Abstract
The superfamily of G protein-coupled receptors (GPCRs) includes at least 800 seven-transmembrane receptors that participate in diverse physiological and pathological functions. GPCRs are the most successful targets of modern medicine, and approximately 36% of marketed pharmaceuticals target human GPCRs. However, the endogenous ligands of more than 140 GPCRs remain unidentified, leaving the natural functions of those GPCRs in doubt. These are the so-called orphan GPCRs, a great source of drug targets. This review focuses on the signaling transduction pathways of the adhesion GPCR family, the LGR subfamily, and the PSGR subfamily, and their potential functions in immunology, development, and cancers. In this review, we present the current approaches and difficulties of orphan GPCR deorphanization and characterization.
Keywords: G protein-coupled receptors, orphan GPCR, deorphanization, Adhesion GPCR family, LGR subfamily, PSGR, Gpr48/Lgr4
Introduction
GPCRs represent the largest superfamily and most diverse group of mammalian transmembrane proteins. The main characteristic feature of these proteins is that they share a common seven-transmembrane (7TM) configuration. GPCRs have attracted a great deal of interest owing to their numerous physiological and pathological roles in transducing extracellular signals into intracellular effector pathways through the activation of heterotrimeric G protein by binding to a broad range of ligands, including proteins1, peptides2, organic compounds3, 4, and eicosanoids5. This makes GPCRs and their signal transduction pathways important specific targets for a variety of physiological functions and therapeutic approaches, ranging from the control of blood pressure, allergic response, kidney function, hormonal disorders, and neurological diseases to the progression of cancer6. Owing to the features of GPCR structure and function, approximately 36% of currently marketed drugs target human GPCRs7. GPCRs have huge potential in biomedical research and drug development.
Human GPCRs can be divided into five main families on the basis of phylogenetic criteria, Glutamate, Rhodopsin, Adhesion, Frizzled/Taste2, and Secretin8. Among the five GPCRs families, Rhodopsin is the most studied. It comprises the largest group of GPCRs. Notably, in recent years, the leucine-rich repeat-containing G-protein coupled receptor (LGR) subfamily, part of Rhodopsin, have displayed enormously important physiological functions in knockout mice studies especially LGR4 and LGR5. Olfactory receptors are also members of Rhodopsin family of GPCRs and are mainly expressed in sensory neurons of olfactory system. These form a multigene family. The PSGR subfamily belongs to the olfactory receptor group. The family has restricted expression in human prostate tissues and is upregulated in prostate cancer. The second largest GPCR family, with 33 members, is the Adhesion family. This family is very special because of its members' secondary structures, with distinctive long N-termini containing adhesion domains8. Limited studies have shown that Adhesion GPCRs are involved in the signaling of cell adhesion, motility, embryonic development, and the immune system. There are still GPCRs for which the natural ligands remain to be identified. These are called orphan GPCRs.
LGRs and PSGR belong to Rhodopsin subfamily and they represent as classical GPCRs in structure and signal transduction. On the other hand, Adhesion GPCRs are novel, and their structures and signal transduction are distinct to the classical GPCRs. In this review, we focused our discussion on LGR subfamily, PSGR subfamily, and Adhesion GPCRs family. We also discussed current screening systems for the deorphanization and characterization of the orphan GPCRs.
Orphan GPCRs
The first GPCR to be identified was rhodopsin in 1878. It was later proven that rhodopsin consists of the GPCR protein opsin and a reversibly covalently bound cofactor, retinal9, 10. After completion of the human genome sequence in 200411, 12, the number of human GPCRs increased to about 800 based on the screening approaches, such as low-stringency hybridization13, PCR-derived methods14, and bioinformatic analyses15. Besides the olfactory receptor family, more than 140 GPCRs have not yet been linked to endogenous ligands. These are the so-called orphan GPCRs (Figure 1)16.
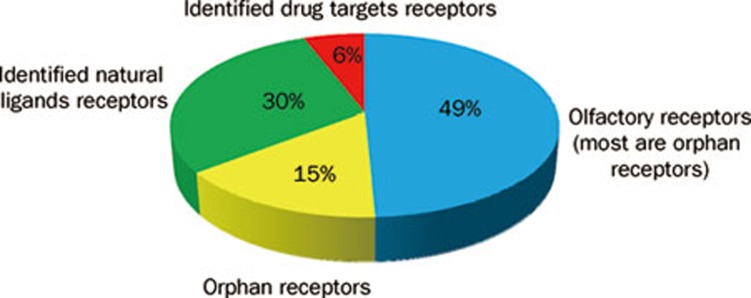
Figure 1.
Percentage of the orphan GPCRs in GPCR superfamily. GPCRs constitute a large transmembrane family of more than 800 members. Among them, 6% are utilized as drug target in clinical applications, and 30% are natural ligand receptors. However, 49% are olfactory receptors (most of them are orphan GPCRs), and 15% are orphan GPCRs. (Data were summarized from a review paper122)
Biological functions of the LGR subfamily
LGRs 4–8 are members of the rhodopsin GPCR family, which can be divided into two groups, LGRs 4, 5, and 6 and LGRs 7 and 8 in terms of their natural ligand. R-spondins have recently been identified as the ligands for LGRs 4, 5, and 617. LGRs 7 and 8 are relaxin family peptide (RXFP) receptors18. According to sequence similarity, LGRs 4, 5, and 6 are closely related to each other, showing almost 50% identities. The three orphan receptors have a substantially large N-terminal extracellular domain (ECD) composed of 17 leucine rich repeats (LRR) (Figure 2)19. Lgr4, also known as Gpr48, has been reported to have many physiological functions by the generation of knockout mice. The loss of Lgr4 results in developmental defects in many areas, including intrauterine growth retardation associated with embryonic and perinatal lethality20, abnormal renal development21, defective postnatal development of the male reproductive tract22, ocular anterior segment dysgenesis23, bone formation and remodeling dysfunction6, impaired hair placode formation24, and defective development of the gall bladder and cystic ducts25. Lgr5 has been proven to be a marker of gastrointestinal tract and hair follicle stem cells26, 27. Knockout of Lgr5 in mice leads to total neonatal lethality accompanied with ankyloglossia and gastrointestinal distension28. Lgr6 also has been shown to be a stem cell marker in hair follicles, and Lgr6-positive stem cells have been found to produce all cell lineages of the skin29. LGR4 and LGR5 are also highly expressed in several types of cancers. LGR5 is up-regulated in human colon and ovarian tumors and promotes cell proliferation and tumor formation in basal cell carcinoma30, 31. Overexpression of LGR4 enhances cervical and colon cancer cell invasiveness and metastasis32. However, despite their critical function in development and cancer, LGR4 and LGR5 will still be considered orphan receptors until R-spondins reported to function as their natural ligands can be proven to regulate Wnt/β-catenin signaling pathway. Some observations of Lgr4 and Lgr5 knockout mice have been strongly relevant to Wnt/β-catenin signaling33, 34. This suggests that LGR4 and LGR5 could be involved in the Wnt pathway. One author stated that R-spondins-Lgr4 induced the signal transduction pathway in a manner independent of G proteins17. However, two independent groups have reported that Lgr4/Gpr48 is associated with the Gαs-cAMP pathway by generating constitutively active forms of Lgr4/Gpr4823, 35. Therefore, the existence of endogenous ligands for the activation of classical G-protein coupled signaling pathways for Lgr4/Gpr48 is still a question. LGR7 and LGR8 share 54% identity. Besides 10 LRRs motif, LGR7 and LGR8 also have an LDL class A (LDLa) motif in the N-terminal, which is an important domain for signal transduction (Figure 2). Traditionally, relaxin/LGR7 has been thought to be a hormone receptor for pregnancy and parturition18. Recently, it has been reported that relaxin/LGR7 also has significant function in non-reproductive organs, such as the heart, and even plays a role in cancer growth and metastasis36. Insulin-like peptide 3 (INSL3), which is a ligand of LGR8, is highly expressed in the Leydig cells of the testis and knocking out Insl3 in mice generates a cryptorchid phenotype. However, reports have been conflicting with respect to LGR8 mutations related to human cryptorchidism18. The role of INSL3 in human adult male is still not clear.
