Abstract
Background
MITE, TRIM and SINEs are miniature form transposable elements (mTEs) that are ubiquitous and dispersed throughout entire plant genomes. Tens of thousands of members cause insertion polymorphism at both the inter- and intra- species level. Therefore, mTEs are valuable targets and resources for development of markers that can be utilized for breeding, genetic diversity and genome evolution studies. Taking advantage of the completely sequenced genomes of Brassica rapa and B. oleracea, characterization of mTEs and building a curated database are prerequisite to extending their utilization for genomics and applied fields in Brassica crops.
Findings
We have developed BrassicaTED as a unique web portal containing detailed characterization information for mTEs of Brassica species. At present, BrassicaTED has datasets for 41 mTE families, including 5894 and 6026 members from 20 MITE families, 1393 and 1639 members from 5 TRIM families, 1270 and 2364 members from 16 SINE families in B. rapa and B. oleracea, respectively. BrassicaTED offers different sections to browse structural and positional characteristics for every mTE family. In addition, we have added data on 289 MITE insertion polymorphisms from a survey of seven Brassica relatives. Genes with internal mTE insertions are shown with detailed gene annotation and microarray-based comparative gene expression data in comparison with their paralogs in the triplicated B. rapa genome. This database also includes a novel tool, K BLAST (Karyotype BLAST), for clear visualization of the locations for each member in the B. rapa and B. oleracea pseudo-genome sequences.
Conclusions
BrassicaTED is a newly developed database of information regarding the characteristics and potential utility of mTEs including MITE, TRIM and SINEs in B. rapa and B. oleracea. The database will promote the development of desirable mTE-based markers, which can be utilized for genomics and breeding in Brassica species. BrassicaTED will be a valuable repository for scientists and breeders, promoting efficient research on Brassica species. BrassicaTED can be accessed at http://im-crop.snu.ac.kr/BrassicaTED/index.php.
Keywords: Brassica, Miniature inverted-repeat transposable element (MITE), Terminal-repeat retrotransposon in miniature (TRIM), Miniature form transposable elements (mTEs), Short interspersed elements (SINEs), TE Database
Findings
Background
Transposable elements (TEs) are ubiquitous and compose a large fraction of most of eukaryotic genomes. For example, more than 80% of the maize genome [1] and 40% of the B. rapa genome are occupied by transposons [2]. TEs can be mobilized and accumulated in the genome through cut and paste mechanism or a replicative transposition mechanism. Transposition of TEs can cause gene knock-out or rearrangement, altered regulation of expression, acquisition of novel function, and/or gene structure modification [3-5]. Although plant genomes have large numbers of TEs, most of them remain as non-autonomous TEs, which cannot transpose without the assistance of a “partner” element providing transposition enzymes, because of accumulation of mutations, incomplete transposition processes or genetic and epigenetic control [4,6]. Miniature form transposable elements (mTEs) are especially small (<800 bp) TEs that are present in high copy numbers. They lack functional coding genes and rely on trans-activation by long and complete counterpart autonomous TEs [3,7-9]. The mTEs include the terminal repeat retrotransposon in miniature (TRIM), short interspersed element (SINE) and miniature inverted-repeat transposable element (MITE) [4,8-10] families.
TRIMs are derivatives of retrotransposons from which the coding domain for transposition has been lost. TRIMs have terminal direct repeats (TDRs) (100–250 bp) flanked by 5-bp target site duplications (TSDs) and contain an internal sequence that begins with the signature region of the primer binding site (PBS) of tRNA-methionine and ends with a polypurine tract (PPT) motif [8,11,12]. Studies have suggested that amplification and mobilization of TRIM elements are controlled by autonomous partner elements like LTR retrotransposons. TRIMs have been identified in various plant species and are present at moderately high levels, with 2000 copies, in the Brassica genome [8,11,13]. TRIM element-based markers have been effective for high quality mapping in Brassica[11].
Short interspersed elements (SINEs) are non-autonomous, non-LTR retrotransposons that are transcribed by RNA polymerase III and widely distributed in eukaryotic organisms [10,14]. SINEs have unique structural features, with so-called head, body and tail sequences, and have been found in higher copy numbers in animals than in plants [15-17]. For instance, the Alu SINE family is present in > 1,500,000 copies in the human genome and covers > 11% of the total genome [14,18,19]. Propagation, maintenance and movement of SINEs are controlled by long-interspersed nucleotide elements (LINEs). To date, 210 SINE families from various plant and animal genomes including 16 SINE families related to Brassicaceae have been reported in SINEBase [20].
MITEs are small, non-autonomous class II DNA transposons that are AT-rich and have DNA structures characteristic of TEs, like terminal inverted repeats (TIRs) and TSD of variable lengths. Based on their TSD, most MITEs have been classified into several superfamilies. Stowaway superfamily members recognize 2-bp (TA) nucleotides and duplicate these at their terminal region via insertion. Similarly, Tourist superfamily members produce unique TAA duplications, hAT superfamily members produce 5-, 6-, or 8-bp TSD, Mutator superfamily members produce 9- or 10-bp TSD, and En/Spm superfamily members produce 3-bp non-unique TSD [4]. Available evidence suggests that MITEs originate through either internal deletion or cross mobilization from autonomous transposable elements [3]. MITEs can amplify into high copy numbers, although the MITEs are thought be mobilized through cut-and-paste mechanisms. Analysis of complete information for MITEs on a genome-wide scale will be important for a better understanding of how MITE transposition can produce high copy numbers, as well as of their influence on genome evolution [21].
mTEs are stably maintained and highly dispersed throughout the genome, often in association with genic regions, where they can alter gene structure and function [22,23]. Insertion of MITEs has been reported to cause changes in pigmentation and flowering time in potato and rice, respectively [24,25]. However, most mTE insertions may not cause phenotypic changes but can alter gene expression and epigenetic patterns [8,26-28]. Their high copy number and close association with gene-rich regions make mTEs a potential source of molecular markers, mTE-based marker systems have been successfully developed in various species for genetic mapping as well as analysis of genetic diversity, genome-wide association and evolution [8,29-34].
