Abstract
A large number of methods are available to deplete ribosomal RNA reads from high throughput RNA sequencing experiments. Such methods are critical for sequencing Drosophila small RNAs between 20 and 30 nucleotides because size selection is not typically sufficient to exclude the highly abundant class of 30 nt 2S rRNA. Here we demonstrate that pre-annealing terminator oligos complimentary to Drosophila 2S rRNA prior to 5′ adapter ligation and reverse transcription efficiently depletes 2S rRNA sequences from the sequencing reaction in a simple and inexpensive way. This depletion is highly specific and is achieved with minimal perturbation of miRNA and piRNA profiles.
Keywords: Drosophila, 2S rRNA, piRNA, miRNA, small RNA sequencing
High-throughput RNA sequencing (RNA-seq) has revolutionized our understanding of RNA biology, but the presence of high abundance RNA species that are not of direct interest can be problematic. For this reason there is great interest in the development of methods that enrich for specific classes of RNA for RNA-seq applications. Historically, the most commonly used method for enriching populations of mRNA has depended on the presence of poly(A) tails. However, this method is not applicable in non-eukaryotes that lack poly-adenylation and is also incapable of enriching for many non-coding RNAs and degraded RNAs that lack poly(A) tails. In contrast to enriching for poly(A), an alternate method is to directly deplete nuisance classes of RNA. A recent survey(1) compared different methods (2–6) to deplete nuisance species of rRNA and favored rRNA hybridization to DNA oligos and removal of RNA-DNA hybrids by RNAse H treatment.
For sequencing small RNAs (miRNAs, siRNAs and piRNAs), enrichment is usually achieved by size selection. Since these classes of RNA are typically between 20 and 30 nt, size selection from total RNA, followed by ligation, PCR and further size selection, typically achieves good results (for review of small RNA sequencing methods see (7)). In Drosophila, however, size selection is not typically sufficient because a highly abundant class of 2S rRNA is 30 nt long. To deal with this problem, two approaches have been developed. One method, using RNAse H, employs hybridization with DNA oligos complementary to 2S RNA and directed degradation (8). A second method involves depletion by hybridization using oligos bound to magnetic beads, from which non-2S rRNA is removed off as supernatant (9). Both of these methods have proven effective in eliminating 2S rRNA reads, but both require additional steps and reagents which, in the case of magnetic beads, can be costly. We therefore sought to determine whether depletion could be achieved during 5′ ligation and cDNA synthesis by specifically blocking 2S RNA products from jointly participating in these two reactions. For 2S RNA, this is expected to yield truncated cDNA products lacking sequences complementary to the 5′ adapter. Without these sequences, library PCR amplification is impossible. Blocking with terminator oligos may thus serve as a simple means to eliminate 2S rRNA from the sequencing reaction without additional enzymatic steps or use of magnetic beads.
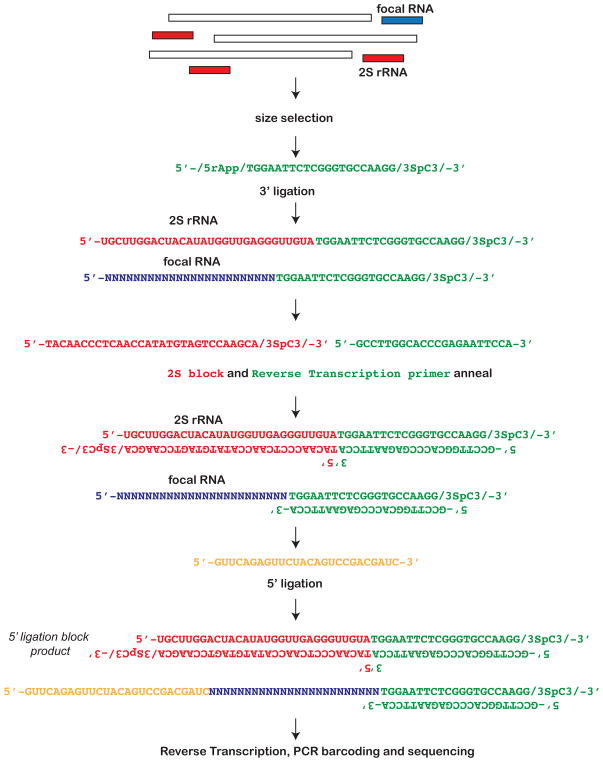
To test this, we performed a modification to a previously published protocol (10), with the intent of precluding 5′ adapter ligation to 2S rRNA. We first performed size selection of 19 to 30 nt RNA with 15% acrylamide/urea gels followed by 3′ adapter ligation. Prior to 5′ adapter ligation, we first pre-annealed a 2S rRNA blocker complementary to the 2S rRNA sequence and ending with a 3′ C3 spacer (Figure 1). Subsequently, we annealed the RT primer complementary to the 3′ adapter. This RT primer was annealed prior to 5′ ligation to eliminate adapter-adapter ligation products (10). After these pre-annealing steps, we performed 5′ ligation and RT followed by PCR-based barcode incorporation and Illumina sequencing (detailed method provided as supplemental materials). For these experiments, two separate size selected small RNA pools (experiment 1 and experiment 2) were extracted from Drosophila virilis ovaries. After gel purification and size selection, the two pools of RNA were each split into no block and block treatments.
