Abstract
Hydrogen sulfide (H2S), generated through various endogenous enzymatic and non-enzymatic pathways, is emerging as a regulator of physiological and pathological events throughout the body. Bacteria in the gastrointestinal tract also produce significant amounts of H2S that regulates microflora growth and virulence responses. However, the impact of the microbiota on host global H2S bioavailability and metabolism remain unknown. To address this question, we examined H2S bioavailability in its various forms (free, acid labile or bound sulfane sulfur), cystathionine gamma lyase (CSE) activity and cysteine levels in tissues from germ free versus conventionally housed mice. Free H2S levels were significantly reduced in plasma and gastrointestinal tissues of germ free mice. Bound sulfane sulfur levels were decreased by 50–80% in germ free mouse plasma, adipose and lung tissues. Tissue CSE activity was significantly reduced in many organs from germ free mice, whereas tissue cysteine levels were significantly elevated compared to conventional mice. These data reveal that the microbiota profoundly regulates systemic bioavailability and metabolism of H2S.
Keywords: sulfide, bacteria, cysteine, microflora, metabolism
Introduction
Hydrogen sulfide has emerged as an important endogenous gasotransmitter in vivo that contributes to numerous physiological and pathological responses of different organs through its ability to modulate oxidative stress, signal transduction and nitric oxide bioavailability (32). Endogenous generation of hydrogen sulfide is complex with enzymatic synthesis occurring through three proteins including cystathione gamma lyase (CSE) and cystathione beta synthase (CBS) that use cysteine as a substrate and 3-mercapto-sulfurtransferase (3-MST) using 3-mercaptopyruvate as a substrate (25,32). Once formed, hydrogen sulfide is very reactive resulting in rapid metabolism into one of three major pools including free, acid labile and bound sulfane sulfur forms (28,29). In this way, hydrogen sulfide bioequivalents may be regulated allowing for conversion and use for various cellular biochemical processes.
Hydrogen sulfide production has long been studied in prokaryotic cells with its generation being important for antioxidant defense, energy production, and cell cycle regulation (1,11,17,20,21). Various species of sulfate reducing bacteria (SRB) typically use thiosulfate to generate hydrogen sulfide, although disproportination of S2O3− to hydrogen sulfide and SO4−2 and decay of S-containing amino acids are also alternative generation pathways (3,17). Studies suggest that gastrointestinal hydrogen sulfide generation plays a critical role in regulating physiological responses such as motility, epithelial cell health and inflammation (4,16,30). Conversely, other reports suggest a pathological role of gastrointestinal hydrogen sulfide generation presumably due to differential microbial colonization contributing to various conditions such as inflammatory bowel disease, colonic nociception and colorectal cancer (18,22). However, the importance of microflora on host hydrogen sulfide formation, bioavailability and metabolism remains unknown, as examination of tissue H2S synthesis has only been performed using conventional mice (13,29). Here we report that the normal microflora profoundly alters H2S bioavailability along with alterations in synthesis enzyme activity and substrate availability.
Materials and Methods
Animals and tissue collection
All animal experiments were approved by the Ethics Committee in Stockholm, Sweden. Eleven to twelve weeks old male germ free C57BL/6J mice (n = 10) and specific pathogen free (conventional) C57BL/6J mice (n = 10) were used. All mice were maintained on autoclaved standard chow (R36; Lactamin, Stockholm, Sweden) and water ad libitum, and kept under controlled 12-h light-dark cycle in 12 h light cycles. The germ free status was checked weekly by culturing faecal samples, both aerobically and anaerobically at +20 and +37 °C for up to 4 weeks (10).
At the day of the experiment, animals were anesthetized by inhalation of 2.2% isoflurane (Forene®, Abbot Scandinavia AB, Kista, Sweden) in air. After blood sampling (inferior vena cava) the animals were sacrificed and tissues rapidly collected. Plasma and tissue samples were immediately homogenized in a stabilization buffer (degassed 100 mM Tris-HCl buffer pH 9.5, 0.1 mM DTPA) to preserve the bioavailable pools of H2S and metabolic proteins. Samples were snap frozen and stored in liquid nitrogen until analyzed.