The genus Brassica (family Brassicaceae) is composed of more than 3700 species including important vegetable and oil-seed crops such as kimchi-cabbage, cabbage, and oilseed rape (B. rapa, B. oleracea and B. napus) [35]. B. rapa was the first Brassica vegetable to be sequenced and annotated, revealing recent genome triplication at 13–17 MYA, after divergence from Arabidopsis thaliana 20 MYA [2]. While only 6% of the A. thaliana genome is occupied by TEs [36,37], around 38-40% of the B. rapa and B. oleracea genome is derived from TEs [2]. Genome analysis has suggested that TEs are important players not only in terms of genome size increases but also for gene and genome evolution, especially in the recently triplicated Brassica genome [2]. Recently, a plant MITE database was created for 41 sequenced plant genomes, and 174 MITE families from B. rapa were identified using in silico MITE identification tools [38]. However, experimental validation of identified mTE families and genome-scale characterization of their members with biological utility for Brassica species have not been developed or included in any databases. Furthermore, there is no resource with information regarding the distribution of TRIM, SINE and MITE families in various genomic regions for the Brassica species. Therefore, we aimed to establish a unique web portal containing detailed characterization of TEs in Brassica species. As a starting point, we have investigated the MITE, TRIM and SINE families of B. rapa and B. oleracea at the whole-genome scale and developed a database, BrassicaTED (Brassica Transposable Elements Database), for effective utilization of MITE, TRIM and SINEs in the Brassica genome.
Construction and content
We developed BrassicaTED as a database to provide comprehensive information about mTEs in the B. rapa and B. oleracea genomes. This database has three-tier architecture, namely a client tier, a middle tier and a database tier (Figure 1). The user-friendly interface, i.e. the client tier, was developed using PHP (v4.3.9) and JavaScript [39-41]. In the database tier, all information related to the mTEs is stored in a MySQL (v4.1.20) database. In the middle tier, an application forces all HTTP requests to be processed by an Apache web server (Figure 1). The database is currently hosted on a CentOS (5.8) Linux operation system. Additionally, we included the NCBI BLAST algorithm (version: 2.2.15) for sequence-based searches and sequence extractor tools.
Figure 1.
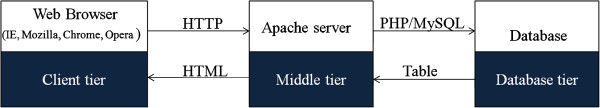
Three-tier architecture of BrassicaTED. The client tier receives input from users and sends the request to an Apache server. The middle tier receives the request, processes it and passes it to the database tier through PHP and MySQL. The user request is extracted from MySQL as a table and displayed on a browser in the html page.
Browse
In the Browse section, BrassicaTED provides a convenient panel in which to browse the structural characteristics, family classification, and sequence information with copy numbers in B. rapa and B. oleracea and A. thaliana for mTEs. It currently contains information for 20 MITE families, 5 TRIM families and 16 SINE families characterized in the whole genomes of B. rapa and B. oleracea. We developed this section to be fully updatable as more mTEs are identified and characterized (Figure 2 [1-I]).
Figure 2.
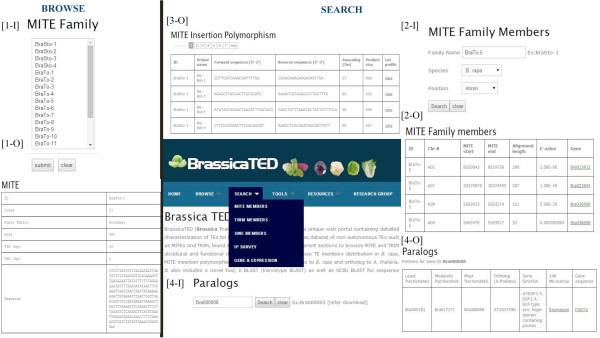
Display of the Browse and Search panels of BrassicaTED. [1-I] The Browse page for the MITE family listing characterized MITEs. [1-O] Output display of the selected MITE family. [2-I] Query page to retrieve MITE family members on each chromosome. [2-O] Result display of MITE member page. [3-O] Display of all MITE families and MITE insertion polymorphism. [4-I] Query box for Paralogs & Expression tab. [4-O] Paralog partners and ortholog gene display for selected genes.
Search
The Search section was developed to allow exploration of annotation and analysis for each mTE family in detail. In this section, there are five sub-categories: MITE Members, TRIM Members, SINE Members, IP Survey, and Gene & Expression (Figure 2).
MITE members
B. rapa pseudo-chromosome sequences with unanchored scaffolds (283 Mb) and gene annotation information (version 1.2) were obtained from the publically available database [42]. We included 20 MITE families that were identified from our studies and others in Brassica[8,30,43-45]. Consensus representative sequences of the 20 MITE families were used to identify their members from 283 Mb B. rapa pseudo-chromosome sequences. The physical position of each MITE on the B. rapa pseudo-chromosome was described using a custom Perl script. A total of 5894 MITEs (Br-members) were recently identified in the B. rapa genome [45]. Among them, 98% could be classified based on the type of genomic sequence in which they were located, such as intergenic region, 5’- or 3’-UTR, exon or coding sequence (CDS), and intron, whereas the remaining 2% were located on unannotated scaffolds of the B. rapa genome.
