Abstract
Engineered zinc finger nucleases (ZFNs) are a tool for genome manipulation that are of great interest to scientists in many fields. To meet the needs of researchers wishing to employ ZFNs, an inexpensive, rapid assembly procedure would be beneficial to laboratories that do not have access to the proprietary reagents often required for ZFN production. Using freely available sequence data derived from the Zinc Finger Targeter database, we developed a protocol for synthesis and directed insertion of user-defined ZFNs into a versatile plasmid expression system. This oligonucleotide-based isothermal DNA assembly protocol was used to determine whether we could generate functional nucleases capable of endogenous gene editing. We targeted the human α-l-iduronidase (IDUA) gene on chromosome 4, mutations of which result in the severe lysosomal storage disease mucopolysaccharidosis type I. In approximately 1 week we were able to design, assemble, and test six IDUA-specific ZFNs. In a single-stranded annealing assay five of the six candidates we tested performed at a level comparable to or surpassing previously reported ZFNs. One of the five subsequently showed nuclease activity at the endogenous genomic IDUA locus. To our knowledge, this is the first demonstration of in silico-designed, oligonucleotide-assembled, synthetic ZFNs, requiring no specialized templates or reagents that are capable of endogenous human gene target site activity. This method, termed CoDA-syn (context-dependent assembly-synthetic), should facilitate a more widespread use of ZFNs in the research community.
Osborn and colleagues report development of a protocol for synthesis and direct insertion of user-defined zinc finger nucleases (ZFN) into versatile expression systems. ZFNs targeting the human α-L-iduronidase (IDUA) gene designed in this manner performed comparably to previously reported ZFNs. This method, termed CoDA-syn (Context Dependent Assembly-synthetic), should facilitate a more widespread utilization of ZFNs in the research community.
Introduction
Zinc finger nucleases (ZFNs) are engineered proteins composed of a DNA-binding domain that confers exquisite specificity for a particular DNA sequence and is tethered to a nonspecific nuclease domain of the FokI endonuclease (Porteus and Carroll, 2005). The DNA-binding domains contain a left array and a right array, each composed of three individual zinc fingers. Each finger contacts 3 bp of DNA so that each array recognizes and binds to a specific 9-bp sequence termed a “half-site” (Porteus and Carroll, 2005). Separating the finger arrays is a 5- to 7-bp “spacer” region that is cleaved by the FokI endonuclease component. Thus, the fully formed complex, by virtue of its 23- to 25-bp target site recognition sequence, is a highly precise DNA-binding protein that holds great promise as a tool for genome engineering in many organisms. Multiple platforms for generating gene-specific ZFNs exist that are available through academic or industrial entities. These platforms include the CompoZr® service and the oligomerized pool engineering (OPEN), modular assembly, and context-dependent assembly (CoDA) methods (Miller et al., 2007; Maeder et al., 2008; Kim et al., 2009, 2011; Porteus, 2010; Sander et al., 2011).
Sangamo BioSciences (Richmond, CA) offers the CompoZr® service (available through Sigma-Aldrich, St. Louis, MO), which relies on a proprietary generation procedure. Investigators may purchase gene-specific ZFNs through this service, although the associated cost of these reagents may be limiting for some (Miller et al., 2007; Meng et al., 2008). The selection-based OPEN platform is a robust system that is less restricted in its availability. However, the procedure requires specialized reagents and expertise that can take months to perfect (Maeder et al., 2008). The modular approach to ZFN generation relies on the assembly of previously characterized ZFN modules into full arrays. The latest generation of this technique appears to show higher rates of success in gene editing than its predecessors but can still require purchase and assembly of the modules by the end user (Kim et al., 2011). An earlier study by Carroll and colleagues, using previously characterized modules, established the ability of ZFNs to be generated by oligonucleotide assembly (Carroll et al., 2006). Although this was an important study, the length of time to assemble the ZFNs was on the order of weeks, the templates for assembly were modular ZFNs that can show a high failure rate (Ramirez et al., 2008), and the final assembled product is not immediately ready for gene editing in cells. Instead, specificity and activity were assessed in a cell-free assay wherein the recombinant ZFN protein was incubated with naked plasmid DNA (Carroll et al., 2006). Thus, the intracellular gene-editing potential of these oligonucleotide-generated ZFNs is not fully known.
A new method known as CoDA has attributes that make it an attractive alternative to the other generation options: it is a more streamlined generation procedure than OPEN, and preserves the critical component of the context dependency of adjacent fingers in an array during construction that is lacking in modular design (Sander et al., 2011). In addition, both the amino acid and nucleotide sequences for individual arrays are publicly available for CoDA-derived proteins on the CoDA Zinc Finger Targeter (ZiFiT) database (Sander et al., 2007). This free dissemination of sequence data gives investigators the template to generate gene-specific ZFNs, although methods for translating the in silico CoDA ZFN data to functional reagents are lacking in the literature.
Therefore, we sought to define a protocol that would allow the end user to rapidly assemble ZFN arrays into a plasmid vector containing heterodimeric FokI nuclease domains (Doyon et al., 2011). Thus, the end result is a single expression plasmid that permits the coordinated expression of candidate left and right ZFN arrays. To determine the efficacy of such an approach we targeted the α-l-iduronidase (IDUA) gene, which is located on the short arm of chromosome 4. Mutations to this gene result in mucopolysaccharidosis type I (MPS I) because of the loss of IDUA enzymatic activity, resulting in a severe phenotype with substantial morbidity and early death (Cleary and Wraith, 1995; Muenzer et al., 2009). IDUA is a highly relevant disease gene that allowed us to validate our assembly method as well as determine whether CoDA proteins, previously shown capable of targeting only zebrafish and plant genes, were also functional for human gene editing.
In 1 week we generated six IDUA-specific ZFNs by this method at comparatively low cost and showed site-specific activity of five of them in a mammalian cell single-stranded annealing assay. To test their activity at an endogenous target locus we used oligonucleotide duplex capture, Surveyor nuclease, and homology-directed integration methods to show, for the first time, the ability of in silico-designed, artificially synthesized CoDA ZFNs to target a human gene, α-l-iduronidase.
Materials and Methods
Zinc finger targeter analysis
The ZiFiT website (http://zifit.partners.org/ZiFiT/ChoiceMenu.aspx) allows users to copy and paste in their sequence of interest and returns the available CoDA ZFN sites as well as the DNA sequences that will encode a particular array. We inserted human IDUA sequence and chose six of the sites that were returned.