Detection of free sulfide, acid-labile sulfide, bound sulfide and cysteine
Concentrations of sulfide and cysteine were measured by RP-HPLC after derivatization with excess monobromobimane (MBB) as stable Sulfide-dibimane and cysteine-S-bimane as we have previously reported (23). Briefly, 30 μl of samples was added into to 70 μl of 100 mM Tris-HCl buffer (pH 9.5, 0.1 mM DTPA), followed by addition of 50 μl of 10 mM MBB and incubated for 30 minutes. The reaction was terminated with 50 μl of 200 mM 5-sulfosalicylic acid and the mixture centrifuged. The resulting supernatant was analyzed by RP-HPLC equipped with a fluorescence detector (λex: 390 nm and λem:475 nm) and an eclipse XDB-C18 column (4.6×250 mm). Typical retention times for bimane adducts of hydrogen sulfide and cysteine were 15.75 and 10.12 min, respectively.
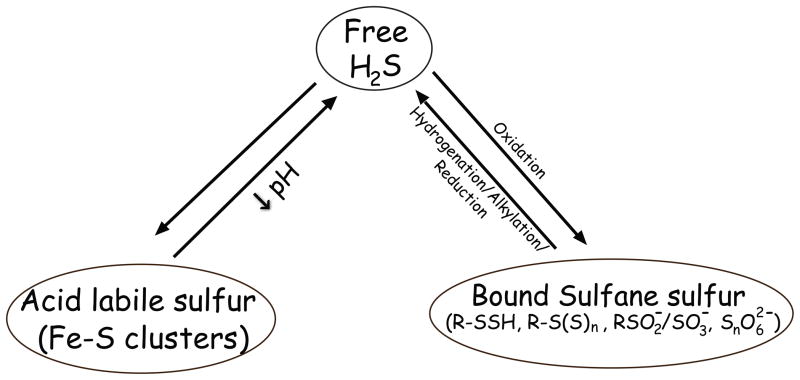
Hydrogen sulfide can exist in many biochemical forms as illustrated in figure 1. Acid-labile sulfide and bound sulfane sulfur was measured as we have previously reported (24). Acid-labile sulfide was released by incubating samples in an acidic solution (pH 2.6, 100 mM phosphate buffer, 0.1 mM DTPA), in an enclosed system to contain volatilized hydrogen sulfide. Volatilized hydrogen sulfide was then trapped in 100 mM Tris-HCl buffer (pH 9.5, 0.1 mM DTPA). The bound sulfane sulfur pool was measured by incubating the sample with 1 mM TCEP in the 100 mM phosphate buffer at pH 2.6 with 0.1 mM DTPA and sulfide measurement performed in a manner analogous to the one described above. The acid-labile pool was determined by subtracting the free hydrogen sulfide value from the value obtained by the acid-liberation protocol. The bound sulfane sulfur pool was determined by subtracting the hydrogen sulfide measurement from the acid-liberation protocol alone compared to that of TCEP plus acidic conditions.
Figure 1. Various biochemical forms of H2S bioavailability.
H2S exists in different biochemical forms within biological systems that can be classified based on chemical properties and/or structure. Freely available H2S represents gaseous H2S and its HS− anion, acid labile sulfide represents iron-sulfur clusters and persulfides, and bound sulfane sulfur represents thiol sulfides, polysulfides, sulfate/sulfite, and bound elemental sulfur.
Cystathionine γ-lyase (CSE) activity measurement
CSE activity was measured as previously reported (12,36). Tissue lysates were incubated with 2 mM cystathionine, 0.25 mM pyridoxal 5′-phosphate in 100 mM Tris-HCl buffer (pH 8.3) for 60 min at 37°C. 10% Trichloroacetic acid was added into reaction mixture. After centrifugation, the supernatant was mixed completely with 1% ninhydrin reagent and incubated for 5 min in a boiling-water bath. After heating, the solution was cooled on ice for 2 minutes and color reaction development measured 20 minutes at 455 nm using a smartSpect Plus spectrophotometer (Bio-Rad). CSE activity was assessed by cystathionine consumption and enzyme activity expressed as nanomoles of cystathionine consumed per mg of total protein per hour of incubation.
Statistical analysis
Resulting hydrogen sulfide species measurements, cysteine and CSE activity levels were statistically compared with Prism software (Graphpad Inc) using an unpaired students t-test between conventional versus germ free mice per organ examined. Distribution of tissue H2S metabolite levels and CSE enzyme activity was also compared within each subject group using one-way ANOVA with Neuman-Keuls multiple comparison test to identify tissues with the most significant differences. A minimum p<0.05 was necessary for significance.
