Abstract
Significance: Modification of cysteine thiols dramatically affects protein function and stability. Hence, the abilities to quantify specific protein sulfhydryl groups within complex biological samples and map disulfide bond structures are crucial to gaining greater insights into how proteins operate in human health and disease. Recent Advances: Many different molecular probes are now commercially available to label and track cysteine residues at great sensitivity. Coupled with mass spectrometry, stable isotope-labeled sulfhydryl-specific reagents can provide previously unprecedented molecular insights into the dynamics of cysteine modification. Likewise, the combined application of modern mass spectrometers with improved sample preparation techniques and novel data mining algorithms is beginning to routinize the analysis of complex protein disulfide structures. Critical Issues: Proper application of these modern tools and techniques, however, still requires fundamental understanding of sulfhydryl chemistry as well as the assumptions that accompany sample preparation and underlie effective data interpretation. Future Directions: The continued development of tools, technical approaches, and corresponding data processing algorithms will, undoubtedly, facilitate site-specific protein sulfhydryl quantification and disulfide structure analysis from within complex biological mixtures with ever-improving accuracy and sensitivity. Fully routinizing disulfide structure analysis will require an equal but balanced focus on sample preparation and corresponding mass spectral dataset reproducibility. Antioxid. Redox Signal. 21, 511–531.
Introduction
Oxidative protein folding refers to the creation of a specific set of protein backbone cross-links via the formation of disulfide bonds between the side chains of cysteine amino-acid residues. This process provides, in effect, a set of molecular staples that stabilize a higher-order protein structure. It is considered an oxidative process, because two electrons are removed from the protein molecule when an intramolecular disulfide bond forms (Fig. 1). Outside of a small proportion of cases (45) where ongoing disulfide interchange plays a direct regulatory role in protein activity, there exists for each protein a unique linkage pattern of disulfide bonds that is consistent between all biologically active or “properly folded” protein molecules. In this regard, disulfide bond structure—defined as the precise set of uniquely linked disulfide bonds in a given protein—represents an extremely consistent (protein molecule-to-protein molecule) post-translational modification. Cysteine residues involved in disulfide bonds (i.e., cystines) are notably the most evolutionarily conserved amino acids and, in 99% of cases, are only replaced in pairs (95, 104). Free cysteine is not particularly conserved (95). When present, however, free cysteine residues tend to play a major role in protein biochemistry—thus, techniques for the analysis sulfhydryl groups are widely applicable.
FIG. 1.
Oxidative protein folding as it generally occurs in the ER (a), the mitochondria (b), and in vitro (84) (c). Oxygen receives electrons in all three cases but not always toward the same molecular fate. ER, endoplasmic reticulum.
Oxidative protein folding within the eukaryotic cell is a rapid, enzyme-mediated process that occurs in the relatively oxidizing environments of the endoplasmic reticulum (ER) and the mitochondria. As summarized by Hu et al. (46), the cytosol maintains a redox potential of −290 mV, but the ER is significantly more oxidizing at a redox potential of −170 to −185 mV; mitochondria redox potentials range from −250 to −280 mV with the matrix generally more reducing than the intermembrane space (56). By way of comparison, the redox potential of bacterial periplasm is −165 mV (49). [The review by Herrmann and Riemer (41a) in this Forum of Antioxidants and Redox Signaling compares and contrasts protein folding in these different environments.] During or after protein biosynthesis, proteins destined for secretion are shuttled to the ER, where protein folding is generally mediated by protein disulfide isomerase (PDI) and ER oxidoreductin 1 (Ero1) enzymes (Fig. 1a). Owing to its unique evolutionary history, a different set of enzymes facilitate mitochondrial protein folding (Fig. 1b) (86). Correct disulfide bonding is critical to ensuring proper protein function and thereby overall health status. For example, altered regulation of the PDI family of enzymes is emerging as an important contributor to a variety of pathological conditions, including neurodegenerative and infectious diseases, cancer, and infertility, with additional roles in hemostasis and lipid homeostasis (3, 8). PDI itself is subject to inhibition by S-nitrosylation of its cysteine residue(s), an event that may lead to neurodegeneration in the forms of Parkinson's and Alzheimer's diseases (96). Independent of enzymes, oxidative stress in the myocardium may contribute to cysteine cross-linking within the protein Titin that produces increased passive tension and hysteresis in heart tissue (32, 61).
Oxidative protein folding can also occur spontaneously in vitro (Fig. 1c). As initially demonstrated by the Nobel Prize-winning work of Anfinsen and colleagues (5, 6, 35, 102, 103), many isolated, unfolded proteins exposed to oxygen are not only capable of forming disulfide bonds spontaneously [vis-à-vis cysteine sulfenic acid (Cys-SOH) intermediates (84)], but they also frequently do so in the correct, native pattern provided the correct buffer conditions are supplied and the polypeptide backbone remains intact (95). However, correct folding ex vivo is never guaranteed. Given, for example, the continual possibility that disulfide scrambled protein isomers may contaminate recombinant proteins intended for pharmaceutical/therapeutic use, the verification of correct disulfide linkage patterns plays a significant role in the development of modern protein-based drugs (40, 64).
The first protein disulfide bond structure was determined for bovine insulin by Sanger and colleagues in 1955 through a logical but laborious process (87). In recent years, the scientific capacity to study protein disulfide bonds in small quantities of material has been accelerated, in large part, by advances in biological mass spectrometry and follow-on creative approaches to sample preparation and data analysis—generating powerful analytical techniques for determining protein disulfide structures.
However, the analysis of fully folded proteins represents only a sub-discipline within the study of oxidative protein folding. Accurately pinpointing,* tracking and quantifying free cysteine residues both within folded proteins and during the folding process are important tasks for many protein chemists. As such, in this review, we discuss modern tools for (and potential sources of error in) tracking, pinpointing, and quantifying free sulfhydryl groups, quantifying the degree of oxidative protein folding over time, and elucidating complete protein disulfide structures. Due to its minimal sample requirements and its flexible and expansive analytical capacity, mass spectrometry has come center stage in recent years, playing an important role in many of the techniques described.
Techniques for Tracking Reduced Cysteine Residues in Proteins
Depending on the question(s) at hand, detecting, quantifying, and determining the degree to which cysteine residue(s) exist in the free thiol form may be carried out in unfractionated biological samples, but they are more routinely carried out in ways that are specific to certain cellular fractions (37), particular proteins (44, 58, 60, 89), or other (semi)purified preparations. The unique chemistry of sulfhydryl groups enables them to serve well as molecular “handles” for self-quantification and for the manipulation and analysis of proteins in general. As such, there are literally hundreds of different thiol-specific reagents that may be employed in a great variety of creative ways to track and quantify sulfhydryl status within proteins (42, 54). We prefer to categorize these probes into two groups: first, as mono-functional small molecules that react specifically with thiol groups under a particular set of conditions (Table 1). Alone, these are often employed to selectively block protein thiol groups without imparting any unique functionality. (Of course, if mass spectrometry is to be used as the analytical endpoint, all sulfhydryl-tagging reagents are bi-functional in the sense that besides blocking thiols, they impart a specific mass shift to each protein sulfhydryl group.) Second, nearly all of the mono-functional thiol reagents in Table 1 can be synthesized as covalent conjugates with a second small molecule or functional group that imparts unique functionality to cysteine residues tagged with the bi-functional reagent (Table 2). Many of the possible combinations of thiol-specific reactants from Table 1 with conjugates from Table 2 have been synthesized and are commercially available (42, 54). Useful example protocols for employing most of these reagents in the laboratory have been compiled by Hermanson (42).
Table 1.
Major Classes of Mono-Functional Thiol-Reactive Reagents
| Major class of thiol blocking reagent | Examples | Positive attributes | Negative attributes |
|---|---|---|---|
| Haloacetamide/haloacetate/haloalkyls | Iodoacetamide, iodoacetate, and variable-length iodo- and bromo-alkyl chains | Highly reactive; results in stable thioether product | Reacts with amines and other nucleophiles if pH is too high |
| Maleimide | Maleimide, N-ethylmaleimide | Very good thiol reactivity and specificity at slightly acidic pH | Suceptible to thiol exchange or ring hydrolysis/opening (2, 63, 90) |
| Disulfide | Cystine, cystamine, glutathione, Ellman's reagent (not used for conjugation), 2,2′-dipyridyl disulfide | Reversible, high reaction specificity | Susceptible to disulfide exchange; should keep acidic pH |
| Thiosulfonate | Methyl methanethiosulfonate (MMTS) | Reversible, fast reaction rate, high reaction specificity | Susceptible to disulfide exchange; should keep acidic pH |
| Metals | Mercury, gold | Can be used to simply block thiols or to create quantum dots, gold nanoparticles, self-assembled monolayers, and other immobilization applications | Hg is highly toxic; Au-S bonds subject to displacement or oxidative disruption |
| Others | Vinyl sulfones, acryloyls, halobenzenes, aziridenes | Useful in specific niche applications | Reactions can be slow, and specificity can be highly pH dependent |
Table 2.
Functional Classes That Can Be Conjugated to One Or More of the Thiol-Reactive Reagents in Table 1
| Conjugatea | Application/utility | Example | Equipment required |
|---|---|---|---|
| Visible spectrum absorbance probes (42, 54) | Quantification of sulfhydryls | Ellman's reagent (5,5′-dithiobis-(2-nitrobenzoic acid) | Spectrophotometer |
| Fluorescent probes (42, 54) | Study of protein structures and interactions; quantification of sulfhydryls | Rhodamine Red™ C2-maleimide | Fluorimeter, fluorescence imager |
| Biotin (1, 42) | Isolation, identification, and quantification of free sulfhydryl-containing proteins | N-[6-(Biotinamido)hexyl]-3'-(2'-pyridyldithio)propionamide | Western blot tools or mass spectrometer |
| ICAT (42), ECAT (42) and isobaric tandem mass tags (1) | Relative quantification of sulfhydryl-containing proteins in complex biological samples | ECAT: (S)-2-(4-(2-bromoacetamido)benzyl)-DOTA lanthanide metal chelate | Mass spectrometer, generally LC-MS |
| Solid-phase materials (36, 43) | Sulfhydryl-based affinity capture/affinity chromatography | Functionalized silica, dextran, sepharose, metals | Application-specific: spectrophotometer and Western blot tools are most common |
In selecting a particular reagent for an experiment, researchers should continually be alert to the fact that solution chemistry—pH in particular—plays a major role in defining the reaction rate as well as the degree of specificity with which most labeling reagents will react with thiols. The relationship of thiol reactivity with pH is, in large part, governed by formation of the highly reactive thiolate anion as solution pH approaches and exceeds the pKa of protein sulfhydryl groups. On average, protein sulfhydryl pKa values range from about 8 to 9.5 (69), but can vary widely from as low as 2.5 (28) to approximately 10 (41)—meaning that the pKa of a single cysteine residue can dramatically affect protein function. The pKa values of protein amino groups tend to fall slightly above those of most sulfhydryls, so there is generally a limited range of solution pH values in which thiol-specific probes will react at a reasonably rapid rate while maintaining specificity for sulfhydryls.
In many cases, the use of protein denaturants, such as detergents, or chaotropic agents, such as urea or guanidine hydrochloride, may also warrant consideration, as they tend to provide improved access to internal protein residues for larger or bi-functional probes. Denaturants should be employed with caution, however, as they may interfere with downstream steps if they are not first removed; mass spectrometry-based analytical methods are often particularly susceptible to failure caused by the presence of denaturants.
