endocytic pathway for internalising large gulps. Immunol. Cell Biol., 89, 836–843 [DOI] [PubMed] [Google Scholar]
- 86. Bar-Sagi D., et al. (1986). Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by ras proteins. Science, 233, 1061–1068 [DOI] [PubMed] [Google Scholar]
- 87. Commisso C., et al. (2013). Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature, 497, 633–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kamphorst J.J., et al. (2013). Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc. Natl. Acad. Sci. U. S. A., 110, 8882–8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Berg J.M., et al. (2002). Biochemistry. 5th edn. W.H. Freeman, New York, NY [Google Scholar]
- 90. Vander Heiden M.G., et al. (2011). Metabolic pathway alterations that support cell proliferation. Cold Spring Harb. Symp. Quant. Biol., 76, 325–334 [DOI] [PubMed] [Google Scholar]
- 91. Von Hoff D.D., et al. (2013). Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med., 369, 1691–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yun J., et al. (2009). Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science, 325, 1555–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gaglio D., et al. (2011). Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol. Syst. Biol., 7, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Racker E., et al. (1985). Glycolysis and methylaminoisobutyrate uptake in rat-1 cells transfected with ras or myc oncogenes. Proc. Natl. Acad. Sci. U. S. A., 82, 3535–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chun S.Y., et al. (2010). Oncogenic KRAS modulates mitochondrial metabolism in human colon cancer cells by inducing HIF-1α and HIF-2α target genes. Mol. Cancer, 9, 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zhao D., et al. (2013). Lysine-5 acetylation negatively regulates lactate dehydrogenase A and is decreased in pancreatic cancer. Cancer Cell, 23, 464–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Icard P., et al. (2012). A global view of the biochemical pathways involved in the regulation of the metabolism of cancer cells. Biochim. Biophys. Acta, 1826, 423–433 [DOI] [PubMed] [Google Scholar]
- 98. Slawson C., et al. (2010). O-GlcNAc signaling: a metabolic link between diabetes and cancer? Trends Biochem. Sci., 35, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hanover J.A., et al. (2010). The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim. Biophys. Acta, 1800, 80–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kornfeld S., et al. (1964). The feedback control of sugar nucleotide biosynthesis in liver. Proc. Natl. Acad. Sci. U. S. A., 52, 371–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wellen K.E., et al. (2010). The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev., 24, 2784–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Mi W., et al. (2011). O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochim. Biophys. Acta, 1812, 514–519 [DOI] [PubMed] [Google Scholar]
- 103. Fuster M.M., et al. (2005). The sweet and sour of cancer: glycans as novel therapeutic targets. Nat. Rev. Cancer, 5, 526–542 [DOI] [PubMed] [Google Scholar]
- 104. Hart G.W., et al. (2010). Glycomics hits the big time. Cell, 143, 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Jänne P.A., et al. (2013). Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol., 14, 38–47 [DOI] [PubMed] [Google Scholar]
- 106. Ghadimi H., et al. (1964). Free amino acids of cord plasma as compared with maternal plasma during pregnancy. Pediatrics, 33, 500–506 [PubMed] [Google Scholar]
- 107. Wise D.R., et al. (2010). Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci., 35, 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. DeBerardinis R.J., et al. (2010). Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene, 29, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Metallo C.M., et al. (2012). Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature, 481, 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wise D.R., et al. (2011). Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. U. S. A., 108, 19611–19616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. DeBerardinis R.J., et al. (2007). Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. U. S. A., 104, 19345–19350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sinha K., et al. (2013). Oxidative stress: the mitochondria-dependent and mitochondria-independent pathways of apoptosis. Arch. Toxicol., 87, 1157–1180 [DOI] [PubMed] [Google Scholar]