Results
Plasma H2S bioavailability in conventional versus germ free mice
Bioavailable H2S can be compartmentalized in various biochemical forms as illustrated in figure 1 (28). Therefore, we employed specific analytical methods that we have developed to measure these various H2S pools between conventional versus germ free mice. Figure 2 illustrates the amount of plasma free H2S, acid labile sulfide and bound sulfane sulfur in conventional and germ free mice (panels A–C, respectively). Germ free mice had significantly reduced plasma free H2S and bound sulfane sulfur levels compared to conventional mice.
Figure 2. Plasma H2S bioavailable pools in conventional and germ free mice.
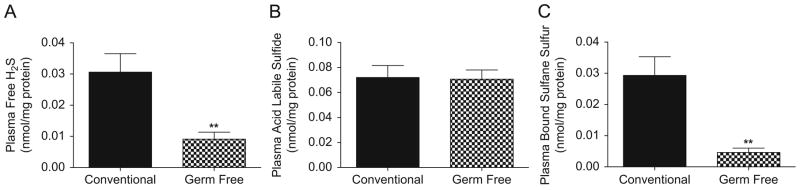
Panels A–C illustrate plasma free H2S, acid labile sulfide and bound sulfane sulfur levels, respectively, between conventional versus germ free mice. The amount of bioavailable H2S pools is normalized to mg total protein. **p<0.01, n=10.
Tissue free H2S levels in conventional versus germ free mice
Figure 3 shows distinct differences regarding the amount of freely available H2S in different organs. In conventional mice, the kidney, stomach and heart showed the highest levels of free H2S while lung and fat tissues were found to have the lowest levels. Germ free mice had significantly less free H2S in cecum and colon compared to conventional mice. These data indicate that the presence of an intestinal microflora significantly contributes to plasma and gastrointestinal organ free H2S levels.
Figure 3. Organ tissue free H2S levels in conventional and germ free mice.
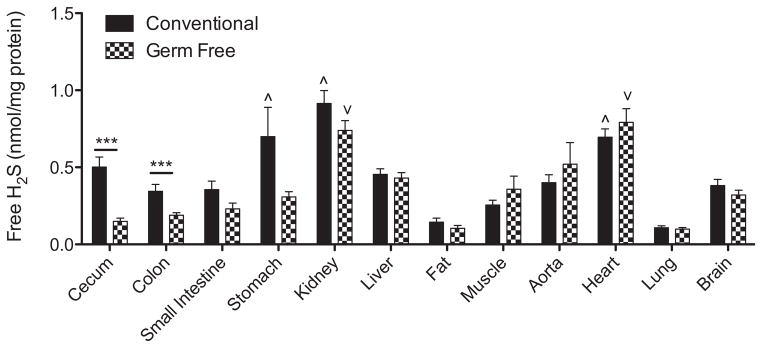
Multiple organs were collected from conventional and germ free mice, and free H2S measured that was normalized to mg total protein. ^p<0.05 tissues with significantly higher amounts of free H2S in conventional mice. ∨p<0.05 tissues with significantly higher amounts of free H2S in germ free mice. ***p<0.001 conventional versus germ free tissue levels. n=10.