The recent availability of 385 Mb of B. oleracea pseudo-chromosome sequences (version 1.0) [46,47] allowed that analysis to expand to the B. oleracea genome. Similar to B. rapa, a total of 6026 members (Bo-Members) were annotated based on physical position and gene annotation information for B. oleracea[42]. This work revealed that compared with Br-members, very few Bo-members (three) are present in the CDS of B. oleracea[45].
TRIM members
In a previous study, we characterized four TRIM families (TRIM of Brassicaceae -1, -2, -3, -4) and their members in 96 Mb of B. rapa BAC end sequences and 434 Mb of B. oleracea shotgun sequences, [8]. Now, we additionally characterized a high copy family, TB-5 (Cassandra), in the pseudo-chromosome sequences of B. rapa and B. oleracea[13]. TRIM element insertion was analyzed based on physical position information and the annotation of the genomes of B. rapa and B. oleracea. In total, 1393 and 1639 copies from the five TRIM families were identified from B. rapa and B. oleracea whole-genome pseudo-chromosome sequences, respectively (Table 1). Distribution analysis of these five TRIM families shows insertion throughout the chromosomes of the B. rapa and B. oleracea genomes. Interestingly, 619 (44%) and 656 (40%) members reside in or within 2 kb of a gene in the B. rapa and B. oleracea genome, respectively. This information will be highly valuable for marker development and characterization of target regions that have TRIM element insertion.
Table 1.
Characteristics of five TRIM families in the B . rapa and B . oleracea genomes
| TRIM family |
No. of TRIM members in
B.
rapa
(283 Mb) genome |
No. of TRIM members in
B.
oleracea
(385 Mb) genome |
Copies in A. thaliana (119 Mb) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Intergenic space a | Near gene b |
Elements in genic regions |
Un mapped d | Total | Intergenic space a | Near gene b |
Elements in genic regions |
||||||||
| 5’ UTR c | CDS | Intron | 3’ UTR c | 5’ UTR c | CDS | Intron | 3’ UTR c | |||||||||
| TB-1 |
206 |
71 |
54 |
12 |
1 |
39 |
17 |
12 |
273 |
164 |
61 |
17 |
0 |
19 |
12 |
65 |
| TB-2 |
319 |
147 |
92 |
15 |
2 |
35 |
18 |
10 |
321 |
162 |
110 |
11 |
0 |
29 |
9 |
16 |
| TB-3 |
52 |
22 |
7 |
4 |
3 |
8 |
4 |
4 |
51 |
25 |
15 |
3 |
1 |
3 |
4 |
0 |
| TB-4 |
351 |
157 |
78 |
14 |
7 |
37 |
14 |
44 |
392 |
235 |
106 |
19 |
2 |
16 |
14 |
0 |
| TB-5 |
465 |
233 |
96 |
8 |
1 |
40 |
13 |
74 |
602 |
397 |
150 |
21 |
0 |
13 |
21 |
96 |
| Total | 1393 | 630 | 327 | 53 | 14 | 159 | 66 | 144 | 1639 | 983 | 442 | 71 | 3 | 80 | 60 | 177 |
aMITE members categorized by their insertion >2 kb from the nearest gene.
bMITE members categorized by their insertion 500-2000 bp from the nearest gene.
cMITE members residing in the 500 bp upstream (5’UTR) or downstream (3’UTR) of the start or stop codon, respectively.
dMITE members found in unanchored scaffolds of the B. rapa genome.
SINE members
We have included the characterization of the members of 16 SINE families from Brassicaceae, which we have named as SINE of Brassicaceae 1–16. The sequence information for 16 families was obtained from SINEBase, and 1270 and 2364 members were retrieved from the B. rapa and B. oleracea genomes [20]. The insertion of SINEs in various genomic locations was characterized based on physical position and genome annotation information, showing that 599 (47.1%) and 1154 (48.8%) of the members from B. rapa and B. oleracea, respectively, were present in close association with genic regions (with < 2 kb of a gene) (Table 2). This will be a valuable resource from which researchers can develop markers for tightly linked genes or regions on the chromosome.
Table 2.
Characteristics of 16 SINE families in the B . rapa and B . oleracea genomes
| SINE family |
No. of SINE members in
B.
rapa
(283 Mb) genome |
No. of SINE members in
B.