- 113. Imlay J.A., et al. (1988). DNA damage and oxygen radical toxicity. Science, 240, 1302–1309 [DOI] [PubMed] [Google Scholar]
- 114. Sosa V., et al. (2013). Oxidative stress and cancer: an overview. Ageing Res. Rev., 12, 376–390 [DOI] [PubMed] [Google Scholar]
- 115. Hurd T.R., et al. (2012). Redox regulation of cell migration and adhesion. Trends Cell Biol., 22, 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Iles K.E., et al. (2002). Macrophage signaling and respiratory burst. Immunol. Res., 26, 95–105 [DOI] [PubMed] [Google Scholar]
- 117. Sena L.A., et al. (2012). Physiological roles of mitochondrial reactive oxygen species. Mol. Cell, 48, 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Fruehauf J.P., et al. (2007). Reactive oxygen species: a breath of life or death? Clin. Cancer Res., 13, 789–794 [DOI] [PubMed] [Google Scholar]
- 119. Cui X. (2012). Reactive oxygen species: the achilles’ heel of cancer cells? Antioxid. Redox Signal., 16, 1212–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Liou G.Y., et al. (2010). Reactive oxygen species in cancer. Free Radic. Res., 44, 479–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Storz P. (2005). Reactive oxygen species in tumor progression. Front. Biosci., 10, 1881–1896 [DOI] [PubMed] [Google Scholar]
- 122. Pani G., et al. (2010). Metastasis: cancer cell’s escape from oxidative stress. Cancer Metastasis Rev., 29, 351–378 [DOI] [PubMed] [Google Scholar]
- 123. Weinberg F., et al. (2010). Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl. Acad. Sci. U. S. A., 107, 8788–8793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Du J., et al. (2013). Regulation of pancreatic cancer growth by superoxide. Mol. Carcinog., 52, 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Romanowska M., et al. (2007). DNA damage, superoxide, and mutant K-ras in human lung adenocarcinoma cells. Free Radic. Biol. Med., 43, 1145–1155 [DOI] [PubMed] [Google Scholar]
- 126. Calvert R.J., et al. (2013). K-ras 4A and 4B mRNA levels correlate with superoxide in lung adenocarcinoma cells, while at the protein level, only mutant K-ras 4A protein correlates with superoxide. Lung Cancer, 80, 263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Vaquero E.C., et al. (2004). Reactive oxygen species produced by NAD(P)H oxidase inhibit apoptosis in pancreatic cancer cells. J. Biol. Chem., 279, 34643–34654 [DOI] [PubMed] [Google Scholar]
- 128. Trachootham D., et al. (2008). Redox regulation of cell survival. Antioxid. Redox Signal., 10, 1343–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Shelton P., et al. (2013). The transcription factor NF-E2-related factor 2 (Nrf2): a protooncogene? FASEB J., 27, 414–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Hayes J.D., et al. (2009). NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem. Sci., 34, 176–188 [DOI] [PubMed] [Google Scholar]
- 131. DeNicola G.M., et al. (2011). Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature, 475, 106–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Lister A., et al. (2011). Nrf2 is overexpressed in pancreatic cancer: implications for cell proliferation and therapy. Mol. Cancer, 10, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Semenza G.L. (2013). HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. Invest., 123, 3664–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Zhao F., et al. (2010). Imatinib resistance associated with BCR-ABL upregulation is dependent on HIF-1alpha-induced metabolic reprograming. Oncogene, 29, 2962–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Dang C. V. (2007). The interplay between MYC and HIF in the Warburg effect. Ernst Schering Found. Symp. Proc, 4, 35–53 [DOI] [PubMed] [Google Scholar]
- 136. Chaika N.V., et al. (2012). MUC1 mucin stabilizes and activates hypoxia-inducible factor 1 alpha to regulate metabolism in pancreatic cancer. Proc. Natl. Acad. Sci. U. S. A., 109, 13787–13792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Chen C., et al. (2009). siRNA targeting HIF-1alpha induces apoptosis of pancreatic cancer cells through NF-kappaB-independent and -dependent pathways under hypoxic conditions. Anticancer Res., 29, 1367–1372 [PubMed] [Google Scholar]