Tissue acid labile sulfide and bound sulfane sulfur levels in conventional versus germ free mice
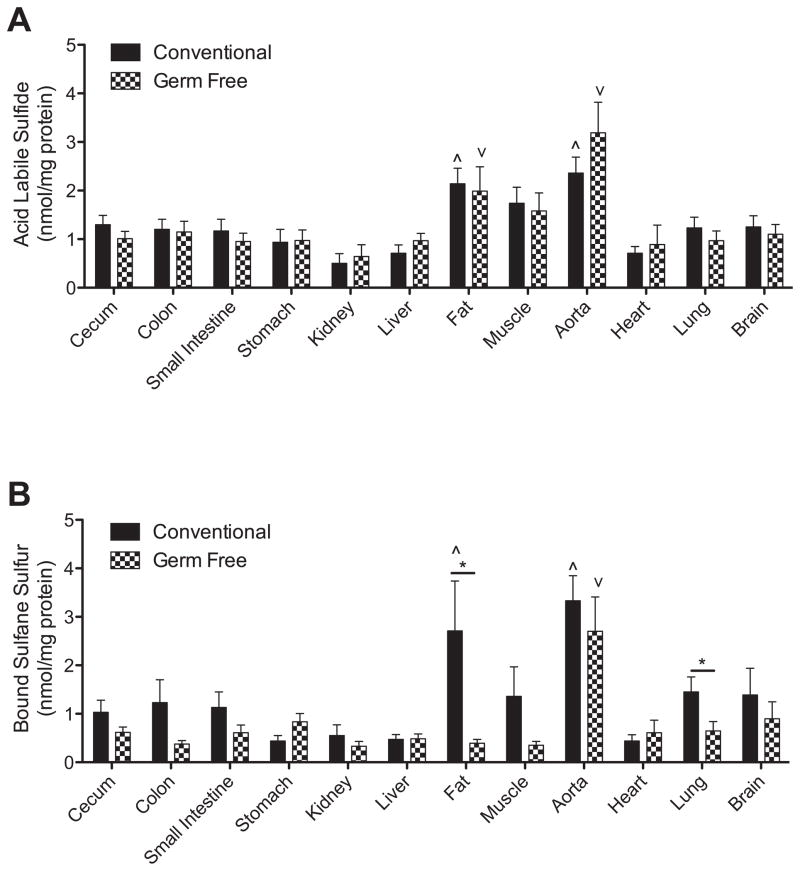
Experiments were performed to measure tissue levels of acid labile sulfide and bound sulfane sulfur. Figure 4, panel A reports various tissue levels of acid labile sulfide in conventional and germ free mice. The absence of a microflora did not significantly affect acid labile H2S pools in any of the organs examined. Although comparisons of tissue acid labile H2S levels in conventional and germ free mice revealed significantly higher amounts in fat and aorta. Figure 4, panel B shows the various tissue levels of bound sulfane sulfur levels (i.e. polysulfides) in conventional and germ free mice. Interestingly, fat and aorta tissues of conventional mice contained the greatest amounts of bound sulfane sulfur pools similar to that of acid labile H2S measurements. However, fat and lung tissue bound sulfane sulfur levels were significantly decreased in germ free mice. These data highlight that the microflora significantly impacts bound sulfane sulfur bioavailability in discrete tissue compartments.
Figure 4. Organ tissue acid labile and bound sulfane sulfur levels in conventional and germ free mice.
Panel A illustrates tissue concentrations of acid labile sulfide levels reported as nanomoles per mg total protein between conventional and germ free mice. ^p<0.05 tissues with significantly higher amounts of acid labile sulfide in conventional mice. ∨p<0.05 tissues with significantly higher amounts of acid labile sulfide in germ free mice. Panel B shows tissue concentrations of bound sulfane sulfur levels reported as nanomoles per mg total protein between conventional and germ free mice. ^p<0.05 tissues with significantly higher amounts of bound sulfane sulfur in conventional mice. ∨p<0.05 tissues with significantly higher amounts of bound sulfane sulfur in germ free mice. *p<0.05 conventional versus germ free bound sulfane sulfur levels. n=10.
