Abstract
Aims: In bacillithiol (BSH)-utilizing organisms, protein S-bacillithiolation functions as a redox switch in response to oxidative stress and protects critical Cys residues against overoxidation. In Bacillus subtilis, both the redox-sensing repressor OhrR and the methionine synthase MetE are redox controlled by S-bacillithiolation in vivo. Here, we identify pathways of protein de-bacillithiolation and test the hypothesis that YphP(BrxA) and YqiW(BrxB) act as bacilliredoxins (Brx) to remove BSH from OhrR and MetE mixed disulfides. Results: We present evidence that the BrxA and BrxB paralogs have de-bacillithiolation activity. This Brx activity results from attack of the amino-terminal Cys residue in a CGC motif on protein BSH-mixed disulfides. B. subtilis OhrR DNA-binding activity is eliminated by S-thiolation on its sole Cys residue. Both the BrxA and BrxB bacilliredoxins mediate de-bacillithiolation of OhrR accompanied by the transfer of BSH to the amino-terminal cysteine of their CGC active site motif. In vitro studies demonstrate that BrxB can restore DNA-binding activity to OhrR which is S-bacillithiolated, but not to OhrR that is S-cysteinylated. MetE is most strongly S-bacillithiolated at Cys719 in vitro and can be efficiently de-bacillithiolated by both BrxA and BrxB. Innovation and Conclusion: We demonstrate that BrxA and BrxB function in the reduction of BSH mixed protein disulfides with two natural substrates (MetE, OhrR). These results provide biochemical evidence for a new class of bacterial redox-regulatory proteins, the bacilliredoxins, which function analogously to glutaredoxins. Bacilliredoxins function in concert with other thiol-disulfide oxidoreductases to maintain redox homeostasis in response to disulfide stress conditions. Antioxid. Redox Signal. 21, 357–367.
Introduction
The cytoplasm is generally a reducing environment that is maintained, in part, by low-molecular-weight (LMW) thiol redox buffers. The most studied LMW thiol is glutathione (GSH), which is present in all eukaryotes and most Gram-negative bacteria, but absent from many Gram-positive bacteria (12, 29, 40). In Firmicutes bacteria, including Bacillus and Staphyloccoccus species, bacillithiol (Cys-GlcN-Malate; BSH) is an abundant LMW thiol (32). BSH functions as a thiol redox buffer in the detoxification of reactive oxygen species (ROS), reactive electrophile species (RES), toxins, and antibiotics (4, 16, 31, 32). Mutants in the BSH synthesis pathway of Bacillus subtilis are especially sensitive to the strong oxidant sodium hypochlorite (NaOCl) and to the electrophilic antibiotic fosfomycin (4, 16).
Innovation.
The BrxA and BrxB proteins have a thioredoxin fold with an active site CGC motif, and they are encoded in genomes of organisms that are known or predicted to synthesize bacillithiol (BSH) as a major low-molecular-weight thiol. We here demonstrate that BrxA and BrxB have bacilliredoxin (Brx) activity and catalyze protein de-bacillithiolation, analogous to protein de-glutathionylation by glutaredoxins (Grx). Brx activity is demonstrated with two physiologically relevant substrates, the OhrR transcriptional repressor that senses reactive oxygen species by S-bacillithiolation on Cys15 and the MetE methionine synthase which is transiently inactivated by S-bacillithiolation on Cys719 and the active site Cys730 in vivo.
The intracellular thiol-disulfide redox balance is affected by ROS, RES, and some thiophilic metals. Bacteria must cope with ROS generated endogenously during respiration or produced during host-pathogen interactions by phagocytic cells of the innate immune system (21, 30, 43). ROS often cause post-translational thiol-modifications in Cys-containing proteins, including intramolecular or intermolecular protein disulfides and mixed disulfides (S-thiolations) between proteins and LMW thiols (2). The oxidation of protein thiols may cause changes in protein structure and activity and can control redox signaling pathways. In eukaryotes, S-glutathionylation is an important redox-regulatory thiol modification, and numerous proteins have been identified as S-glutathionylated using large-scale redox proteomics studies (7, 22, 23, 26). S-glutathionylation serves to protect critical Cys residues against overoxidation and acts as a mechanism for the redox control of protein function in eukaryotes and some bacteria (7).
It was recently shown that BSH is used for S-thiolations in BSH-producing Firmicutes bacteria (4, 5). In B. subtilis, S-bacillithiolation functions as a redox-regulatory mechanism in response to cumene hydroperoxide (CHP) and hypochlorite stress (4, 24). The redox-controlled OhrR repressor is S-bacillithiolated after CHP and NaOCl stress (4, 24). This BSH-modified repressor (OhrR-SSB) is inactive, which leads to the induction of the OhrA peroxiredoxin (14, 15). The methionine synthase MetE is S-bacillithiolated in NaOCl-treated cells at the active site Cys730 and the non-conserved Cys719 in vivo. This leads to enzyme inactivation and a transient methionine auxotrophy (4).
The pathways that catalyze specific de-glutathionylation in eukaryotes and Escherichia coli involve glutaredoxins (Grx), classified into di-thiol Grx with CPTC active site motifs and monothiol Grx with a typical CGPS active site (25). Grx proteins have a basic thioredoxin (Trx)-fold with a four-stranded β sheet that is surrounded by three α-helices with the active site motif in the loop between β-sheet 1 and α-helix 1 (25). The di-thiol Grx1 and Grx3 proteins of E. coli have a redox potential of −233 and −198 mV, which is higher than that of thioredoxins (−270 mV) (3). Most di-thiol Grx proteins catalyze protein de-glutathionylation via a monothiol mechanism in which the N-terminal Cys thiolate attacks the S-glutathionylated protein, resulting in reduction of the mixed disulfide and release of an S-glutathionylated Grx (Grx-SSG) intermediate. This Grx-SSG intermediate is reduced by GSH, resulting in the formation of oxidized glutathione disulfide (GSSG), which is subsequently reduced by the GSSG reductase with NADPH as an electron donor (1, 17, 36). Alternatively, the Grx-SSG intermediate may be resolved by the second active site Cys residue, thereby leading to a Grx intramolecular disulfide (44). This di-thiol mechanism is often involved in the reduction of inter- or intramolecular disulfides and is less efficient for protein de-glutathionylation, and the Grx intramolecular disulfide can even divert the catalytic cycle and hinder Grx activity (1, 25).
The identification of many conserved S-bacillithiolated proteins among various Firmicutes bacteria (4, 5) suggests that there should be also conserved Grx-like enzymes which function as bacilliredoxins (Brx) in protein de-bacillithiolation (16). Here, we show that YphP(BrxA) and YqiW(BrxB) function in the de-bacillithiolation of two important substrates, OhrR and MetE.
Results
OhrR and MetE as model substrates for bacilliredoxin activity
S-bacillithiolation inactivates the redox-controlled OhrR repressor after oxidative stress (4, 24). In vitro, repressor activity is restored by thiol reductants, but the pathways mediating protein re-activation in vivo are not yet known. In response to NaOCl stress, the methionine synthase MetE is strongly S-bacillithiolated on both Cys719 and the active site residue Cys730 and, as a result, cells experience a transient methionine auxotrophy (4). This indicates that MetE oxidation can be a growth-limiting event in NaOCl-treated cells. As for OhrR, the pathways that might serve to reactivate S-bacillithiolated MetE are unknown.