oleracea
(385 Mb) genome |
Copies in A. thaliana (119 Mb) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Intergenic space a | Near gene b |
Elements in genic regions |
Un mapped d | Total | Intergenic space a | Near gene b |
Elements in genic regions |
||||||||
| 5’ UTR c | CDS | Intron | 3’ UTR c | 5’ UTR c | CDS | Intron | 3’ UTR c | |||||||||
| SB-1 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
98 |
49 |
32 |
7 |
0 |
4 |
6 |
0 |
| SB-2 |
43 |
2 |
0 |
2 |
0 |
38 |
1 |
0 |
59 |
8 |
4 |
0 |
0 |
45 |
2 |
132 |
| SB-3 |
134 |
70 |
36 |
6 |
0 |
12 |
6 |
4 |
278 |
156 |
80 |
20 |
0 |
9 |
13 |
4 |
| SB-4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
38 |
| SB-5 |
155 |
72 |
54 |
6 |
0 |
11 |
7 |
5 |
375 |
205 |
99 |
23 |
0 |
18 |
30 |
3 |
| SB-6 |
248 |
134 |
71 |
12 |
0 |
7 |
14 |
10 |
394 |
213 |
132 |
21 |
0 |
7 |
21 |
28 |
| SB-7 |
372 |
202 |
106 |
17 |
0 |
11 |
20 |
16 |
355 |
197 |
119 |
16 |
0 |
2 |
21 |
3 |
| SB-8 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
80 |
28 |
26 |
14 |
0 |
2 |
10 |
0 |
| SB-9 |
141 |
62 |
49 |
7 |
0 |
6 |
12 |
5 |
201 |
102 |
69 |
11 |
0 |
7 |
12 |
0 |
| SB-10 |
16 |
5 |
10 |
0 |
0 |
0 |
0 |
1 |
134 |
67 |
48 |
7 |
0 |
6 |
6 |
0 |
| SB-11 |
4 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
56 |
22 |
25 |
3 |
0 |
2 |
4 |
0 |
| SB-12 |
10 |
5 |
3 |
0 |
0 |
2 |
0 |
0 |
101 |
46 |
37 |
7 |
0 |
5 |
6 |
0 |
| SB-13 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
95 |
43 |
30 |
7 |
0 |
7 |
8 |
0 |
| SB-14 |
142 |
69 |
43 |
9 |
0 |
12 |
8 |
1 |
137 |
74 |
37 |
10 |
0 |
6 |
10 |
0 |
| SB-15 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| SB-16 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
4 |
| Total | 1270 | 629 | 373 | 59 | 0 | 99 | 68 | 42 | 2364 | 1210 | 738 | 146 | 1 | 120 | 149 | 212 |
aMITE members categorized by their insertion >2 kb from the nearest gene.
bMITE members categorized by their insertion 500–2000 bp from the nearest gene.
cMITE members resid in the 500 bp upstream (5’UTR) or downstream (3’UTR) of the start or stop codon, respectively.
dMITE members found in unanchored scaffolds of the B. rapa genome.
Insertion polymorphism (IP) survey
Insertion polymorphism inter- and intra- Brassica species due to insertion or absence of a mTE in specific accessions was surveyed by PCR using flanking primers [48]. For MITE insertion polymorphism (MIP) analysis, 187 and 145 MITE targets were surveyed from B. rapa and B. oleracea members, respectively [30,45]. Among them, primers for 162 Br-members and 127 Bo-members produced clear amplicons, of which over than 52% (150) showed MIP in the tested Brassica relatives and their MIP information were summarized such as Figure 3. BrassicaTED includes information for the primers used and polymorphism profiles with the tested accessions. Data for additional insertion polymorphism surveys of more MITE members and for TRIM and SINE members will be added in the future.
Figure 3.
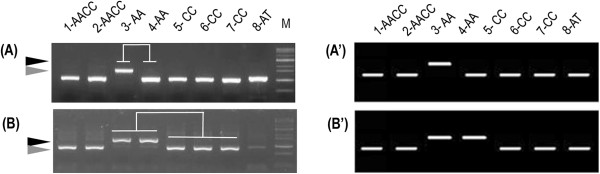
MITE insertion polymorphism analysis shows intra- and inter- species polymorphism between B. rapa and B. oleracea. Representative analysis of intra (A)- and inter (B)- species polymorphism between B. rapa and B. oleracea. The original gels are shown in A and B, whereas (A’) and (B’) show the corresponding images generated based on the gel scores. The accessions used for the MIP survey are the same as those in [30].
Gene and expression analysis
BrassicaTED includes information regarding paralogous genes in B. rapa and their orthologs from A. thaliana, obtained from the B. rapa genome database [42]. Single, duplicate, and triplicate paralogs and their orthologs in A. thaliana are listed according to the gene order in A. thaliana. The expression level of B. rapa genes with internal MITE insertion was surveyed against the B. rapa 24 k microarray database, which was generated from four different stress treatments: cold (4°C), salt (250 mM NaCl), drought (air-dry) and ABA (100 μM) [49].
Tools
This section includes tools for basic bioinformatics analysis to visualize or map the query sequence onto the chromosome region and perform similarity searches. A sequence extractor tool is also provided to extract target sequences for further analysis, such as for MIP primer development or related gene structure identification (Figure 4).
Figure 4.
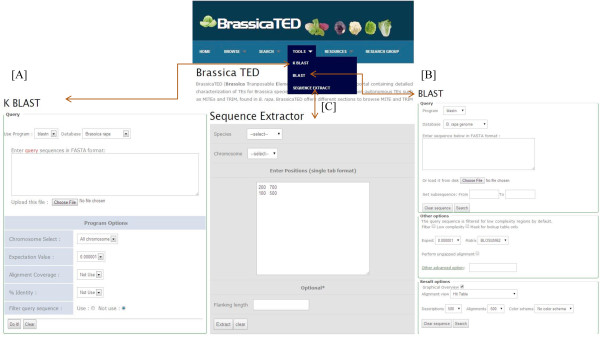
Display of tools panel. Display of K BLAST [A] and NCBI BLAST [B] and Sequence Extractor [C].
K BLAST
We developed a new tool, which we call K BLAST (Karyotype BLAST), to map the unique query sequence (s) onto homologous chromosomal regions and visualize the resulting distribution pattern. Users can visualize the locations of many different query sequences on each chromosome in different colors using K BLAST. It is a straightforward approach to inspect the distribution or dispersion of any DNA sequences (e.g. genes or repeats) on the chromosomes of B. rapa and B. oleracea. We have also included A. thaliana genome sequence for use in K BLAST.
BLAST
The tool panel offers the standard NCBI BLAST program for sequence similarity searches against the genome sequences. Users can match nucleotide or protein sequence (s) against database sequences such as whole-genome sequences of B. rapa, B. oleracea, and A. thaliana.