CSE activity and cysteine levels in conventional versus germ free mice
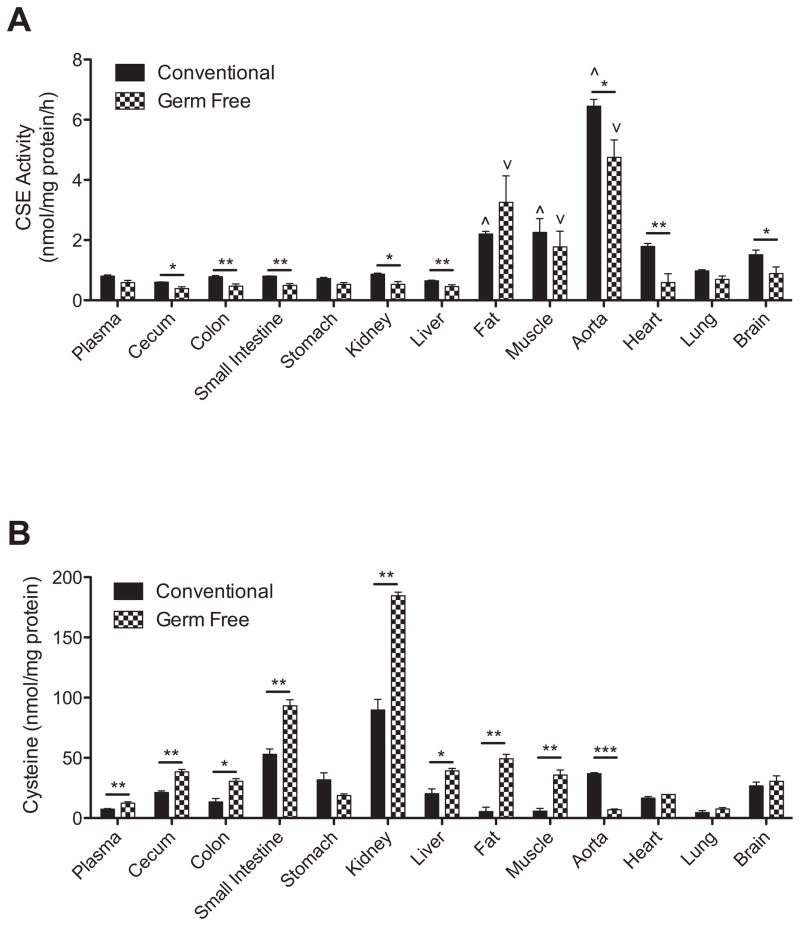
Figure 5, panel A, reports the amount of CSE activity amongst various tissues in conventional and germ free mice. Comparison of tissue CSE activity in conventional mice revealed that aorta, fat and muscle tissue contain the most abundant enzyme activity. Absence of a microflora elicited a significant decrease in tissue CSE activity in multiple organs including the cecum, colon, small intestine, kidney, liver, aorta, heart and brain; although, the aorta, fat and muscle tissues still displayed the greatest CSE activity. Figure 5, panel B, illustrates the amount of cysteine measured in various tissues from conventional and germ free mice. In germ free mice, cysteine levels were significantly increased in the plasma, cecum, colon, small intestine, kidney, liver, fat and muscle compared with conventional mice. Interestingly, only aorta tissue from germ free mice showed a significant decrease in cysteine levels. These data reveal that the presence of a microflora significantly impacts CSE activity and cysteine bioavailability.
Figure 5. Organ tissue CSE activity and cysteine levels in conventional and germ free mice.
Panel A reports differences in tissue CSE enzyme activity levels as reported as nanomoles of substrate consumed per mg of total protein. ^p<0.05 tissues with significantly higher amounts of CSE enzyme activity in conventional mice. ∨p<0.05 tissues with significantly higher amounts of CSE enzyme activity in germ free mice. *p<0.05 and **p<0.01 conventional versus germ free tissue CSE enzyme activity. Panel B illustrates tissue cysteine levels as nanomoles per mg of total protein between conventional and germ free mice. *p<0.05, **p<0.01 and ***p<0.001 conventional versus germ free tissue cysteine levels. n=10.
Discussion
It is increasingly apparent that the gut microbiota plays a key role in modulating health and disease of its host (2). Numerous studies demonstrate that gut commensal bacteria participate in regulating gastrointestinal neurophysiology, mucosal immunity, epithelial health and survival as well as overall metabolic activity (5,26). These responses are most likely due to the copious amount of bacterial products that are produced and released within the host that influence various physiological and pathological responses involving chronic inflammation and metabolic dysfunction (8,19,31,33). Of these metabolites, sulfur/sulfate reducing bacteria generate significant amounts of H2S luminally that has been posited to be involved in gastrointestinal pathophysiology (18,22). However, other findings suggest that the role of microbial H2S production may not be completely deleterious with possible benefits conveyed to the host (16,30).
The contribution of the microbiota to host H2S metabolism and regulation of its bioavailability in different biochemical forms is poorly understood. A recent study by Flannigan and colleagues found no differences in colonic tissue H2S synthesis rates between germ free mice versus mice colonized with altered Schaedler flora (6). However, in our study we found significantly differences in H2S biochemical pool bioavailability coupled with altered synthesis enzyme activity and substrate levels using sensitive analytical HPLC based methods that allow for accurate measurement of H2S metabolism compared to the methylene blue detection method that we and others have reported to be subject to experimental artifact (9,23,24). Our findings highlight the importance of the microbiota in contributing to the regulation of H2S metabolism and synthesis that was hereto unknown. Intriguingly, our results demonstrate that the gut mictobiota regulates H2S homeostasis not only locally in the gut but also systemically in various tissues.