BrxA and BrxB are putative bacilliredoxins
We postulated that BrxA (formerly YphP) and BrxB (formerly YqiW) might function in protein de-bacillithiolation (Fig. 1). As previously noted (16), these two Trx-like proteins (DUF1094 protein family) display a high co-occurrence across bacterial genomes with BSH biosynthesis genes (Fig. 2A). Moreover, previous structural studies demonstrate that BrxA has a typical Trx-fold (Fig. 2B), exhibits a high redox potential (E0′=−118 mV), and displays weak protein disulfide isomerase activity in vitro (8). Similar to BrxA, the BrxB paralogue (53% identity, 70% similarity) contains a potentially redox-active CGC motif. These features led us to postulate that these proteins represent a new family of redox enzymes, the bacilliredoxins (Brx family) (16, 18). Recently, it was shown that BrxA is bacillithiolated at its active site Cys after NaOCl treatment in Bacillus and Staphylococcus species (4, 5). The presence of BrxA S-bacillithiolated on Cys53 in vivo is consistent with a possible role in the reduction of protein-BSH mixed disulfides (4).
FIG. 1.
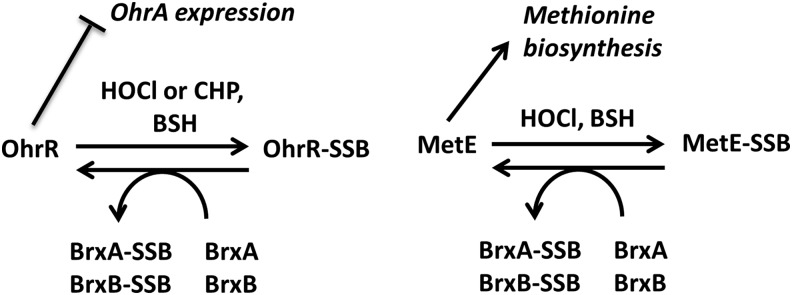
Model for S-bacillithiolation and de-bacillithiolation of OhrR and MetE indicating possible roles for the bacilliredoxins BrxA and BrxB. OhrR is a dimeric repressor that represses the transcription of ohrA (13). In response to oxidative stress, Bacillus subtilis OhrR is inactivated by S-bacillithiolation at its sole Cys residue (Cys15), leading to up-regulation of the OhrA peroxiredoxin that confers CHP and NaOCl resistance (4, 14). Oxidative stress also leads to S-bacillithiolation of the methionine synthase MetE at Cys730 and Cys719, resulting in methionine auxotrophy (4). The S-bacillithiolated OhrR-SSB and MetE-SSB proteins can be de-bacillithiolated in vitro by the BrxA and BrxB bacilliredoxins. Pathways for recycling of the Brx-SSB intermediate are not yet resolved. CHP, cumene hydroperoxide; NaOCl, sodium hypochlorite.
FIG. 2.
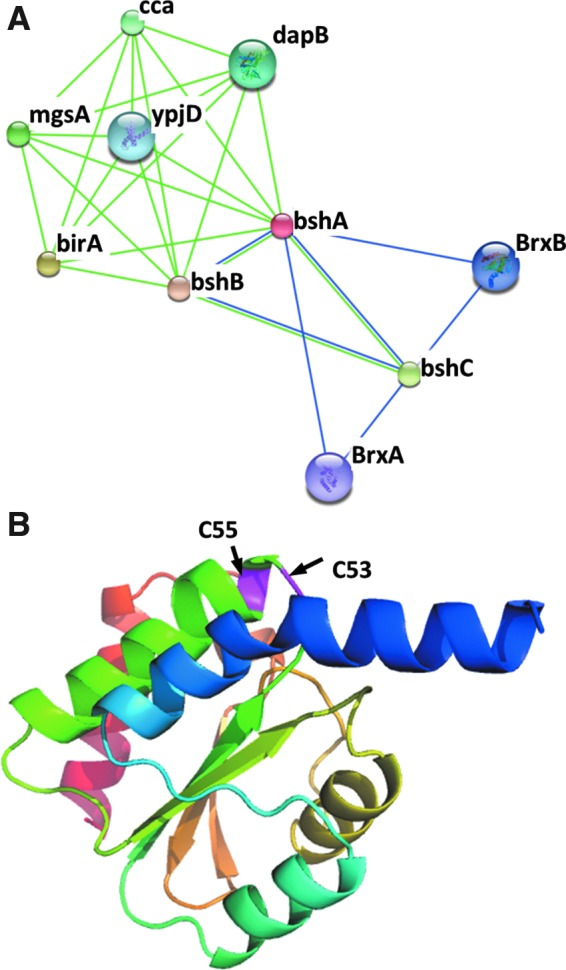
Phylogenomic profiling and structural studies suggest that BrxA and BrxB may function as bacilliredoxins. (A) The genes encoding bshA, B, and C co-occur across sequenced bacterial genomes with those encoding two putative Brxs (BrxA and BrxB) as revealed using the EMBL/Strings websearch interface (15). (B) Structure of BrxA (8) showing two conserved cysteines (C53 and C55). The BrxB paralogue is predicted to have a very similar structure.
S-thiolation by BSH inactivates OhrR repressor function in vitro
B. subtilis OhrR contains a single Cys residue that is S-bacillithiolated in vivo after CHP and NaOCl treatment, leading to inactivation of the repressor (24). In our previous in vitro studies of OhrR modification, before the discovery and characterization of BSH, we used l-Cys to monitor the effects of mixed disulfides on OhrR function (24). The availability of pure, chemically synthesized BSH (35) encouraged us to examine the effects of protein S-bacillithiolation directly. OhrR DNA-binding was monitored using a fluorescence anisotropy (FA)-based assay with an ohrA DNA probe representing the operator site as defined by the high-resolution structure of the OhrR:ohrA complex (20) (Fig. 3). In this assay, DNA binding by OhrR is evident as an increase in the FA signal as described (24, 37–39). The functional inactivation of OhrR by the addition of CHP, and subsequent reactivation by the addition of a thiol reductant (DTT), can be followed by monitoring the FA signal in real time (24).
FIG. 3.
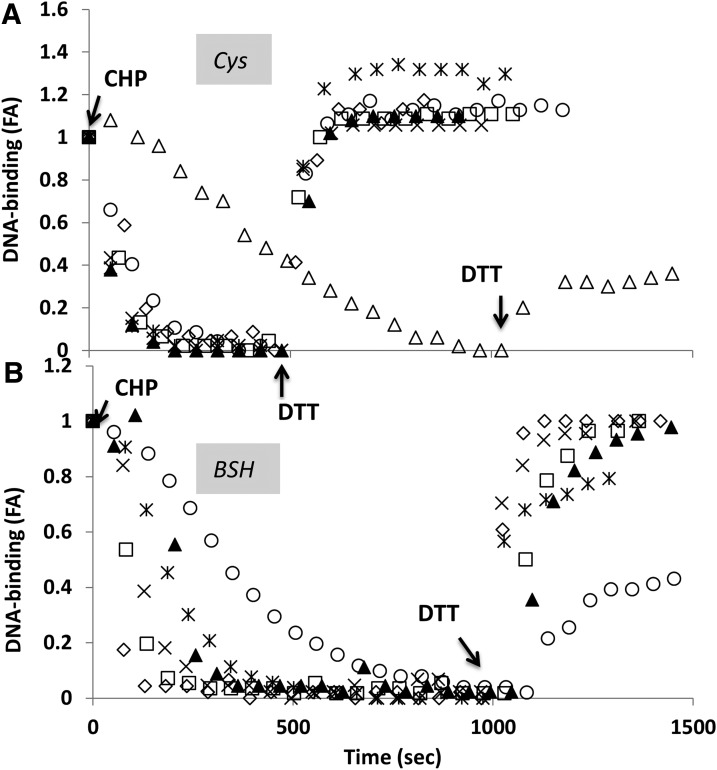
OhrR can be inactivated by mixed disulfide formation with BSH after CHP treatment. OhrR binding to ohrA probe was detected using FA assay. Anisotropy was measured for the free probe (corresponding to 0 on the y axis) and after the addition of 300 nM of OhrR (corresponding to 1 on the y axis). Three micromolar CHP was added in the absence of thiol (△) or 10 μM (◯), 50 μM (★), 0.1 mM (×) 0.25 mM (▲), 0.5 mM (□), and 1 mM (◆) l-Cys (A) and BSH (B). Inactivation of WT OhrR by CHP treatment in the presence of BSH and restoration of DNA-binding activity by the addition of 10 mM DTT (times of addition are indicated by the arrows). BSH, bacillithiol; FA, fluorescence anisotropy.