Sequence extract
We incorporated a sequence extractor tool using Python script. This can be utilized by entering single or multiple start and end positions in column tab-delimited format in the input query box for specific regions on the selected chromosome of B. rapa and B. oleracea. This tool also provides an optional input box to add flanking region length for input start and end positions.
Utility and discussion
TEs are important factors affecting genome size and genome evolution. Among those, miniature form TEs (mTEs) are valuable targets for genomics and phylogenetic studies. Comprehensive analysis and tools are desirable for effective utilization of mTEs for molecular studies, and were lacking. We developed BrassicaTED web as an interface for extensive characterization of MITEs, TRIM and SINEs in the Brassica genome. We will keep adding newly identified mTE elements to our database.BrassicaTED provides three sections (Browse, Search, and Tools) with user-friendly navigation. First, there are three parts of the Browse section: one each for MITEs, TRIMs and SINEs. Users can choose any MITE family in the list box and click ‘submit’ to enter the area pertaining to that family (Figure 2 [1-I]). This tab retrieves a summary of all MITE families included, such as MITE element classification, sequence information, structure, total members of a particular family with navigation to view the members and their closely related members, etc. (Figure 2 [1-O]). Similarly, TRIM and SINE family members can also be queried to obtain the family characterization by choosing any element from that family section. The Browse panel thus allows users to find summaries of complete information about families of MITEs, TRIMs and SINEs.In the Search panel, users can type the name of any family in the search box to find its members. Using the position drop-down option, users can filter the members returned by choosing the required genomic location (intergenic, CDS, intron, UTR) and clicking ‘Search’ for retrieval (Figure 2 [2-I]). This retrieves members of the selected family in each chromosome, their start and end positions on the chromosome, the length of the query alignment and e-value threshold, obtained from annotation (Figure 2 [2-O]). When ‘intergenic’ is chosen, the names of the two genes flanking the element are returned. When other options like CDS, intron, 5′UTR and 3′UTR are chosen, the member located in the corresponding genic regions is given with the gene name.
Brassica genomes contain triplicated homologous counterparts relative to the genome of A. thaliana[2,50]. Recent investigation of BraSto-2 suggested that MITEs may have important roles in triplicated Brassica genome evolution [30]. BrassicaTED also provides navigation tools to determine paralogous partners and microarray-based gene expression profiles of individual mTE-inserted genes (Figure 2 [2-I]). Transposition of MITEs, TRIM and SINEs in various genomic locations can alter the gene structure and also gene expression patterns [8,30,44]. mTEs associated with genes may cause gene silencing, sub-functionalization or neo-functionalization. The information regarding mTE position and annotation of genes harboring mTE insertion will be helpful for association studies and candidate gene approaches by comparison of their functional diversity among a range of accessions.
The Search panel also includes IP survey, which is a valuable source for genomics, genetic diversity studies, and evolutionary analysis [31,51,52]. Currently, there are about 18349 IP targets from 41 mTEs known in the Brassica genome and >50-85% of them are related to genic regions. The accumulated IP marker information from 289 MITE targets (Figure 2 [3-O]) will be effectively used for Brassica genomics studies. We surveyed 289 MIP targets that provided good genetic diversity based on presence or absence of a MITE in the target region among different accessions. MIP survey profiles in seven different Brassica derivatives were scored as 1, 2 and 3 for insertion as a full site, non-insertion as an empty site and both insertion and non-insertion as corresponding full and empty sites, respectively. The gel analysis has also been included as a link and users can view the gel profile with graphical images and accession information by clicking the link (Figure 3). This information will help researchers choose MIP primers for diversity studies using their own population. We will continue to update our database with more IP analysis of mTE targets. We also welcome other researchers to submit their data regarding mTEs of Brassica species.
The next sub-section of the Search panel is ‘Gene & Expression’. As a mesopolyploid species, the B. rapa genome has been estimated to contain 41,174 protein coding genes in triplicated form compared to the genome of A. thaliana[2]. Our study on BraMi-1 MITE insertion suggests that MITEs may modify the structure of one copy among such triplicated genes and eventually its expression pattern [8,30]. The ‘Gene & Expression’ analysis provides information regarding paralogous genes and 24 k microarray expression profiles for any gene of B. rapa. Under this sub-section, users can find differential expression patterns of mTE-inserted genes via comparison with expression of their paralogs. Users can obtain the available paralogous partner and gene sequences in FASTA format for any B. rapa gene by entering the gene ID (Figure 2 [4-I]). Clicking the ‘Expression’ link navigates to another page where the user can select expression profiles derived from four stress treatments: cold, salt, drought and ABA (Figure 2 [4-O]). The expression level of the selected genes will be shown with output as a graph or a table. This feature will be updated with more microarray datasets and can be used to analyze expression differences for any gene of interest.
In the Tools section using K BLAST, users can visualize the distribution of query sequences with user-defined specific options. This program contains options similar to NCBI BLAST but it has been updated with several new features (Figure 4 [A]). Users can perform K BLAST searches against all of the chromosomes or any particular chromosome of B. rapa, B. oleracea, or A. thaliana by selecting the chromosome number in the ‘Chromosome Select’ option. Furthermore, users can filter the output based on alignment coverage and sequence identity from each query by choosing a range between 60 and 100%. In K BLAST, up to 25 queries can be visualized on chromosome sequences simultaneously. An example shows the K BLAST output using three different mTE families (BraSto-3 TB-1, and SB-10) (Figure 5). The NCBI BLAST algorithm retrieves the matched database sequences in a flat file format, and even though it displays a graphical representation, it cannot show the overall positions precisely, unlike K BLAST. In addition, K BLAST offers options for filtering the query coverage and the percentage of sequence identity, which can be highly useful for research.BrassicaTED also offers a sequence extractor tool. With this tool, the user can select the chromosome number in the drop-down menu, input the start and end position(s) in the input text box, and click ‘Extract’ to retrieve the sequences. This returns an output of extracted sequences in FASTA format with defined positions. We have provided an example of the input position on the program as reference for the users (Figure 4 [C]). This tool will be useful for users wanting to extract target region(s) with flanking sequence for designing primers or further analyses.