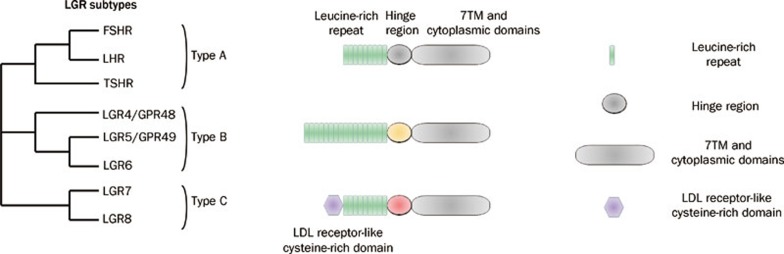
Figure 2.
LGR subfamily GPCRs. The Type A LGRs includes the follicle-stimulating hormone receptor (FSHR), the luteinizing hormone receptor (LHR) and the thyroid-stimulating hormone receptor (TSHR). The Type B LGR comprises three members, Gpr48/LGR4, LGR5, and LGR6 which remain orphan GPCRs at the present time. By contrast, Type C LGRs have only two members, LGR7 and LGR8 which have been demonstrated to be the relaxin family receptors. Type A contains 9 LRRs in the ectodomain, whereas Type B contains 17 LRRs. By contrast, Type C has an N-terminal LDL receptor-like cysteine-rich domain not found in other LGRs. 7TM, seven-transmembrane; LDL, low-density lipoprotein; LRR, leucine-rich repeat; LGR, leucine-rich repeat-containing G-protein-coupled receptor; FSHR, follicle-stimulating hormone receptor; LHR, luteinizing hormone receptor; TSHR, thyroid-stimulating hormone receptor.
PSGRs subfamily in prostate cancer
Mammalian olfactory receptors, which are the members of the Rhodopsin family of GPCRs and mainly expressed in sensory neurons of the olfactory epithelium in the nose, are used to sense the chemical environment37. Recently, some olfactory receptors have also been found in other organs. For example, MOR23 is expressed both in the olfactory epithelium and in sperm and functions as a chemosensing receptor during sperm-egg communication, thereby modulating fertilization in the reproductive system38. The new olfactory receptor family members PSGR1 and PSGR2 have been found to have restricted expression in human prostate tissues, as shown by Northern blot and real-time PCR analysis of over 20 different human tissue types39, 40, 41. PSGR subfamily expression increases significantly in the epithelial cells of prostate intraepithelial neoplasia (PIN) patients and in prostate cancer patients relative to non-cancerous controls and benign prostatic hyperplasia tissues, suggesting that the PSGR subfamily may play an important role in early prostate cancer development42. The PSGR subfamily has been proven to be strongly associated with the clinical parameters (clinical stages, Gleason scores, recurrence status, and metastasis) and its members could serve as biomarkers for prostate cancer42, 43. PSGR subfamily transcripts even can be used as diagnostic markers in urine44. It has also been reported that PSGR expression detection together with the well-known prostate cancer marker prostate-specific antigen (PSA), prostate cancer gene 3 (PCA3), and α-methylacyl-CoA racemase (AMACR) can increases diagnostic specificity in the detection of prostate cancer43, 44, 45. Recently, Neuhaus EM et al reported that through intracellular Ca2+ flux using a bank of steroid hormones and through odorant-related compound screening, certain steroids and β-ionone have been proven to be active ligands for PSGR46. PSGR-induced Ca2+signaling was found to require the involvement of endogenous Ca2+-selective transient receptor potential vanilloid type 6 (TRPV6) channels47. Incubation of prostate cancer cells with β-ionone inhibits cell proliferation46. This suggests that PSGR signaling is also involved in prostate cancer cell progression.
Adhesion GPCR family
GPCRs in the Adhesion family have a relative long N-terminal domain, which contains many so-called adhesion domains (Figure 3). These adhesion domains only existed in some adhesion molecules, such as integrins, cadherins, and selectins; and the domains are thought to have adhesive properties. Another striking characteristic of all the Adhesion GPCRs is that there is a GPS (GPCR proteolytic site) domain linking the 7TM region to the extracellular domain, which acts as an autocatalytic site48, 49. As a novel GPCRs family, most of the members are orphan and only a few of them have been identified as having natural ligands and functions.
Figure 3.
Schematic diagram of the extracellular N-terminal domain within the Adhesion GPCRs. The extracellular N-terminal domains of 33 Adhesion GPCRs was predicted by the RPS-BLAST against the conserved domain database (CCD). CA, cadherin domain; calx-beta, domain found in Na+–Ca2+ exchangers; CUB, resembles the structure of immunoglobins; EAR, epilepsy-associated repeat; EGF-Lam, laminin EGF-like domain; EGF, epidermal growth factor domain; HBD, hormone-binding domain; herpes-gp2, resembles the equine herpes virus glycoprotein gp2 structure; GBL, galactose-binding lectin domain; Ig, immunoglobulin domain; OLF, olfactomedin domain; LamG, laminin G domain; LRR, leucine-rich repeat domain; PTX, pentraxin domain; Puf, displays structural similarity to RNA-binding protein from the Puf family; SEA, domain found in sea-urchin sperm protein; SIN, resembles the primary structure of the SIN component of the histone deacetylase complex; TSP1, thrombospondin domain. C-type lectin, similar to the C-type lectin or carbohydrate-recognition domain; GPS, GPCR proteolytic site domain.