As previously reported (24), in the absence of any LMW thiol, the oxidation of OhrR by CHP leads to only a slow loss of DNA-binding activity (Fig. 3A, open triangles). This occurs despite the rapid oxidation of OhrR to form the sulfenic acid derivative (which retains DNA-binding activity), and it is correlated with the slow processes of condensation to form a cyclic sulfenamide derivative or overoxidation of the initial sulfenic acid derivative to the sulfonic acid. The addition of DTT partially restores DNA-binding activity by the slow re-reduction of the sulfenamide derivative, but cannot restore activity to the overoxidized (sulfonic acid-containing) protein (24). Importantly, the presence of an LMW thiol in the reaction captures the initial sulfenic acid derivative as a mixed disulfide and rapidly triggers the allosteric changes that result in the loss of DNA-binding activity. As previously shown, even low concentrations of l-Cys (10 μM) suffice to rapidly inactivate the OhrR repressor with formation of the S-cysteinylated product, OhrR-SSCys (Fig. 3A). BSH also facilitates the rapid release of OhrR from DNA, although somewhat higher concentrations of BSH were needed to achieve the same rate of OhrR dissociation at the pH (8.0) of these in vitro reactions (Fig. 3B). Nevertheless, BSH (rather than free Cys) is the major cofactor for mixed disulfide formation in vivo, likely due to the increased thiol acidity of BSH whose reactive thiolate anion concentrations are 10–50-fold higher than those of free Cys thiolate at physiological pH in B. subtilis (34). The addition of DTT to either OhrR-SSCys or OhrR-SSB rapidly and fully restored OhrR DNA-binding activity (Fig. 3A, B).
OhrR binds DNA as a homodimer, and we have previously shown that S-cysteinylation of a single Cys residue (per dimer) is sufficient for derepression (10). This was done by constructing a single-chain OhrR (scOhrR) in which the two protein chains (protomers) were connected by a short amino-acid linker and encoded by a single gene. Using this construct, it was possible to selectively mutate the active site Cys residue in only one of the two “monomer” domains. To study whether bacillithiolation of a single Cys residue is also sufficient to inactivate OhrR, two single chain OhrR variants, WT-WT and WT-C15S, were purified as described (10). In the latter molecule, the active site Cys residue in the second encoded monomer domain is replaced by Ser, resulting in an scOhrR with only a single Cys residue. As reported (10), the oxidation of OhrR with CHP in the presence of Cys led to rapid dissociation of both the WT-WT and WT-C15S scOhrR variants from the DNA operator (Supplementary Fig. S1A; Supplementary Data are available online at www.liebertpub.com/ars). In parallel, we now show that BSH can also mediate the dissociation of the WT-WT and WT-C15S single chain OhrR variants after CHP treatment (Supplementary Fig. S1B). These data indicate that, similar to S-cysteinylation, S-bacillithiolation of a single subunit in the scOhrR (and presumably also in the native OhrR dimer) is sufficient for inactivation.
BrxA and BrxB de-bacillithiolate OhrR-SSB
In initial studies, we noted that, in contrast to the strong thiol-reducing agent DTT (Fig. 3), DNA-binding activity could not be restored to OhrR-SSB by the addition of either purified BrxA or BrxB (data not shown). Since these assays required that the Brx proteins be in buffers lacking thiol-reducing agents, we hypothesized that the CGC motif might have been oxidized before the assay. Indeed, both Cys residues in BrxA and BrxB were oxidized as judged by their inability to be modified by 4-acetamido-4′-maleimidylstilbene-2, 2′-disulfonic acid (AMS) treatment (data not shown).
To obtain reduced BrxA and BrxB, we purified the proteins in the presence of EDTA and incubated the proteins with a 50-fold molar excess of NaBH4 as reductant, as previously described for studies of Grxs (9). To prevent modification of the BrxA or BrxB proteins by free BSH, we first purified OhrR-SSB away from any residual BSH using three cycles of buffer exchange with desalting spin columns. Incubation of reduced BrxA or BrxB with OhrR-SSB resulted in de-bacillithiolation of OhrR and transfer of the BSH moiety to the bacilliredoxin protein as monitored by immunoblotting with anti-BSH antibodies (5) (Fig. 4A). Control experiments demonstrated that buffer alone (treated with NaBH4 under identical incubation conditions) failed to de-bacillithiolate OhrR.
FIG. 4.
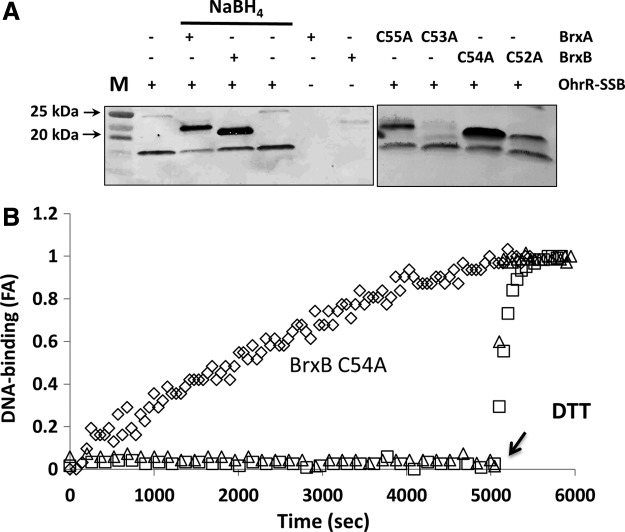
De-bacillithiolation of OhrR-SSB by BrxA and BrxB. (A) OhrR-SSB was generated in vitro using CHP and purified BSH, and the level of protein S-bacillithiolation was monitored by immunoblot analysis using anti-BSH antibodies. Left panel: Reduced BrxA and BrxB proteins (7.5 μM) (and a no protein control) were prepared by incubation with NaBH4 and tested for their ability to de-bacillithiolate OhrR (7.5 μM) as evidenced by a loss of OhrR-SSB signal and the appearance of S-bacillithiolated BrxA-SSB and BrxB-SSB. It should be noted that BrxA and BrxB are not detected unless they are S-bacillithiolated by incubation with OhrR-SSB. Right panel: Incubation of OhrR-SSB with BrxA C55A, BrxA C53A, BrxB C54A, or BrxB C52A indicates that thiol transfer occurs efficiently with those proteins which retain the active site (amino-terminal) Cys residue and inefficiently, if at all, with those retaining only the resolving Cys residue. (B) Ability of Brx proteins to restore DNA-binding activity of OhrR-SSB as monitored by FA analysis. Purified OhrR-SSB (300 nM) was incubated with fluorescently labeled ohrA operator DNA, and no binding (increased anisotropy) was observed. At time zero, various BrxA and BrxB proteins (3 μM) were added, and the regeneration of active (DNA-binding OhrR) was monitored over time. Under these conditions, BrxB C54A slowly reactivates OhrR-SSB (◊), but not S-cysteinylated OhrR (△). WT BrxA (not shown), BrxB (×), and BrxB C52A (□) were unable to reactivate OhrR.