Pinpointing and Tracking Reduced Cysteine Residues Within Proteins
Protein sequence alone is inadequate for determining whether cysteine residues exist in fully reduced, free thiol-bearing form. The identification of such cysteinyl residues is often necessary for adequate protein characterization. This often starts with recognizing and pinpointing cysteinyl residues, which may be accomplished via several different approaches that rely on (i) sulfhydryl tagging, (ii) proteolysis, and (iii) the subsequent generation of analytical signatures that are unique to the tagged peptides. For example, simple isotope-coded affinity tag (ICAT)-based approaches (33, 91) that pinpoint cysteinyl residues within complex (impure) protein samples are based on splitting a protein sample and labeling half with a light sulfhydryl-reactive ICAT reagent and the other half with a heavy, stable isotope-labeled sulfhydryl-reactive ICAT reagent, mixing, proteolytically digesting, and then analyzing by mass spectrometry. This results in readily identifiable pairs of mass spectral peaks in single-stage mass spectra that are separated by the mass difference between the light and heavy ICAT tags. Mass mapping and, if necessary, analysis of the tagged peptides by tandem mass spectrometry (MS/MS) can reveal the identity of the labeled cysteine residue(s). (More complex ICAT-based schemes for relative quantification are described next.)
If a protein is purified and investigators wish to skip directly to the identification of cysteine residues without the need to first pinpoint them, a cyanylation/cleavage strategy may be found useful: As studied in depth by the Watson group, cyanylation of cysteinyl residues (51, 109) followed by a reaction with ammonia (110, 111) or (more efficiently) with other primary amines (27) facilitates proteolytic cleavage N-terminal to cysteine residues. Subsequent analysis by mass spectrometry and mass mapping readily reveals the loci of cysteinyl residues in the original protein.
Alternatively, after proteolysis, protein cysteinyl residues labeled with 4-dimethylaminophenylazophenyl-4′-maleimide can be tracked by their absorbance in the visible spectrum. This readily facilitates the isolation of cysteine-containing peptides by high-performance liquid chromatography (HPLC). If this is insufficient, subsequent analysis of labeled peptides by matrix-assisted laser desorption ionization (MALDI)-MS produces a unique in-source decay-based signature pattern in single-stage mass spectra, which, in combination with mass mapping, readily reveals what cysteine residues existed in the free thiol form in the original intact protein (12). This approach has been applied to tyrosine hydroxylase, the rate-limiting enzyme in dopamine synthesis, to investigate the susceptibility of particular cysteine residues to modification by S-glutathionylation (10).
Quantification of Reduced Cysteine Residues
Given the number of molecular probes that are available today (Tables 1 and 2), there are dozens, if not hundreds, of possible approaches to sulfhydryl quantification. Broadly speaking, this may be accomplished on either absolute or relative terms. Some of the most widely applicable approaches that are likely to be employed well into the future are described next.
Absolute quantification
The most common approach to absolute molar quantification of total in-solution protein sulfhydryls involves a reaction with Ellman's reagent [5,5′-dithiobis-(2-nitrobenzoic acid) or DTNB] (24). The utility of DTNB is derived from the red-shifted absorbance spectrum of the free 2-nitro-5-thiobenzoic acid (TNB) species that is released on a reaction of DTNB with protein thiols. At pH 8.0, TNB facilitates rapid spectrophotometric quantification without the removal of residual DTNB, provided measurements from proper negative control sample(s) are subtracted out (34). Such use of controls is important, because the molar absorptivity of TNB at 412 nm (which is around 14,000 M−1 cm−1) can vary not only with pH, but with salt and denaturant concentration as well (85). Selecting the appropriate control(s) is highly dependent on experimental context, but they should match the samples in as many biophysical aspects as possible without introducing additional sulfhydryl groups. Preisolation of protein away from sulfhydryl-containing small molecules (such as cysteine or glutathione) is generally required before protein-specific sulfhydryl quantification with DTNB.
Relative quantification
Relative quantification of protein thiols requires at least two different samples or conditions—for example, a “before-and-after” or a “with-and-without x treatment” set of samples—and consists of parallel processing and analysis followed by calculating the ratio of the analytical signal measured for each sample individually. In most experiments, changes over multiple orders of magnitude do not occur and the assumption is appropriately made that instrument response remains linear over the range of concentrations encountered. Thus, calibration curves are frequently not employed.
There are several different classes of thiol-specific reagent conjugates (with dozens of examples each) that may be employed for relative quantification of sulfhydryls (Table 2). These are based on unique physical separation and/or detection properties. Relative quantification can be carried out on a whole sample #1 versus whole sample #2 basis (e.g., for a comparison of total thiol-specific probe incorporation). This approach has been used to show that protein thiols represent a larger active redox pool than glutathione—implying a role for protein thiols as a defense against cellular oxidative stress (37). On the other hand, if there are specific target proteins of interest, comparisons may be made on a specific-protein(s)-in-sample #1 versus specific-protein(s)-in-sample #2 basis (44, 59). In this case, in addition to means for detection or separation based on the thiol-specific probe, some form of protein separation is also necessary. Two-dimensional gels are frequently employed for this purpose, where it is possible to quantify the relative abundance of sulfhydryls within specific proteins in at least two samples at a time using difference gel electrophoresis (DIGE) (62) or related techniques. This general approach has proved useful for generating a comprehensive view of the changes to all cellular proteins in response to oxidative insults (44, 59). Though it is a powerful technique for global protein relative quantification, two-dimensional DIGE is limited with regard to throughput and the analysis of proteins at the extremes of molecular weight, isoelectric point, solubility, and copy number.
If protein site-specific quantification of thiols relative to their reversible oxidation product(s) is desired, oxidation-based isotope-coded affinity tag (OxICAT)-based approaches (58, 89) [derived from the ICAT technique (33)] may be found useful (Fig. 2). For example, one of the original applications of OxICAT was to help identify a specific group of redox-regulated proteins that protect Escherichia coli under conditions of oxidative stress (58). In general, OxICAT enables the detection of most reversible oxidative thiol modifications (Fig. 3), but if specificity toward certain oxidative modifications is desired, tris(2-carboxyethyl)phosphine (TCEP) can be substituted with a modification-specific reducing agent, such as ascorbate, arsenite, glutaredoxin, or hydroxylamine [reviewed elsewhere (71)]. As reviewed and diagramed by Murray and Van Eyk (71), these highly targeted approaches are beginning to be employed in a variety of in vivo applications vis-à-vis the popular and highly flexible “biotin switch” technique (52, 53) (Fig. 4).
FIG. 2.
Protein site-specific relative quantification of sulfhydryls using the OxICAT technique. In general, the ICAT reagent consists of a thiol-reactive probe (typically iodoacetamide—though conceptually any of the thiol probes in Table 1 could be utilized), a light or heavy labeled linker, and a biotin group for affinity purification (33, 91). OxICAT may be employed for (a) within-sample determination of site-specific sulfhydryl status relative to a reversible cysteine modification or, in modified form, for (b) relative quantification of the degree to which specific cysteine residues are modified after oxidative treatment. Only one sulfhydryl group per protein is shown for the sake of simplicity; in practice, additional sulfhydryl groups would generate tryptic fragments with distinct masses, and each protein sulfhydryl group would be analyzed independently by the mass spectrometer. Information about modification of multiple cysteines within a single protein molecule can only be derived from analysis of the intact protein in a separate experiment. OxICAT, oxidation-based isotope-coded affinity tag.
FIG. 3.
Biologically encountered cysteine modifications. Numbers adjacent to sulfur atoms indicate oxidation state. Reversibility status refers to general dynamic reversibility under biological conditions (regardless of whether the modification can be reduced in vitro). The difference between a structural disulfide bond and an allosteric disulfide bond lies in its redox potential and whether or not it is below (out of reach of) the redox potentials of biologically relevant oxidoreductases (20, 107). Notably, all common chemical reducing reagents readily reduce both allosteric and structural disulfides. *The generation of cysteine sulfinic acid is enzymatically reversible in at least some cases (105, 106).
FIG. 4.
Generic biotin switch-based strategy (52, 53) for the analysis of selectively reversible cysteine modifications. Besides facilitating detection, this approach may be combined with the ICAT strategy for relative quantification of reversible protein thiol modifications (108, 121). -NO, nitrosylation; -OH, sulfenylation; -SG, glutathionylation; S-Palm, palmitoylation; ICAT, isotope coded affinity tag.
Monitoring Protein Thiols During Oxidative Protein Folding
Reduced proteins undergo spontaneous oxidative folding in vitro. Under the appropriate buffer conditions, this process results in formation of the correct disulfide bond structure and restoration of protein activity (5, 35, 102, 103). Though not necessarily representative of the enzyme-mediated in vivo folding process (Fig. 1a, b), observation of the spontaneous in vitro oxidative protein folding process can lend insights into the structure(s) of folding intermediates, which, in turn, may be able to shed light on the molecular etiology of diseases in which protein misfolding is involved such as Alzheimer's, Huntington's, Creutzfeldt-Jakob's, and Parkinson's.
The experimental process generally starts with a reduction of prepurified protein and subsequent purification away from reducing reagent. This can be accomplished through the use of column chromatography (5), protein precipitation, and subsequent dialysis (97), or (more rapidly) using spin filters (84). Conceptually, oxidative protein folding can then be monitored by tagging sulfhydryls at specific time points with a reagent that facilitates their quantification relative to maximum and minimum signal boundaries defined by parallel samples of completely reduced and completely oxidized protein, respectively. Key to such experiments is the ability to take a molecular “snapshot” of the folding process. This entails a rapid reaction of free cysteine residues with a reagent that renders them unable to participate further in the folding process. Such “capping” reactions should take place on a timescale that is negligible with regard to the protein folding process of interest. Depending on the nature of the particular experiment, this timescale may be in the order of either milliseconds or minutes. Cysteinyl tagging as a means of monitoring the protein folding process was briefly discussed in a review by Konermann and Simmons in 2003 (57). As they point out, millisecond time-scale labeling can be accomplished with reagents such as methyl methanethiosulfonate (MMTS) and DTNB, followed by spectrophotometric measurements that assess the degree of labeling (34).
However, such fast reactivity is not always needed, and in many cases, traditional alkylating reagents may be used when folding is not mediated by enzymes—provided the pH is optimized for rapid reactivity while maintaining sulfhydryl specificity. This approach has been illustrated by the early studies of Anfinsen and colleagues (5, 6, 35, 103) and by others more recently (55, 83, 84). The option to employ traditional alkylating reagents provides greater flexibility in experimental design and has been taken advantage of in several studies of oxidative protein folding where mass spectrometric analysis of tagged, intact proteins has served as the final analytical modality (55, 83, 84). The advantage of a mass spectrometry-based analytical approach is that it provides the precise fractional distribution of protein molecules with 0, 2, 4, 6…n free cysteine residues and, hence, detailed kinetic information on these particular subclasses of oxidative folding intermediates (Fig. 5)—enabling researchers to determine the rates at which folding states with discrete numbers of disulfide bonds appear and disappear. If desired, this detailed data set can then be processed to provide aggregate-type folding kinetics data such as “Percent Folded” (as might be obtained with spectrophotometry-based methods) and “Percent in the Fully Folded State” (as might be obtained with activity-based measurements of folding) (Fig. 5).
FIG. 5.