The physiological and pathological importance of the various H2S biochemical pools (free, acid labile and bound sulfane sulfur) remains poorly understood (24,28). Our detailed examination of these different pools in the major organs reveals new information regarding basal and microbial modulation of tissue H2S bioavailability and synthesis rates. Interestingly, we found that aorta and adipose tissue contained the highest amount of H2S bioavailable equivalents primarily due to large greater amounts of acid labile and bound sulfane sulfur forms. Our observation of abundant aortic H2S bioavailability affirms recent findings from Levitt et al that found the aorta contained the largest amount of H2S (free and acid labile forms) as measured using gas chromatography-chemiluminescent techniques (13). With the use of our selective H2S pool liberation methods coupled with HPLC analysis, we further identified adipose tissue as an equally important biological reservoir for H2S bioequivalents that was not previously examined (13). These observations are consistent with numerous studies documenting the importance of H2S for cardiovascular health and endocrine and metabolic disease such as diabetes (32).
It is particularly interesting that the presence of a microbiota significantly altered adipose tissue bound sulfane sulfur, CSE enzyme activity and cysteine levels given the fact that germ free mice have lesser fat mass compared to conventional mice, and that the type of enteric flora significantly impacts fat pad mass (7,27). It is possible that the presence of a microflora differentially effects distal H2S metabolism in the adipocyte that modulates subsequent adipose metabolism responses. This notion is supported by two reports demonstrating 1) alterations in plasma H2S levels in obese patients, and 2) that the H2S donor diallyl trisulfide can suppress 3T3-L1 adipogenesis stimulation (15,34). Additional studies are clearly needed to determine whether the microbiota may contribute to adipocyte function and obesity due to alterations in H2S metabolism.
To our surprise, the absence of a microflora was associated with a significantly reduced CSE enzyme activity in numerous tissues and coincident with an increase in tissue cysteine levels. These observations suggest an interesting hypothesis that bacterial products could possibly influence CSE activity or expression. Alternatively, calcium/calmodulin has been reported to modulate CSE activity, and the enzyme may also be targeted for sumoylation, both of which might be altered in germ free animals (25,35). Tissue cysteine levels were found to be significantly elevated in many of the same tissues with blunted CSE activity. This may simply reflect less utilization of substrate due to decreased enzyme activity or it could indicate enhanced cellular cysteine uptake and shunting of H2S synthesis through cystathione beta synthase activity or altered regulation of redox status as many of these tissues did not manifest abundant deficiency of H2S bioavailability. While a clear explanation for these differences is not readily apparent, future studies will address these questions in greater detail. It will also be necessary to better understand exact mechanistic and signaling pathways involved in induction of systemic H2S synthesis by the gut microbiota. One possibility is that bacterial products leak into the blood stream and induce H2S generating enzymes systemically. Indeed, bacterial lipopolysaccharide (LPS) has been shown to up regulate CSE in certain cell types (14). It is also possible that a portion of H2S bioequivalents found in blood and tissues of conventional mice come from gut lumen by sulpur metabolizing bacteria.
In conclusion, we have shown that the host microbiota serves an important role in controlling host tissue H2S bioavailability and metabolism. These data highlight the possibility that several effects of the microflora on physiological or pathological responses could involve its modulation of H2S bioavailability.
Acknowledgments
This work was funded in part by NIH grant HL113303 to C.G.K.