Adhesion GPCRs in immunology
Immune response is coordinated by an assortment of membrane receptors, including TLRs, integrins, lectins, the Ig superfamily, selectins, and GPCRs, which are found on leukocytes10, 50, 51. The first Adhesion GPCR to be discovered, epidermal growth factor-like module containing mucin-like receptor protein 1 (EMR1, F4/80 receptor), which is an epidermal growth factor (EGF)-seven transmembrane (7M) receptor, have a predominantly leukocyte-restricted expression pattern52. Though the expression of Emr1 is restricted, the function of this receptor remained unknown until the generation of Emr1 knock-out mice. The mouse model indicates that Emr1 is critical to the induction of CD8+ regulatory T-cells in peripheral tolerance53. Besides EMR1, the EGF-TM7 subfamily includes EMR2, EMR3, EMR4, and CD97, all of which belong to the Adhesion GPCR family. Unlike the highly specific expression of EMR1, the other EGF-TM7 receptors are expressed largely in myeloid cells (monocytes, macrophages, neutrophils, and dendritic cells) and in some lymphoid cells (T and B cells)54. Chondroitin sulfate has recently been identified as the ligand for EMR2 and CD97, which mediate cell attachment55. CD97, the leukocyte activation antigen, also has been shown to bind to the complement regulatory protein DAF/CD55 (decay accelerating factor) and the longest splice variant of CD97 has the highest capacity to bind to CD55-expressing cells. Although CD97 and EMR2 differ by only 3 amino acids (in the EGF domain), the activity of EMR2 binding to CD55 is significantly weaker56, 57. The precise function of the CD97-CD55 interaction is still not fully understood. Using knock-out mice and x-ray crystallography, Abbott RJ et al demonstrated that T cells and complement regulatory activities of CD55 occur on opposite faces of the molecule, suggesting that the CD97-CD55 complex might simultaneously regulate both the innate and adaptive immune responses58, 59. EMR3 has been reported as a marker for mature granulocytes, and it can interact with the ligand that expresses at the surface of monocyte-derived macrophages and activated human neutrophils60, 61. EMR4 has been reported to interact with a cell surface protein as a ligand on A20 B-lymphoma cells62.
Adhesion GPCRs in development
The most extensively studied Adhesion GPCRs in embryonic development are the so-called 7TM-cadherin subfamily (Celsr/Flamingo/Starry night). All the members of this subfamily posses extracellular domains containing nine atypical cadherin repeats which have linked the combination of EGF-like and laminin G-like domains63. The 7TM-cadherins are an evolutionarily conserved gene subfamily with homologues discovered from ascidians to mammals63. In mammals, the subfamily comprises 3 genes, Celsr1, Celsr2, and Celsr3. There are 4 genes (fmila, fmilb, fmi2, and fmi3) in zebrafish and only one homologue, called flamingo and starry night, in Drosophila. Drosophila studies provide us with a distinct function view of Flamingo/Starry night as a core planar polarity protein64. Its functions include regulating dendrite extension from sensory neurons65, 66, modulating target selection by photoreceptor axons67, accelerating axon advance from sensory and motor neurons68, and limiting ectopic neuromuscular junction formation and maintenance of motor axon terminals69. Gene knockout and knockdown of 7TM-cadherins has also confirmed this observation in vertebrates. 7TM-cadherins regulate morphogenetic movements, neural tube closure, orientation of sensory hair cells in inner ear, and hair follicle patterning63, 70, 71, 72, 73. Recently, Adhesion GPCRs Gpr124 and Gpr126, which are not 7TM-cadherins, have been shown to regulate the development of different tissues in mice. Gpr124 affects CNS-specific angiogenesis and Gpr126 affects Schwann cells to initiate myelination74, 75, 76, 77. This suggests that more members of this family may be involved in development and that this may be due to the adhesive or other properties of N-terminal domains.
Adhesion GPCRs in cancers
Because cell adhesion molecules have a vital role in cancer progression, it is reasonable to speculate that Adhesion GPCRs also play important functions in cancer progression and metastasis. Leukocyte Adhesion GPCR EMR2 has been proven to be overexpressed in human breast cancer and is associated with patient survival78. CD97 is involved in tumor-environment interactions and mediates tumor invasion79. It has been reported that the 7TM-cadherin receptors may also be involved in human cancers, such as gastric cancer, lung cancer, and melanoma80. Interestingly, unlike other Adhesion GPCRs, GPR56 has been shown to suppress some cancer cell growth and metastasis through interacting with tissue transglutaminase (TG2)81, 82.
Signal transduction mediated by Adhesion GPCRs
Most Adhesion GPCRs are orphan receptors, which is the main reason whether or not Adhesion GPCRs are involved in G protein signaling. In addition, the complicated structure of Adhesion GPCRs, comprising both largely ECD and 7TM domains, make it possible for Adhesion GPCRs to go through the signaling pathway in a G-protein-independent manner83. For example, Gpr124 regulates angiogenic sprouting into neural tissues through TGF-beta pathway in mouse76. BAI1 can function as an engulfment receptor in response to “eat me” signal phosphatidylserine, which leads to BAI1 directly bind and activate the ELMO/DOCK180/RAC module84. It has been reported that GPR124 and GPR125 can interact with several viral oncoproteins by its cytoplasmic PDZ domain. And the rat Ig-Hepta (GPR116) has been shown to form a homodimer that is linked by disulphide bonds. Moreover, this receptor undergoes two proteolytic cleavages, and cleaved product in the SEA domain might act as a ligand to bind to GPR11685, 86, 87. Therefore, these 7TM receptors may mediate G-protein independent signaling pathway in cellular functions.
Though some Adhesion GPCRs go through G-protein- independent pathways, others have been proven to go through the classic G-protein-dependent pathway. Lectomedin receptor-1 was co-purified with the Gαo88. Also, GPR56 has been shown to form a complex with Gq/11 and G12/13 in the neural progenitor cells89, 90. Gpr126 modulates Schwann cells, initiating myelination by classic cAMP pathway74. Latrophilin, which is activated by the ligand LTX, can transduce the intercellular Ca2+ signal pathway. These observations indicate that this family can transmit signals through both classical G-protein-dependent and G-protein-independent mechanisms.
Deorphanization strategy
GPCRs are the most prominent family of pharmacological targets in biomedicine91. The deorphanization of orphan GPCRs is one of the most important missions in orphan GPCR research. Deorphanization is the process of identifying ligands that are highly selective for orphan GPCRs. In general, the standard assays are radio-ligand binding, calcium flux, GTPγ binding, and modulation of cAMP levels92, 93, 94, 95, 96, 97, 98.
With the development of molecular technology, several lines of approaches have been used for deorphanization. The first, according to the sequence and function similarity, ligands of the identified receptors are used to examine GPCRs with identical sequences or domains. This sequence similarity strategy resulted in the identification of the ligands of Edg3 and Edg5, whose sequences are similar to that of the S1P receptors, with >50% amino acid identity99, 100, 101. The function similarity strategy lead to the identification of the ligands of Lgr5 homologues, R-spondins, which stimulate the growth of intestinal stem cells17. However, this approach must be carefully evaluated because its predictions are not always accurate. For example, alkyl imidazole functions as dual histamine H3/H4 receptor ligands, while histamine H3/H4 receptors share very little sequence identity102, 103. Although the EGF domain of CD97 and EMR2 share 97% identity, only CD97 shows high affinity with CD55 but not with EMR2. The second strategy used to identify natural ligands works by determining the expression profile relationship between receptor and the putative ligand. This technique led to the identification of the receptors of RDC7 and RDC8 as adenosine A1 and A2A receptors, all of which are highly transcribed in the brain cortex, thyroid follicular cells, and testis104, 105. The third technique is used to identify GPCRs that have specific expression profiles and distinct cytoplasm signal pathways. This method uses extracts of tissues that contain potential ligands to screen by the GPCRs mediated signaling assays. Some hormone proteins, such as nociptin, orexins, apelin, prolactin, and ghrelin, were successfully identified using this strategy106, 107, 108, 109. The fourth strategy has been used successfully to deorphanize Adhesion GPCRs. It involves engineering recombinant soluble extracellular regions of Adhesion GPCRs with an Fc-fragment in N-terminal and biotinylation signal at the C-terminal. This acts as probe to screen the extracellular matrix components. This led to the identification of certain ligands for myeloid cell Adhesion GPCR51, 110. In recent years, the so-called reverse pharmacology strategy has also been used to identify the ligands of orphan GPCRs98. This is carried out by expressing these orphan GPCRs in eukaryotic cells by DNA transfection and then coupling them to ligands to examine the binding affinity of the cells and ligands111, 112. With this approach, many peptide hormones, including ghrelin, which stimulates hunger; kisspeptin and metastin, which are involved in puberty development and cancer metastasis; orexin and hypocretin, which mediate food intake and induce wakefulness and energy expenditure, have been discovered within the last decade113. However, the successful application of reverse pharmacology method depends on three major elements: sufficient orphan receptor expression, high-quality ligands and robust screening assays to detect receptor activation114, 115. Fortunately, with development of membrane protein expression and purification techniques, neuropeptides and synthetic ligands have been applied to large-scale screening116. Of the three elements outlined above, choosing an appropriate detection assay is the most problematic.