Oxidative protein folding monitored by mass spectrometric analysis of intact protein. In vitro protein folding monitored via sulfhydryl alkylation with maleimide at pH 5 and analysis of the intact protein (ribonuclease A) by electrospray ionization-mass spectrometry (a). Charge deconvoluted mass spectral peak areas were used to calculate the relative quantities of protein in each oxidative folding state over time (b). Percent of total folding progress (such as might be reported with a chromo- or fluorophoric probe) as well as the proportion of protein in the fully folded state (c) were calculated in an analogous manner as described elsewhere [(84), data from oxygen atmosphere folding experiment; Fig. 3 of indicated reference]. Percent of total folding progress subtracts from 100% any remaining protein sulfhydryls without distinction between different folding states. (In this case, however, since we directly measured the relative abundance of individual folding states, partially folded states were weighted in proportion to the remaining number of sulfhydryl groups within them. This weighted value was then subtracted from 100%.) Percent of total folding progress is an arbitrary form of measurement that is not molecularly specific. It is often used because spectrophotometric approaches to measuring protein folding cannot quantify individual protein states and read the sample as an aggregate. Percent of protein in the fully folded state is generally not equal to the former measurement due to the existence of partially folded intermediates that represent a form of “folding progress.”
Besides direct sulfhydryl-labeling techniques, there are a couple of indirect methods by which to monitor the overall progress of oxidative protein folding in vitro. One of the oldest of such approaches relies on measuring protein activity as a surrogate of completely folded protein. (Naturally, this approach only works for proteins that possess some function which can be monitored.) This was one of the major approaches adopted by Anfinsen and colleagues back in the 1960s in their seminal work on the oxidative protein folding of Ribonuclease A (6, 35, 103), but it can still be useful even when modern techniques and instrumentation are available to help reveal the order in which disulfide bonds form (97).
A second common indirect approach involves halting the oxidative protein folding process by quenching with acid [or rapid alkylator (50)], then profiling the qualitatively distinct, partially folded intermediates by reversed-phase-HPLC (14, 15, 26, 75, 97). Since most oxidative folding intermediates can be separated by HPLC, the process can be monitored as chromatograms transition from a single peak representing the fully reduced protein into a myriad of other peaks and then (ideally) into a final peak representative of the fully folded protein into which all of the partially folded forms eventually coalesce. At any point during the folding process, individual folding intermediates may be isolated by HPLC with fraction collection and, subsequently, characterized (e.g., by MS and MS/MS) to ascertain which disulfide bonds formed first (97). After identification, it may be of interest to quantitatively track one or more intermediates by HPLC in subsequent experiments to gain a better understanding of their role in the oxidative protein folding process. This approach has been employed to demonstrate major differences in folding pathways for proteins that are structurally homologous with regard to cysteine content (15). Others have extended research in this arena by using this approach to show that amino-acid sequence rather than cysteine pattern determines the in vitro folding mechanisms (of at least conotoxins) (26).
Of related interest, Happersberger et al. in 1998 developed a method to monitor protein folding in which arsonous acid derivatives were employed to trap (lock in place) spatially neighboring but disconnected cysteine residues (38). This permitted mass spectrometry-based structural characterization of unique but merely transient protein conformations. In turn, they employed the technique to elucidate the folding pathway of recombinant human macrophage-colony stimulating-factor β (39).
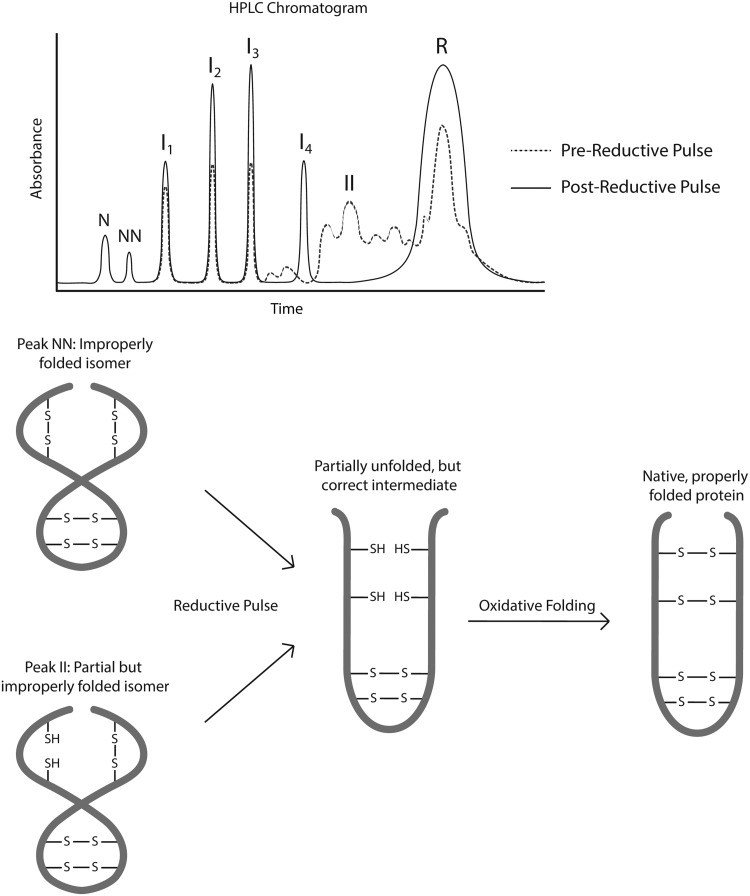
Several years later, Welker et al. developed the use of reductive pulses of 5–10 mM dithiothreitol (DTT) for 0.5–2 min to reduce only those structured intermediates whose disulfide bonds were exposed to solvent; intermediates with buried disulfide bonds were not reduced (Fig. 6). The technique was found to be useful in the characterization of oxidative folding pathways (101).
FIG. 6.
Reductive pulses to facilitate correct oxidative protein folding, monitored by HPLC. As described by Scheraga and colleagues (72, 101), reductive pulses of dithiothreitol can be employed to eliminate improperly folded intermediates and fully folded non-native (incorrect) structures, enabling them to refold in the correct manner. I, properly folded intermediate; II, improperly folded intermediate; N, native, properly folded form; NN, non-native fully folded form; R, fully reduced form. Chromatograms adapted with permission from Welker et al. (101). Copyright 2004 American Chemical Society. HPLC, high-performance liquid chromatography.
It should be noted that in some cases the in vitro folding process may end with a mixture of native and disulfide-scrambled fully oxidized protein molecules. At this point, neither labeling techniques nor HPLC alone can identify the properly folded protein structure. In 2008, Narayan et al. (72) reported a technique based on the combination of a 2-min reductive pulse with DTT to reduce “…disulfide bonds not protected by stable, three-dimensional structure” (Fig. 6), with analysis by high-resolution mass spectrometry to detect the remaining fully folded (and, therefore, properly disulfide bonded) protein molecules. The robust technique was demonstrated with a mixture of seven different proteins (72).
In vivo, under enhanced cellular oxidative stress, the oxidative protein folding process may result in the formation of abnormal interprotein disulfide bonds. These are most readily identified using diagonal electrophoresis or redox gels (22, 92) in which proteins are separated under nonreducing conditions, then reduced, and separated along a second dimension. Proteins that do not form intermolecular disulfides run at the same molecular weight in both dimensions, resulting in their deposition along a diagonal. Off-diagonal proteins should have experienced a significant change in molecular weight after reduction and, by virtue of being off the diagonal, are readily pinpointed. Coupled with modern proteomics techniques for protein identification, this represents a powerful approach to solve the otherwise nearly intractable problem of identifying novel disulfide-based protein heterodimers. Among other applications, this technique has been applied to mammalian neurons (22) and cardiac myocytes (13) to ascertain the widespread effects of intracellular oxidative stress in disease.
Sources of Error in the Analysis of Protein Thiols and Related Modifications
The conclusions drawn from most experiments that employ techniques for tracking the status of cysteine residues in proteins are based on the assumed specificity, completeness, and/or (in some cases) irreversibility of sulfhydryl reactivity with particular reagents. Under certain circumstances, these underlying assumptions may become invalid. As such, it is important for researchers to be aware of such situations and either avoid them or verify that they do not affect their investigations.
Lack of labeling specificity (over-labeling/nonspecific labeling)
Most alkylating reagents that react with sulfhydryl groups may also react with other protein nucleophiles (such as primary amino groups) at an appreciable rate if the pH of the reaction solution approaches the pKa of the nucleophile in question (42). Experiments, for example in which a bi-functional sulfhydryl probe is employed to label sulfhydryl-containing protein(s) followed by spectroscopic or antibody-based detection, are particularly susceptible to a potentially false assumption of labeling specificity. Control experiments (e.g., with sulfhydryl-free proteins or proteins with sulfhydryls blocked with a dissimilar reagent or proteins containing known numbers of free cysteine residues) and/or verification by mass spectrometry (if feasible) are vital to ensuring the integrity of such experiments.
Incomplete labeling
Similar to “over-labeling” or “non-specific labeling,” under-labeling of cysteinyl residues may lead to false conclusions in the interpretation of experimental results. False-negative conclusions are most easily envisioned under such circumstances, but these conclusions may be inverted or amplified depending on experimental design. For example, under-labeling during oxidative protein folding experiments (in which an alkylating reagent is employed to tag and “count” remaining sulfhydryl groups) may be misinterpreted as an increased state of folding. Otherwise, in more complicated interpretation schemes such as the negative signature mass algorithm developed by the Watson group for elucidation of protein disulfide bond structures (80) (described next), under-labeling may result in ruling out a correct conclusion and ending up with no valid conclusion whatsoever. In such cases, additional steps may be required to validate and ensure comprehensive labeling of cysteinyl residues (11).
Label reversibility
Incorrectly assuming label irreversibility can lead to pitfalls similar to those of incomplete labeling. Two situations, in particular, warrant mention. First, disulfide bonds generated by labeling reactions are susceptible to disulfide exchange or inadvertent reduction. As such, experimental workflows involving such labeling reactions should involve subsequent acidification (to prevent formation of reactive thiolate anions) and extreme precaution (coupled with verification) to prevent reduction and/or disulfide exchange. When acidification is not an option, it is generally necessary to remove all traces of residual reducing reagent as well as all sample thiols; in many cases, this may entail excess addition of a nondisulfide-based alkylating reagent. Second, and less well known, is the fact that protein-maleimide bonds may be susceptible to hydrolysis or exchange—particularly in biological environments (2, 63, 90). Shen et al. (90) have shown that maleimide ring opening via hydrolysis results in ring-opened adducts with greater stability. If the label has a specific binding affinity (e.g., biotin) or spectral properties, purification of the labeled protein by affinity column, gel electrophoresis, spin column, or HPLC can be used to verify label retention. If necessary, mass spectrometry can be used to verify the retention of nearly any label.
Sulfhydryl instability
Protein sulfhydryl groups are not intrinsically stable ex vivo in the presence of oxygen. The same chemical principles that mediate protein folding in vitro (4, 5, 35, 102, 103) also facilitate in-solution dimerization of single cysteine-containing peptides (84) and, in some cases, protein polymerization due to spontaneous intermolecular disulfide bond formation when the concentration of protein is too high (6). Such disulfide bond formation is an oxidation reaction: Two thiols do not spontaneously react with one another in the absence of an oxidizing reagent that, ultimately, receives the lost electrons. In vitro, the oxidizing reagent is oxygen, and the oxidized thiol intermediate is Cys-SOH (84).