Figure 5.
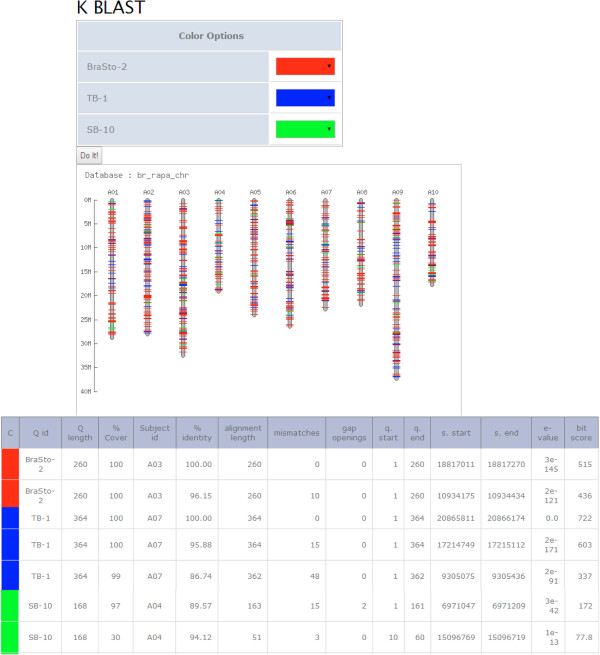
Output display of the K BLAST tool using three mTE families.
In sum, we have characterized members of 41 mTE families using the whole-genome pseudo-chromosome sequences of B. rapa and B. oleracea, which allowed us to develop a database to promote effective utilization of TEs. Previously developed databases for B. rapa such as BRAD mostly handle distribution and mining of whole-genome data [42]. Similarly, the B. oleracea genome database (Bolbase) [46] was designed to provide genomic data, which mainly includes whole genome sequence, annotation and synteny comparison with B. rapa and A. thaliana. Though recently reported database for plant MITEs, P-MITE, has data for MITE families from 41 plant species, it does not provide specific information regarding MITE insertion positions in the B. rapa and B. oleracea genomes. By contrast, BrassicaTED is built to establish an interactive web platform for all types of mTEs and includes in-depth analysis of 41 mTE families in B. rapa and B. oleracea, as well as in A. thaliana. In addition, BrassicaTED provides tools to extract gene sequences and compare the expression of paralogous genes using the 24 k microarray datasets.
Conclusion
BrassicaTED is a new database exclusively made with mTEs of Brassica species, especially from B. rapa and B. oleracea. This will be an important repository to promote the utilization of mTEs and the elucidation of the effects of mTEs on genome evolution in Brassica species. Unlike other publically available Brassica databases, BrassicaTED has a unique user-friendly visualization tool (K-BLAST) and a microarray expression data comparison tool for B. rapa. BrassicaTED will be a valuable storehouse for scientists and breeders who work on Brassica species and will be continuously updated as more data is uncovered.
Availability and requirements
Datasets in BrassicaTED are freely accessible for research purposes for non-profit and academic organizations at http://im-crop.snu.ac.kr/BrassicaTED/index.php. The database is optimized for Internet Explorer, Mozilla Firefox, Google Chrome and Safari.
Competing interests
The authors declare that they have no competing interests.
Author’s contributions
PS and TJY developed the methodology and conducted the study. JM, PS, SCL and BSC participated in the database and bioinformatics tool development. JM, PS, NS, SL and TJY drafted the manuscript, which was revised by all authors. All authors read and approved the final manuscript.
Contributor Information
Jayakodi Murukarthick, Email: murukarthick@snu.ac.kr.
Perumal Sampath, Email: sampathsnu@snu.ac.kr.
Sang Choon Lee, Email: sclee0923@snu.ac.kr.
Beom-Soon Choi, Email: bschoi@phyzen.com.
Natesan Senthil, Email: senthil_natesan@tnau.ac.in.
Shengyi Liu, Email: liusy@oilcrops.cn.
Tae-Jin Yang, Email: tjyang@snu.ac.kr.
Acknowledgments
This work was supported by Golden Seed Project (Center for Horticultural Seed Development, No. 309008-05-1-cg000), Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA) and Korea Forest Service (KFS), and a grant from the Next-Generation BioGreen 21 Program (No. PJ009076), Rural Development Administration, Republic of Korea. Perumal Sampath was supported by a Korean Government Scholarship (KGSP) 2008–2013 from the National Institute for International Education (NIIED), Ministry of Education, Science, and Technology, Republic of Korea.