The rate of GPCR deorphanization decreased drastically at the turn of the century, suggesting some gap the processes exit. Herein, we discuss several factors that may account for the problem. The greatest challenge in deorphanization of the receptors is the limited knowledge about them, especially with respect to their physiological functions and their roles as transmitters of signal pathways. Thus, experimental design is rendered difficult by the lack of signal transduction assays and positive controls113. Second, the majority of approaches to deorphanization rely on monitoring changes at the second messenger level, which is regulated by G proteins. However, GPCRs can transduce signal pathways diversely, sometimes even beyond G proteins. In this case, identifying the relevant signaling pathway is key point to deorphanization. For example, some orphan GPCRs require accessory proteins for their activity. This working model has been shown in calcitonin GPCRs, which require RAMPs (receptor activity-modifying proteins) for their activation. To identify the ligand of this kind of GPCR, new screening assays for specific accessory proteins must be set up117, 118. Third, there is a possibility that some transmitters are only expressed at a particular time during the life span or at certain specific conditions9. Although it is risky and challenging, it is necessary to find more effective transmitters for deorphanization and put them to use. Lastly, some orphan GPCRs can form heterodimers with other GPCRs and function in a ligand-independent manner, and there is no outcome for the identification of the ligands of this kind of orphan GPCRs. For example, GABABR1 and GABABR2 form well-known heterodimer receptors and GABABR1 is involved in ligand-binding, whereas GABABR2 only acts as the signaling unit. GABABR2 is an orphan receptor in the heterodimer complex without any known ligand119, 120, 121.
Perspectives in the research of orphan GPCRs
In recent years, the numbers of new orphan GPCRs have increased and several members have been relatively well characterized. However, the progress of orphan GPCR function research has been hampered by the lack of identified ligands and by the unique structures of the GPCR themselves. Further investigation of their signaling pathways is valuable to understand the physiological and pathological roles of these new orphan GPCRs. The development of orphan GPCR knockout mice has also been shown to be a successful method for the characterization of their physiological and pathological functions. The knockout approach for orphan GPCRs are essential for our understanding of these receptor functions and their potential pathways. Functional and specific antibodies can serve probes not only for the ligands, but also for developing therapies for tumors and genetic disorders in which orphan GPCRs are involved. Although progress is very difficult, searching for the ligands of orphan GPCRs and identifying their physiological functions will continue. With recent discoveries of more and more orphan GPCR signaling pathways, understanding of their particular physiological functions and deorphanization for therapeutic purposes should accelerate in the coming years.
Acknowledgments
This work is partially supported by grants from National Basic Research Program of China (2012CB910400), National Natural Science Foundation of China (30800653, 30930055, and 81071437), the Science and Technology Commission of Shanghai Municipality (11DZ2260300), and by grants from NIH (5R01CA134731) and DOD (W81XWH-10-10147).
References
- Parmentier M, Libert F, Maenhaut C, Lefort A, Gerard C, Perret J, et al. Molecular cloning of the thyrotropin receptor. Science. 1989;246:1620–2. doi: 10.1126/science.2556796. [DOI] [PubMed] [Google Scholar]
- Masu Y, Nakayama K, Tamaki H, Harada Y, Kuno M, Nakanishi S. cDNA cloning of bovine substance-K receptor through oocyte expression system. Nature. 1987;329:836–8. doi: 10.1038/329836a0. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Kobilka BK, Strader DJ, Benovic JL, Dohlman HG, Frielle T, et al. Cloning of the gene and cDNA for mammalian beta-adrenergic receptor and homology with rhodopsin. Nature. 1986;321:75–9. doi: 10.1038/321075a0. [DOI] [PubMed] [Google Scholar]
- Kobilka BK, MacGregor C, Daniel K, Kobilka TS, Caron MG, Lefkowitz RJ. Functional activity and regulation of human beta 2-adrenergic receptors expressed in Xenopus oocytes. J Biol Chem. 1987;262:15796–802. [PubMed] [Google Scholar]
- Felder CC, Briley EM, Axelrod J, Simpson JT, Mackie K, Devane WA. Anandamide, an endogenous cannabimimetic eicosanoid, binds to the cloned human cannabinoid receptor and stimulates receptor-mediated signal transduction. Proc Natl Acad Sci U S A. 1993;90:7656–60. doi: 10.1073/pnas.90.16.7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Zhou W, Zhou X, Li D, Weng J, Yi Z, et al. Regulation of bone formation and remodeling by G-protein-coupled receptor 48. Development. 2009;136:2747–56. doi: 10.1242/dev.033571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask-Andersen M, Almen MS, Schioth HB. Trends in the exploitation of novel drug targets. Nat Rev Drug Discov. 2011;10:579–90. doi: 10.1038/nrd3478. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–72. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Chung S, Funakoshi T, Civelli O. Orphan GPCR research. Br J Pharmacol. 2008;153:S339–46. doi: 10.1038/sj.bjp.0707606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DY, Kim K, Kwon HB, Seong JY. Cellular and molecular biology of orphan G protein-coupled receptors. Int Rev Cytol. 2006;252:163–218. doi: 10.1016/S0074-7696(06)52003-0. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Bunzow JR, Van Tol HH, Grandy DK, Albert P, Salon J, Christie M, et al. Cloning and expression of a rat D2 dopamine receptor cDNA. Nature. 1988;336:783–7. doi: 10.1038/336783a0. [DOI] [PubMed] [Google Scholar]
- Libert F, Parmentier M, Lefort A, Dinsart C, Van Sande J, Maenhaut C, et al. Selective amplification and cloning of four new members of the G protein-coupled receptor family. Science. 1989;244:569–72. doi: 10.1126/science.2541503. [DOI] [PubMed] [Google Scholar]
- Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, et al. The G protein-coupled receptor repertoires of human and mouse. Proc Natl Acad Sci U S A. 2003;100:4903–8. doi: 10.1073/pnas.0230374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levoye A, Dam J, Ayoub MA, Guillaume JL, Jockers R. Do orphan G-protein-coupled receptors have ligand-independent functions? New insights from receptor heterodimers. EMBO Rep. 2006;7:1094–8. doi: 10.1038/sj.embor.7400838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A. 2011;108:11452–7. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong RC, Shilling PJ, Lobb DK, Gooley PR, Bathgate RA. Membrane receptors: structure and function of the relaxin family peptide receptors. Mol Cell Endocrinol. 2010;320:1–15. doi: 10.1016/j.mce.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Hsu SY, Kudo M, Chen T, Nakabayashi K, Bhalla A, van der Spek PJ, et al. The three subfamilies of leucine-rich repeat-containing G protein-coupled receptors (LGR): identification of LGR6 and LGR7 and the signaling mechanism for LGR7. Mol Endocrinol. 2000;14:1257–71. doi: 10.1210/mend.14.8.0510. [DOI] [PubMed] [Google Scholar]
- Mazerbourg S, Bouley DM, Sudo S, Klein CA, Zhang JV, Kawamura K, et al. Leucine-rich repeat-containing, G protein-coupled receptor 4 null mice exhibit intrauterine growth retardation associated with embryonic and perinatal lethality. Mol Endocrinol. 2004;18:2241–54. doi: 10.1210/me.2004-0133. [DOI] [PubMed] [Google Scholar]
- Kato S, Matsubara M, Matsuo T, Mohri Y, Kazama I, Hatano R, et al. Leucine-rich repeat-containing G protein-coupled receptor-4 (LGR4, Gpr48) is essential for renal development in mice. Nephron Exp Nephrol. 2006;104:e63–75. doi: 10.1159/000093999. [DOI] [PubMed] [Google Scholar]
- Mendive F, Laurent P, Van Schoore G, Skarnes W, Pochet R, Vassart G. Defective postnatal development of the male reproductive tract in LGR4 knockout mice. Dev Biol. 2006;290:421–34. doi: 10.1016/j.ydbio.2005.11.043. [DOI] [PubMed] [Google Scholar]
- Weng J, Luo J, Cheng X, Jin C, Zhou X, Qu J, et al. Deletion of G protein-coupled receptor 48 leads to ocular anterior segment dysgenesis (ASD) through down-regulation of Pitx2. Proc Natl Acad Sci U S A. 2008;105:6081–6. doi: 10.1073/pnas.0708257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri Y, Kato S, Umezawa A, Okuyama R, Nishimori K. Impaired hair placode formation with reduced expression of hair follicle-related genes in mice lacking Lgr4. Dev Dyn. 2008;237:2235–42. doi: 10.1002/dvdy.21639. [DOI] [PubMed] [Google Scholar]
- Yamashita R, Takegawa Y, Sakumoto M, Nakahara M, Kawazu H, Hoshii T, et al. Defective development of the gall bladder and cystic duct in Lgr4-hypomorphic mice. Dev Dyn. 2009;238:993–1000. doi: 10.1002/dvdy.21900. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40:1291–9. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- Morita H, Mazerbourg S, Bouley DM, Luo CW, Kawamura K, Kuwabara Y, et al. Neonatal lethality of LGR5 null mice is associated with ankyloglossia and gastrointestinal distension. Mol Cell Biol. 2004;24:9736–43. doi: 10.1128/MCB.24.22.9736-9743.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327:1385–9. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- McClanahan T, Koseoglu S, Smith K, Grein J, Gustafson E, Black S, et al. Identification of overexpression of orphan G protein-coupled receptor GPR49 in human colon and ovarian primary tumors. Cancer Biol Ther. 2006;5:419–26. doi: 10.4161/cbt.5.4.2521. [DOI] [PubMed] [Google Scholar]
- Tanese K, Fukuma M, Yamada T, Mori T, Yoshikawa T, Watanabe W, et al. G-protein-coupled receptor GPR49 is up-regulated in basal cell carcinoma and promotes cell proliferation and tumor formation. Am J Pathol. 2008;173:835–43. doi: 10.2353/ajpath.2008.071091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Kitagawa K, Hiramatsu Y, Kikuchi H, Isobe T, Shimada M, et al. Up-regulation of GPR48 induced by down-regulation of p27Kip1 enhances carcinoma cell invasiveness and metastasis. Cancer Res. 2006;66:11623–31. doi: 10.1158/0008-5472.CAN-06-2629. [DOI] [PubMed] [Google Scholar]
- Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15:701–6. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–8. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Kitagawa K, Shimada M, Uchida C, Hattori T, Oda T, et al. Generation of a constitutively active mutant of human GPR48/LGR4, a G-protein-coupled receptor. Hokkaido Igaku Zasshi. 2006;81:101–5. [PubMed] [Google Scholar]
- Feng S, Agoulnik IU, Truong A, Li Z, Creighton CJ, Kaftanovskaya EM, et al. Suppression of relaxin receptor RXFP1 decreases prostate cancer growth and metastasis. Endocr Relat Cancer. 2010;17:1021–33. doi: 10.1677/ERC-10-0073. [DOI] [PubMed] [Google Scholar]
- Spehr M, Munger SD. Olfactory receptors: G protein-coupled receptors and beyond. J Neurochem. 2009;109:1570–83. doi: 10.1111/j.1471-4159.2009.06085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda N, Yomogida K, Okabe M, Touhara K. Functional characterization of a mouse testicular olfactory receptor and its role in chemosensing and in regulation of sperm motility. J Cell Sci. 2004;117:5835–45. doi: 10.1242/jcs.01507. [DOI] [PubMed] [Google Scholar]
- Xu LL, Stackhouse BG, Florence K, Zhang W, Shanmugam N, Sesterhenn IA, et al. PSGR, a novel prostate-specific gene with homology to a G protein-coupled receptor, is overexpressed in prostate cancer. Cancer Res. 2000;60:6568–72. [PubMed] [Google Scholar]
- Weng J, Wang J, Hu X, Wang F, Ittmann M, Liu M. PSGR2, a novel G-protein coupled receptor, is overexpressed in human prostate cancer. Int J Cancer. 2006;118:1471–80. doi: 10.1002/ijc.21527. [DOI] [PubMed] [Google Scholar]
- Xia C, Ma W, Wang F, Hua S, Liu M. Identification of a prostate-specific G-protein coupled receptor in prostate cancer. Oncogene. 2001;20:5903–7. doi: 10.1038/sj.onc.1204803. [DOI] [PubMed] [Google Scholar]
- Weng J, Wang J, Cai Y, Stafford LJ, Mitchell D, Ittmann M, et al. Increased expression of prostate-specific G-protein-coupled receptor in human prostate intraepithelial neoplasia and prostate cancers. Int J Cancer. 2005;113:811–8. doi: 10.1002/ijc.20635. [DOI] [PubMed] [Google Scholar]
- Wang J, Weng J, Cai Y, Penland R, Liu M, Ittmann M. The prostate-specific G-protein coupled receptors PSGR and PSGR2 are prostate cancer biomarkers that are complementary to alpha-methylacyl-CoA racemase. Prostate. 2006;66:847–57. doi: 10.1002/pros.20389. [DOI] [PubMed] [Google Scholar]