The implications of this are primarily twofold. First, experiments carried out on sulfhydryl-containing proteins in vitro should be carried out on “fresh” samples—not on samples that have been stored in solution for prolonged periods of time without particular attention paid to their redox status. Thus, for example, when DTNB is employed to quantify sulfhydryls, absorbance should be read without delay, as TNB can re-oxidize to DTNB when exposed to air. Second, in vitro experiments designed to elucidate proteins that are biologically modified by Cys-SOH should be carried out with great caution to avoid false-positive findings, as every sulfhydryl group is likely to be susceptible to this form of artifactual Cys-SOH formation—including simple, free-cysteine containing peptides (84). Sample processing in an oxygen-free glove box, exclusion of metals and/or addition of chelators, or, if possible, the addition of excess “sacrificial” sulfur-containing compounds can help investigators avoid such artifactual sulfhydryl oxidation.
Techniques for Elucidating Protein Disulfide Structures
The techniques covered up to this point are largely designed to interrogate whether protein cysteine residues exist in the free thiol form, disulfide-bound (cystinyl) form, or in what ratios (reduced vs. oxidized) redox-regulated proteins exist under different conditions. It has been recognized for decades that the activity of proteins which contain multiple disulfide bridges depends not only on the formation of disulfides, but also on the correct bridging pattern or “disulfide structure” (35). Hence, the determination of disulfide bond structure is as integral to understanding protein structure-function relationships as is the knowledge of amino-acid sequence (the latter generally being required for elucidation of the former).
Most of the modern techniques for solving protein disulfide structures involve techniques such as mass spectrometry, NMR, or X-ray crystallography that require advanced analytical instrumentation. Besides the general requirement for significantly greater quantities of purified protein, NMR and crystallographic approaches are generally employed for the purpose of gaining structural information well beyond that of disulfide structure alone—that is, disulfide structures are often a “bonus feature” of NMR or crystallographic datasets. As such, this review will focus on mass spectrometric approaches that enable the targeted determination of disulfide structures with limited quantities of protein which would generally contra-indicate the use of NMR or crystallography.
Broadly speaking, there are two general sample preparation approaches in which mass spectrometry may be employed to elucidate protein disulfide bond structures: the “Direct” approach in which disulfide linkages are kept intact and analyzed directly (such as outlined in Fig. 7), and “Indirect” partial reduction-based strategies in which disulfide linkages are logically deduced based on the interpretation of mass spectral data through the lens of sample preparation. The former was the only mainstream approach available till the early 1990s when partial reduction-based strategies came online and offered an attractive alternative. Since the mid-2000s, however, the Direct approach has received renewed interest and research focus vis-à-vis its coupling with MS/MS and novel data mining algorithms. For most investigators, the decision of which technique(s) to use often comes down to practical matters involving target protein characteristics and available instrumentation.
FIG. 7.
The Direct approach to solving disulfide bond structures. Adapted with permission from Tang and Speicher (94).
Irrespective of which general approach is taken, the care and precise manner in which samples are prepared are critically important to an investigator's ability to glean valid, useful information. The need for such care and precaution begins even before formal steps to elucidate disulfide structures: During all preliminary protein purification steps, for example, it is important to minimize opportunities for disulfide scrambling by maintaining a buffer pH that is at least slightly acidic. This minimizes the formation of thiolate anions on cysteinyl residues that may potentially displace a half-cystine in an existing disulfide bond. In many cases, it may be useful to block cysteine residues with an alkylating reagent as the first analytical step (120). Thorough protein isolation before analysis and, when necessary, the use of proteases that are immobilized will also help minimize ambiguous or difficult-to-interpret results due to proteinaceous contaminants. Notably, both the Direct and Indirect approaches require foreknowledge of the protein sequence and the total number of disulfide bonds; the latter can be determined using techniques described earlier.
The Direct Approach
The classic, commonly employed Direct approach to the determination of protein disulfide structures (Fig. 7) is explained in complete detail by Tang and Speicher (94). In modern terms, the procedure consists of (i) proteolytic cleavage; (ii) separation of the resulting peptides by HPLC before and after sample reduction to facilitate the pinpointing of disulfide-containing peptides on the basis of chromatographic peak shifts; (iii) single-stage mass spectral mass mapping analysis and identification of the peptides represented by HPLC peaks of interest; and (iv) further peptide isolation, partial reduction, and iterative sample processing as needed to map all the disulfide bonds within the original protein structure.
As illustrated in Figure 7, the ideal scenario after proteolysis is one in which all peptides joined by a disulfide bond contain just a single cystine. If these polypeptides can be accurately mass mapped (i.e., identified by virtue of highly accurate mass measurements) and their identities confirmed by Edman sequencing or MS/MS (or MSn—i.e., MS/MS/MS or more), disulfide connectivity is obvious and no additional work is necessary. If, on the other hand, there is more than one cystine and the protein backbone cannot be cleaved between critical cysteine residues, additional iterative workflow(s) become necessary. In unmodified form, 10–50 nmol of purified protein are recommended and likely to be required for use with this approach (94). Modifications that scale down the chromatography, couple it with mass spectrometry and eliminate Edman sequencing, however, can significantly decrease these sample requirements.
Unambiguously pinpointing and then identifying† chromatographic peaks (or mass spectral ions) representative of intra- or inter-chain disulfide-linked peptides (without necessarily identifying disulfide connectivities in cases where more than one cystine is present) is the first major challenge in all workflows that seek Direct identification and connectivity assignment of disulfide linkages. Depending on proteolytic cleavage patterns and the resulting linkage types encountered (Fig. 8), the identification of disulfide-linked peptides may comprise well more than half of the information required to elucidate a complete disulfide structure. Indeed, complete disulfide structures may be solved in one pass if all of the identified intra- and inter-chain peptides contain only a single disulfide bond—making cleavage between all cysteine/cystine residues a major goal of proteolysis in the Direct approach. Thus, both bench-based and MSn/informatics-driven approaches have been developed to meet the challenge of disulfide-bridged peptide identification.
FIG. 8.

The 10 unique types of intra- and interchain disulfide bond structures that can be derived from a protein/peptide with two cystines. The first generation represents intact proteins. Subsequent generations represent substructures (denoted by lower case letters in boxes) that result when cleavage has taken place in the preceding generation at corresponding capital letter sites—that is, capital letters (logically read as “OR” when present more than once within a single structure) indicate cut positions which result in their lowercase counterpart structures. Single cystine-containing peptides “a” and “g” are the most desirable products in the Direct approach, because disulfide linkages can be assigned immediately once they are identified. However, as illustrated, the production of single cystine-containing peptides requires thrice the number of backbone cleavages when disulfide bonds overlap one another along the original protein/peptide.
Unique bench chemistry-focused approaches to pinpointing and identifying disulfide-linked peptides
In 2001, Wallis et al. (98) described a clever technical maneuver involving proteolysis in 50% H218O to facilitate the pinpointing and, therefore, eventual identification and connectivity mapping of proteolytic fragments containing inter-chain disulfide bonds. The technique results in unique isotopic signatures in single-stage (i.e., “full scan”/non-MS/MS) mass spectra and obviates the need for analysis by HPLC before and after reduction to facilitate the pinpointing of disulfide-linked peptides. The approach has been reviewed in detail elsewhere (30).
In 2009, Pompach et al. (78) reported that the inclusion of 200 μM cystamine during initial protein purification and during proteolytic digestion facilitated digestion at normal, slightly alkaline pH without disulfide bond scrambling—making it easier, in general, to achieve all-important cleavage of the protein backbone between as many cysteine residues as possible. HPLC-based separation of the resulting peptides was carried out in line with ultra-high resolution/high mass accuracy Fourier transform mass spectrometry and semi-automated data analysis using the Automated Spectrum Assignment Program (119) to generate a library of single-chain and disulfide cross-linked peptides. The approach was utilized to successfully characterize the disulfide structures of hen egg lysozyme, human CD69, mouse leukocyte receptor NKR-P1A, and β-N-acetylhexosaminidases from Aspergillus oryzae and Penicillium oxalicum. Only several micrograms of each protein were required under this approach. It was noted, of course, that if proteolytic cleavage was not sufficient between cysteine residues (as evidenced by a lack of protease-specific cleavage sites or simply the inability to assign all disulfide linkages), then accurate mass measurement alone would not suffice the complete assignment of all disulfide bonds and the implementation of additional workflow(s) would become necessary.
Building on their own earlier work, Huang et al. (47) recently published a novel technique in which protease-derived peptides are dimethylated to significantly enhance a1 ion formation during mass spectral collision-induced dissociation (CID)‡. After one-pot sample preparation, low microgram quantities of sample are analyzed by LC-MS/MS, during which inter-chain disulfide-containing peptides are readily pinpointed when two abundant a1 ions are observed in an MS2 spectrum. Identification of the linked peptides is then provided by (i) the mass of the precursor ion, (ii) the identities of the amino acids at the N-termini (given by the masses of the a1 ions), and (iii) the masses of additional b and y ions. To automate data interpretation using this custom technique, Huang et al. developed an accompanying software package known as RADAR that takes all of the peptide features mentioned earlier into account, providing rapid disulfide connectivity information for dipeptides linked by a single cystine. (As of October, 2013 a free trial of RADAR software may be accessed at www.mass-solutions.com.tw/Index_Eng.aspx) The approach was demonstrated on the monoclonal antibodies bevacizumab (Avastin) and trastuzumab (Herceptin), with a note from the authors that with the aid of multiple proteases the connectivity of all bevacizumab disulfide bonds can be solved automatically. As with most other Direct approaches, cleavage between cysteine residues constitutes a key hurdle to deriving complete solutions of disulfide connectivity.
Gas-phase fragmentation, MSn, and informatics-focused approaches for identifying disulfide-linked peptides
Besides creative bench-based chemistries, numerous unique sample and time-saving mass spectrometric approaches have been developed to facilitate the critical task of rapidly pinpointing inter-chain disulfides. As described earlier, the identification of such peptides is the first major hurdle in Direct approaches to disulfide structure elucidation. Thus, many MS-based techniques are now being complemented by software algorithms that help pinpoint, identify, and even map the connectivity of disulfide-linked peptide chains. Offsetting the sample and time-saving advantages of these techniques is the fact that proteases and follow-on gas-phase approaches to fragmenting peptide ions often fail to break apart polypeptides in ways which reveal desired information on disulfide connectivities. Thus, as with all mass spectrometry-based methods for solving disulfide structures, there is no routine way of ensuring the acquisition of complete data sets in a single experiment that will provide all desired disulfide connectivities; as such, additional follow-up experiments using complementary approaches are often necessary.
Matrix-assisted laser desorption ionization
Depending on experimental conditions employed, the laser energy imparted to disulfide-linked polypeptides during MALDI can lead to in-source cleavage of C-S and S-S bonds (21, 76, 81, 122), generating triplet peak sets separated by 32 Da (the mass of a sulfur atom), which can be useful toward pinpointing and eventually mapping disulfide bond connectivities (21, 81, 88) (Fig. 9). [Notably, such triplets have also been observed in high-energy CID of disulfide-linked peptides (7).] The MALDI matrix employed plays a critical role in generating the triplet pattern (21, 48, 76) and, if desired, can be adjusted to suppress the pattern altogether (48).
FIG. 9.
MALDI-induced cleavage of cystines can generate peak patterns in single-stage (“full scan”) mass spectra that readily pinpoint disulfide bridged peptides. The matrix 2,5′-dihydroxybenzoic acid, high laser fluence, and relatively long time spans before mass analysis (microseconds or more) favor generation of the peaks observed in the upper spectrum (76, 81). The matrix α-cyano-4-hydroxycinnamic acid, minimal laser fluence, and rapid mass analysis (e.g., time-of-flight) favor generation of the peaks observed in the lower spectrum (88). MALDI, matrix-assisted laser desorption ionization.