Oligonucleotide design, generation, and purification
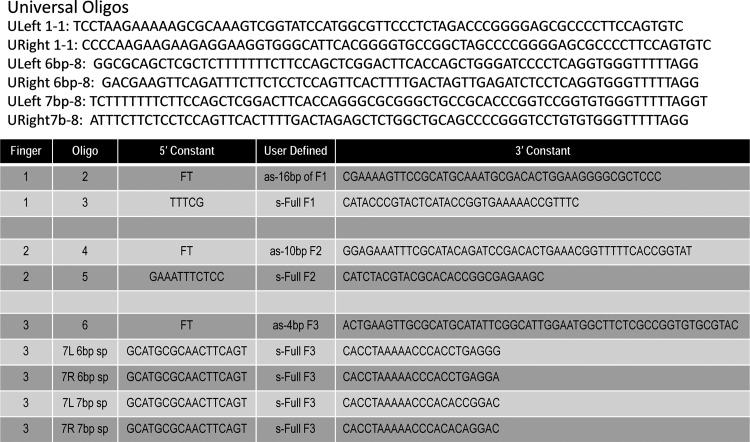
For the present study, oligonucleotides (oligos) were ordered from Integrated DNA Technologies (Coralville, IA). Each array contained eight oligos and, for the left array fingers, all the fingers used the Universal Left 1-1 primer. Depending on the spacer sequence a Universal Left 6bp primer (also used when 5-bp spacers were present) or Universal Left 7bp primer was used. For the right arrays a Universal Right 1-1 primer was used and a Universal Right 6bp or Universal Right 7bp primer was used. The universal primers were ordered on the 100-nmol scale and purified by polyacrylamide gel electrophoresis (PAGE) whereas the remaining oligos were ordered on the 10-nmol scale with standard desalting. The entire primer set used in this work is presented in Supplementary Table S1 (supplementary data are available online at www.liebertonline.com/hum).
Overlapping extension PCR
The eight oligos for a left or right array were resuspended in TE (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA) to a concentration of 50 μM and then pooled to a final per-oligo concentration of 200 nM. For the left arrays the mixture was composed of the Universal Left 1-1 and Universal Left 6bp or 7bp eighth (1-8) oligos, depending on the spacer sequence. For right arrays the Universal Right 1 and Universal Right 6bp or 7bp eight oligos were used. One microliter of the primer mix was used in a 100-μl left or right finger PCR containing 1× high-fidelity (HF) buffer (New England BioLabs [NEB], Ipswich, MA), 200 μM dNTPs, 2 μl of 50 mM MgCl2, 2 U of Phusion DNA polymerase (NEB), and 0.2 μM of primer (Universal Left assembly Forward [5′-TCCT AAG AAA AAG CGC AAA GTC GGT-3′] and Left 6bp assembly Reverse [5′-GGCGCAGCTCGCTCTTTTTTTC-3′] or Left 7bp assembly Reverse [5′-CTTTTTTTCTTCCAGCTCGGAC-3′] for the left 6- or 7-bp arrays and Right PCR assembly Forward [5′-CCCAAGAAGAAGAGGAAGGTGGGCATTC-3′] and Right PCR assembly Reverse [5′-TTTCTTCTCCTCCAGTTCAC-3′]). The reaction conditions were as follows: 98°C for 30 sec; 34 cycles of 98°C for 10 sec, 60°C for 30 sec, and 72°C for 30 sec; and 72°C for 10 min.
Vector digestion and isothermal DNA assembly
Two sets of digests were required for each assembly. One generated the “vector backbone” and the second created a large quantity of the “middle fragment.” For the pMJO-6 vector 1 μg of the vector was digested with BglII and XbaI (NEB) to create the backbone and 10 μg was digested with BamHI and NheI to generate the middle fragment. For pMJO-7 the vector backbone was generated by a PstI and XbaI digest whereas the middle fragment was created by an NheI/RsrII digest (NEB). The PCR fragments and vector digests were resolved on a 1.5% agarose gel and the top band of the backbone digest, the smaller band of the middle digest, and each PCR product were purified with a QIAquick gel extraction kit (Qiagen, Hilden, Germany) with the plasmid fragments eluted in 10 μl and the PCR products eluted in 50 μl of water. Two microliters each of the backbone and middle fragments were pooled and to this was added 1.5 μl each of the PCR fragments. Five microliters of this was then added to a 15-μl aliquot of the isothermal DNA assembly mixture and incubated immediately at 50°C for 30 min as described (Gibson et al., 2009) and in the online supplement.
Colony PCR and DNA sequencing
Candidate colonies were picked with a pipette tip, placed in 50 μl of water, boiled for 5 min, placed on ice for 2 min, and spun at high speed for 1 min. Five microliters of the lysate was then used for colony PCR, using 0.2 μM each of the left assembly forward and right assembly reverse primers, described previously, under the following conditions: 98°C for 5 min; 34 cycles of 98°C for 40 sec, 57°C for 40 sec, and 72°C for 70 sec; and 72°C for 10 min with 200 μM dNTPs, 1× CoralLoad PCR buffer, and 2.5 U of Taq DNA polymerase (Qiagen). After confirmation of left and right finger inserts the colonies were grown overnight and plasmid DNA was isolated with a Wizard Plus SV mini-prep kit (Promega, Madison, WI) and submitted for sequencing with the left sequencing primer (5′-CACCATGGATTATAAGGATCACGATGG-3′) and the right sequencing primer (5′-CGCAAGTTCAACAATGGTG-3′).
Single-stranded annealing target plasmid construction
The template plasmid for single-stranded annealing (SSA) assembly was the pGL3 Control Vector from Promega. To generate the SSA targeting vector, in which the ZFN site is inserted between two halves of the luciferase gene, the following primers were used for the left half: pGL3 HindIII Forward (5′-TTCCAGAAGTAGTGAGGAGG-3′) and SSA Reverse (5′-XXXXXXXXXXXXXXXXXXXXXXXXXX(X)TCATCACATAGGACCTCTCACACACAGTTCG-3′), where the “X” portion represents the 26-bp sequence obtained from ZiFiT for a particular 6-bp spacer ZFN site [note: for 7-bp sites the target site comprises 27 bp and this extra base is denoted as (X)] and was incorporated into the primer in the antisense orientation, and the italicized portion corresponds to pGL3. To generate the right half, SSA forward primer 5′-TGATGAXXXXXXXXXXXXXXXXXXXXXXXXXX(X)CGACATTTATAATGAACGTGAATTGC-3′ was used This primer contains a 5′ double stop codon followed by 26 or 27 bp of the ZFN target site in the sense orientation followed by the pGL3 sequence (an example of SSA primers is shown in Supplementary Fig. S1B). The pGL3-specific reverse primer (pGL XbaI Reverse, 5′-ATGTATCTTATCATGTCTGC-3′) was used to generate the right half of the SSA reporter. To reduce the number of plasmids we included two target sites per plasmid. The PCR conditions for the left/right reactions were as follows: 0.2 μM of primer with 1 ng of pGL3 (Promega), 1× high-fidelity (HF) buffer (NEB), 200 μM dNTPs, 2 μl of 50 mM MgCl2, and 2 U of Phusion DNA polymerase (NEB), and the reaction was performed at 98°C for 30 sec; 34 cycles of 98°C for 10 sec, 60°C for 30 sec, and 72°C for 120 sec; and 72°C for 10 min.
Simultaneously, as the PCR is underway, 1 μg of the pGL3 vector was digested with HindIII and XbaI. The PCRs and vector digest were resolved on a 1% agarose gel and the upper band of the vector and the PCR products were gel purified as described previously and eluted in 20 μl (vector) and 100 μl (PCR) of water. Three microliters of the vector was mixed with 1.5 μl each of the PCR products and 5 μl was used for isothermal DNA assembly and transformation as described previously. Colonies were digested with SphI to confirm the presence of an insert and then sequenced with SSA sequencing primer 5′-TTATCGGAGTTGCAGTTGCG-3′.