References
- 1.Alexander M. Microbial formation of environmental pollutants. Adv Appl Microbiol. 1974;18:1–73. doi: 10.1016/s0065-2164(08)70569-0. [DOI] [PubMed] [Google Scholar]
- 2.Backhed F. Host responses to the human microbiome. Nutr Rev. 2012;70 (Suppl 1):S14–7. doi: 10.1111/j.1753-4887.2012.00496.x. [DOI] [PubMed] [Google Scholar]
- 3.Barrett EL, Clark MA. Tetrathionate reduction and production of hydrogen sulfide from thiosulfate. Microbiol Rev. 1987;51:192–205. doi: 10.1128/mr.51.2.192-205.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blachier F, Davila AM, Mimoun S, Benetti PH, Atanasiu C, Andriamihaja M, Benamouzig R, Bouillaud F, Tome D. Luminal sulfide and large intestine mucosa: friend or foe? Amino Acids. 2010;39:335–47. doi: 10.1007/s00726-009-0445-2. [DOI] [PubMed] [Google Scholar]
- 5.Delzenne NM, Neyrinck AM, Backhed F, Cani PD. Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat Rev Endocrinol. 2011;7:639–46. doi: 10.1038/nrendo.2011.126. [DOI] [PubMed] [Google Scholar]
- 6.Flannigan KL, McCoy KD, Wallace JL. Eukaryotic and prokaryotic contributions to colonic hydrogen sulfide synthesis. Am J Physiol Gastrointest Liver Physiol. 2011;301:G188–93. doi: 10.1152/ajpgi.00105.2011. [DOI] [PubMed] [Google Scholar]
- 7.Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci U S A. 2012;109:594–9. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamer HM, De Preter V, Windey K, Verbeke K. Functional analysis of colonic bacterial metabolism: relevant to health? Am J Physiol Gastrointest Liver Physiol. 2012;302:G1–9. doi: 10.1152/ajpgi.00048.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes MN, Centelles MN, Moore KP. Making and working with hydrogen sulfide: The chemistry and generation of hydrogen sulfide in vitro and its measurement in vivo: a review. Free Radic Biol Med. 2009;47:1346–53. doi: 10.1016/j.freeradbiomed.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 10.Jansson EA, Huang L, Malkey R, Govoni M, Nihlen C, Olsson A, Stensdotter M, Petersson J, Holm L, Weitzberg E, Lundberg JO. A mammalian functional nitrate reductase that regulates nitrite and nitric oxide homeostasis. Nat Chem Biol. 2008;4:411–7. doi: 10.1038/nchembio.92. [DOI] [PubMed] [Google Scholar]
- 11.Kadota H, Ishida Y. Production of volatile sulfur compounds by microorganisms. Annu Rev Microbiol. 1972;26:127–38. doi: 10.1146/annurev.mi.26.100172.001015. [DOI] [PubMed] [Google Scholar]
- 12.Kashiwamata S, Greenberg DM. Studies on cystathionine synthase of rat liver. Properties of the highly purified enzyme. Biochim Biophys Acta. 1970;212:488–500. doi: 10.1016/0005-2744(70)90255-x. [DOI] [PubMed] [Google Scholar]
- 13.Levitt MD, Abdel-Rehim MS, Furne J. Free and acid-labile hydrogen sulfide concentrations in mouse tissues: anomalously high free hydrogen sulfide in aortic tissue. Antioxid Redox Signal. 2011;15:373–8. doi: 10.1089/ars.2010.3525. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Whiteman M, Moore PK. Dexamethasone inhibits lipopolysaccharide-induced hydrogen sulphide biosynthesis in intact cells and in an animal model of endotoxic shock. J Cell Mol Med. 2009;13:2684–92. doi: 10.1111/j.1582-4934.2008.00610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lii CK, Huang CY, Chen HW, Chow MY, Lin YR, Huang CS, Tsai CW. Diallyl trisulfide suppresses the adipogenesis of 3T3-L1 preadipocytes through ERK activation. Food Chem Toxicol. 2012;50:478–84. doi: 10.1016/j.fct.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 16.Linden DR, Levitt MD, Farrugia G, Szurszewski JH. Endogenous production of H2S in the gastrointestinal tract: still in search of a physiologic function. Antioxid Redox Signal. 2010;12:1135–46. doi: 10.1089/ars.2009.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lloyd D. Hydrogen sulfide: clandestine microbial messenger? Trends Microbiol. 2006;14:456–62. doi: 10.1016/j.tim.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Medani M, Collins D, Docherty NG, Baird AW, O’Connell PR, Winter DC. Emerging role of hydrogen sulfide in colonic physiology and pathophysiology. Inflamm Bowel Dis. 2011;17:1620–5. doi: 10.1002/ibd.21528. [DOI] [PubMed] [Google Scholar]