Coupled with the right matrix and rapid mass analysis (i.e., Time-of-Flight), the MALDI phenomenon of in-source decay-mediated disulfide scission of polypeptide chains into two separate peptides enabled Schnaible et al. (88) to develop an alternate but widely applicable pattern-based method for the screening of disulfide bonded polypeptides in single-stage MALDI mass spectra (Fig. 9). Subsequent LIFT-TOF/TOF-MS§ (a form of MS/MS) of the disulfide bonded peptides facilitated confirmation and mapping of disulfide bond connectivities. This technique represents an efficient combination of in-source fragmentation and directed MSn for disulfide structure determination.
Collision-induced dissociation
Low-energy CID (also known as collisionally activated dissociation) of protonated gas phase peptides is currently the most commonly employed form of biological MS/MS. It routinely provides peptide backbone cleavage and, therefore, sequence information in MSn spectra, but has a reputation for not disrupting disulfide bonds in high abundance (67, 70, 123, 124). Notably, however, Clark et al. have found that double backbone cleavages of disulfide-bonded, double-chain peptides are common (but not predictable) under CID conditions (19). In fortuitous cases, such cleavages may provide cystinyl linkage patterns in multi-chain peptides containing more than one disulfide bond—suggesting that such cleavages ought to be considered in algorithms that seek to solve disulfide structures based on MS/MS data (discussed next). For example, cleavage at both “F” sites in the second-generation “b” structure (Fig. 8) instantly jumps the complexity down two levels—that is, to the point of two fourth-generation “g” structures that readily provide information on disulfide connectivity.
Electron capture dissociation and electron transfer dissociation
Electron capture dissociation (ECD) and electron transfer dissociation (ETD) involve the acquisition of a destabilizing low-energy electron by a multiply protonated gas-phase polypeptide. Cleavage at disulfide linkages is the most common dissociation pathway for inter-chain disulfide-containing peptide ions under both ECD (124) and ETD (17) conditions. Such cleavages help pinpoint these important peptides.
In a new technique that illustrates the utility of this phenomenon, Clark et al. (18) provide a way to take advantage of it in a common-sense, highly efficient manner (Fig. 10): After collection of LC-ETD-MS2 datasets from tryptic digests, they plot the theoretically unmodified/unlinked, sequence-and-cleavage-site predicted extracted ion chromatograms (XICs)** for all cysteine-containing peptides in their target protein. Since ETD causes cleavage between the sulfur atoms of a disulfide bond, any two of these different XICs that produce perfectly aligned HPLC retention times represent candidate disulfide-linked peptide chains which were linked together before ETD. To confirm a pairing, the summed masses of the partners are verified to match with those of the precursor ion for the MS2 spectrum. Assigned pairs are additionally confirmed from CID fragmentation data collected in a separate analysis. The authors demonstrate that special cases such as multi-chain peptides and intra-chain disulfides can be dealt with in relatively simple ways without the need for additional experimentation. As demonstrated by its utility in assigning the disulfide bond structure of a recombinant monomer of the HIV envelope protein gp120, this insightful data analysis technique represents an excellent starting point (if not ending point as well) for ETD-based attempts to directly characterize protein disulfide structures.
FIG. 10.
Clark et al.'s LC-ETD-MS2 XIC-based technique for assigning disulfide bond connectivities (18) in tryptic peptides. Shown is a figure that illustrates the assignment of peptides with one disulfide bond. XICs are constructed to show where peptide mass marker ions of cysteine-containing peptides elute. Co-eluting peptides are assigned as disulfide pairs after further scrutiny, as described in the text. The inset shows an example mass spectrum of the peptides GYSLGNWVCAAK and CK that were disulfide bonded and dissociated in ETD. Data from lysozyme are shown. Reprinted with permission from Clark et al. (18). Copyright 2013 American Chemical Society. ETD, electron transfer dissociation; XIC, extracted ion chromatograms.
ETD-mediated cleavage at disulfide linkages nicely complements the tendency of CID to cause fragmentation along the peptide backbone—as recognized and capitalized on several years ago by Wu et al. (114), who combined ETD and CID within the same instrument for in-line sequential mixed MSn experiments in which one and then the other fragmentation technique was employed to identify cystine-containing peptides and map disulfide connectivities therein. The next year, the same group followed up this initial study with the multi-enzyme-mediated complete disulfide mapping of recombinant tissue plasminogen activator, which contains 17 cystines and an unpaired cysteine (113). After this feat, they reported the disulfide structures of three monoclonal antibodies, including the elucidation of scrambled disulfide linkages (99). Most recently, they have used the technique to map the disulfide structure of recombinant human Arylsulfatase A (73), a glycolipid metabolizing protein with a cystine knot and a pair of nested disulfides. Obviously, this constitutes a powerful approach, one that has been caught on to by other groups as well (65, 66, 68, 74). Notably, Pike et al. (77) have employed ECD MS2 and CID MS2 data of proteolytically generated peptides to map seven out of nine disulfide bonds in an engineered viral glycoprotein.
Algorithms and software for use with the direct approach
A couple of algorithms for use with particular disulfide mapping strategies have already been mentioned. Widely employed and readily available software tools designed to help users sift through single-stage mass spectral data to pinpoint and identify disulfide-linked peptides include GPMAW (www.gpmaw.com), PROWL's PeptideMap (http://prowl.rockefeller.edu/prowl/peptidemap.html), and Protein Prospector's MS-Bridge (http://prospector.ucsf.edu). Notably, most algorithms designed for use with the Direct approach are largely focused on the identification of inter-chain disulfide bonds; single-chain peptides with one or more disulfides are readily identifiable by common mass mapping approaches and, if they generate adequate sequence information, can be found by most MS/MS search algorithms such as Sequest and MASCOT.
Two additional, readily available search algorithms of note that were specifically designed for identifying disulfide-containing peptides include MassMatrix (115) and DBond (16). Both of these algorithms are designed to search tandem mass spectra from low-energy CID datasets for the presence of intra- and inter-chain disulfide bonds. DBond reports peptides with single cystines, and MassMatrix reports peptide fragments with up to two cystines. DBond, however, takes into account fragmentation patterns that are specific to disulfide bonds such as cysteine thioaldehyde (−2 Da), cysteine persulfide (+32 Da), and dehydroalanine (−34 Da). Both programs employ scoring models to assign confidence levels to putative identifications. Neither algorithm, however, is able to assign disulfide connectivities in cases where there is more than one disulfide bond.
The recently introduced algorithm DisConnect (9) takes a major step forward by automating the interpretation of MSn data that can often carry information on disulfide connectivity in peptides linked by more than one disulfide. This approach promises access into some of the perceived advantages of CID-based MS/MS for disulfide structure analysis that up until now had been largely unrealized in practice—including minimal sample requirements and the potential to eliminate iterative bench-based sample preparation loops in the disulfide structure-solving process (Figs. 7 and 8).
Indirect, Partial Reduction-Based Strategies
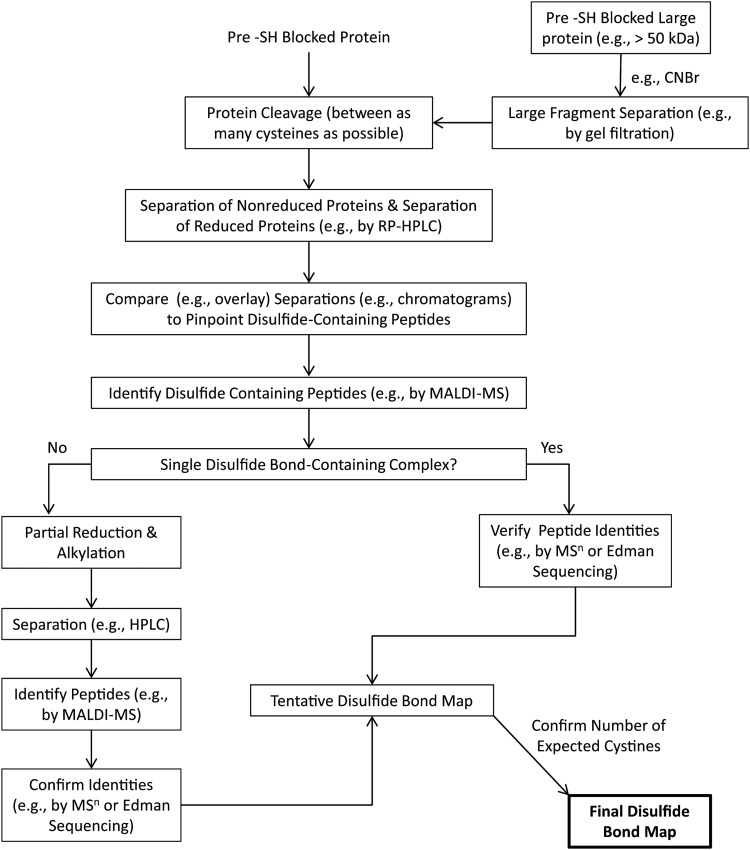
As an alternative to the direct identification of intact disulfide linkages, partial reduction approaches seek to reduce a single disulfide bond within a folded protein molecule, then alkylate and identify the resulting tagged cysteine residues. Partial reduction-based strategies to assign disulfide bond linkages have existed for more than 30 years (25, 82). Introduction into the workflow of reducing reagents such as TCEP (31) that are reactive at acidic pH—and hence prevent disulfide exchange during reduction—were key to the further development and widespread implementation of partial reduction-based approaches.
In the mid-1990s, the Watson group coupled partial reduction strategies with the old discovery (51) that cyanylated cysteine residues may be employed to induce site-specific protein backbone cleavage immediately N-terminal to cysteine residues (110). This resulted in a routinized laboratory workflow (Fig. 11) for the elucidation of oxidative protein folding intermediates (100, 112, 117) and complex disulfide structures (11, 79)—including proteins with adjacent cysteine residues (116). Other strategically complementary partial reduction strategies based on differential alkylation have also been developed (29, 118). Notably, the technique developed by Goransson and Craik (29) alkylates cysteine residues such that they become tryptic cleavage sites.
FIG. 11.
The Wu and Watson (110) partial reduction/cyanylation strategy for solving disulfide bond structures. Ideally, for a protein with n cystines, all n unique singly reduced isoforms would be produced and isolated by HPLC, providing the complete disulfide structure. When this is not possible, the negative signature mass algorithm (80) (see text) may be employed to rule out potential disulfide linkages. TCEP, tris(2-carboxyethyl)phosphine; CDAP, 1-cyano-4-dimethylaminopyridinium tetrafluoroborate; itz, iminothiazolidine group.
Different proteins follow divergent pathways of partial reduction, many of which are not ideally suited to provide optimal information content under the partial reduction/cyanylation approach (i.e., do not produce copious quantities of all possible singly reduced protein isoforms that are readily separable by HPLC). In addition, proteins with complex disulfide structures generate numerous partially reduced, often-inseparable isoforms and large quantities of mass spectral data in need of interpretation. These problems gave rise to the conceptual development and software-based implementation of the “Negative Signature Mass Algorithm” for use with the partial reduction/cyanylation approach (80). This approach operates on the principle of ruling out (rather than ruling in) possible disulfide linkages based on the observations of specific cyanylation-induced cleavage products. In short, the algorithm is driven by the fact that, given complete cyanylation (11, 80), the internal cysteine residues of cyanylation-induced cleavage products cannot logically be connected to the cysteines at the peptide termini. Thus, in Figure 11, for example, without fractionation of partially reduced isomers, the observed itz71-150 fragment enables an arrival at the conclusion that Cys71 and Cys 151 may (but not necessarily) be connected to one another, but Cys71 cannot be connected to Cys102 or Cys130—otherwise, there would have been cleavage at the connected residue [or at least mass-based evidence of side-chain β-elimination (110)]. The Negative Signature Mass Algorithm is compatible with all stages of protein partial reduction as well as with the lack of physical separation of partially reduced isoforms. Notably, the study of Echterbille et al. (23) suggests that, in the future, ion mobility mass spectrometry may serve as an alternate technique to HPLC when it comes to separating the different partially reduced isoforms of peptides and proteins. This approach may already be feasible with top-down compatible fragmentation strategies such as ECD and ETD.