- Rigau M, Morote J, Mir MC, Ballesteros C, Ortega I, Sanchez A, et al. PSGR and PCA3 as biomarkers for the detection of prostate cancer in urine. Prostate. 2010;70:1760–7. doi: 10.1002/pros.21211. [DOI] [PubMed] [Google Scholar]
- Rigau M, Ortega I, Mir MC, Ballesteros C, Garcia M, Llaurado M, et al. A three-gene panel on urine increases PSA specificity in the detection of prostate cancer. Prostate. 2011;71:1736–45. doi: 10.1002/pros.21390. [DOI] [PubMed] [Google Scholar]
- Neuhaus EM, Zhang W, Gelis L, Deng Y, Noldus J, Hatt H. Activation of an olfactory receptor inhibits proliferation of prostate cancer cells. J Biol Chem. 2009;284:16218–25. doi: 10.1074/jbc.M109.012096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spehr J, Gelis L, Osterloh M, Oberland S, Hatt H, Spehr M, et al. G protein-coupled receptor signaling via Src kinase induces endogenous human transient receptor potential vanilloid type 6 (TRPV6) channel activation. J Biol Chem. 2011;286:13184–92. doi: 10.1074/jbc.M110.183525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnoperov VG, Bittner MA, Beavis R, Kuang Y, Salnikow KV, Chepurny OG, et al. alpha-Latrotoxin stimulates exocytosis by the interaction with a neuronal G-protein-coupled receptor. Neuron. 1997;18:925–37. doi: 10.1016/s0896-6273(00)80332-3. [DOI] [PubMed] [Google Scholar]
- Baud V, Chissoe SL, Viegas-Pequignot E, Diriong S, N'Guyen VC, Roe BA, et al. EMR1, an unusual member in the family of hormone receptors with seven transmembrane segments. Genomics. 1995;26:334–44. doi: 10.1016/0888-7543(95)80218-b. [DOI] [PubMed] [Google Scholar]
- Taylor PR, Martinez-Pomares L, Stacey M, Lin HH, Brown GD, Gordon S. Macrophage receptors and immune recognition. Annu Rev Immunol. 2005;23:901–44. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- Yona S, Lin HH, Stacey M. Immunity and adhesion-GPCRs. Adv Exp Med Biol. 2010;706:121–7. doi: 10.1007/978-1-4419-7913-1_10. [DOI] [PubMed] [Google Scholar]
- Austyn JM, Gordon S. F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur J Immunol. 1981;11:805–15. doi: 10.1002/eji.1830111013. [DOI] [PubMed] [Google Scholar]
- Lin HH, Faunce DE, Stacey M, Terajewicz A, Nakamura T, Zhang-Hoover J, et al. The macrophage F4/80 receptor is required for the induction of antigen-specific efferent regulatory T cells in peripheral tolerance. J Exp Med. 2005;201:1615–25. doi: 10.1084/jem.20042307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwakkenbos MJ, Pouwels W, Matmati M, Stacey M, Lin HH, Gordon S, et al. Expression of the largest CD97 and EMR2 isoforms on leukocytes facilitates a specific interaction with chondroitin sulfate on B cells. J Leukoc Biol. 2005;77:112–9. doi: 10.1189/jlb.0704402. [DOI] [PubMed] [Google Scholar]
- Stacey M, Chang GW, Davies JQ, Kwakkenbos MJ, Sanderson RD, Hamann J, et al. The epidermal growth factor-like domains of the human EMR2 receptor mediate cell attachment through chondroitin sulfate glycosaminoglycans. Blood. 2003;102:2916–24. doi: 10.1182/blood-2002-11-3540. [DOI] [PubMed] [Google Scholar]
- Hamann J, Vogel B, van Schijndel GM, van Lier RA. The seven-span transmembrane receptor CD97 has a cellular ligand (CD55, DAF) J Exp Med. 1996;184:1185–9. doi: 10.1084/jem.184.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann J, Stortelers C, Kiss-Toth E, Vogel B, Eichler W, van Lier RA. Characterization of the CD55 (DAF)-binding site on the seven-span transmembrane receptor CD97. Eur J Immunol. 1998;28:1701–7. doi: 10.1002/(SICI)1521-4141(199805)28:05<1701::AID-IMMU1701>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Abbott RJ, Spendlove I, Roversi P, Fitzgibbon H, Knott V, Teriete P, et al. Structural and functional characterization of a novel T cell receptor co-regulatory protein complex, CD97–CD55. J Biol Chem. 2007;282:22023–32. doi: 10.1074/jbc.M702588200. [DOI] [PubMed] [Google Scholar]
- Capasso M, Durrant LG, Stacey M, Gordon S, Ramage J, Spendlove I. Costimulation via CD55 on human CD4+ T cells mediated by CD97. J Immunol. 2006;177:1070–7. doi: 10.4049/jimmunol.177.2.1070. [DOI] [PubMed] [Google Scholar]
- Matmati M, Pouwels W, van Bruggen R, Jansen M, Hoek RM, Verhoeven AJ, et al. The human EGF-TM7 receptor EMR3 is a marker for mature granulocytes. J Leukoc Biol. 2007;81:440–8. doi: 10.1189/jlb.0406276. [DOI] [PubMed] [Google Scholar]
- Stacey M, Lin HH, Hilyard KL, Gordon S, McKnight AJ. Human epidermal growth factor (EGF) module-containing mucin-like hormone receptor 3 is a new member of the EGF-TM7 family that recognizes a ligand on human macrophages and activated neutrophils. J Biol Chem. 2001;276:18863–70. doi: 10.1074/jbc.M101147200. [DOI] [PubMed] [Google Scholar]
- Stacey M, Chang GW, Sanos SL, Chittenden LR, Stubbs L, Gordon S, et al. EMR4, a novel epidermal growth factor (EGF)-TM7 molecule up-regulated in activated mouse macrophages, binds to a putative cellular ligand on B lymphoma cell line A20. J Biol Chem. 2002;277:29283–93. doi: 10.1074/jbc.M204306200. [DOI] [PubMed] [Google Scholar]
- Formstone CJ. 7TM-Cadherins: developmental roles and future challenges. Adv Exp Med Biol. 2010;706:14–36. doi: 10.1007/978-1-4419-7913-1_2. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Casal J, Struhl G. Cell interactions and planar polarity in the abdominal epidermis of Drosophila. Development. 2004;131:4651–64. doi: 10.1242/dev.01351. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–78. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- Sweeney NT, Li W, Gao FB. Genetic manipulation of single neurons in vivo reveals specific roles of flamingo in neuronal morphogenesis. Dev Biol. 2002;247:76–88. doi: 10.1006/dbio.2002.0702. [DOI] [PubMed] [Google Scholar]
- Chen PL, Clandinin TR. The cadherin Flamingo mediates level-dependent interactions that guide photoreceptor target choice in Drosophila. Neuron. 2008;58:26–33. doi: 10.1016/j.neuron.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinel MC, Whitington PM. The atypical cadherin Flamingo is required for sensory axon advance beyond intermediate target cells. Dev Biol. 2009;327:447–57. doi: 10.1016/j.ydbio.2008.12.026. [DOI] [PubMed] [Google Scholar]
- Bao H, Berlanga ML, Xue M, Hapip SM, Daniels RW, Mendenhall JM, et al. The atypical cadherin flamingo regulates synaptogenesis and helps prevent axonal and synaptic degeneration in Drosophila. Mol Cell Neurosci. 2007;34:662–78. doi: 10.1016/j.mcn.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin JA, Quint E, Tsipouri V, Arkell RM, Cattanach B, Copp AJ, et al. Mutation of Celsr1 disrupts planar polarity of inner ear hair cells and causes severe neural tube defects in the mouse. Curr Biol. 2003;13:1129–33. doi: 10.1016/s0960-9822(03)00374-9. [DOI] [PubMed] [Google Scholar]