Figure 1.
Overall small RNA library preparation scheme with 2S block reaction.
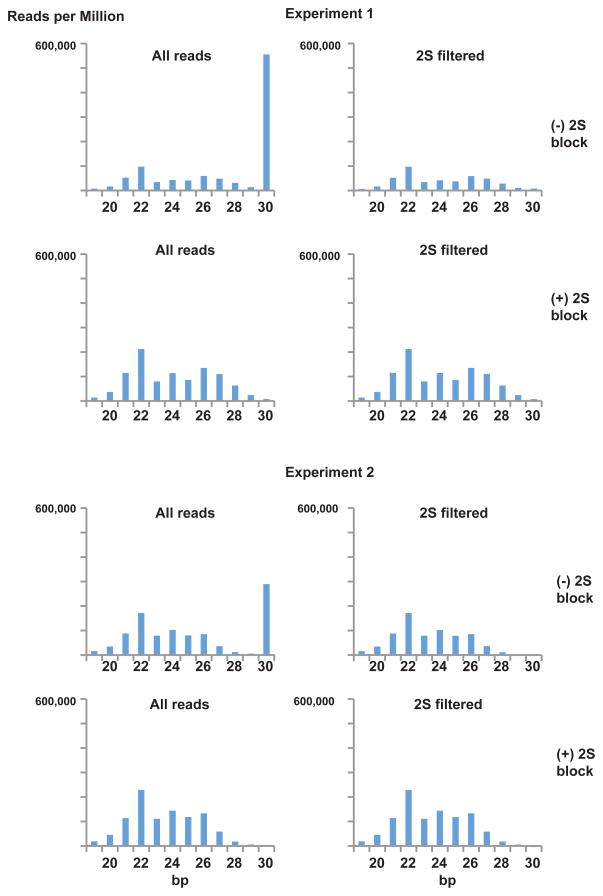
To measure depletion of 2S rRNA, we quantified the proportion of 19 to 30 bp reads mapping to 2S rRNA. Table 1 indicates that 2S rRNA was nearly entirely eliminated when the 2S block was utilized. Since the number of uniqe small RNA reads is greater for the blocked samples in both experiments, we can conclude that the block is not leading to PCR compression of library complexity. Examining the size distribution of reads (Figure 2), one can see that the 2S block reaction largely eliminates the 30 bp class of RNA without perturbing the underlying size distribution. In all cases, a 22 nt peak of miRNA is evident against a background of presumptive piRNA and siRNA.
Table 1.
2S mapped reads from 2S block (−) and (+) small RNA sequence runs. The percent genome mapping in total reads is indicated in parentheses. The percentage of total reads in 2S reads in also in parentheses.
| Experiment 1 | Experiment 2 | |||
|---|---|---|---|---|
| 2S block (−) | 2S block (+) | 2S block (−) | 2S block (+) | |
| Total Reads (% Mapped) | 919686 (99) | 689333 (98) | 1556883 (98) | 1186377 (98) |
| Unique Reads | 107102 | 154663 | 206277 | 221156 |
| 2S Reads (% Total Reads) | 515483 (56) | 437 (<0.1) | 454481 (29) | 496 (<0.1) |
Figure 2.
Size distribution of small RNA reads from two experiments, with and without 2S block treatment. 2S filtered distributions were obtained after removal of 2S mapping sequences.
To determine whether non-specific 2S block hybridization perturbed signatures of miRNA expression from ovaries, we mapped non-2S rRNA small RNA reads against the annotated miRNA library from Drosophila virilis. Comparison of profiles was achieved by measuring the proportion of the non-2S reads that mapped to these miRNAs. miRNA profiles were very similar between blocked and non-blocked libraries (Figure 3A, C). In experiment one, there was some difference in the expression profile among two of the most highly expressed miRNAs. However, this was not the case in experiment two. To directly compare the miRNA profiles between experiments, we used Pearson’s r as a metric (Figure 3C). Among all pairwise comparisons and considering all miRNAs, the pair with most similar results was Experiment 2 (no 2S block) and Experiment 2 (2S block) (r=0.999). Overall, results indicate that Experiment 1 (no 2S block) is an outlier since its Pearson’s r value is lower with all other samples. However, its miRNA profile is most similar to its own block treatment pair (r=0.965). Since block pairs and no block pairs were never most similar to each other, we can conclude that blocking treatment did not result in a systematic bias in miRNA profile signatures. Excluding the four highly expressed RNAs, the most similar two pairwise results were achieved within RNA extraction rather than within block/no block treatment.