- 138. Chen J., et al. (2003). Dominant-negative hypoxia-inducible factor-1 alpha reduces tumorigenicity of pancreatic cancer cells through the suppression of glucose metabolism. Am. J. Pathol., 162, 1283–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Wilkinson S., et al. (2009). Hypoxia-selective macroautophagy and cell survival signaled by autocrine PDGFR activity. Genes Dev., 23, 1283–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Zhang H., et al. (2008). Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem., 283, 10892–10903 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 141. Rausch V., et al. (2012). Autophagy mediates survival of pancreatic tumour-initiating cells in a hypoxic microenvironment. J. Pathol., 227, 325–335 [DOI] [PubMed] [Google Scholar]
- 142. Guerra C., et al. (2013). Genetically engineered mouse models of pancreatic adenocarcinoma. Mol. Oncol., 7, 232–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Levine A.J., et al. (2010). The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science, 330, 1340–1344 [DOI] [PubMed] [Google Scholar]
- 144. Harris C.C. (1996). p53 tumor suppressor gene: from the basic research laboratory to the clinic–an abridged historical perspective. Carcinogenesis, 17, 1187–1198 [DOI] [PubMed] [Google Scholar]
- 145. Kruse J.P., et al. (2009). Modes of p53 regulation. Cell, 137, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Levine A.J., et al. (1991). The p53 tumour suppressor gene. Nature, 351, 453–456 [DOI] [PubMed] [Google Scholar]
- 147. Aylon Y., et al. (2007). Living with p53, dying of p53. Cell, 130, 597–600 [DOI] [PubMed] [Google Scholar]
- 148. Scarpa A., et al. (1993). Pancreatic adenocarcinomas frequently show p53 gene mutations. Am. J. Pathol., 142, 1534–1543 [PMC free article] [PubMed] [Google Scholar]
- 149. Oshima M., et al. (2013). Immunohistochemically detected expression of 3 major genes (CDKN2A/p16, TP53, and SMAD4/DPC4) strongly predicts survival in patients with resectable pancreatic cancer. Ann. Surg., 258, 336–346 [DOI] [PubMed] [Google Scholar]
- 150. Pérez-Mancera P.A., et al. (2012). What we have learned about pancreatic cancer from mouse models. Gastroenterology, 142, 1079–1092 [DOI] [PubMed] [Google Scholar]
- 151. Gottlieb E., et al. (2010). p53 regulation of metabolic pathways. Cold Spring Harb. Perspect. Biol., 2, a001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Shen L., et al. (2012). The fundamental role of the p53 pathway in tumor metabolism and its implication in tumor therapy. Clin. Cancer Res., 18, 1561–1567 [DOI] [PubMed] [Google Scholar]
- 153. Illuzzi J.L., et al. (2011). Modifications of p53 and the DNA damage response in cells expressing mutant form of the protein huntingtin. J. Mol. Neurosci., 45, 256–268 [DOI] [PubMed] [Google Scholar]
- 154. Johnson R.F., et al. (2012). Nuclear factor-κB, p53, and mitochondria: regulation of cellular metabolism and the Warburg effect. Trends Biochem. Sci., 37, 317–324 [DOI] [PubMed] [Google Scholar]
- 155. Madan E., et al. (2011). Regulation of glucose metabolism by p53: emerging new roles for the tumor suppressor. Oncotarget, 2, 948–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Matoba S., et al. (2006). p53 regulates mitochondrial respiration. Science, 312, 1650–1653 [DOI] [PubMed] [Google Scholar]
- 157. Suzuki S., et al. (2010). Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc. Natl. Acad. Sci. U. S. A., 107, 7461–7466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Hu W., et al. (2010). Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc. Natl. Acad. Sci. U. S. A., 107, 7455–7460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Kondoh H., et al. (2005). Glycolytic enzymes can modulate cellular life span. Cancer Res., 65, 177–185 [PubMed] [Google Scholar]
- 160. Schwartzenberg-Bar-Yoseph F., et al. (2004). The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res., 64, 2627–2633 [DOI] [PubMed] [Google Scholar]
- 161. Bensaad K., et al. (2006). TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell, 126, 107–120 [DOI] [PubMed] [Google Scholar]