Single-stranded annealing assay
Twenty-four hours before gene transfer, HEK 293 cells were seeded in quadruplicate in a 24-well dish at a density of 200,000 cells per well in complete RPMI medium. Doses of 50, 100, 250, and 500 ng of the appropriate nuclease and 25 ng of the SSA reporter were transfected into 293 cells, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. As a comparative control a separate group into which only the SSA plasmid was transfected was included. As a normalization control all reactions also contained 4 ng of the pRL-TK Renilla luciferase plasmid (Promega). Twenty-four hours after gene transfer the cells were lysed in 300 μl of 1× passive lysis buffer (Promega). Intracellular luminescence was measured with a Dual-Luciferase reporter assay system (Promega) according to the manufacturer's instructions. Firefly luciferase values were normalized to Renilla luciferase values and then that value was divided by the SSA reporter-alone treatment group to give the fold activation of a candidate ZFN.
Oligonucleotide duplex capture
Oligo capture was performed with, as described (Miller et al., 2011), the following annealed primers: top (5′-X*X*XXGTACGGATCCAAGCTTCGTCGACCTAGCC-3′) and bottom (5′-X*X*XXGGCTAGGTCGACGAAGCTTGGATCCGTAC-3′), where “X” refers to the target site-specific overhang and the asterisks (*) refer to a phosphorothioate linkage (primer pairs are listed in Supplementary Table S1 as Top/Bottom in the “Oligo Duplex ss oligos” section). Five microliters of a 40 μM oligo mix±the nuclease were transfected by Lipofection (Invitrogen) into 293 cells and genomic DNA was harvested 72 hr later. PCR was performed with the oligo-specific primer 5′-GTACGGATCCAAGCTTCGTCGACCTAGCC-3′ and the gene-specific endogenous primer (“endo”-designated primers in Supplementary Table S1) with Platinum Taq DNA polymerase high fidelity (Invitrogen) and 1× Q solution (Qiagen) at 94°C for 120 sec, followed by 35 cycles of 94°C for 30 sec, 60°C for 30 sec, and 68°C for 60 sec; and 68°C for 10 min. PCR products were cloned into the TOPO TA cloning vector (Invitrogen) and sequenced.
Surveyor nuclease assay
HEK 293 cells were transfected by lipofection (Invitrogen) with the individual nucleases or a GFP control plasmid. Genomic DNA was harvested 72 hr later and PCR was performed with the Endo Forward and Endo Reverse primers from Supplementary Table S1 under the same conditions as the oligonucleotide duplex capture assay. The PCR products were purified with a QIAquick PCR purification kit (Qiagen) and the Surveyor nuclease procedure was performed as described by Guschin and colleagues (2010).
Homology-directed integration
Minimal donor arms that flank the N170 cut site on either side of the spacer region extending 40 bp into the genomic locus were included in PCR primers: HDI short arm donor F (5′-C*G*CTGCCACACAGCCAGGCTGACCAGTACGTCCTCAGCTGGGACCAGCAGCTCAACCTCACCGGGTAGGGGAGGCGCTTTTCC-3′) and HDI short arm reverse: (5′-*C*AGCAGCCAGTGGGTCCGGACCTGCTTGATGCCGCGGTGAGGGACGGCGCCCAC AGCTGGTTCTTTCCGCCTCAGAAGC-3′), where the asterisks (*) refer to a phosphorothioate linkage. PCR conditions to generate the donor were as follows: 0.2 μM of primer with 1 ng of target plasmid, 1× high-fidelity (HF) buffer (NEB), 200 μM dNTPs, 2 μl of 50 mM MgCl2, and 2 U of Phusion DNA polymerase (NEB), and PCR was performed at 98°C for 30 sec; 34 cycles of 98°C for 10 sec, 60°C for 30 sec, and 72°C for 120 sec; and 72°C for 10 min. The entire PCR product was resolved on a 1% agarose gel, gel purified as described previously, and resuspended in 10 μl of water. To this was added 500 ng of nuclease 170 (N170) and the entire mixture was nucleofected (solution V, program Q-001; Lonza, Basel, Switzerland). At 72 hr postnucleofection genomic DNA was harvested from these cells (or cells receiving the donor in the absence of nuclease) and PCR was performed with three primers: two forward primers (N170 Endo F, 5′-GGCTTGAACGTGTGTGTCAG-3′; and the donor-specific HR screen F, 5′-CATCGCATTGTCTGAGTAGG-3′) with N170 Endo Reverse (5′-ACACACACAGGGATGCTCAC-3′). The PCR conditions were as follows: 1× buffer, 1× Q solution (Qiagen), a 0.2 μM concentration of each primer, 0.2 mM dNTPs, and 5 U of Taq DNA polymerase (Qiagen) at 94°C for 120 sec; 40 cycles of 94°C for 30 sec, 60°C for 30 sec, and 72°C for 40 sec; and 72°C for 10 min. PCR products were resolved on a 1% agarose gel for densitometric analysis as well as cloned into the TOPO TA cloning vector (Invitrogen) and sequenced.
Densitometry
Ethidium bromide-stained DNA gels were photographed and saved as TIFF files for analysis with ImageJ (Rasband, 2011). The density of the bands was determined and the frequency of gene modification was determined according to the following equation: 100×(1−(1−fraction cleaved)1/2) as described (Guschin et al., 2010).
Results and Discussion
In silico gene target site selection and oligonucleotide design
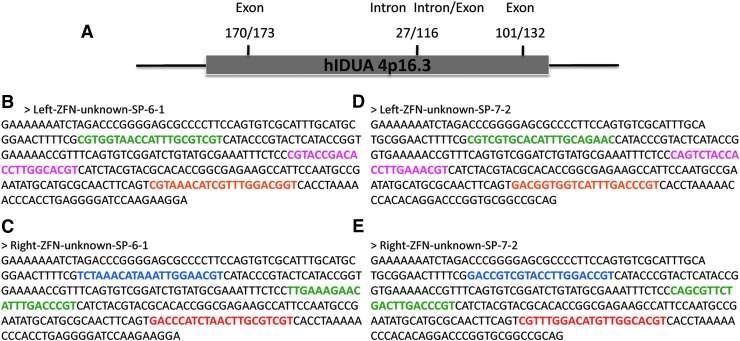
The ZiFiT version 3.3 website contains options for target gene analysis for ZFN generation by the modular, OPEN, and CoDA platforms. We analyzed sequence from the IDUA gene on human chromosome 4, using the CoDA ZiFiT database. We targeted the 5′, middle, and 3′ portions of the gene and each target site is within 200 bp of a base pair that when mutated results in mucopolysaccharidosis type I (MPS I). Our target sites were named 170, 173, 27, 116, 101, and 132. Targets 170 and 173 are near the 5′ portion of the gene and are both in exons; target 27 is within an intron, and target 116 is at an intron/exon boundary, in the central portion of the gene; and targets 101 and 132 are 3′ proximal and are both in exons (Fig. 1A). We chose these loci as they are clinically relevant and limited our candidate list to these in order to optimize and validate the assembly procedure. Of note, the relationship between these selected sites and the CompoZr® knockout ZFN kit targeting IDUA (CKOZFN10448; Sigma-Aldrich) is currently unknown.
FIG. 1.