To gain insights into which of the Cys residues in each Brx protein are required for bacilliredoxin activity, we also purified the single Cys to Ala substitution mutants BrxA C55A, BrxA C53A, BrxB C54A, and BrxB C52A. Using these proteins, de-bacillithiolation was only observed for the BrxA C55A and BrxB C54A proteins and no prior reduction with NaBH4 was required. These results confirm that both BrxA and BrxB can de-bacillithiolate OhrR-SSB and further indicate that the BSH moiety is transferred from OhrR Cys15 to the amino-terminal Cys residue of the bacilliredoxin. In control experiments, we generated S-bacillithiolated BrxB C54A and incubated this protein with OhrR. The lack of transfer from BrxB-SSB to OhrR indicates that the equilibrium of this thiol transfer reaction strongly favors the debacillithiolation of OhrR (Supplementary Fig. S2).
BrxB C54A reduces OhrR-SSB to regenerate active OhrR
Next, we sought to determine whether the addition of Brx proteins was sufficient to restore DNA-binding activity to OhrR-SSB in an FA assay. Under these much more dilute conditions (OhrR-SSB is at a 25-fold lower concentration than in the direct thiol transfer assays), neither of the wild-type proteins was able to restore DNA binding to OhrR-SSB even after NaBH4 reduction. However, the addition of BrxB C54A (but not the C52A) protein restored OhrR DNA-binding activity (Fig. 4B), which was consistent with the high activity noted for this protein in the direct thiol transfer assays (Fig. 4A). Remarkably, BrxB C54A could not reactivate OhrR-SSCys (Fig. 4B), despite the fact that this inactive repressor is readily reactivated by DTT (Fig. 3A). This indicates that BrxB acts as a bacilliredoxin; this activity requires the active site Cys52 residue, and it shows specificity for the BSH moiety. Further studies will be required to determine whether BrxA and/or BrxB are required for de-bacillithiolation of OhrR in vivo.
BrxA and BrxB de-bacillithiolate MetE-SSB
In addition to OhrR, the MetE methionine synthase has been identified as a physiologically important target of S-bacillithiolation in NaOCl-exposed cells (4). Previous mass spectrometry (MS) analyses of tryptic peptides identified S-bacillithiolation in vivo at Cys719 and the active site Cys730 (4). To determine whether BrxA or BrxB proteins could de-bacillithiolate these MetE-SSB isoforms, we generated cell extracts from cells treated for 10 min with 75 μM NaOCl. Consistent with earlier studies, NaOCl treatment led to the appearance of a prominent band of ∼90 kDa molecular mass as detected using anti-BSH antibodies. As noted in earlier studies, this MetE-SSB band is only detected in NaOCl-treated wild-type cells, and it is not observed in cells that do not synthesize BSH (5).
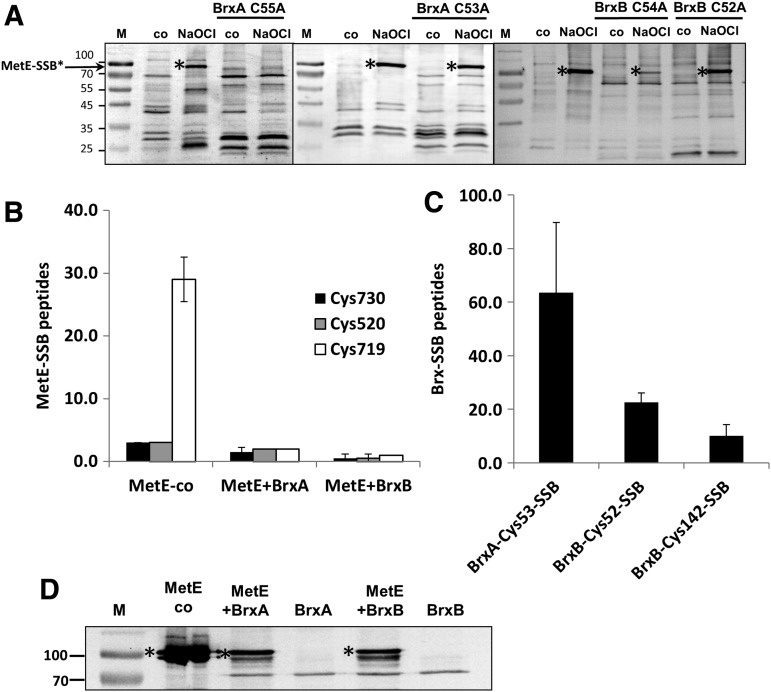
Due to their greater stability under our assay conditions, we initially tested the ability of BrxA C55A (containing the active site C53 residue) and BrxB C54A (containing the active site C52 residue) to de-bacillithiolate MetE-SSB generated in vivo by NaOCl stress. The intensity of the MetE-SSB band significantly decreased on incubation with the BrxA C55A and BrxB C54A proteins, but not after incubation with the active site mutants BrxA C53A and BrxB C52A (Fig. 5A). Thus, BrxA C55A and BrxB C54A proteins can de-bacillithiolate the MetE-SSB isoforms generated in vivo, and this reaction requires the active site (amino-terminal) Cys residue.
FIG. 5.
De-bacillithiolation of MetE-SSB by BrxA and BrxB. (A) S-bacillithiolated MetE (MetE-SSB) was generated in vivo by the treatment of cells with NaOCl for 10 min. Cell extracts were prepared, and the presence of MetE-SSB (*) was monitored by immunoblotting with anti-BSH antibodies as previously described (4, 5). The prominent ∼90 kDa band in NaOCl-treated cells corresponds to MetE-SSB, and other bands are other S-bacillithiolated proteins as previously documented by MS (4, 5). Incubation of purified BrxA C55A (∼15 μM) for 30 min with crude cell extracts eliminated the MetE-SSB signal, and BrxB C54A greatly reduced the signal intensity. In contrast, BrxA C53A and BrxB C52A did not detectably de-bacillithiolate MetE-SSB. The left lane in each panel contains size markers. (B–D) To test the ability of native BrxA and BrxB proteins to de-bacillithiolate MetE-SSB, and to determine which site(s) of modification are affected by Brx proteins, we prepared MetE-SSB in vitro by the incubation of MetE with oxidized BSH (BSSB). MetE-SSB modification was monitored by MS and was found to be predominantly on Cys719 with lesser amounts of modification on Cys520 and Cys730. Incubation with 15 μM BrxA and BrxB (after reduction with NaBH4) led to nearly complete de-bacillithiolation of MetE-SSB as revealed by the quantitation of spectral counts in the MS analysis (B) and by BSH immunoblots (D). Concomitant with MetE-SSB de-bacillithiolation, both BrxA and BrxB became S-bacillithiolated at their active site Cys residues with a minor amount of S-bacillithiolation also detected on BrxB Cys142 (C). BSSB, oxidized bacillithiol disulfide; MS, mass spectrometry.
MetE can also be S-bacillithiolated in vitro by treatment with oxidized bacillithiol disulfide (BSSB) as monitored using either non-reducing BSH-specific immunoblots or Orbitrap Velos liquid chromatography (LC)-MS/MS analysis (Fig. 5B and Supplementary Table S1). MS most strongly revealed S-bacillithiolation of MetE at Cys719 with this oxidant, with relatively few spectral counts for the BSH-modified Cys520 and Cys730 peptides (Fig. 5B). Incubation of MetE-SSB with wild-type BrxA or BrxB, after prior reduction with NaBH4, led to substantial de-bacillithiolation of Cys719 (Fig. 5B). As expected, analysis of the BrxA and BrxB proteins after incubation with MetE-SSB revealed transfer of the BSH moiety to the active site residues of the bacilliredoxins, with a small amount of bacillithiolation also detected on BrxB Cys142 (Fig. 5C and Supplementary Tables S2 and S3). No bacillithiolation was detected on BrxA Cys55 or on BrxB Cys54.