- Carreira-Barbosa F, Kajita M, Morel V, Wada H, Okamoto H, Martinez Arias A, et al. Flamingo regulates epiboly and convergence/extension movements through cell cohesive and signalling functions during zebrafish gastrulation. Development. 2009;136:383–92. doi: 10.1242/dev.026542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruna B, Jenny A, Lee D, Mlodzik M, Schier AF. Planar cell polarity signalling couples cell division and morphogenesis during neurulation. Nature. 2006;439:220–4. doi: 10.1038/nature04375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravni A, Qu Y, Goffinet AM, Tissir F. Planar cell polarity cadherin Celsr1 regulates skin hair patterning in the mouse. J Invest Dermatol. 2009;129:2507–9. doi: 10.1038/jid.2009.84. [DOI] [PubMed] [Google Scholar]
- Monk KR, Naylor SG, Glenn TD, Mercurio S, Perlin JR, Dominguez C, et al. A G protein-coupled receptor is essential for Schwann cells to initiate myelination. Science. 2009;325:1402–5. doi: 10.1126/science.1173474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnert F, Mancuso MR, Shamloo A, Wang HT, Choksi V, Florek M, et al. Essential regulation of CNS angiogenesis by the orphan G protein-coupled receptor GPR124. Science. 2010;330:985–9. doi: 10.1126/science.1196554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KD, Pan L, Yang XM, Hughes VC, Walls JR, Dominguez MG, et al. Angiogenic sprouting into neural tissue requires Gpr124, an orphan G protein-coupled receptor. Proc Natl Acad Sci U S A. 2011;108:2807–12. doi: 10.1073/pnas.1019761108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk KR, Oshima K, Jors S, Heller S, Talbot WS. Gpr126 is essential for peripheral nerve development and myelination in mammals. Development. 2011;138:2673–80. doi: 10.1242/dev.062224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JQ, Lin HH, Stacey M, Yona S, Chang GW, Gordon S, et al. Leukocyte adhesion-GPCR EMR2 is aberrantly expressed in human breast carcinomas and is associated with patient survival. Oncol Rep. 2011;25:619–27. doi: 10.3892/or.2010.1117. [DOI] [PubMed] [Google Scholar]
- Galle J, Sittig D, Hanisch I, Wobus M, Wandel E, Loeffler M, et al. Individual cell-based models of tumor-environment interactions: Multiple effects of CD97 on tumor invasion. Am J Pathol. 2006;169:1802–11. doi: 10.2353/ajpath.2006.060006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. WNT/PCP signaling pathway and human cancer (review) Oncol Rep. 2005;14:1583–8. [PubMed] [Google Scholar]
- Xu L, Begum S, Hearn JD, Hynes RO. GPR56, an atypical G protein-coupled receptor, binds tissue transglutaminase, TG2, and inhibits melanoma tumor growth and metastasis. Proc Natl Acad Sci U S A. 2006;103:9023–8. doi: 10.1073/pnas.0602681103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Begum S, Barry M, Crowley D, Yang L, Bronson RT, et al. GPR56 plays varying roles in endogenous cancer progression. Clin Exp Metastasis. 2010;27:241–9. doi: 10.1007/s10585-010-9322-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno N, Itoh H. Signal transduction mediated through adhesion-GPCRs. Adv Exp Med Biol. 2010;706:157–66. doi: 10.1007/978-1-4419-7913-1_14. [DOI] [PubMed] [Google Scholar]
- Park D, Tosello-Trampont AC, Elliott MR, Lu M, Haney LB, Ma Z, et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450:430–4. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- Abe J, Fukuzawa T, Hirose S. Cleavage of Ig-Hepta at a “SEA” module and at a conserved G protein-coupled receptor proteolytic site. J Biol Chem. 2002;277:23391–8. doi: 10.1074/jbc.M110877200. [DOI] [PubMed] [Google Scholar]
- Abe J, Suzuki H, Notoya M, Yamamoto T, Hirose S. Ig-hepta, a novel member of the G protein-coupled hepta-helical receptor (GPCR) family that has immunoglobulin-like repeats in a long N-terminal extracellular domain and defines a new subfamily of GPCRs. J Biol Chem. 1999;274:19957–64. doi: 10.1074/jbc.274.28.19957. [DOI] [PubMed] [Google Scholar]
- Fukuzawa T, Hirose S. Multiple processing of Ig-Hepta/GPR116, a G protein-coupled receptor with immunoglobulin (Ig)-like repeats, and generation of EGF2-like fragment. J Biochem. 2006;140:445–52. doi: 10.1093/jb/mvj170. [DOI] [PubMed] [Google Scholar]
- Lelianova VG, Davletov BA, Sterling A, Rahman MA, Grishin EV, Totty NF, et al. Alpha-latrotoxin receptor, latrophilin, is a novel member of the secretin family of G protein-coupled receptors. J Biol Chem. 1997;272:21504–8. doi: 10.1074/jbc.272.34.21504. [DOI] [PubMed] [Google Scholar]
- Little KD, Hemler ME, Stipp CS. Dynamic regulation of a GPCR-tetraspanin-G protein complex on intact cells: central role of CD81 in facilitating GPR56-Galpha q/11 association. Mol Biol Cell. 2004;15:2375–87. doi: 10.1091/mbc.E03-12-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi T, Sakata K, Yoshizaki K, Tago K, Mizuno N, Itoh H. Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a G alpha 12/13 and Rho pathway. J Biol Chem. 2008;283:14469–78. doi: 10.1074/jbc.M708919200. [DOI] [PubMed] [Google Scholar]
- Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat Rev Drug Discov. 2011;10:47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]
- Crouch MF, Osmond RI. New strategies in drug discovery for GPCRs: high throughput detection of cellular ERK phosphorylation. Comb Chem High Throughput Screen. 2008;11:344–56. doi: 10.2174/138620708784534806. [DOI] [PubMed] [Google Scholar]
- Bikkavilli RK, Tsang SY, Tang WM, Sun JX, Ngai SM, Lee SS, et al. Identification and characterization of surrogate peptide ligand for orphan G protein-coupled receptor mas using phage-displayed peptide library. Biochem Pharmacol. 2006;71:319–37. doi: 10.1016/j.bcp.2005.10.050. [DOI] [PubMed] [Google Scholar]
- Overton HA, Babbs AJ, Doel SM, Fyfe MC, Gardner LS, Griffin G, et al. Deorphanization of a G protein-coupled receptor for oleoylethanolamide and its use in the discovery of small-molecule hypophagic agents. Cell Metab. 2006;3:167–75. doi: 10.1016/j.cmet.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Pausch MH. G-protein-coupled receptors in Saccharomyces cerevisiae: high-throughput screening assays for drug discovery. Trends Biotechnol. 1997;15:487–94. doi: 10.1016/S0167-7799(97)01119-0. [DOI] [PubMed] [Google Scholar]