Conclusions
The unique chemistry of cysteine side chains enables them to serve as molecular handles for protein manipulation and has facilitated the development of dozens of unique probes and corresponding approaches to track cysteine residue status within proteins. Given its continually increasing resolving power and analytical sensitivity, mass spectrometry is employed to handle an increasing share of research effort on this front. Nevertheless, as approaches to probing cysteine status become more and more advanced, the fundamental concern for labeling specificity, labeling completeness and potential reversibility, and consideration of sulfhydryl instability in the presence of oxygen should continue to be respected, as these features often underpin assumptions that are generally taken for granted when experimental results are interpreted.
As a related discipline, disulfide structure analysis is now largely oriented toward direct mapping of intact disulfide bonds using a bottom-up mass spectrometric approach in combination with CID as well as other modern fragmentation techniques such as ETD. The complex datasets generated from such experiments require greater reliance on algorithms and software-based tools for data interpretation. Thus, perhaps the greatest hurdle to truly routinized workflows for disulfide structure analysis lies in improving the reproducibility of sample preparation and mass spectral datasets. Ultimately, continued progress in this direction will mean a significantly decreased need for iterative experimental workflows and, therefore, faster resolution of complex disulfide structures to meet the quality control demands of modern biological therapeutics and other challenging cystinyl proteins.
Abbreviations Used
- CDAP
1-cyano-4-dimethylaminopyridinium tetrafluoroborate
- CID
collision-induced dissociation
- Cys-SOH
cysteine sulfenic acid
- DIGE
difference gel electrophoresis
- DTNB
5,5′-dithiobis-(2-nitrobenzoic acid)
- DTT
dithiothreitol
- ECD
electron capture dissociation
- ER
endoplasmic reticulum
- Ero1
ER oxidoreductin 1
- ETD
electron transfer dissociation
- HPLC
high-performance liquid chromatography
- ICAT
isotope-coded affinity tag
- itz
iminothiazolidine group
- MALDI
matrix-assisted laser desorption ionization
- MMTS
methyl methanethiosulfonate
- MS/MS
tandem mass spectrometry
- MSn
tandem mass spectrometry with two or more stages of fragmentation
- OxICAT
oxidation-based isotope-coded affinity tag
- PDI
protein disulfide isomerase
- TCEP
tris(2-carboxyethyl)phosphine
- TNB
2-nitro-5-thiobenzoic acid
- XIC
extracted ion chromatograms
Footnotes
As used in this review, the term “pinpointing” refers merely to obtaining knowledge that an analytical signal represents a free cysteine residue. Thus, the term “pinpointing” does not provide any information on peptide or protein identity; the term “identifying” is used for this purpose.
As used in this section, the term “pinpointing” refers merely to obtaining knowledge that a chromatographic or mass spectral signal represents an intra- or interchain disulfide-linked peptide; “pinpointing” does not provide any information on peptide identity. The term “identifying” refers to the ascertainment of the intra- or interchain peptide sequence(s), but it does not provide a disulfide connectivity pattern if more than one cystine is present.
CID is a means of fragmenting gas-phase ions by which the ions are accelerated into a region of the mass spectrometer containing an inert gas. Here, they become vibrationally excited and, subsequently, decompose into fragments that can be analyzed in an ensuing stage of mass spectrometry. a1 ions are generated by N-terminal fragmentation of the first amino-acid residue where cleavage occurs between the α-carbon and the carbonyl carbon. Since the side chain is retained on the ion, its mass can (in most cases) provide the identity of the N-terminal amino acid.
LIFT is not an acronym but it refers to the process of raising the potential energy of product ions for acceleration after they have been selected for MS/MS and fragmented (88, 93.)
XICs are chromatograms consisting of continuous ion abundance measurements from a single nominal mass. They are analogous to chromatograms corresponding to a single wavelength from an HPLC run monitored by a diode array detector.
Acknowledgments
The authors gratefully acknowledge support from the Virginia G. Piper Charitable Trust and NIH grants R01DK082542 and R24DK090958.
References
- 1.ThermoFisher Scientific. Available at www.piercenet.com
- 2.Alley SC, Benjamin DR, Jeffrey SC, Okeley NM, Meyer DL, Sanderson RJ, and Senter PD. Contribution of linker stability to the activities of anticancer immunoconjugates. Bioconjugate Chem 19: 759–765, 2008 [DOI] [PubMed] [Google Scholar]
- 3.Andreu CI, Woehlbier U, Torres M, and Hetz C. Protein disulfide isomerases in neurodegeneration: from disease mechanisms to biomedical applications. FEBS Lett 586: 2826–2834, 2012 [DOI] [PubMed] [Google Scholar]
- 4.Anfinsen CB. Principles that govern the folding of protein chains. Science 181: 223–230, 1973 [DOI] [PubMed] [Google Scholar]
- 5.Anfinsen CB. and Haber E. Studies on the reduction and re-formation of protein disulfide bonds. J Biol Chem 236: 1361–1363, 1961 [PubMed] [Google Scholar]
- 6.Anfinsen CB, Haber E, Sela M, and White FH, Jr., The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc Natl Acad Sci U S A 47: 1309–1314, 1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bean MF. and Carr SA. Characterization of disulfide bond position in proteins and sequence-analysis of cystine-bridged peptides by tandem mass-spectrometry. Anal Biochem 201: 216–226, 1992 [DOI] [PubMed] [Google Scholar]
- 8.Benham AM. The protein disulfide isomerase family: key players in health and disease. Antioxid Redox Signal 16: 781–789, 2012 [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharyya M, Gupta K, Gowd KH, and Balaram P. Rapid mass spectrometric determination of disulfide connectivity in peptides and proteins. Mol Biosyst 9: 1340–1350, 2013 [DOI] [PubMed] [Google Scholar]
- 10.Borges CR, Geddes T, Watson JT, and Kuhn DM. Dopamine biosynthesis is regulated by S-glutathionylation. Potential mechanism of tyrosine hydroxylast inhibition during oxidative stress. J Biol Chem 277: 48295–48302, 2002 [DOI] [PubMed] [Google Scholar]
- 11.Borges CR, Qi J, Wu W, Torng E, Hinck AP, and Watson JT. Algorithm-assisted elucidation of disulfide structure: application of the negative signature mass algorithm to mass-mapping the disulfide structure of the 12-cysteine transforming growth factor beta type II receptor extracellular domain. Anal Biochem 329: 91–103, 2004 [DOI] [PubMed] [Google Scholar]
- 12.Borges CR. and Watson JT. Recognition of cysteine-containing peptides through prompt fragmentation of the 4-dimethylaminophenylazophenyl-4'-maleimide derivative during analysis by MALDI-MS. Protein Sci 12: 1567–1572, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brennan JP, Wait R, Begum S, Bell JR, Dunn MJ, and Eaton P. Detection and mapping of widespread intermolecular protein disulfide formation during cardiac oxidative stress using proteomics with diagonal electrophoresis. J Biol Chem 279: 41352–41360, 2004 [DOI] [PubMed] [Google Scholar]
- 14.Buczek O, Green BR, and Bulaj G. Albumin is a redox-active crowding agent that promotes oxidative folding of cysteine-rich peptides. Biopolymers 88: 8–19, 2007 [DOI] [PubMed] [Google Scholar]
- 15.Chang JY. and Li L. Divergent folding pathways of two homologous proteins, BPTI and tick anticoagulant peptide: compartmentalization of folding intermediates and identification of kinetic traps. Arch Biochem Biophys 437: 85–95, 2005 [DOI] [PubMed] [Google Scholar]
- 16.Choi S, Jeong J, Na S, Lee HS, Kim HY, Lee KJ, and Paek E. New algorithm for the identification of intact disulfide linkages based on fragmentation characteristics in tandem mass spectra. J Proteome Res 9: 626–635, 2010. Available at http://prix.hanyang.ac.kr/download/dbond.jsp [DOI] [PubMed] [Google Scholar]
- 17.Chrisman PA, Pitteri SJ, Hogan JM, and McLuckey SA. SO2- electron transfer ion/ion reactions with disulfide linked polypeptide ions. J Am Soc Mass Spectrom 16: 1020–1030, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark DF, Go EP, and Desaire H. Simple approach to assign disulfide connectivity using extracted ion chromatograms of electron transfer dissociation spectra. Anal Chem 85: 1192–1199, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark DF, Go EP, Toumi ML, and Desaire H. Collision induced dissociation products of disulfide-bonded peptides: ions result from the cleavage of more than one bond. J Am Soc Mass Spectrom 22: 492–498, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cook KM. and Hogg PJ. Post-translational control of protein function by disulfide bond cleavage. Antioxid Redox Signal 18: 1987–2015, 2013 [DOI] [PubMed] [Google Scholar]
- 21.Crimmins DL, Saylor M, Rush J, and Thoma RS. Facile, in-situ matrix-assisted laser-desorption ionization mass-spectrometry analysis and assignment of disulfide pairings in heteropeptide molecules. Anal Biochem 226: 355–361, 1995 [DOI] [PubMed] [Google Scholar]
- 22.Cumming RC, Andon NL, Haynes PA, Park M, Fischer WH, and Schubert D. Protein disulfide bond formation in the cytoplasm during oxidative stress. J Biol Chem 279: 21749–21758, 2004 [DOI] [PubMed] [Google Scholar]
- 23.Echterbille J, Quinton L, Gilles N, and De Pauw E. Ion mobility mass spectrometry as a potential tool to assign disulfide bonds arrangements in peptides with multiple disulfide bridges. Anal Chem 85: 4405–4413, 2013 [DOI] [PubMed] [Google Scholar]
- 24.Ellman GL. Tissue sulfhydryl groups. Arch Biochem Biophys 82: 70–77, 1959 [DOI] [PubMed] [Google Scholar]
- 25.Fryklund L. and Eaker D. The complete covalent structure of a cardiotoxin from the venom of Naja nigricollis (African black-necked spitting cobra). Biochemistry 14: 2865–2871, 1975 [DOI] [PubMed] [Google Scholar]
- 26.Fuller E, Green BR, Catlin P, Buczek O, Nielsen JS, Olivera BM, and Bulaj G. Oxidative folding of conotoxins sharing an identical disulfide bridging framework. FEBS J 272: 1727–1738, 2005 [DOI] [PubMed] [Google Scholar]
- 27.Gallegos-Perez JL, Rangel-Ordonez L, Bowman SR, Ngowe CO, and Watson JT. Study of primary amines for nucleophilic cleavage of cyanylated cystinyl proteins in disulfide mass mapping methodology. Anal Biochem 346: 311–319, 2005 [DOI] [PubMed] [Google Scholar]
- 28.Gan ZR. and Wells WW. Identification and reactivity of the catalytic site of pig liver thioltransferase. J Biol Chem 262: 6704–6707, 1987 [PubMed] [Google Scholar]