α-l-Iduronidase (IDUA) sequence analysis and oligonucleotide design. (A) Three separate regions of the human IDUA gene were targeted for the development of six nucleases. Above each numerically designated site is its location (intron or exon). The ZiFiT CoDA Design Zinc Finger Nucleases option reveals target sites and spacer sequence and displays the DNA sequence to be assembled. (B) Shown are examples of the nucleotide sequence required to encode a 6-bp left or (C) right array, and a (D) 7-bp left or (E) right ZFN array. Color-coded regions in the left array sequences correspond to Finger One (green), Finger Two (pink), and Finger Three (orange). In the right arrays Finger One is in blue, Finger Two is in green, and Finger Three is in red.
ZiFiT also returns the DNA sequence required to encode a particular nuclease candidate. Shown are nucleotide sequences for a representative left and right three-fingered ZFN array that require either a 6-bp (Fig. 1B and C) or 7-bp spacer (Fig. 1D and E) region. The length of the spacer region, in which the double-stranded DNA break occurs, is critical from a design standpoint as the amino acid linker tethering the ZFN to the nuclease domain differs depending on the spacer size (Handel et al., 2009). The highlighted bases in the sequences comprise the unique DNA-binding portions of the array. A framework for artificial synthesis was derived from this sequence and the oligo design rules are represented in Fig. 2 and an example oligo assembly pool is listed in Supplementary Fig. 1A. This strategy relies on eight oligos; with the first and eighth oligos serving as universal primers that can be used for any left or right assembly for a particular spacer sequence. As such, these oligos are slightly longer than the others and therefore we recommend PAGE purification to ensure full-length products. The remaining six oligos, defined by the user on the basis of the sequence of the left or right array they wish to generate from the ZiFiT database, overlaps its neighbor by 21 bp and contains 39 bases of new sequence. These oligos were ordered from Integrated DNA Technologies at the 10-nmol scale with standard desalting at a per-oligo cost of approximately $12.
FIG. 2.
Context-dependent assembly-synthetic (CoDA-syn) eight-oligonucleotide design strategy. Top: Universal primers: The system is designed such that each array will use a universal left or right 5′ primer (ULeft 1-1, URight 1-1) and, depending on the spacer sequence, a universal eighth oligo will be used for all left or right arrays (Universal Left 6bp spacer, ULeft 6bp-8; Universal Right 6bp spacer, URight 6bp-8; Universal Left 6bp spacer, ULeft 7bp-8; Universal Right 6bp spacer, URight 7bp-8). Bottom: The remaining six oligos constitute the DNA sequence that encodes a particular user-defined left or right array. Oligo 2 will contain the first 16 bp of Finger 1 in the antisense orientation and is at the 5′ terminal end of the oligo. Oligo 3 contains the full Finger One sequence in the sense orientation. Oligo 4 contains the first 10 bp of Finger Two in the antisense orientation. Oligo 5 will contain the entire Finger Two sequence in the sense orientation. Oligo 6 will have the first 4 bp of Finger Three in the antisense orientation. Oligo 7 will have the entire Finger Three sequence in the sense orientation. The oligos in position 7 are unique for left/right/6/7-bp spacers. FT, finger terminal; as, antisense; s, sense; F, finger.
CoDA-syn DNA assembly method
We developed an assembly procedure in which four separate double-stranded DNA (dsDNA) fragments are simultaneously combined to form an intact expression plasmid. We created two entry expression plasmids: the pMJO-6 vector, which allows for expression of any nuclease with a 5- or 6-bp spacer, and the pMJO-7 vector, which is used for nucleases requiring 7-bp spacers. Each vector is under the control of the cytomegalovirus (CMV) promoter and also contains a T7 RNA polymerase-binding site that allows ZFNs to be delivered as either DNA or RNA. Each vector contains a perfect Kozak consensus sequence and mammalian and zebrafish codon-optimized heterodimeric ZFN monomers containing a simian virus 40 (SV40) nuclear localization signal. The nucleases are separated from one another by a self-cleaving picornaviral T2A peptide sequence that mediates equimolar ZFN left and right array expression from a single open reading frame (Osborn et al., 2005). In addition, the FokI nuclease domains have been modified to encode (+) and (–) obligate heterodimers that significantly reduce ZFN toxicity (Doyon et al., 2011).
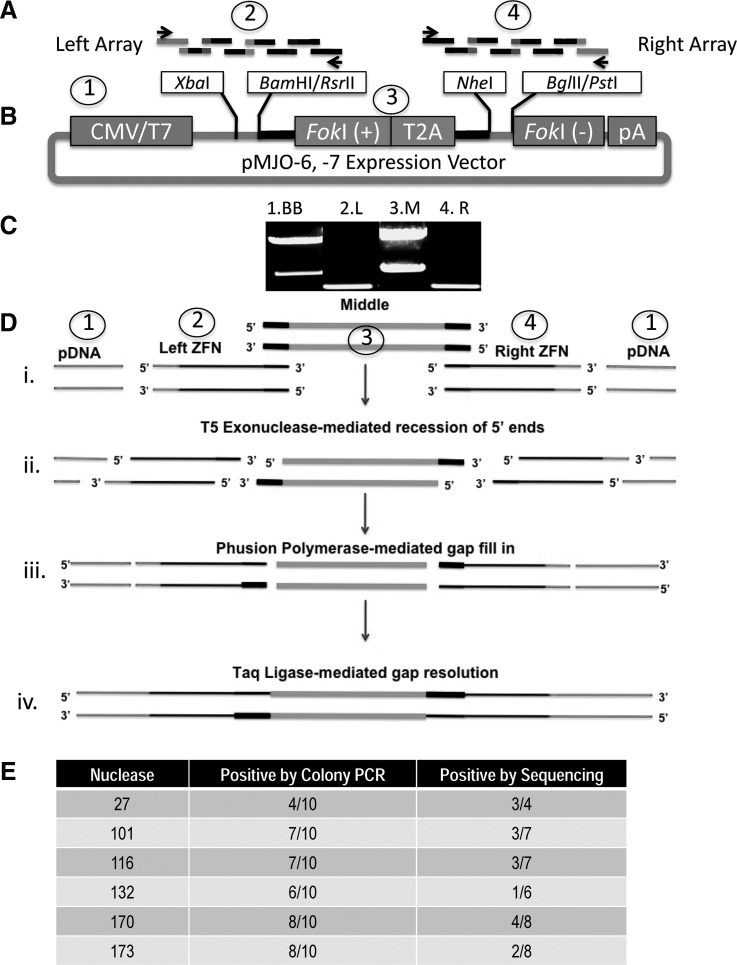
The single-stranded oligos that form the left and right ZFN arrays were pooled at an equimolar concentration and subjected to overlapping extension PCR using terminal flanking PCR primers to generate left and right array dsDNA products (Fig. 3A). Thus, the left array dsDNA served as fragment two and shared overlap regions with fragments one (expression plasmid backbone downstream of the CMV promoter) and three [upstream of the FokI (+) heterodimer] (Fig. 3A and B). Similarly, the right ZFN PCR product overlapped with the region upstream of the FokI (–) heterodimer domain of the fragment 1 plasmid backbone (Fig. 3A and B). To generate plasmid sequences homologous to the PCR products, and to avoid the potential for PCR-introduced errors in vector sequence, a restriction digest was used to liberate vector backbone DNA containing sequences required for bacterial propagation (Fig. 3B, fragment 1) and a “middle” fragment containing the FokI (+) nuclease dimer (Fig. 3B, fragment 3). Figure 3C shows the four dsDNA assembly fragments: the backbone, ZFN left, middle fragment, and ZFN right. All of these were then pooled in one reaction (Fig. 3D, i) with a isothermal DNA assembly mixture that contained T5 exonuclease, Phusion DNA polymerase, and Taq ligase, which work in concert to connect the DNA fragments (Gibson et al., 2009). The T5 exonuclease served to recess the overlap regions between the multiple DNA fragments, creating single-stranded overhangs that can anneal (Fig 3D, ii). The reaction was performed at 50°C, which results in slow inactivation of the heat-labile T5 enzyme and allows for homologous region fill-in by Phusion polymerase (Fig. 3D, iii). Last, the intact expression plasmid was completed after Taq ligase-mediated covalent linkage of the gaps (Fig. 3D, iv).