- 162. Jiang P., et al. (2011). p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol., 13, 310–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Rosenfeldt M.T., et al. (2013). p53 status determines the role of autophagy in pancreatic tumour development. Nature, 504, 296–300 [DOI] [PubMed] [Google Scholar]
- 164. Lizcano J.M., et al. (2004). LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J., 23, 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Lüttges J., et al. (2004). Rare ductal adenocarcinoma of the pancreas in patients younger than age 40 years. Cancer, 100, 173–182 [DOI] [PubMed] [Google Scholar]
- 166. Morton J.P., et al. (2010). LKB1 haploinsufficiency cooperates with Kras to promote pancreatic cancer through suppression of p21-dependent growth arrest. Gastroenterology, 139, 586–97, 597.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Yuan H.X., et al. (2013). Nutrient sensing, metabolism, and cell growth control. Mol. Cell, 49, 379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Mihaylova M.M., et al. (2011). The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol., 13, 1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Jeon S.M., et al. (2012). AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature, 485, 661–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Egan D.F, et al. (2011). Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science, 331, 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Guo J.Y., et al. (2013). Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev., 27, 1447–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Zadra G., et al. (2013). The fat side of prostate cancer. Biochim. Biophys. Acta, 1831, 1518–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Ma X., et al. (2011). The metabolic features of normal pancreas and pancreatic adenocarcinoma: preliminary result of in vivo proton magnetic resonance spectroscopy at 3.0 T. J. Comput. Assist. Tomogr., 35, 539–543 [DOI] [PubMed] [Google Scholar]
- 174. Yabushita S., et al. (2013). Metabolomic and transcriptomic profiling of human K-ras oncogene transgenic rats with pancreatic ductal adenocarcinomas. Carcinogenesis, 34, 1251–1259 [DOI] [PubMed] [Google Scholar]
- 175. Zhang G., et al. (2013). Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clin. Cancer Res., 19, 4983–4993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Mohammed A., et al. (2012). Endogenous n-3 polyunsaturated fatty acids delay progression of pancreatic ductal adenocarcinoma in Fat-1-p48(Cre/+)-LSL-Kras(G12D/+) mice. Neoplasia, 14, 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Philip B., et al. (2013). A high-fat diet activates oncogenic Kras and COX2 to induce development of pancreatic ductal adenocarcinoma in mice. Gastroenterology, 145, 1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Wang F., et al. (2009). Increased lipid metabolism and cell turnover of MiaPaCa2 cells induced by high-fat diet in an orthotopic system. Metabolism., 58, 1131–1136 [DOI] [PubMed] [Google Scholar]
- 179. Pierre A.S., et al. (2013). Trans-10, cis-12 conjugated linoleic acid induced cell death in human colon cancer cells through reactive oxygen species-mediated ER stress. Biochim. Biophys. Acta, 1831, 759–768 [DOI] [PubMed] [Google Scholar]
- 180. Di Stefano M., et al. (2013). Diversification of NAD biological role: the importance of location. FEBS J., 280, 4711–4728 [DOI] [PubMed] [Google Scholar]
- 181. Chiarugi A., et al. (2012). The NAD metabolome–a key determinant of cancer cell biology. Nat. Rev. Cancer, 12, 741–752 [DOI] [PubMed] [Google Scholar]
- 182. Garten A., et al. (2009). Nampt: linking NAD biology, metabolism and cancer. Trends Endocrinol. Metab., 20, 130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Billington R.A., et al. (2008). NAD depletion by FK866 induces autophagy. Autophagy, 4, 385–387 [DOI] [PubMed] [Google Scholar]
- 184. Chini C.C.S., et al. (2013). Targeting of NAD metabolism in pancreatic cancer cells: potential novel therapy for pancreatic tumors. Clin. Cancer Res., 20, 120–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. von Heideman A., et al. (2010). Safety and efficacy of NAD depleting cancer drugs: results of a phase I clinical trial of CHS 828 and overview of published data. Cancer Chemother. Pharmacol., 65, 1165–1172 [DOI] [PubMed] [Google Scholar]