References
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du F, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B. et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326(5956):1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, Huang S, Li X, Hua W, Wang J, Wang X, Freeling M, Pires JC, Paterson AH, Chalhoub B, Wang B, Hayward A, Sharpe AG, Park BS, Weisshaar B, Liu B, Li B, Liu B, Tong C, Song C, Duran C. et al. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011;43(10):1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Jiang N, Wessler SR. Plant transposable elements: where genetics meets genomics. Nat Rev Genet. 2002;3(5):329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell AJ, Pearce SR, Kumar A. Plant transposable elements and the genome. Curr Opin Genet Dev. 1994;4(6):838–844. doi: 10.1016/0959-437x(94)90068-x. [DOI] [PubMed] [Google Scholar]
- Lisch D. Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol. 2009;60:43–66. doi: 10.1146/annurev.arplant.59.032607.092744. [DOI] [PubMed] [Google Scholar]
- Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9(5):397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TJ, Kwon SJ, Choi BS, Kim JS, Jin M, Lim KB, Park JY, Kim JA, Lim MH, Kim HI, Lee HJ, Lim YP, Paterson AH, Park BS. Characterization of terminal-repeat retrotransposon in miniature (TRIM) in Brassica relatives. Theor Appl Genet. 2007;114(4):627–636. doi: 10.1007/s00122-006-0463-3. [DOI] [PubMed] [Google Scholar]
- Witte C-P, Le QH, Bureau T, Kumar A. Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc Natl Acad Sci U S A. 2001;98(24):13778–13783. doi: 10.1073/pnas.241341898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov DA, Vassetzky NS. Origin and evolution of SINEs in eukaryotic genomes. Heredity (Edinb) 2011;107(6):487–495. doi: 10.1038/hdy.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SJ, Kim DH, Lim MH, Long Y, Meng JL, Lim KB, Kim JA, Kim JS, Jin M, Kim HI, Ahn SN, Wessler SR, Yang TJ, Park BS. Terminal repeat retrotransposon in miniature (TRIM) as DNA markers in Brassica relatives. Mol Genet Genomics. 2007;278(4):361–370. doi: 10.1007/s00438-007-0249-6. [DOI] [PubMed] [Google Scholar]
- Kalendar R, Tanskanen J, Chang W, Antonius K, Sela H, Peleg O, Schulman AH. Cassandra retrotransposons carry independently transcribed 5S RNA. Proc Natl Acad Sci USA. 2008;105(15):5833–5838. doi: 10.1073/pnas.0709698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P, Yang TJ. Comparative analysis of Cassandra TRIMs in three Brassicaceae genomes. Plant Genet Resour: Characterization Util. 2014. in press.
- Kramerov DA, Vassetzky NS. Short retroposons in eukaryotic genomes. Int Rev Cytol. 2005;247:165–221. doi: 10.1016/S0074-7696(05)47004-7. [DOI] [PubMed] [Google Scholar]
- Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, Deragon JM, Westerman RP, Sanmiguel PJ, Bennetzen JL. Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 2009;5(11):e1000732. doi: 10.1371/journal.pgen.1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenke T, Dobel T, Sorensen TR, Junghans H, Weisshaar B, Schmidt T. Targeted identification of short interspersed nuclear element families shows their widespread existence and extreme heterogeneity in plant genomes. Plant Cell. 2011;23(9):3117–3128. doi: 10.1105/tpc.111.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David S, Yaakov B, Kashkush K. Genome-wide analysis of short interspersed nuclear elements SINES revealed high sequence conservation, gene association and retrotranspositional activity in wheat. Plant J. 2013;76(2):201–210. doi: 10.1111/tpj.12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov VN, Leem S-H, Solomon G, Mullokandov M, Chae J-Y, Yoon Y-H, Shin Y-S, Kouprina N, Larionov V. A novel strategy for analysis of gene homologues and segmental genome duplications. J Mol Evol. 2003;56(6):702–710. doi: 10.1007/s00239-002-2442-x. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3(5):370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Vassetzky NS, Kramerov DA. SINEBase: a database and tool for SINE analysis. Nucleic Acids Res. 2013;41(Database issue):D83–D89. doi: 10.1093/nar/gks1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G. MITE Digger, an efficient and accurate algorithm for genome wide discovery of miniature inverted repeat transposable elements. BMC bioinformatics. 2013;14:186. doi: 10.1186/1471-2105-14-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta JM, Santiago N. Plant LTR-retrotransposons and MITEs: control of transposition and impact on the evolution of plant genes and genomes. Gene. 2003;311:1–11. doi: 10.1016/s0378-1119(03)00557-2. [DOI] [PubMed] [Google Scholar]
- Lu C, Chen J, Zhang Y, Hu Q, Su W, Kuang H. Miniature inverted-repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol Biol Evol. 2012;29(3):1005–1017. doi: 10.1093/molbev/msr282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose M, Abe Y, Ozeki Y. Miniature inverted-repeat transposable elements of Stowaway are active in potato. Genetics. 2010;186(1):59–66. doi: 10.1534/genetics.110.117606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12(12):2473–2483. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, Okumoto Y, Tanisaka T, Wessler SR. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461(7267):1130–1134. doi: 10.1038/nature08479. [DOI] [PubMed] [Google Scholar]
- Fattash I, Rooke R, Wong A, Hui C, Luu T, Bhardwaj P, Yang G. Miniature inverted-repeat transposable elements: discovery, distribution, and activity1. Genome. 2013;56(9):475–486. doi: 10.1139/gen-2012-0174. [DOI] [PubMed] [Google Scholar]