FIG. 3.
CoDA-syn DNA assembly. (A) Overlapping extension PCR. The left (fragment 1) or right (fragment 4) ZFN arrays containing eight overlapping oligonucleotides were mixed together for use in an overlapping extension PCR to generate dsDNA. (B) Expression vector preparation. To generate the backbone fragment of the 6-bp spacer plasmid (pMJO-6) the vector was digested with BglII and XbaI (fragment 1). To generate the middle portion (fragment 3) this plasmid was digested with BamHI and NheI. For the 7-bp spacer (pMJO-7) the backbone/fragment 1 was generated with PstI and XbaI, and the middle/3 fragment was produced by digesting with NheI and RsrII. (C) Agarose gel analysis of dsDNA fragments. Fragment 1, plasmid backbone (upper band); fragment 2, left ZFN array; fragment 3, middle fragment (lower band); fragment 4, right ZFN array. (D) Isothermal DNA assembly. (i) Overlapping fragments 1–4 were mixed together and assembled. (ii) T5 exonuclease creates ssDNA complementary overhangs that allow complementary base pair annealing. (iii) Phusion polymerase fills in the gaps, which are then sealed by Taq ligase (iv). (E) Colony PCR and sequence analysis of assembled nuclease candidates.
After bacterial transformation of the postassembly mixture, we consistently observed >100 transformants per reaction. Prescreening by colony PCR showed that 40–80% of the candidates contained the desired left-middle-right fragments (Fig. 3E). This procedure is based on the homologous sequences of the left and right fingers for the middle fragment as well as the corresponding vector sequences (Fig. 3). However, because the architecture for any ZFN is highly similar, a left–right fusion product (i.e., with the exclusion of the middle fragment) can also be present (data not shown). We have observed that the amount of middle fragment present is critical for the recovery of a fully competent expression cassette. Therefore, the middle fragment generation digest uses an excess of plasmid (Fig. 3C) to generate this segment for the reaction, and decreasing its input results in a reduction of the desired left-middle-right finger insertions (data not shown).
We sequenced all of the candidates identified in the colony PCR and observed that between 16 and 75% of the plasmids showed fully correct left and right finger sequence (Fig. 3E). Consistent with previous findings, the errors identified by sequencing were small deletions as opposed to misassembled DNA fragments (Gibson et al., 2011). The Gibson group showed fully correct sequence in their assembly strategy at rates of 0–85.7%, which encompasses the range seen in our results of 16–75% (Gibson et al., 2011). Of note, their strategy typically assembled eight oligos into a plasmid and then determined assembly accuracy by sequencing. We assembled two pools of eight oligos each and assembled them simultaneously into a single plasmid and considered the sequence to be correct only if the entire 16-oligo synthetic sequence was correct. The source of the error was most likely the inherent defects of oligonucleotide synthesis, which include truncated oligos or oligos with internal deletions (Xiong et al., 2008). These errors can be minimized by having the oligos purified by PAGE, although this would add significantly to the cost and time frame for synthesis. Despite the imperfections of oligo chemistry we were able to obtain at least one sequence confirmed ZFN for each of our candidates during the initial screening process. In addition, if required (which we did not find necessary in this work), more transformants can be screened by colony PCR and sequenced. These data highlight the speed, efficiency, and applicability of the DNA isothermal assembly method for the rapid generation of ZFNs.
Single-stranded annealing assay
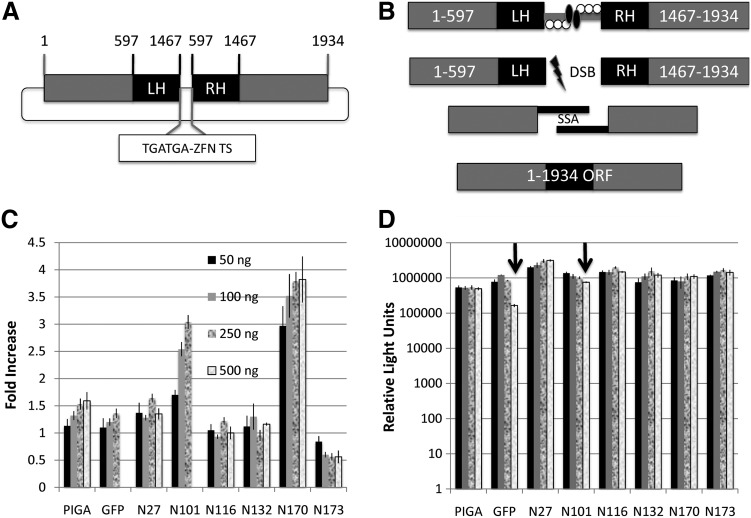
To assess candidate ZFN activity we employed the SSA assay, whereby a ZFN target sequence was inserted between two duplicated halves of the firefly luciferase gene (Fig. 4A). In this form the luciferase reporter gene does possess some basal luciferase activity and establishing this level is important, as it can vary between different SSA targeting plasmids. When ZFNs are cointroduced with this reporter they can bind to the specific target sequence and introduce a double-stranded DNA break, resulting in cellular repair in which the duplicated halves are used as templates that reconstitute the full open reading frame (Fig. 4B). The six IDUA ZFNs were transfected at four different concentrations into HEK 293 cells along with the appropriate SSA target. As an internal transfection control and a marker for ZFN toxicity we also included a Renilla luciferase plasmid and determined dual luciferase expression, using the Dual-Luciferase reporter assay system, which allows for sequential measurement of firefly and Renilla luciferase in the same sample. For comparison, we included a previously characterized, OPEN-generated nuclease to the phosphatidylinositol glycan anchor biosynthesis, class A (PIGA) gene (Zou et al., 2009) as well as a ZFN pair for the green fluorescent protein (GFP) gene (Alwin et al., 2005; Bhakta and Segal, 2010). Five of our candidates (27, 101, 116, 132, and 170) showed activity that equaled or surpassed that of the PIGA and GFP nucleases and one candidate (170) exceeded each of the previously characterized ZFNs by at least a 2-fold margin (Fig. 4C). PIGA, GFP, and nucleases 101 and 170 showed higher activity as the dose of plasmid increased (Fig. 4C). Measurement of the cotransfected Renilla luciferase highlights the importance of including the Renilla plasmid. First, it serves as a normalizing transfection control; and second, it is an indicator of toxicity due to nonspecific ZFN cleavage. A decrement in Renilla luciferase expression is likely due to genotoxicity caused by off-target nuclease activity and this was observed for the GFP and 101 nucleases as the dose increased (Fig. 4D). As the ZFN dose increased Renilla expression decreased for these two nucleases and we set an arbitrary “toxicity limit” defined as a ≥1.5-fold decrease in Renilla luciferase for a particular treatment dose versus the lowest dose ZFN group. Any value exceeding this resulted in classification of the nuclease as toxic and the SSA data were discarded because the low Renilla levels resulted in an artificially inflated SSA fold increase value. The PIGA, 27, 116, 132, 170, and 173 nucleases did not appear to exhibit any associated toxicity (Fig. 4D). However, because nuclease 173 failed to activate at the level of PIGA or GFP (Fig. 4C) it was classified as inactive and was not tested further. These data show the usefulness of the SSA to rapidly quantify ZFN activity in a user-defined cell type and underscores the importance of testing the nucleases over a range of concentrations to determine whether toxicity is evident.