Roles of BrxA and BrxB in ROS resistance in vivo
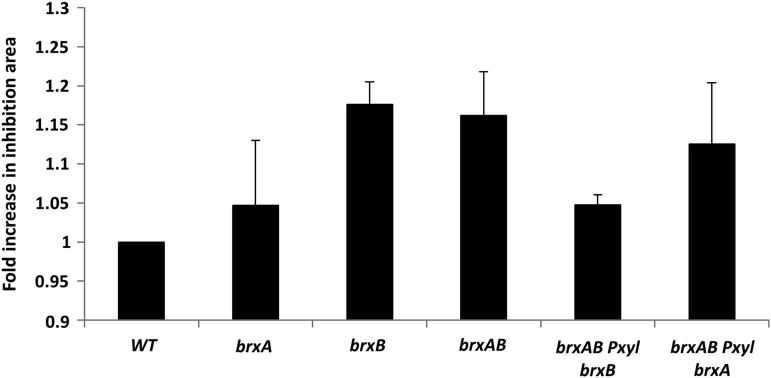
Protein S-bacillithiolation arises in cells exposed to CHP, as first noted for OhrR (24). Chemical sensitivity studies using a zone of growth inhibition assay demonstrate that BrxB contributes to fitness in cells exposed to CHP (Fig. 6). Compared with the brxB mutant, the brxA mutant had a comparatively mild phenotype. The sensitivity of the brxB (and brxA brxB double mutant) strain was complemented by the expression of brxB from a xylose-inducible promoter. The identity of the protein(s) that are oxidized by CHP, thereby resulting in growth inhibition, are not known, but these results lead us to speculate that BrxB preferentially de-bacillithiolates these key proteins.
FIG. 6.
BrxB is required for optimal resistance to CHP. Zone of inhibition assays were done using WT, brxA, brxB, and brxA brxB double-mutant (brxAB) strains. The area of inhibition was determined for each condition and is reported as average and standard deviation relative to the wild-type control (n>3). The sensitivity phenotype was complemented by the expression of brxB (but not by brxA) after induction from a xylose-dependent promoter.
Since BrxA and BrxB are able to de-bacillithiolate MetE-SSB in vitro, and MetE oxidation is correlated with a transient cessation of growth after NaOCl treatment (4), we next tested brxA and brxB single and brxA brxB double mutants for growth differences on challenge with NaOCl. However, no significant differences in growth lag or recovery were noted relative to the wild-type strain, nor was there a dramatic increase in the observed level of MetE S-bacillithiolation (data not shown). We also failed to detect an increased sensitivity to a variety of other thiol reactive agents in the brxA brxB double mutant, including H2O2, t-butyl peroxide, and diamide (data not shown). We, therefore, speculate that the thioredoxin system, or other unknown thiol-disulfide oxidoreductases, can substitute for BrxA and BrxB in the de-bacillithiolation of MetE-SSB and likely other proteins in vivo. This is not unexpected, as analogous studies of Grx mutants in E. coli have revealed only mild and strain-dependent chemical sensitivity for singly mutant strains, whereas multiply mutant strains can sometimes display much more severe phenotypes (42).
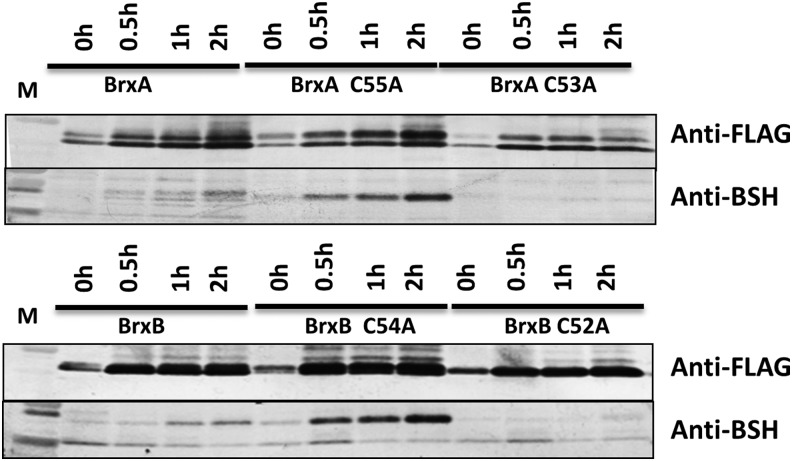
If the relevant in vivo activity of BrxA and BrxB is to de-bacillithiolate oxidized proteins, we predict that exposure to CHP should lead to at least transient S-bacillithiolation of BrxA and BrxB. To test this notion, we expressed FLAG-tagged BrxA, BrxB, and the related Cys mutant proteins in a brxB brxA double mutant background. The FLAG-tagged proteins were recovered by immunoprecipitation after the treatment of cells with CHP, and the recovered proteins were analyzed for reactivity with anti-BSH antibodies. Analysis of BrxA and BrxB protein levels, as monitored using the anti-FLAG antibody, revealed that both proteins were up-regulated (relative to total cell protein) within 30 min of CHP treatment. Further studies will be required to determine the factor(s) that might regulate the expression or accumulation of BrxA and BrxB in response to oxidative stress. However, when relative levels of protein S-bacillithiolation were monitored using anti-BSH antibodies, it was apparent that S-bacillithiolated BrxA and BrxB accumulated in those strains expressing mutations in the resolving Cys residue (BrxA C55A and BrxB C54A) (Fig. 7). This is consistent with a model in which Brx proteins act to de-bacillithiolate client proteins and where oxidation to the disulfide can lead to the release of BSH (a dithiol mechanism).
FIG. 7.
BrxA and BrxB are S-bacillithiolated in vivo in CHP-treated cells. FLAG-tagged versions of BrxA, BrxA C55A, BrxA C53A, BrxB, BrxB C54A, and BrxB C52A were expressed in the background of the brxA brxB double mutant. Cells were treated with 50 μM CHP, and samples were collected at the times indicated. The BrxA and BrxB proteins were immunoprecipitated using anti-FLAG antibodies and analyzed by Western blotting with either anti-BSH or anti-FLAG antibodies.
Increased S-bacillithiolation of the BrxA C55A and BrxB C54A proteins was also apparent after the treatment of cells with NaOCl. Previously, BrxA was found to be S-bacillithiolated in vivo in both control and NaOCl-treated cells at the active site Cys53 (4). FLAG-tagged BrxA C55A and BrxB C54A proteins were purified from B. subtilis cells after NaOCl stress. Analysis using Orbitrap LC-MS/MS identified peptides with the BrxA Cys53-SSB and BrxB Cys52-SSB modifications, confirming that both are trapped as S-bacillithiolated bacilliredoxins in vivo (Supplementary Table S4).
Discussion
Post-translational modifications are ubiquitous in biology and serve major roles in metabolic and genetic regulation. Transient protein modification, and in particular S-thiolation, may also serve a protective role under oxidative stress conditions. In the presence of ROS, cysteine residues can be oxidized to the sulfenic (RSOH), sulfinic (RSO2H), and sulfonic (RSO3H) acid derivatives (6). While sulfinate and sulfonate are often irreversible modifications, sulfenic acids can serve as an intermediate leading to either intra- or intermolecular disulfide bond formation. The capture of a Cys-sulfenate by an LMW thiol to form a mixed disulfide mixture protects the modified Cys residue against overoxidation. In eukaryotic cells and many Gram-negative bacteria, S-glutathionylation is a predominant mechanism for reversible cysteine modification (7). S-glutathionylation is a predominant mechanism for reversible cysteine modification and can also function to regulate protein activity (2, 7). Several criteria were proposed to define S-glutathionylation as a regulatory event: S-glutathionylation should change protein function, it should occur in response to a physiological stimulus and lead to a specific cellular response, and there should be a mechanism for reversal by de-glutathionylation (36). De-glutathionylation is carried out by specific GSH-mixed disulfide oxidoreductases known as Grxs (13, 17).