Figure 3.
Comparison of Drosophila virilis ovary small RNA profiles between no block and 2S block treatments. A) log10-log10 miRNA profiles from two experiments, comparing no block and 2S block treatments, after 2S rRNA sequence filtering. Measurements are in Reads Per Million Mapped (RPMM) - where Mapped (M) is measured from the total number of reads mapping to the D. virilis genome. B) log10-log10 TE mapping small RNA profiles from two experiments, comparing non block and 2S block treatments. Measurements are the RPMM of non-2S RNA/non-miRNA mapping reads that map to each of the annotated Drosophila virilis TEs. C) miRNA profile similarity comparisons as measured by Pearson’s r. Values in parentheses are with highly expressed miRNAs excluded. Red indicates most similar pair among a set of comparisons. D) TE small RNA profile similarity. All results, block or no block, are highly similar.
In addition, we also determined whether 2S blocking perturbed the signature of small RNAs targeting transposable elements. We mapped non-2S/non-miRNA reads against the 10 well annotated TEs from Drosophila virilis. The vast majority of those mapping TEs were 23 – 30 nt in length, indicating them as piRNAs. TE mapping small RNA profiles are remarkably robust across all treatments and there is no evidence 2S blocking perturbs these profiles (Figure 3B). Interestingly, small RNA profiles from TEs are more robust to experimental variation than miRNA profiles (Figure 3D). This might be explained by the fact that each TE represents many different piRNAs. In contrast to single miRNA measurements, pools of diverse piRNAs may be robust to stochastic variation in cloning, PCR and sequencing efficiency.
To conclude, we have demonstrated a simple and inexpensive method to limit rRNA reads from small RNA sequence reactions. This is most likely achieved by limiting 5′ adapter ligation to the double-stranded rRNA/oligonucleotide duplex. Previous studies have shown that 5′ adapter/3′ adapter ligation events are minimized in the presence of duplex DNA (10). In addition, the presence of a 3′ C3 spacer may further contribute to ligation blocking. Since rRNA excess is a general problem for RNA-seq, similar blocking methods that limit adapter ligation and full length cDNA synthesis may prove generally simple and useful, limiting the use of magnetic beads and additional enzymatic steps.
Supplementary Material
Acknowledgments
Funding for this work was provided by NSF Award MCB-1022165 and NIH P20GM103638.
Footnotes
Competing Interests Statement
The authors declare no competing interests.
Refererences
- 1.Adiconis X, Borges-Rivera D, Satija R, Deluca DS, Busby MA, Berlin AM, Sivachenko A, Thompson DA, et al. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat Methods. 2013;10:623–629. doi: 10.1038/nmeth.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morlan JD, Qu K, Sinicropi DV. Selective depletion of rRNA enables whole transcriptome profiling of archival fixed tissue. PLoS One. 2012;7:e42882. doi: 10.1371/journal.pone.0042882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang R, Jaritz M, Guenzl P, Vlatkovic I, Sommer A, Tamir IM, Marks H, Klampfl T, et al. An RNA-Seq strategy to detect the complete coding and non-coding transcriptome including full-length imprinted macro ncRNAs. PLoS One. 2011;6:e27288. doi: 10.1371/journal.pone.0027288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yi H, Cho YJ, Won S, Lee JE, Jin Yu H, Kim S, Schroth GP, Luo S, Chun J. Duplex-specific nuclease efficiently removes rRNA for prokaryotic RNA-seq. Nucleic Acids Res. 2011;39:e140. doi: 10.1093/nar/gkr617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tariq MA, Kim HJ, Jejelowo O, Pourmand N. Whole-transcriptome RNAseq analysis from minute amount of total RNA. Nucleic Acids Res. 2011;39:e120. doi: 10.1093/nar/gkr547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhuang F, Fuchs RT, Robb GB. Small RNA expression profiling by high-throughput sequencing: implications of enzymatic manipulation. J Nucleic Acids. 2012;2012:360358. doi: 10.1155/2012/360358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li CJ, V, Vagin V, Lee SH, Xu J, Ma SM, Xi HL, Seitz H, Horwich MD, et al. Collapse of Germline piRNAs in the Absence of Argonaute3 Reveals Somatic piRNAs in Flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5′ precision of both MicroRNAs and their miRNA* strands in flies. Current Biology. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vigneault F, Ter-Ovanesyan D, Alon S, Eminaga S, CCD, Seidman JG, Eisenberg E, MCG High-throughput multiplex sequencing of miRNA. Curr Protoc Hum Genet. 2012;Chapter 11(Unit 11 12):11–10. doi: 10.1002/0471142905.hg1112s73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.