FIG. 4.
CoDA-syn candidate dosage activity and toxicity: single-stranded annealing (SSA) reporter. (A) The specific ZFN target site was introduced downstream of a double-stop codon (TGATGA-ZFN TS) luciferase reporter that is composed of nucleotides (nt) 1–597 and then a duplicated region of nt 597–1467, followed by the remaining nucleotide sequence of the luciferase gene (nt 1467–1937). (B) SSA mechanism. ZFN binding to the target site results in a double-stranded break (lightning bolt) with recession and repair by the cellular SSA pathway, using the duplicated regions as the template. The fully active open reading frame is the result of repair. (C) Candidate ZFN SSA activity. The six synthetic IDUA ZFNs were cotransfected at four different concentrations with the appropriate SSA reporter into HEK 293 cells and luminescence was measured 24 hr later and normalized to Renilla luciferase expression. For comparison we included the human PIGA nuclease as well as a GFP-specific set of ZFNs. (D) Renilla luciferase activity. Renilla luciferase was measured with the Dual-Luciferase reporter assay system, using the same cells as in (C). Arrows indicate a 1.5-fold or greater decrease in Renilla activity due to dose-dependent ZFN toxicity. Data are represented as fold increase relative to cells transfected with the SSA reporter alone. n=4 replicates, mean and SD are shown. DSB, double-strand break; GFP, green fluorescent protein; LH/RH, left and right half (nt 597–1467) SSA duplicated regions; ORF, open reading frame; PIGA, phosphatidylinositol glycan anchor biosynthesis, class A; TGATGA-ZFN TS, double-stop codon zinc finger nuclease target site.
ZFN endogenous locus activity
An important consideration for the employment of ZFNs is the epigenetic profile of the endogenous target locus. Chromatin condensation, DNA structure, or competing endogenous binding factors can impact ZFN accessibility to the target (Bhakta and Segal, 2010). These phenomena are likely not accounted for in the SSA because the target plasmid is an extrachromosomal episome that is incubated for only 24 hr and, thus, may not be subject to the same epigenetic patterns of the genomic locus.
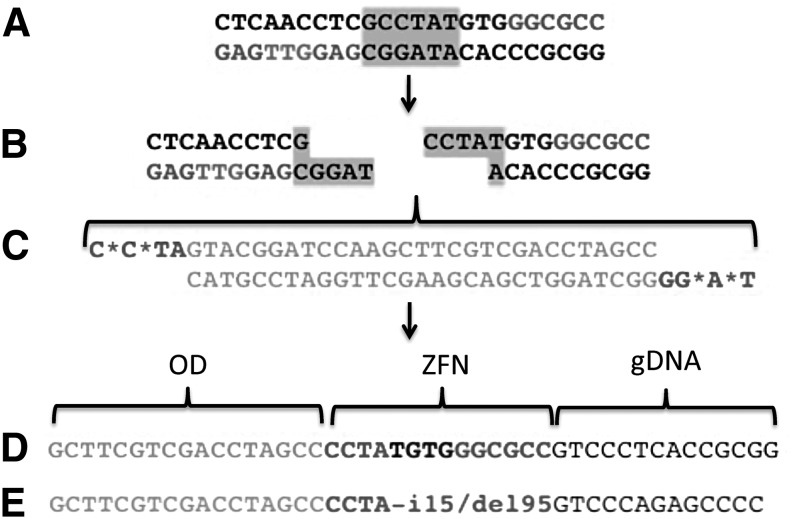
To assess synthetic ZFN activity at the endogenous target we employed the oligonucleotide duplex (OD) assay (Orlando et al., 2010). The principle behind this assay is that nuclease cutting results in a staggered, 4-bp 5′ overhang into which a target-specific OD can be ligated (Orlando et al., 2010). We tested nucleases 27, 116, 101, 132, and 170, using site-specific ODs for each, and then PCR-amplified the genomic DNA from treated cells, using an OD-specific forward primer and a genomic locus-specific reverse primer. Only nuclease 170/OD-treated cells yielded a PCR product; cells treated with nuclease 27/OD, 116/OD, 101/OD, and 132/OD and, importantly, cells transfected with an OD in the absence of a nuclease failed to show a PCR product (data not shown). Figure 5A shows the nuclease 170 target that, when cleaved by the nuclease, is staggered (Fig. 5B) and contains complementary overhangs into which an OD (Fig. 5C) can be inserted. We sequenced the PCR products from the nuclease 170/OD treatment group and observed two events: A perfect ligation of the OD into the target site (Fig. 5D) as well as the presence of an insertion of 15 bp followed by a deletion of 95 bp at the site (Fig. 5E). Both events are consistent with nuclease activity at the target site, the former being characteristic of a cut and perfect repair-mediated insertion and the latter being common for resection and insertion of the OD by the error-prone nonhomologous end joining pathway (Orlando et al., 2010; Miller et al., 2011). These results show that nucleases 27, 116, 101, and 132, despite their activity in the SSA (Fig. 3C), were not functional at the endogenous target locus, suggesting that the genomic landscape of these ZFN targets may be unfavorable for these candidates (Remy et al., 2010). These results also show that nuclease 170 was able to modify the endogenous target site as evidenced by the insertion of the OD at the target site (Fig. 5D and E). These data are the first evidence of functionality of a CoDA-based protein in human gene editing.
FIG. 5.
CoDA-syn nuclease activity at the cellular IDUA locus. (A and B) Nuclease 170 target site (A) before and (B) after nuclease cleavage resulting in staggered 5′, 4-bp overhangs. (C) Artificial oligonucleotide duplex (OD) with overhang regions compatible with a specific ZFN cleavage product and also containing phosphorothioate linkages (*) to prevent intracellular nuclease degradation. Detection of nuclease-dependent endogenous target site modification. The OD insertion event can be detected by using OD and genomic DNA-specific primers and characterized by DNA sequencing. Sequence analysis of genome editing. (D) OD insertion into the target site by perfect ligation. (E) OD target site insertion after DNA recession and repair by nonhomologous end joining resulting in a 15-bp insertion and a 95-bp deletion (i15/del95) followed by genomic sequence.