- Bates B, Zhang L, Nawoschik S, Kodangattil S, Tseng E, Kopsco D, et al. Characterization of Gpr101 expression and G-protein coupling selectivity. Brain Res. 2006;1087:1–14. doi: 10.1016/j.brainres.2006.02.123. [DOI] [PubMed] [Google Scholar]
- Suga H, Haga T. Ligand screening system using fusion proteins of G protein-coupled receptors with G protein alpha subunits. Neurochem Int. 2007;51:140–64. doi: 10.1016/j.neuint.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Howard AD, McAllister G, Feighner SD, Liu Q, Nargund RP, Van der Ploeg LH, et al. Orphan G-protein-coupled receptors and natural ligand discovery. Trends Pharmacol Sci. 2001;22:132–40. doi: 10.1016/s0165-6147(00)01636-9. [DOI] [PubMed] [Google Scholar]
- Okamoto H, Takuwa N, Yatomi Y, Gonda K, Shigematsu H, Takuwa Y. EDG3 is a functional receptor specific for sphingosine 1-phosphate and sphingosylphosphorylcholine with signaling characteristics distinct from EDG1 and AGR16. Biochem Biophys Res Commun. 1999;260:203–8. doi: 10.1006/bbrc.1999.0886. [DOI] [PubMed] [Google Scholar]
- An S, Zheng Y, Bleu T. Sphingosine 1-phosphate-induced cell proliferation, survival, and related signaling events mediated by G protein-coupled receptors Edg3 and Edg5. J Biol Chem. 2000;275:288–96. doi: 10.1074/jbc.275.1.288. [DOI] [PubMed] [Google Scholar]
- Pyne S, Pyne NJ. Sphingosine 1-phosphate signalling in mammalian cells. Biochem J. 2000;349:385–402. doi: 10.1042/0264-6021:3490385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovenberg TW, Roland BL, Wilson SJ, Jiang X, Pyati J, Huvar A, et al. Cloning and functional expression of the human histamine H3 receptor. Mol Pharmacol. 1999;55:1101–7. [PubMed] [Google Scholar]
- Oda T, Morikawa N, Saito Y, Masuho Y, Matsumoto S. Molecular cloning and characterization of a novel type of histamine receptor preferentially expressed in leukocytes. J Biol Chem. 2000;275:36781–6. doi: 10.1074/jbc.M006480200. [DOI] [PubMed] [Google Scholar]
- Maenhaut C, Van Sande J, Libert F, Abramowicz M, Parmentier M, Vanderhaegen JJ, et al. RDC8 codes for an adenosine A2 receptor with physiological constitutive activity. Biochem Biophys Res Commun. 1990;173:1169–78. doi: 10.1016/s0006-291x(05)80909-x. [DOI] [PubMed] [Google Scholar]
- Libert F, Schiffmann SN, Lefort A, Parmentier M, Gerard C, Dumont JE, et al. The orphan receptor cDNA RDC7 encodes an A1 adenosine receptor. EMBO J. 1991;10:1677–82. doi: 10.1002/j.1460-2075.1991.tb07691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lecea L, Kilduff TS, Peyron C, Gao X, Foye PE, Danielson PE, et al. The hypocretins: hypothalamus-specific peptides with neuroexcitatory activity. Proc Natl Acad Sci U S A. 1998;95:322–7. doi: 10.1073/pnas.95.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinuma S, Habata Y, Fujii R, Kawamata Y, Hosoya M, Fukusumi S, et al. A prolactin-releasing peptide in the brain. Nature. 1998;393:272–6. doi: 10.1038/30515. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Amemiya A, Ishii M, Matsuzaki I, Chemelli RM, Tanaka H, et al. Orexins and orexin receptors: a family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell. 1998;92:573–85. doi: 10.1016/s0092-8674(00)80949-6. [DOI] [PubMed] [Google Scholar]
- Tatemoto K, Hosoya M, Habata Y, Fujii R, Kakegawa T, Zou MX, et al. Isolation and characterization of a novel endogenous peptide ligand for the human APJ receptor. Biochem Biophys Res Commun. 1998;251:471–6. doi: 10.1006/bbrc.1998.9489. [DOI] [PubMed] [Google Scholar]
- Lin HH, Chang GW, Huang YS, Hsiao CC, Stacey M, Gordon S. Multivalent protein probes for the identification and characterization of cognate cellular ligands for myeloid cell surface receptors. Methods Mol Biol. 2009;531:89–101. doi: 10.1007/978-1-59745-396-7_7. [DOI] [PubMed] [Google Scholar]
- Libert F, Vassart G, Parmentier M. Current developments in G-protein-coupled receptors. Curr Opin Cell Biol. 1991;3:218–23. doi: 10.1016/0955-0674(91)90142-l. [DOI] [PubMed] [Google Scholar]
- Mills A, Duggan MJ. Orphan seven transmembrane domain receptors: reversing pharmacology. Trends Biotechnol. 1994;12:47–9. doi: 10.1016/0167-7799(94)90099-X. [DOI] [PubMed] [Google Scholar]
- Ozawa A, Lindberg I, Roth B, Kroeze WK. Deorphanization of novel peptides and their receptors. AAPS J. 2010;12:378–84. doi: 10.1208/s12248-010-9198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese A, George SR, Kolakowski LF, Jr, Lynch KR, O'Dowd BF. Novel GPCRs and their endogenous ligands: expanding the boundaries of physiology and pharmacology. Trends Pharmacol Sci. 1999;20:370–5. doi: 10.1016/s0165-6147(99)01366-8. [DOI] [PubMed] [Google Scholar]
- Lerner MR. Tools for investigating functional interactions between ligands and G-protein-coupled receptors. Trends Neurosci. 1994;17:142–6. doi: 10.1016/0166-2236(94)90087-6. [DOI] [PubMed] [Google Scholar]
- Wise A, Jupe SC, Rees S. The identification of ligands at orphan G-protein coupled receptors. Annu Rev Pharmacol Toxicol. 2004;44:43–66. doi: 10.1146/annurev.pharmtox.44.101802.121419. [DOI] [PubMed] [Google Scholar]
- McLatchie LM, Fraser NJ, Main MJ, Wise A, Brown J, Thompson N, et al. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature. 1998;393:333–9. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- Hay DL, Poyner DR, Sexton PM. GPCR modulation by RAMPs. Pharmacol Ther. 2006;109:173–97. doi: 10.1016/j.pharmthera.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Galvez T, Duthey B, Kniazeff J, Blahos J, Rovelli G, Bettler B, et al. Allosteric interactions between GB1 and GB2 subunits are required for optimal GABAB receptor function. EMBO J. 2001;20:2152–9. doi: 10.1093/emboj/20.9.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA, Borowsky B, Tamm JA, Craig DA, Durkin MM, Dai M, et al. GABAB receptors function as a heteromeric assembly of the subunits GABABR1 and GABABR2. Nature. 1998;396:674–9. doi: 10.1038/25348. [DOI] [PubMed] [Google Scholar]
- Robbins MJ, Calver AR, Filippov AK, Hirst WD, Russell RB, Wood MD, et al. GABAB2 is essential for g-protein coupling of the GABAB receptor heterodimer. J Neurosci. 2001;21:8043–52. doi: 10.1523/JNEUROSCI.21-20-08043.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–57. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]