- 29.Goransson U. and Craik DJ. Disulfide mapping of the cyclotide kalata B1. Chemical proof of the cystic cystine knot motif. J Biol Chem 278: 48188–48196, 2003 [DOI] [PubMed] [Google Scholar]
- 30.Gorman JJ, Wallis TP, and Pitt JJ. Protein disulfide bond determination by mass spectrometry. Mass Spectrom Rev 21: 183–216, 2002 [DOI] [PubMed] [Google Scholar]
- 31.Gray WR. Disulfide structures of highly bridged peptides: a new strategy for analysis. Protein Sci 2: 1732–1748, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grutzner A, Garcia-Manyes S, Kotter S, Badilla CL, Fernandez JM, and Linke WA. Modulation of titin-based stiffness by disulfide bonding in the cardiac titin N2-B unique sequence. Biophys J 97: 825–834, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, and Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol 17: 994–999, 1999 [DOI] [PubMed] [Google Scholar]
- 34.Ha JH. and Loh SN. Changes in side chain packing during apomyoglobin folding characterized by pulsed thiol-disulfide exchange. Nat Struct Biol 5: 730–737, 1998 [DOI] [PubMed] [Google Scholar]
- 35.Haber E. and Anfinsen CB. Side-chain interactions governing the pairing of half-cystine residues in ribonuclease. J Biol Chem 237: 1839–1844, 1962 [PubMed] [Google Scholar]
- 36.Hage DS. (Ed) Handbook of Affinity Chromatography. Boca Raton, FL: CRC Press Taylor & Francis Group, 2006 [Google Scholar]
- 37.Hansen RE, Roth D, and Winther JR. Quantifying the global cellular thiol-disulfide status. Proc Natl Acad Sci U S A 106: 422–427, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Happersberger HP, Przybylski M, and Glocker MO. Selective bridging of bis-cysteinyl residues by arsonous acid derivatives as an approach to the characterization of protein tertiary structures and folding pathways by mass spectrometry. Anal Biochem 264: 237–250, 1998 [DOI] [PubMed] [Google Scholar]
- 39.Happersberger HP, Stapleton J, Cowgill C, and Glocker MO. Characterization of the folding pathway of recombinant human macrophage-colony stimulating-factor beta (rhM-CSF beta) by bis-cysteinyl modification and mass spectrometry. Proteins Suppl 2: 50–62, 1998 [PubMed] [Google Scholar]
- 40.Harris RJ, Shire SJ, and Winter C. Commercial manufacturing scale formulation and analytical characterization of therapeutic recombinant antibodies. Drug Dev Res 61: 137–154, 2004 [Google Scholar]
- 41.Held JM. and Gibson BW. Regulatory control or oxidative damage? Proteomic approaches to interrogate the role of cysteine oxidation status in biological processes. Mol Cell Proteomics 11: R111 013037 (pgs. 1–14), 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Herrmann and Riemer Three approaches to one problem: Protein folding in the periplasm, the endoplasmic reticulum, and the intermembrane space. Antioxid Redox Signal 21: 438–456, 2014 [DOI] [PubMed] [Google Scholar]
- 42.Hermanson GT. Bioconjugate Techniques. San Diego, CA: Academic Press, 2008 [Google Scholar]
- 43.Hermanson GT, Krishna Mallia A, and Smith PK. Immobilized Affinity Ligand Techniques. San Diego, CA: Academic Press, 1992 [Google Scholar]
- 44.Hochgrafe F, Mostertz J, Albrecht D, and Hecker M. Fluorescence thiol modification assay: oxidatively modified proteins in Bacillus subtilis. Mol Microbiol 58: 409–425, 2005 [DOI] [PubMed] [Google Scholar]
- 45.Hogg PJ. Contribution of allosteric disulfide bonds to regulation of hemostasis. J Thromb Haemost 7Suppl 1: 13–16, 2009 [DOI] [PubMed] [Google Scholar]
- 46.Hu JJ, Dong LX, and Outten CE. The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J Biol Chem 283: 29126–29134, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang SY, Hsieh YT, Chen CH, Chen CC, Sung WC, Chou MY, and Chen SF. Automatic disulfide bond assignment using a(1) ion screening by mass spectrometry for structural characterization of protein pharmaceuticals. Anal Chem 84: 4900–4906, 2012 [DOI] [PubMed] [Google Scholar]
- 48.Huwiler KG, Mosher DF, and Vestling MM. Optimizing the MALDI-TOF-MS observation of peptides containing disulfide bonds. J Biomol Tech 14: 289–297, 2003 [PMC free article] [PubMed] [Google Scholar]
- 49.Ito K. and Inaba K. The disulfide bond formation (Dsb) system. Curr Opin Struct Biol 18: 450–458, 2008 [DOI] [PubMed] [Google Scholar]
- 50.Iwaoka M, Juminaga D, and Scheraga HA. Regeneration of three-disulfide mutants of bovine pancreatic ribonuclease A missing the 65–72 disulfide bond: characterization of a minor folding pathway of ribonuclease A and kinetic roles of Cys65 and Cys72. Biochemistry 37: 4490–4501, 1998 [DOI] [PubMed] [Google Scholar]
- 51.Jacobson GR, Schaffer MH, Stark GR, and Vanaman TC. Specific chemical cleavage in high yield at the amino peptide bonds of cysteine and cystine residues. J Biol Chem 248: 6583–6591, 1973 [PubMed] [Google Scholar]
- 52.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, and Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol 3: 193–197, 2001 [DOI] [PubMed] [Google Scholar]
- 53.Jaffrey SR. and Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001: PL1, 2001 [DOI] [PubMed] [Google Scholar]
- 54.Johnson I. and Spence MTZ. The Molecular Probes Handbook: A Guide to Fluorescent Probes and Labeling Technologies. USA: Life Technologies Corporation, 2010 [Google Scholar]
- 55.Karala AR, Lappi AK, Saaranen MJ, and Ruddock LW. Efficient peroxide-mediated oxidative refolding of a protein at physiological pH and implications for oxidative folding in the endoplasmic reticulum. Antioxid Redox Signal 11: 963–970, 2009 [DOI] [PubMed] [Google Scholar]
- 56.Kojer K, Bien M, Gangel H, Morgan B, Dick TP, and Riemer J. Glutathione redox potential in the mitochondrial intermembrane space is linked to the cytosol and impacts the Mia40 redox state. EMBO J 31: 3169–3182, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Konermann L. and Simmons DA. Protein-folding kinetics and mechanisms studied by pulse-labeling and mass spectrometry. Mass Spectrom Rev 22: 1–26, 2003 [DOI] [PubMed] [Google Scholar]
- 58.Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, Strahler JR, Andrews PC, and Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A 105: 8197–8202, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leichert LI. and Jakob U. Protein thiol modifications visualized in vivo. PLoS Biol 2: 1723–1737, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leichert LI. and Jakob U. Global methods to monitor the thiol-disulfide state of proteins in vivo. Antioxid Redox Signal 8: 763–772, 2006 [DOI] [PubMed] [Google Scholar]
- 61.Lewinter MM. and Granzier HL. Cardiac titin and heart disease. J Cardiovasc Pharmacol 2013. [Epub ahead of print]; DOI: 10.1097/FJC.0000000000000007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lilley KS. and Friedman DB. All about DIGE: quantification technology for differential-display 2D-gel proteomics. Expert Rev Proteomics 1: 401–409, 2004 [DOI] [PubMed] [Google Scholar]
- 63.Lin D, Saleh S, and Liebler DC. Reversibility of covalent electrophile-protein adducts and chemical toxicity. Chem Res Toxicol 21: 2361–2369, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu H, Gaza-Bulseco G, Chumsae C, and Newby-Kew A. Characterization of lower molecular weight artifact bands of recombinant monoclonal IgG1 antibodies on non-reducing SDS-PAGE. Biotechnol Lett 29: 1611–1622, 2007 [DOI] [PubMed] [Google Scholar]
- 65.Lutz E, Wieser H, and Koehler P. Identification of disulfide bonds in wheat gluten proteins by means of mass spectrometry/electron transfer dissociation. J Agric Food Chem 60: 3708–3716, 2012 [DOI] [PubMed] [Google Scholar]
- 66.Medzihradszky KF. and Bohlen CJ. Partial de novo sequencing and unusual CID fragmentation of a 7 kDa, disulfide-bridged toxin. J Am Soc Mass Spectrom 23: 923–934, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JE, Shabanowitz J, and Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochim Biophys Acta 1764: 1811–1822, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mo J, Tymiak AA, and Chen G. Characterization of disulfide linkages in recombinant human granulocyte-colony stimulating factor. Rapid Commun Mass Spectrom 27: 940–946, 2013 [DOI] [PubMed] [Google Scholar]
- 69.Moran LA, Scrimgeour KG, Horton HR, Och RS, and Rawn JD. Biochemistry. Englewood Cliffs, NJ: Neil Patterson Publishers/Prentice Hall, 1994 [Google Scholar]
- 70.Mormann M, Eble J, Schwoppe C, Mesters RM, Berdel WE, Peter-Katalinic J, and Pohlentz G. Fragmentation of intra-peptide and inter-peptide disulfide bonds of proteolytic peptides by nanoESI collision-induced dissociation. Anal Bioanal Chem 392: 831–838, 2008 [DOI] [PubMed] [Google Scholar]
- 71.Murray CI. and Van Eyk JE. Chasing cysteine oxidative modifications: proteomic tools for characterizing cysteine redox status. Circ Cardiovasc Genet 5: 591, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Narayan M, Welker E, Zhai H, Han X, Xu G, McLafferty FW, and Scheraga HA. Detecting native folds in mixtures of proteins that contain disulfide bonds. Nat Biotechnol 26: 427–429, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ni W, Lin M, Salinas P, Savickas P, Wu SL, and Karger BL. Complete mapping of a cystine knot and nested disulfides of recombinant human arylsulfatase A by multi-enzyme digestion and LC-MS analysis using CID and ETD. J Am Soc Mass Spectrom 24: 125–133, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nili M, Mukherjee A, Shinde U, David L, and Rotwein P. Defining the disulfide bonds of insulin-like growth factor-binding protein-5 by tandem mass spectrometry with electron transfer dissociation and collision-induced dissociation. J Biol Chem 287: 1510–1519, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pantoja-Uceda D, Arolas JL, Aviles FX, Santoro J, Ventura S, and Sommerhoff CP. Deciphering the structural basis that guides the oxidative folding of leech-derived tryptase inhibitor. J Biol Chem 284: 35612–35620, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Patterson SD. and Katta V. Prompt fragmentation of disulfide-linked peptides during matrix-assisted laser desorption ionization mass spectrometry. Anal Chem 66: 3127–3132, 1994 [DOI] [PubMed] [Google Scholar]
- 77.Pike GM, Madden BJ, Melder DC, Charlesworth MC, and Federspiel MJ. Simple, automated, high resolution mass spectrometry method to determine the disulfide bond and glycosylation patterns of a complex protein: subgroup A avian sarcoma and leukosis virus envelope glycoprotein. J Biol Chem 286: 17954–17967, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pompach P, Man P, Kavan D, Hofbauerova K, Kumar V, Bezouska K, Havlicek V, and Novak P. Modified electrophoretic and digestion conditions allow a simplified mass spectrometric evaluation of disulfide bonds. J Mass Spectrom 44: 1571–1578, 2009 [DOI] [PubMed] [Google Scholar]