By analogy, S-bacillithiolation also likely serves both a protective function and a redox switch in cells that are exposed to ROS. S-bacillithiolation transiently inactivates the function of both OhrR and MetE in vivo (4, 24). Both proteins are S-bacillithiolated in vivo after oxidative stress, and here, we demonstrate that S-bacillithiolation prevents DNA binding by OhrR. De-bacillithiolation of OhrR-SSB occurs preferentially with BrxB (as observed using the BrxB C54A mutant), restores OhrR DNA-binding activity, and results in transfer of the BSH moiety to the active site BrxB Cys52 residue. Remarkably, this restoration of DNA binding is specific for the S-bacillithiolated protein and is ineffective with OhrR that has been inactivated by S-cysteinylation. S-bacillithiolation of MetE occurs at multiple sites both in vivo and in vitro, including Cys520, Cys719, and the active site residue Cys730. As shown here, de-bacillithiolation of MetE-SSB can be catalyzed by both BrxA and BrxB proteins with near quantitative transfer of the BSH moiety to the active site Cys residues of the bacillliredoxins.
The catalytic mechanisms of the BrxA and BrxB proteins are not yet understood in detail, but the conserved CGC motif is clearly important. With wild-type protein, the S-bacillithiolated Brx proteins presumably represent an intermediate in the reduction pathway. Preliminary evidence indicates that 2 mM BSH does not efficiently reduce S-bacillithiolated BrxB C54A protein, arguing that BrxB is more likely to function by a dithiol mechanism (data not shown), which is consistent with the propensity of the purified proteins to form intramolecular disulfides and the observed accumulation of S-bacillithiolated Brx proteins in mutants lacking the second (resolving) Cys residue. The oxidized BrxB protein is presumably reduced by an unknown thiol/disulfide oxidoreductase.
The proposed physiological role of Brx proteins is to facilitate the de-bacillithiolation of client proteins that are generated in response to oxidative stress. Support for this notion is provided by the observation that growth inhibition by CHP is aggravated in cells lacking BSH, and a similar effect is noted for cells lacking BrxA and BrxB. This supports a model in which the major role of BSH in protecting cells against the toxic effects of this peroxide is to modify vulnerable Cys residues that can then be regenerated by Brx proteins once the chemical threat is neutralized (by, e.g., peroxidases). At present, this protective role of S-bacillithiolation and bacilliredoxins has been most clearly visualized with CHP. In contrast, the brxA brxB double mutant was relatively unaffected in resistance to several other redox stress conditions that were shown to cause strong S-bacillithiolations (e.g., NaOCl and diamide stress) (5). This is consistent with the presence in the cell of numerous other thiol-disulfide oxidoreductases, including the essential thioredoxin (Trx) system, and the general notion that these systems are, at least, partially redundant in function. Interestingly, the standard thiol redox potentials of BSH (−221 mV) (34) and BrxA (−118 mV) (8) are notably higher than those of GSH (−240 mV) and typical Grx proteins (−200 mV) (3). It will be interesting to see whether these biophysical differences between the BSH and GSH redox systems are counterbalanced by differences in the relative kinetic properties of Brx and Grx enzymes. Ongoing studies, including genetic, physiological, and redox proteomics, will seek to tease apart these overlapping functions and clarify the roles of BSH, Brx proteins, and BSH-utilizing enzymes in cellular responses to redox stress.
Materials and Methods
Bacterial strains and growth conditions
B. subtilis strains were cultivated under vigorous agitation at 37°C in Belitsky minimal medium (BMM) as previously described (41) or in LB. The antibiotics were used at the following concentrations: 1 μg/ml erythromycin, 25 μg/ml lincomycin, 5 μg/ml chloramphenicol, 10 μg/ml kanamycin, and 100 μg/ml spectinomycin. NaOCl (15% stock solution) and CHP were purchased from Sigma-Aldrich. For NaOCl stress, cells were grown in BMM to an optical density at 500 nm (OD500) of 0.4 and treated with 75 μM NaOCl freshly diluted in distilled water. To determine the effects of bshC, brxA, and brxB mutations on ROS susceptibility, sensitivity was measured using both growth assays in minimal medium to detect methionine auxotrophy (4) and disk diffusion assays to detect overall levels of chemical sensitivity as described (16).
DNA manipulations
Routine molecular biology procedures were done according to (33). Restriction enzymes, DNA ligase, Klenow fragment, and DNA polymerase were used according to the manufacturer's instructions (New England Biolabs). Site-directed mutagenesis was done by polymerase chain reaction (PCR) and overlap extension according to (19). Mutants were constructed using long-flanking-homology PCR as described (28).
Expression and purification of recombinant His-tagged MetE, BrxA, BrxB, BrxA C53A, BrxA C55A, BrxB C52A, and BrxB C54A proteins
E. coli BL21(DE3)pLysS (Invitrogen) was used for overproduction of His-tagged, BrxA, BrxB, BrxA C53A, BrxA C55A, BrxB C52A, and BrxB C54A proteins. Expression plasmids for protein overproduction were constructed using pET16b as follows: Coding sequences were amplified by PCR with B. subtilis chromosomal DNA as template, and the products were cloned in the NdeI-BamHI sites of pET16B. The resulting plasmids were confirmed by DNA sequencing and transformed into E. coli BL21(DE3)pLysS. To avoid problems with protein aggregation, His-tagged MetE was overexpressed using an MBP fusion and in vitro cleavage system as described (11).
For protein expression, E. coli BL21(DE3)pLysS strains carrying expression plasmids were cultured in 1 L LB medium, and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added at mid-log phase (OD600 of 0.5) for 2 h. Recombinant His-tagged proteins were purified using PrepEase™ His-Tagged High Yield purification Resin (USB) under native conditions according to the instructions of the manufacturer. His-tagged proteins were eluted in 50 mM NaH2PO4, 300 mM NaCl, 1 mM EDTA, and 250 mM imidazole pH 8.0. A second step of purification was performed, when needed, using a size exclusion Superdex 200 column. The proteins were dialysed with 10 mM Tris-HCl pH 8.0, 100 mM NaCl, and 1 mM EDTA 50% glycerol (v/v) and stored at −80°C.
Fluorescence anisotropy
A 6-carboxyfluorescein-(6F-)-labeled ohrA operator DNA fragment was generated by annealing 5′-6F-TACAATTAAATTGTATACAATTAAATTGTA-3′ (Integrated DNA Technologies) and its unlabeled complement. It should be noted that the resulting duplex is identical to that used to determine the structure of the OhrR:ohrA operator complex (20) with the exception that the 30 bp fragment is fully duplex. FA measurements (λex=495 nm; slit width=15 nm, λem=520 nm; slit width=20 nm, integration time=1 s) were performed with 50 nM DNA and 300 nM OhrR (monomer) in 3 ml of 20 mM Tris (pH 8.0) containing 150 mM NaCl and 5% (v/v) glycerol. FA values were recorded automatically every 10 s with an LS55 luminescence spectrometer (PerkinElmer).
Preparation of OhrR-SSB
Bacillithiolation or cysteinylation of OhrR was done by treating 50 μM OhrR monomer with 10×molar excess of CHP in the presence of 1 mM BSH. In parallel, S-cysteinylated OhrR was prepared using l-Cys in the place of BSH. After 20 min of incubation at room temperature, buffer exchange was done at least twice using micro-spin columns (Bio-Rad) to remove excess thiol and CHP.
Bacilliredoxin assays using in vitro generated OhrR-SSB
Brx assays for OhrR were done by incubating 7.5 μM of OhrR-SSB with equal molar concentrations of BrxB, BrxA, or their cysteine mutant derivatives at room temperature (22°C). After 20 min, non-reducing sodium dodecyl sulfate (SDS) loading dye was added, and the samples were boiled for 10 min. Proteins were separated by 15% non-reducing SDS-polyacrylamide gel electrophoresis (PAGE), and bacillithiolated proteins were detected using anti-BSH polyclonal rabbit antiserum (5) at 1:500 dilution, followed by incubation with goat anti-rabbit antibodies that were coupled to alkaline phosphatase (Sigma Chemical Co.) and colorimetric detection.