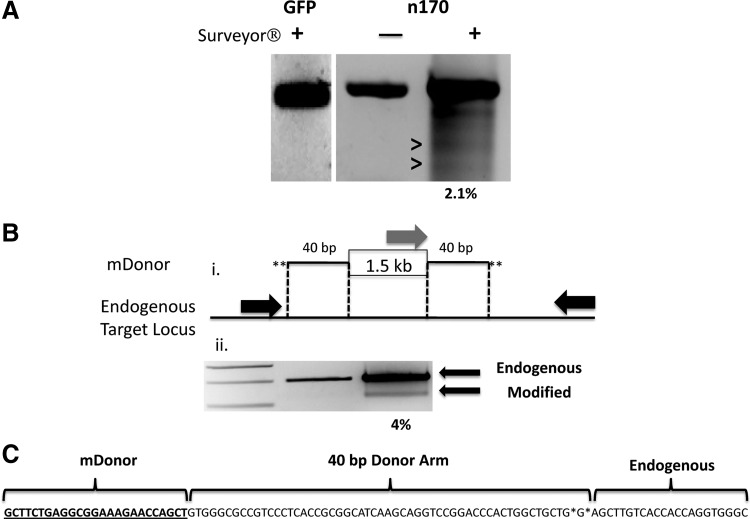
The OD assay is a qualitative measurement of nuclease activity at a target site and, to determine the frequency of endogenous gene cutting, we performed the Surveyor nuclease procedure. This assay uses a member of the CEL family of proteins that are mismatch-specific nucleases capable of detecting nuclease-dependent nonhomologous end-joining (NHEJ) events at the target locus (Guschin et al., 2010). We treated HEK 293 cells with a GFP plasmid or nuclease 170, harvested the DNA, and performed the Surveyor assay and observed no bands in the GFP control or nuclease 170-treated cells that were not incubated with the Surveyor enzyme (Fig. 6A). However, nuclease 170-treated cells that were subjected to Surveyor enzyme showed the production of two bands of a size predicted if NHEJ occurred (Fig. 6A). Densitometric quantification showed a nuclease cutting frequency of approximately 2% (Fig. 6A). Because the Surveyor procedure can require gene-specific optimization and can be difficult to interpret in an unambiguous manner (Voskarides and Deltas, 2009) we also performed a homology-directed insertion (HDI) assay. The principle of this assay is that minimal donor arms (mDonor) can be added to a sequence of choice by PCR and then introduced into cells in the presence of a nuclease, resulting in site-specific insertion of the cargo DNA. Therefore, we amplified a 1.5-kb fragment of plasmid DNA with PCR primers that added 40 bp of mDonor sequence, derived from the IDUA ZFN locus, and then transfected this with nuclease 170 into HEK 293 cells. Using a three-primer PCR strategy including locus- and donor-specific forward primers and a locus-specific reverse primer (Fig. 6B, i), we were able to simultaneously amplify the endogenous and modified alleles (Fig. 6B, ii). Densitometry showed a modification rate of 4% and sequencing showed a perfect junction between the plasmid cargo, the mDonor arm, and the endogenous IDUA locus (Fig. 6C). Thus, we have documented successful IDUA gene modification with our synthetic nuclease. Despite the slight differences in nuclease cutting frequency determined by the Surveyor and HDI assays, each is within the previously reported range for CoDA ZFN activity: <1–18% (Sander et al., 2011).
FIG. 6.
Quantitative CoDA-syn IDUA nuclease activity. surveyor nuclease assay for quantification of CoDA-syn nuclease. (A) GFP or CoDA-syn 170 nuclease-treated cells were assayed by amplifying the endogenous IDUA locus. Lanes 1 and 2 show GFP or N170 treated cells±Surveyor that does not show a fragmentation band. Lane 3 is N170-treated cells also treated with Surveyor that shows the generation of fragments (>) consistent with imperfect nonhomologous end joining after nuclease cutting. (B) Minimal donor (mDonor) arm integration and quantification of allele modification. (i) A 1.5-kb plasmid DNA fragment was PCR amplified with primers with phosphorothioate linkages (*) containing an additional 40 bp homologous to the endogenous IDUA locus that flanks the N170 target sequence (dashed lines). (ii) This PCR fragment was cotransfected with the N170 nuclease into 293 cells and a three-primer PCR was performed. The two black primers are unmodified IDUA locus-specific primers that yield an ∼500-bp product (lane 2; and lane 3, upper band). The gray primer is specific for the mDonor and when site-specific integration and allele modification occurs a PCR product of ∼400 bp is observed (lane 3, lower band) Lane 1, molecular weight standards. (C) Minimal donor sequence analysis. Flanking sequence analysis showed a perfect insertion of the mDonor into the IDUA locus. At left is the mDonor sequence, the middle is the 40-bp donor arm, and at right is the endogenous IDUA sequence.
We report two major findings from this work: first, we describe the CoDA-syn method, whereby single-stranded oligos are assembled into left or right ZFN arrays and simultaneously inserted into a ready-to-use, broadly applicable expression plasmid. These plasmids, pMJO-6 and −7, are freely distributed through the University of Minnesota Center for Genome Engineering (Minneapolis, MN) via Addgene (Cambridge, MA) and can be employed for gene editing in zebrafish and mammals. In addition, our strategy is also compatible with a plant expression plasmid (Zhang et al., 2010). Second, we successfully applied this strategy and have, for the first time, documented human gene editing using a CoDA-derived protein. This expands the usefulness of CoDA proteins, which previously were assessed for activity only in plants and zebrafish (Sander et al., 2011). The recovery of one active nuclease pair is consistent with a common gene-targeting strategy for ZFNs, that is, to target multiple sites in the gene in order to obtain a functional nuclease (Miller et al., 2007, 2011; Maeder et al., 2008; Pruett-Miller et al., 2008; Kim et al., 2009; Sander et al., 2011). An advantage of our system is that the speed, and amenability to scaling up, with which ZFNs can be generated and tested allow for a multiple target redundancy scheme to maximize the chances of recovering a functional nuclease.
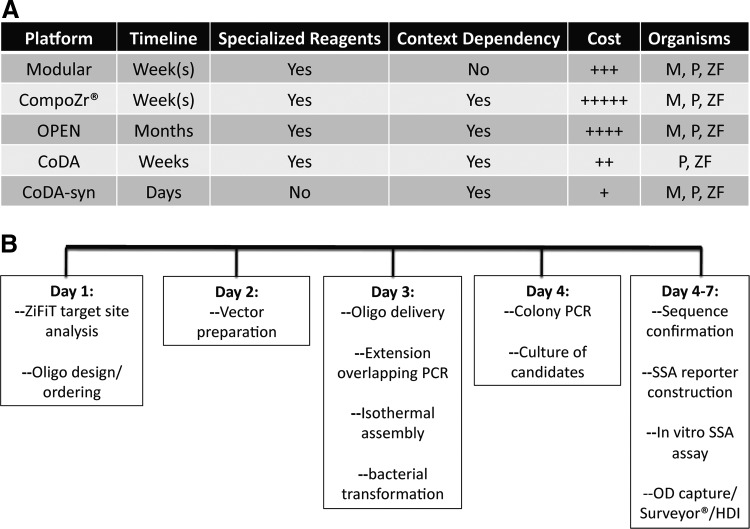
Thus, our method is cost-effective, ensures context dependency, requires no specialized reagents, can be performed in approximately 1 week by users with basic molecular biology skills, and can be used in multiple model organisms (Fig. 7). Last, our method complements a report detailing the rapid generation of a new class of genome-modifying reagents: the transcription activator-like effector nucleases (TALENs) (Christian et al., 2010; Cermak et al., 2011). These reagents share similar properties with ZFNs and have exhibited high rates of activity, thus showing great promise for use in gene editing (Christian et al., 2010; Miller et al., 2011). To date, no comprehensive comparison of the two classes of proteins has been performed and our rapid ZFN synthesis method and the TALEN assembly procedure, both able to be performed in approximately 1 week, should facilitate the widespread use of both reagents and further the working knowledge of each.