- 19.Oldstone MB. Molecular mimicry, microbial infection, and autoimmune disease: evolution of the concept. Curr Top Microbiol Immunol. 2005;296:1–17. doi: 10.1007/3-540-30791-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ortenberg R, Beckwith J. Functions of thiol-disulfide oxidoreductases in E. coli: redox myths, realities, and practicalities. Antioxid Redox Signal. 2003;5:403–11. doi: 10.1089/152308603768295140. [DOI] [PubMed] [Google Scholar]
- 21.Poole LB. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch Biochem Biophys. 2005;433:240–54. doi: 10.1016/j.abb.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Rowan FE, Docherty NG, Coffey JC, O’Connell PR. Sulphate-reducing bacteria and hydrogen sulphide in the aetiology of ulcerative colitis. Br J Surg. 2009;96:151–8. doi: 10.1002/bjs.6454. [DOI] [PubMed] [Google Scholar]
- 23.Shen X, Pattillo CB, Pardue S, Bir SC, Wang R, Kevil CG. Measurement of plasma hydrogen sulfide in vivo and in vitro. Free Radic Biol Med. 2011;50:1021–31. doi: 10.1016/j.freeradbiomed.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen X, Peter EA, Bir S, Wang R, Kevil CG. Analytical measurement of discrete hydrogen sulfide pools in biological specimens. Free Radic Biol Med. 2012;52:2276–83. doi: 10.1016/j.freeradbiomed.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh S, Banerjee R. PLP-dependent H(2)S biogenesis. Biochim Biophys Acta. 2011;1814:1518–27. doi: 10.1016/j.bbapap.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tlaskalova-Hogenova H, Stepankova R, Kozakova H, Hudcovic T, Vannucci L, Tuckova L, Rossmann P, Hrncir T, Kverka M, Zakostelska Z, Klimesova K, Pribylova J, Bartova J, Sanchez D, Fundova P, Borovska D, Srutkova D, Zidek Z, Schwarzer M, Drastich P, Funda DP. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol Immunol. 2011;8:110–20. doi: 10.1038/cmi.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 28.Ubuka T. Assay methods and biological roles of labile sulfur in animal tissues. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;781:227–49. doi: 10.1016/s1570-0232(02)00623-2. [DOI] [PubMed] [Google Scholar]
- 29.Vitvitsky V, Kabil O, Banerjee R. High turnover rates for hydrogen sulfide allow for rapid regulation of its tissue concentrations. Antioxid Redox Signal. 2012;17:22–31. doi: 10.1089/ars.2011.4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallace JL, Ferraz JG, Muscara MN. Hydrogen sulfide: an endogenous mediator of resolution of inflammation and injury. Antioxid Redox Signal. 2012;17:58–67. doi: 10.1089/ars.2011.4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang D, Xia M, Yan X, Li D, Wang L, Xu Y, Jin T, Ling W. Gut microbiota metabolism of anthocyanin promotes reverse cholesterol transport in mice via repressing miRNA-10b. Circ Res. 2012;111:967–81. doi: 10.1161/CIRCRESAHA.112.266502. [DOI] [PubMed] [Google Scholar]
- 32.Wang R. Physiological implications of hydrogen sulfide: a whiff exploration that blossomed. Physiol Rev. 2012;92:791–896. doi: 10.1152/physrev.00017.2011. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whiteman M, Gooding KM, Whatmore JL, Ball CI, Mawson D, Skinner K, Tooke JE, Shore AC. Adiposity is a major determinant of plasma levels of the novel vasodilator hydrogen sulphide. Diabetologia. 2010;53:1722–6. doi: 10.1007/s00125-010-1761-5. [DOI] [PubMed] [Google Scholar]
- 35.Yang G, Wu L, Jiang B, Yang W, Qi J, Cao K, Meng Q, Mustafa AK, Mu W, Zhang S, Snyder SH, Wang R. H2S as a physiologic vasorelaxant: hypertension in mice with deletion of cystathionine gamma-lyase. Science. 2008;322:587–90. doi: 10.1126/science.1162667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zavaczki E, Jeney V, Agarwal A, Zarjou A, Oros M, Katko M, Varga Z, Balla G, Balla J. Hydrogen sulfide inhibits the calcification and osteoblastic differentiation of vascular smooth muscle cells. Kidney Int. 2011;80:731–9. doi: 10.1038/ki.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]