FA assays were used to monitor the DNA-binding activity of OhrR as described (10, 24, 38). 6-FAM labeled ohrA probe (50 nM) was mixed with 300 nM of OhrR, OhrR-SSB, or OhrR-S-Cys. For reactivation of S-thiolated OhrR, 3 μM of BrxB, BrxB C52A, or BrxB C54A proteins were added and FA values were recorded every 10 s. As a negative control, the same reactions were repeated in the absence of OhrR and showed that BrxB, BrxB C52A, or BrxB C54A were unable to bind to the probe.
Immunoprecipitation of BrxA and BrxB
The FLAG-tagged BrxA, BrxA C55A, BrxA C53A BrxB, BrxB C52A, and BrxB C54A proteins were purified from brxA brxB mutant strain with Anti-FLAG M2 magnetic beads (Sigma) as described (27) except that elution of the proteins from the beads was done using non-reducing SDS-loading dye. Protein samples were resolved by 15% SDS-PAGE, and immunoblot analyses were done using anti-BSH or anti-FLAG antibodies.
Bacilliredoxin assays using in vitro generated MetE-SSB and in vivo bacillithiolated cellular protein extracts
For preparation of MetE-SSB, purified MetE was reduced with 5 mM DTT for 10 min at room temperature, and then, excess DTT was removed by desalting with a PD10 column (GE Healthcare Life Sciences) in potassium phosphate buffer (100 mM KH2PO4, pH 7.2). MetE was S-bacillithiolated by incubation with 10 mM BSSB at 30°C for 1 h and desalted using PD10 column. Fractions containing MetE were determined using UV absorbance at 280 nm and were pooled and concentrated to ∼90 μM using an Amicon spin column (Millipore) and tested for mBBr labeling to ensure complete removal of released BSH. Before the Brx assay, the BrxA and BrxB proteins were reduced using NaBH4 for 2 h as previously described for Grxs (9). For the Brx assays, in vitro generated MetE-SSB was incubated with BrxA or BrxB proteins at 37°C for 30 min. The Brx reaction was stopped with 20 mM iodoacetamide (IAM) and separated using non-reducing 13% SDS-PAGE. The SDS gel was stained with Coomassie Brilliant Blue or subjected to BSH-specific immunoblot analysis.
For experiments using in vivo S-bacillithiolated MetE, cells were harvested before and 10 min after exposure to 75 μM NaOCl, lysed by sonication, and reactive thiols were alkylated in TE-buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA) with 100 mM IAM. Unreacted IAM was removed from the extracts using Bio-Rad microspin columns. Purified BrxA C55A, BrxA C53A, BrxB C54A, and BrxB C52A proteins were added to 25 μg alkylated crude protein extract in potassium phosphate buffer (100 mM KH2PO4, pH 7.2) and incubated at 37°C for 30 min. The reaction was stopped by the addition of 20 mM IAM and subjected to non-reducing SDS-PAGE and BSH-specific immunoblot analysis.
Linear trap quadrupole-Orbitrap Velos MS
The MetE-SSB, BrxA, and BrxB bands were cut from Coomassie-stained non-reducing SDS gels and tryptic in-gel digested. Tryptic peptides were subjected to reversed-phase column chromatography (self packed C18 column, 100-μm inside diameter×200 mm) on an Easy-nLC II system (Thermo Fisher Scientific). Elution was performed by a binary gradient of buffer A [0.1% (v/v) acetic acid] and B [99.9% (v/v) ACN, 0.1% (v/v) acetic acid] over a period of 100 min with a flow rate of 300 nl/min. MS and MS/MS data were acquired with the linear trap quadrupole (LTQ)-Orbitrap-Velos mass spectrometer (Thermo Fisher Scientific) that was equipped with a nanoelectrospray ion source as described (4).
Post-translational thiol-modifications of proteins were identified by searching all MS/MS spectra in “dta” format against a B. subtilis 168 target-decoy protein sequence database extracted from UniprotKB release 12.7 (UniProt Consortium, Nucleic acids research 2007, 35, D193-197) using Sorcerer™-SEQUEST® (Sequest v. 2.7 rev. 11, Thermo Electron, including Scaffold_3_00_02; Proteome Software, Inc.). The Sequest search was carried out with the following parameters: a parent ion mass tolerance of 10 ppm and a fragment ion mass tolerance of 1.00 Da. Approximately two tryptic miscleavages were allowed. Methionine oxidation (+15.9949 Da), cysteine carbamidomethylation (+57.0215 Da), and S-bacillithiolations (+396.0839 Da for C13H22N2O10S) were set as variable post-translational modifications in the Sequest search. Proteins were identified by at least two peptides applying a stringent Sequest filter. Sequest identifications required at least ΔCn scores of greater than 0.10 and XCorr scores of greater than 2.2, 3.3, and 3.75 for doubly, triply, and quadruply charged peptides, respectively.
Quantification of bacillithiolated peptides
Abundance of bacillithiolated peptides was estimated using Scaffold (Sequest v. 2.7 rev. 11 Thermo Electron, including Scaffold_3_00_05; Proteome Software, Inc.) to perform spectral count analysis. After fragmentation of each peptide, scaffold quantifies the protein amounts according to the number of MS/MS fragment ion spectra, which is defined as the spectral count for that peptide. Relative abundances of specific bacillithiolated peptide can be estimated by the ratio of the spectral counts. All of the samples were processed in parallel, and equal amounts of tryptic digest were loaded onto the LC column for LC/MS/MS analysis.