- 79.Qi J, Wu J, Somkuti GA, and Watson JT. Determination of the disulfide structure of sillucin, a highly knotted, cysteine-rich peptide, by cyanylation/cleavage mass mapping. Biochemistry 40: 4531–4538, 2001 [DOI] [PubMed] [Google Scholar]
- 80.Qi J, Wu W, Borges CR, Hang D, Rupp M, Torng E, and Watson JT. Automated data interpretation based on the concept of “negative signature mass” for mass-mapping disulfide structures of cystinyl proteins. J Am Soc Mass Spectrom 14: 1032–1038, 2003 [DOI] [PubMed] [Google Scholar]
- 81.Qin J. and Chait BT. Identification and characterization of posttranslational modifications of proteins by MALDI ion trap mass spectrometry. Anal Chem 69: 4002–4009, 1997 [DOI] [PubMed] [Google Scholar]
- 82.Reeve JR, Cheng KW, and Pierce JG. Partial reduction of disulfide bonds in the hormone-specific subunits of TSH and LH. Biochem Biophys Res Commun 67: 149–155, 1975 [DOI] [PubMed] [Google Scholar]
- 83.Regaya I, Andreotti N, Di Luccio E, De Waard M, and Sabatier JM. Effect of Cu2+ on the oxidative folding of synthetic maurotoxin in vitro. J Biomol Struct Dyn 26: 75–82, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rehder DS. and Borges CR. Cysteine sulfenic acid as an intermediate in disulfide bond formation and nonenzymatic protein folding. Biochemistry 49: 7748–7755, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Riddles PW, Blakeley RL, and Zerner B. Reassessment of Ellman's reagent. Methods Enzymol 91: 49–60, 1983 [DOI] [PubMed] [Google Scholar]
- 86.Riemer J, Bulleid N, and Herrmann JM. Disulfide formation in the ER and mitochondria: two solutions to a common process. Science 324: 1284–1287, 2009 [DOI] [PubMed] [Google Scholar]
- 87.Ryle AP, Sanger F, Smith LF, and Kitai R. The disulphide bonds of insulin. Biochem J 60: 541–556, 1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schnaible V, Wefing S, Resemann A, Suckau D, Bucker A, Wolf-Kummeth S, and Hoffmann D. Screening for disulfide bonds in proteins by MALDI in-source decay and LIFT-TOF/TOF-MS. Anal Chem 74: 4980–4988, 2002 [DOI] [PubMed] [Google Scholar]
- 89.Sethuraman M, McComb ME, Heibeck T, Costello CE, and Cohen RA. Isotope-coded affinity tag approach to identify and quantify oxidant-sensitive protein thiols. Mol Cell Proteomics 3: 273–278, 2004 [DOI] [PubMed] [Google Scholar]
- 90.Shen BQ, Xu K, Liu L, Raab H, Bhakta S, Kenrick M, Parsons-Reponte KL, Tien J, Yu SF, Mai E, Li D, Tibbitts J, Baudys J, Saad OM, Scales SJ, McDonald PJ, Hass PE, Eigenbrot C, Nguyen T, Solis WA, Fuji RN, Flagella KM, Patel D, Spencer SD, Khawli LA, Ebens A, Wong WL, Vandlen R, Kaur S, Sliwkowski MX, Scheller RH, Polakis P, and Junutula JR. Conjugation site modulates the in vivo stability and therapeutic activity of antibody-drug conjugates. Nat Biotechnol 30: 184–189, 2012 [DOI] [PubMed] [Google Scholar]
- 91.Shiio Y. and Aebersold R. Quantitative proteome analysis using isotope-coded affinity tags and mass spectrometry. Nat Protoc 1: 139–145, 2006 [DOI] [PubMed] [Google Scholar]
- 92.Sommer A. and Traut RR. Diagonal polyacrylamide-dodecyl sulfate gel electrophoresis for the identification of ribosomal proteins crosslinked with methyl-4-mercaptobutyrimidate. Proc Natl Acad Sci U S A 71: 3946–3950, 1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Suckau D, Resemann A, Schuerenberg M, Hufnagel P, Franzen J, and Holle A. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal Bioanal Chem 376: 952–965, 2003 [DOI] [PubMed] [Google Scholar]
- 94.Tang HY. and Speicher DW. Determination of disulfide-bond linkages in proteins. Curr Protoc Protein Sci Chapter 11: Unit 11.11.1–11.11.20, 2004 [DOI] [PubMed] [Google Scholar]
- 95.Thornton JM. Disulfide bridges in globular-proteins. J Mol Biol 151: 261–287, 1981 [DOI] [PubMed] [Google Scholar]
- 96.Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, and Lipton SA. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature 441: 513–517, 2006 [DOI] [PubMed] [Google Scholar]
- 97.van den Berg B, Chung EW, Robinson CV, and Dobson CM. Characterisation of the dominant oxidative folding intermediate of hen lysozyme. J Mol Biol 290: 781–796, 1999 [DOI] [PubMed] [Google Scholar]
- 98.Wallis TP, Pitt JJ, and Gorman JJ. Identification of disulfide-linked peptides by isotope profiles produced by peptic digestion of proteins in 50% O-18 water. Protein Sci 10: 2251–2271, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang Y, Lu Q, Wu SL, Karger BL, and Hancock WS. Characterization and comparison of disulfide linkages and scrambling patterns in therapeutic monoclonal antibodies: using LC-MS with electron transfer dissociation. Anal Chem 83: 3133–3140, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Watson JT, Yang Y, and Wu J. Capture and identification of folding intermediates of cystinyl proteins by cyanylation and mass spectrometry. J Mol Graph Model 19: 119–128, 2001 [DOI] [PubMed] [Google Scholar]
- 101.Welker E, Hathaway L, and Scheraga HA. A new method for rapid characterization of the folding pathways of multidisulfide-containing proteins. J Am Chem Soc 126: 3720–3721, 2004 [DOI] [PubMed] [Google Scholar]
- 102.White FH, Jr., Regeneration of native secondary and tertiary structures by air oxidation of reduced ribonuclease. J Biol Chem 236: 1353–1360, 1961 [PubMed] [Google Scholar]
- 103.White FH, Jr., and Anfinsen CB. Some relationships of structure to function in ribonuclease. Ann N Y Acad Sci 81: 515–523, 1959 [DOI] [PubMed] [Google Scholar]
- 104.Wong JWH, Ho SYW, and Hogg PJ. Disulfide bond acquisition through eukaryotic protein evolution. Mol Biol Evol 28: 327–334, 2011 [DOI] [PubMed] [Google Scholar]
- 105.Woo HA, Chae HZ, Hwang SC, Yang KS, Kang SW, Kim K, and Rhee SG. Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science 300: 653–656, 2003 [DOI] [PubMed] [Google Scholar]
- 106.Woo HA, Kang SW, Kim HK, Yang KS, Chae HZ, and Rhee SG. Reversible oxidation of the active site cysteine of peroxiredoxins to cysteine sulfinic acid. Immunoblot detection with antibodies specific for the hyperoxidized cysteine-containing sequence. J Biol Chem 278: 47361–47364, 2003 [DOI] [PubMed] [Google Scholar]
- 107.Wouters MA, Fan SW, and Haworth NL. Disulfides as redox switches: from molecular mechanisms to functional significance. Antioxid Redox Signal 12: 53–91, 2010 [DOI] [PubMed] [Google Scholar]
- 108.Wu C, Parrott AM, Liu T, Jain MR, Yang Y, Sadoshima J, and Li H. Distinction of thioredoxin transnitrosylation and denitrosylation target proteins by the ICAT quantitative approach. J Proteomics 74: 2498–2509, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu J, Gage DA, and Watson JT. A strategy to locate cysteine residues in proteins by specific chemical cleavage followed by matrix-assisted laser desorption/ionization time- of-flight mass spectrometry. Anal Biochem 235: 161–174, 1996 [DOI] [PubMed] [Google Scholar]
- 110.Wu J. and Watson JT. A novel methodology for assignment of disulfide bond pairings in proteins. Protein Sci 6: 391–398, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu J. and Watson JT. Optimization of the cleavage reaction for cyanylated cysteinyl proteins for efficient and simplified mass mapping. Anal Biochem 258: 268–276, 1998 [DOI] [PubMed] [Google Scholar]
- 112.Wu J, Yang Y, and Watson JT. Trapping of intermediates during the refolding of recombinant human epidermal growth factor (hEGF) by cyanylation, and subsequent structural elucidation by mass spectrometry. Protein Sci 7: 1017–1028, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu SL, Jiang H, Hancock WS, and Karger BL. Identification of the unpaired cysteine status and complete mapping of the 17 disulfides of recombinant tissue plasminogen activator using LC-MS with electron transfer dissociation/collision induced dissociation. Anal Chem 82: 5296–5303, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu SL, Jiang HT, Lu QZ, Dai SJ, Hancock WS, and Karger BL. Mass Spectrometric determination of disulfide linkages in recombinant therapeutic proteins using online LC-MS with electron-transfer dissociation. Anal Chem 81: 112–122, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xu H, Zhang LW, and Freitas MA. Identification and characterization of disulfide bonds in proteins and peptides from tandem MS data by use of the MassMatrix MS/MS search engine. J Proteome Res 7: 138–144, 2008. Available at www.massmatrix.net/mm-cgi/home.py [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yang Y, Wu J, and Watson JT. Disulfide mass mapping in proteins containing adjacent cysteines is possible with cyanylation/cleavage methodology. J Am Chem Soc 120: 5834–5835, 1998 [Google Scholar]
- 117.Yang Y, Wu J, and Watson JT. Probing the folding pathways of long R(3) insulin-like growth factor-I (LR(3)IGF-I) and IGF-I via capture and identification of disulfide intermediates by cyanylation methodology and mass spectrometry. J Biol Chem 274: 37598–37604, 1999 [DOI] [PubMed] [Google Scholar]
- 118.Yen TY, Yan H, and Macher BA. Characterizing closely spaced, complex disulfide bond patterns in peptides and proteins by liquid chromatography/electrospray ionization tandem mass spectrometry. J Mass Spectrom 37: 15–30, 2002 [DOI] [PubMed] [Google Scholar]
- 119.Young MM, Tang N, Hempel JC, Oshiro CM, Taylor EW, Kuntz ID, Gibson BW, and Dollinger G. High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc Natl Acad Sci U S A 97: 5802–5806, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang W, Marzilli LA, Rouse JC, and Czupryn MJ. Complete disulfide bond assignment of a recombinant immunoglobulin G4 monoclonal antibody. Anal Biochem 311: 1–9, 2002 [DOI] [PubMed] [Google Scholar]
- 121.Zhang X, Huang B, Zhou X, and Chen C. Quantitative proteomic analysis of S-nitrosated proteins in diabetic mouse liver with ICAT switch method. Protein Cell 1: 675–687, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhou J, Ens W, Poppeschriemer N, Standing KG, and Westmore JB. Cleavage of interchain disulfide bonds following matrix-assisted laser-desorption. Int J Mass Spectrom 126: 115–122, 1993 [Google Scholar]
- 123.Zubarev RA. Electron-capture dissociation tandem mass spectrometry. Curr Opin Biotechnol 15: 12–16, 2004 [DOI] [PubMed] [Google Scholar]
- 124.Zubarev RA, Kruger NA, Fridriksson EK, Lewis MA, Horn DM, Carpenter BK, and McLafferty FW. Electron capture dissociation of gaseous multiply-charged proteins is favored at disulfide bonds and other sites of high hydrogen affinity. J Am Chem Soc 121: 2857–2862, 1999 [Google Scholar]