FIG. 7.
CoDA-syn procedure comparison to alternate ZFN generation methods. (CoDA-syn procedure comparison to alternate ZFN generation methods. (A) Multiple platforms exist for generating gene-specific ZFNs. A generalized time line, the necessity for acquiring/purchasing specific starting materials, whether context dependency is inherent to the generation, a generalized cost analysis, and ability to modify genes in multiple organisms are provided. (B) CoDA-syn assembly schema. The in silico gene analysis and oligonucleotide design and ordering can be accomplished in 1 day. Before oligo delivery the expression plasmid vector and middle fragments can be prepared by restriction digest and gel purification and archived. After oligo delivery, they are resuspended, pooled, and subjected to overlapping extension PCR that yields a 300-bp product. The purified vector and PCR fragments are added to the isothermal DNA assembly mixture and incubated at 50°C for 30 min and then transformed into Epi300 bacteria. On day 4, 10 colonies are chosen for colony PCR followed by overnight culture. Plasmids are sequenced while the SSA reporter is generated. Once full sequence confirmation is received the in vitro analysis of synthetic ZFN candidates can be performed in the cell type of choice using the SSA, OD, Surveyor, and Homology Directed Integration (HDI) assays. M, mammalian; P, plant; ZF, zebrafish.
Supplementary Material
Acknowledgments
The authors are grateful to J. Keith Joung and Morgan L. Maeder (Harvard University), and to Daniel F. Voytas, Nikunj Somia, and Kathleen Conklin (University of Minnesota), for helpful discussions; and to John Oleksowicz for manuscript preparation. This work was supported by the Children's Cancer Research Fund (M.J.O., B.R.B., and J.T.) and the American Cancer Society (IRG-58-001-49-IRG to M.J.O.).
Author Disclosure Statement
No competing financial interests exist.
References
- Alwin S. Gere M.B. Guhl E., et al. Custom zinc-finger nucleases for use in human cells. Mol. Ther. 2005;12:610–617. doi: 10.1016/j.ymthe.2005.06.094. [DOI] [PubMed] [Google Scholar]
- Bhakta M.S. Segal D.J. The generation of zinc finger proteins by modular assembly. Methods Mol. Biol. 2010;649:3–30. doi: 10.1007/978-1-60761-753-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Morton J.J. Beumer K.J. Segal D.J. Design, construction and in vitro testing of zinc finger nucleases. Nat. Protoc. 2006;1:1329–1341. doi: 10.1038/nprot.2006.231. [DOI] [PubMed] [Google Scholar]
- Cermak T. Doyle E.L. Christian M., et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M. Cermak T. Doyle E.L., et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary M.A. Wraith J.E. The presenting features of mucopolysaccharidosis type IH (Hurler syndrome) Acta Paediatr. 1995;84:337–339. doi: 10.1111/j.1651-2227.1995.tb13640.x. [DOI] [PubMed] [Google Scholar]
- Doyon Y. Vo T.D. Mendel M.C., et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat. Methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- Gibson D.G. Smith H.O. Hutchison C.A., III, et al. Chemical synthesis of the mouse mitochondrial genome. Nat. Methods. 2011;7:901–903. doi: 10.1038/nmeth.1515. [DOI] [PubMed] [Google Scholar]
- Gibson D.G. Young L. Chuang R.Y., et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Guschin D.Y. Waite A.J. Katibah G.E., et al. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- Handel E.M. Alwin S. Cathomen T. Expanding or restricting the target site repertoire of zinc-finger nucleases: The inter-domain linker as a major determinant of target site selectivity. Mol. Ther. 2009;17:104–111. doi: 10.1038/mt.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J. Lee H.J. Kim H., et al. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Lee M.J. Kim H., et al. Preassembled zinc-finger arrays for rapid construction of ZFNs. Nat. Methods. 2011;8:7. doi: 10.1038/nmeth0111-7a. [DOI] [PubMed] [Google Scholar]
- Maeder M.L. Thibodeau-Beganny S. Osiak A., et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X. Noyes M.B. Zhu L.J., et al. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.C. Holmes M.C. Wang J., et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Miller J.C. Tan S. Qiao G., et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Muenzer J. Wraith J.E. Clarke L.A. Mucopolysaccharidosis I: Management and treatment guidelines. Pediatrics. 2009;123:19–29. doi: 10.1542/peds.2008-0416. [DOI] [PubMed] [Google Scholar]
- Orlando S.J. Santiago Y. Dekelver R.C., et al. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res. 2010;38:e152. doi: 10.1093/nar/gkq512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn M.J. Panoskaltsis-Mortari A. McElmurry R.T., et al. A picornaviral 2A-like sequence-based tricistronic vector allowing for high-level therapeutic gene expression coupled to a dual-reporter system. Mol. Ther. 2005;12:569–574. doi: 10.1016/j.ymthe.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Porteus M. Creating zinc finger nucleases using a modular-assembly approach. Cold Spring Harbor Protoc. 2010. 2010. pdb.prot5530. [DOI] [PubMed]
- Porteus M.H. Carroll D. Gene targeting using zinc finger nucleases. Nat. Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- Pruett-Miller S.M. Connelly J.P. Maeder M.L., et al. Comparison of zinc finger nucleases for use in gene targeting in mammalian cells. Mol. Ther. 2008;16:707–717. doi: 10.1038/mt.2008.20. [DOI] [PubMed] [Google Scholar]
- Ramirez C.L. Foley J.E. Wright D.A., et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband W.S. ImageJ. U.S. National Institutes of Health; Bethesda, MD: 1997–2011. [Jun;2011 ]. [Google Scholar]
- Remy S. Tesson L. Menoret S., et al. Zinc-finger nucleases: A powerful tool for genetic engineering of animals. Transgenic Res. 2010;19:363–371. doi: 10.1007/s11248-009-9323-7. [DOI] [PubMed] [Google Scholar]
- Sander J.D. Dahlborg E.J. Goodwin M.J., et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat. Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J.D. Zaback P. Joung J.K., et al. Zinc Finger Targeter (ZiFiT): An engineered zinc finger/target site design tool. Nucleic Acids Res. 2007;35:W599–W605. doi: 10.1093/nar/gkm349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskarides K. Deltas C. Screening for mutations in kidney-related genes using SURVEYOR nuclease for cleavage at heteroduplex mismatches. J. Mol. Diagn. 2009;11:311–318. doi: 10.2353/jmoldx.2009.080144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong A.S. Peng R.H. Zhuang J., et al. Chemical gene synthesis: Strategies, softwares, error corrections, and applications. FEMS Microbiol. Rev. 2008;32:522–540. doi: 10.1111/j.1574-6976.2008.00109.x. [DOI] [PubMed] [Google Scholar]
- Zhang F. Maeder M.L. Unger-Wallace E., et al. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12028–12033. doi: 10.1073/pnas.0914991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J. Maeder M.L. Mali P., et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.