Supplementary Material
Abbreviations Used
- AMS
4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid
- BMM
Belitsky minimal medium
- Brx
bacilliredoxin
- BSH
bacillithiol
- BSSB
oxidized bacillithiol disulfide
- CHP
cumene hydroperoxide
- Cys
cysteine
- FA
fluorescence anisotropy
- Grx
glutaredoxin
- GSH
glutathione
- GSSG
oxidized glutathione disulfide
- IAM
iodoacetamide
- IPTG
isopropyl-β-d-thiogalactopyranoside
- LC
liquid chromatography
- LMW
low molecular weight
- LTQ
linear trap quadrupole
- Met
methionine
- MS
mass spectrometry
- NaOCl
sodium hypochlorite
- OD
optical density
- PAGE
polyacrylamide gel electrophoresis
- PCR
polymerase chain reaction
- protein-SSB
BSH protein mixed disulfide
- RES
reactive electrophile species
- ROS
reactive oxygen species
- scOhrR
single-chain OhrR
- SDS
sodium dodecyl sulfate
Acknowledgments
The authors thank Sebastian Grund for technical assistance in the LC-MS/MS analysis. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (AN746/3-1) to H.A., by a grant from the Biotechnology and Biological Science Research Council (BB/H013504/1) to C.J.H., and by a grant from the National Science Foundation (MCB-1020481) to J.D.H.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Allen EM. and Mieyal JJ. Protein-thiol oxidation and cell death: regulatory role of glutaredoxins. Antioxid Redox Signal 17: 1748–1763, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antelmann H. and Helmann JD. Thiol-based redox switches and gene regulation. Antioxid Redox Signal 14: 1049–1063, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aslund F, Berndt KD, and Holmgren A. Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J Biol Chem 272: 30780–30786, 1997 [DOI] [PubMed] [Google Scholar]
- 4.Chi BK, Gronau K, Mader U, Hessling B, Becher D, and Antelmann H. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol Cell Proteomics 10: M111.009506, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chi BK, Roberts AA, Huyen TT, Basell K, Becher D, Albrecht D, Hamilton CJ, and Antelmann H. S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in Firmicutes bacteria. Antioxid Redox Signal 18: 1273–1295, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claiborne A, Yeh JI, Mallett TC, Luba J, Crane EJ, 3rd, Charrier V, and Parsonage D. Protein-sulfenic acids: diverse roles for an unlikely player in enzyme catalysis and redox regulation. Biochemistry 38: 15407–15416, 1999 [DOI] [PubMed] [Google Scholar]
- 7.Dalle-Donne I, Rossi R, Colombo G, Giustarini D, and Milzani A. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem Sci 34: 85–96, 2009 [DOI] [PubMed] [Google Scholar]
- 8.Derewenda U, Boczek T, Gorres KL, Yu M, Hung LW, Cooper D, Joachimiak A, Raines RT, and Derewenda ZS. Structure and function of Bacillus subtilis YphP, a prokaryotic disulfide isomerase with a CXC catalytic motif. Biochemistry 48: 8664–8671, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckers E, Bien M, Stroobant V, Herrmann J, and Deponte M. Biochemical characterization of dithiol glutaredoxin 8 from Saccharomyces cerevisiae: the catalytic redox mechanism redux. Biochemistry 48: 1410–1423, 2009 [DOI] [PubMed] [Google Scholar]
- 10.Eiamphungporn W, Soonsanga S, Lee JW, and Helmann JD. Oxidation of a single active site suffices for the functional inactivation of the dimeric Bacillus subtilis OhrR repressor in vitro. Nucleic Acids Res 37: 1174–1181, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eschenfeldt W, Lucy S, Millard C, Joachimiak A, and Mark I. A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol Biol 498: 105–115, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fahey RC. Novel thiols of prokaryotes. Annu Rev Microbiol 55: 333–356, 2001 [DOI] [PubMed] [Google Scholar]
- 13.Fernandes AP. and Holmgren A. Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid Redox Signal 6: 63–74, 2004 [DOI] [PubMed] [Google Scholar]
- 14.Fuangthong M, Atichartpongkul S, Mongkolsuk S, and Helmann JD. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J Bacteriol 183: 4134–4141, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuangthong M. and Helmann JD. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc Natl Acad Sci U S A 99: 6690–6695, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaballa A, Newton GL, Antelmann H, Parsonage D, Upton H, Rawat M, Claiborne A, Fahey RC, and Helmann JD. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc Natl Acad Sci U S A 107: 6482–6486, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallogly MM. and Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr Opin Pharmacol 7: 381–391, 2007 [DOI] [PubMed] [Google Scholar]
- 18.Helmann JD. Bacillithiol, a new player in bacterial redox homeostasis. Antioxid Redox Signal 15: 123–133, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho SN, Hunt HD, Horton RM, Pullen JK, and Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59, 1989 [DOI] [PubMed] [Google Scholar]
- 20.Hong M, Fuangthong M, Helmann JD, and Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol Cell 20: 131–141, 2005 [DOI] [PubMed] [Google Scholar]
- 21.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol 57: 395–418, 2003 [DOI] [PubMed] [Google Scholar]
- 22.Jortzik E, Wang L, and Becker K. Thiol-based posttranslational modifications in parasites. Antioxid Redox Signal 17: 657–673, 2012 [DOI] [PubMed] [Google Scholar]
- 23.Kehr S, Jortzik E, Delahunty C, Yates JR, 3rd, Rahlfs S, and Becker K. Protein S-glutathionylation in malaria parasites. Antioxid Redox Signal 15: 2855–2865, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JW, Soonsanga S, and Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci U S A 104: 8743–8748, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lillig CH, Berndt C, and Holmgren A. Glutaredoxin systems. Biochim Biophys Acta 1780: 1304–1317, 2008 [DOI] [PubMed] [Google Scholar]
- 26.Lindahl M, Mata-Cabana A, and Kieselbach T. The disulfide proteome and other reactive cysteine proteomes: analysis and functional significance. Antioxid Redox Signal 14: 2581–2642, 2011 [DOI] [PubMed] [Google Scholar]
- 27.Luo Y. and Helmann JD. A sigmaD-dependent antisense transcript modulates expression of the cyclic-di-AMP hydrolase GdpP in Bacillus subtilis. Microbiology 158:2732–2741, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mascher T, Margulis NG, Wang T, Ye RW, and Helmann JD. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol 50: 1591–1604, 2003 [DOI] [PubMed] [Google Scholar]
- 29.Masip L, Veeravalli K, and Georgiou G. The many faces of glutathione in bacteria. Antioxid Redox Signal 8: 753–762, 2006 [DOI] [PubMed] [Google Scholar]
- 30.Mishra S. and Imlay J. Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch Biochem Biophys 525: 145–160, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newton GL, Fahey RC, and Rawat M. Detoxification of toxins by bacillithiol in Staphylococcus aureus. Microbiology 158: 1117–1126, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newton GL, Rawat M, La Clair JJ, Jothivasan VK, Budiarto T, Hamilton CJ, Claiborne A, Helmann JD, and Fahey RC. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat Chem Biol 5: 625–627, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J. and Russel DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory, 2001 [Google Scholar]
- 34.Sharma SV, Arbach M, Roberts AA, Macdonald CJ, Groom M, and Hamilton CJ. Biophysical features of bacillithiol: the glutathione surrogate of Bacillus subtilis and other Firmicutes. ChemBioChem 14: 2160–2168, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma SV, Jothivasan VK, Newton GL, Upton H, Wakabayashi JI, Kane MG, Roberts AA, Rawat M, La Clair JJ, and Hamilton CJ. Chemical and chemoenzymatic syntheses of bacillithiol: a unique low-molecular-weight thiol amongst low G+C Gram-positive bacteria. Angew Chem Int Ed Engl 50: 7101–7104, 2011 [DOI] [PubMed] [Google Scholar]
- 36.Shelton MD, Chock PB, and Mieyal JJ. Glutaredoxin: role in reversible protein S-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid Redox Signal 7: 348–366, 2005 [DOI] [PubMed] [Google Scholar]
- 37.Soonsanga S, Fuangthong M, and Helmann JD. Mutational analysis of active site residues essential for sensing of organic hydroperoxides by Bacillus subtilis OhrR. J Bacteriol 189: 7069–7076, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soonsanga S, Lee JW, and Helmann JD. Conversion of Bacillus subtilis OhrR from a 1-Cys to a 2-Cys peroxide sensor. J Bacteriol 190: 5738–5745, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soonsanga S, Lee JW, and Helmann JD. Oxidant-dependent switching between reversible and sacrificial oxidation pathways for Bacillus subtilis OhrR. Mol Microbiol 68: 978–986, 2008 [DOI] [PubMed] [Google Scholar]
- 40.Stout J, De Vos D, Vergauwen B, and Savvides SN. Glutathione biosynthesis in bacteria by bifunctional GshF is driven by a modular structure featuring a novel hybrid ATP-grasp fold. J Mol Biol 416: 486–494, 2012 [DOI] [PubMed] [Google Scholar]
- 41.Stulke J, Hanschke R, and Hecker M. Temporal activation of beta-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J Gen Microbiol 139: 2041–2045, 1993 [DOI] [PubMed] [Google Scholar]
- 42.Vlamis-Gardikas A, Potamitou A, Zarivach R, Hochman A, and Holmgren A. Characterization of Escherichia coli null mutants for glutaredoxin 2. J Biol Chem 277: 10681–10868, 2002 [DOI] [PubMed] [Google Scholar]
- 43.Winterbourn CC. and Kettle AJ. Redox reactions and microbial killing in the neutrophil phagosome. Antioxid Redox Signal 18: 642–660, 2013 [DOI] [PubMed] [Google Scholar]
- 44.Zaffagnini M, Bedhomme M, Lemaire SD, and Trost P. The emerging roles of protein glutathionylation in chloroplasts. Plant Sci 185–186: 86–96, 2012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.