- Piriyapongsa J, Jordan IK. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS One. 2007;2(2):e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K, Cho E, Yang G, Campbell MA, Yano K, Okumoto Y, Tanisaka T, Wessler SR. Dramatic amplification of a rice transposable element during recent domestication. Proc Natl Acad Sci USA. 2006;103(47):17620–17625. doi: 10.1073/pnas.0605421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P, Lee S-C, Lee J, Izzah NK, Choi B-S, Jin M, Park B-S, Yang TJ. Characterization of a new high copy Stowaway family MITE, BRAMI-1 in Brassica genome. BMC Plant Biol. 2013;13(1):56. doi: 10.1186/1471-2229-13-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monden Y, Naito K, Okumoto Y, Saito H, Oki N, Tsukiyama T, Ideta O, Nakazaki T, Wessler SR, Tanisaka T. High potential of a transposon mPing as a marker system in japonica x japonica cross in rice. DNA Res. 2009;16(2):131–140. doi: 10.1093/dnares/dsp004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatout C, Warwick S, Lenoir A, Deragon J-M. SINE insertions as clade markers for wild crucifer species. Mol Biol Evol. 1999;16(11):1614. [Google Scholar]
- Shedlock AM, Okada N. SINE insertions: powerful tools for molecular systematics. Bioessays. 2000;22(2):148–160. doi: 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Prieto J, Pouilly N, Jenczewski E, Deragon J, Chevre A. Development of crop-specific transposable element (SINE) markers for studying gene flow from oilseed rape to wild radish. Theor Appl Genet. 2005;111(3):446–455. doi: 10.1007/s00122-005-2017-5. [DOI] [PubMed] [Google Scholar]
- Al-Shehbaz I, Beilstein M, Kellogg E. Systematics and phylogeny of the Brassicaceae (Cruciferae): an overview. Plant Syst Evol. 2006;259(2–4):89–120. [Google Scholar]
- Kapitonov VV, Jurka J. Molecular paleontology of transposable elements from Arabidopsis thaliana. Genetica. 1999;107(1–3):27–37. [PubMed] [Google Scholar]
- Zhang X, Wessler SR. Genome-wide comparative analysis of the transposable elements in the related species Arabidopsis thaliana and Brassica oleracea. Proc Natl Acad Sci USA. 2004;101(15):5589–5594. doi: 10.1073/pnas.0401243101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Hu Q, Zhang Y, Lu C, Kuang H. P-MITE: a database for plant miniature inverted-repeat transposable elements. Nucleic Acids Res. 2014;42:D1176–D1181. doi: 10.1093/nar/gkt1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakodi M. A web accessible resource for investigating cassava phenomics and genomics information: BIOGEN BASE. Bioinformation. 2011;6(10):391. doi: 10.6026/97320630006391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murukarthick J, Senthil N, Raveendran M, Prabhakaran P. Biogen Base–An Interactive Maize Database for Phenomics Platform. Int J Comput Appl. 2011;2:56–61. [Google Scholar]
- Senthil N, Murukarthick J, Pandiyan M, Karthikeyan A, Sudha M, Raveendran M, Jayamani P, Kalaiselvi S, Nagarajan P. Article: Paithumbase-Biometrical Traits based Query System for Studying Mungbean [Vigna radiata (L.) Wilczek] Phenomics. Int J Appl. 2012;4:36–39. [Google Scholar]
- Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Li P, Hua W, Wang X. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 2011;11:136. doi: 10.1186/1471-2229-11-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Wessler SR. MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 2010;38(22):e199. doi: 10.1093/nar/gkq862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarilar V, Marmagne A, Brabant P, Joets J, Alix K. BraSto, a Stowaway MITE from Brassica: recently active copies preferentially accumulate in the gene space. Plant Mol Biol. 2011;77(1–2):59–75. doi: 10.1007/s11103-011-9794-9. [DOI] [PubMed] [Google Scholar]
- Sampath P, Murukarthick J, Izzah NK, Lee J, Choi HI, Shirasawa K, Choi BS, Liu S, Nou IS, Yang TJ. Genome-Wide Comparative Analysis of 20 Miniature Inverted-Repeat Transposable Element Families in Brassica rapa and B. oleracea. PLoS One. 2014;9(4):e94499. doi: 10.1371/journal.pone.0094499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zhao M, Wang X, Tong C, Huang S, Tehrim S, Liu Y, Hua W, Liu S. Bolbase: a comprehensive genomics database for Brassica oleracea. BMC Genomics. 2013;14(1):664. doi: 10.1186/1471-2164-14-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin I, Zhao M, Yu J, Huang S, Wang X, Yue Z, Li H, Yang L, Wu J, Zhou Q, Wang W, King GJ, Pires JC, Lu C, Wu Z, Sampath P, Wang Z, Guo H, Pan S, Yang L, Min J, Zhang D, Jin D, Li W, Belcram H. et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaakov B, Ceylan E, Domb K, Kashkush K. Marker utility of miniature inverted-repeat transposable elements for wheat biodiversity and evolution. Theor Appl Genet. 2012;124(7):1365–1373. doi: 10.1007/s00122-012-1793-y. [DOI] [PubMed] [Google Scholar]
- Lee SC, Lim MH, Kim JA, Lee SI, Kim JS, Jin M, Kwon SJ, Mun JH, Kim YK, Kim HU. Transcriptome analysis in Brassica rapa under the abiotic stresses using Brassica 24 K oligo microarray. Mol Cells. 2008;26(6):595–605. [PubMed] [Google Scholar]
- Yang TJ, Kim JS, Kwon SJ, Lim KB, Choi BS, Kim JA, Jin M, Park JY, Lim MH, Kim HI, Lim YP, Kang JJ, Hong JH, Kim CB, Bhak J, Bancroft I, Park BS. Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell. 2006;18(6):1339–1347. doi: 10.1105/tpc.105.040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casa AM, Brouwer C, Nagel A, Wang L, Zhang Q, Kresovich S, Wessler SR. The MITE family heartbreaker (Hbr): molecular markers in maize. Proc Natl Acad Sci USA. 2000;97(18):10083–10089. doi: 10.1073/pnas.97.18.10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons M, Cardle L, Rostoks N, Waugh R, Flavell AJ. Isolation, analysis and marker utility of novel miniature inverted repeat transposable elements from the barley genome. Mol Genet Genomics. 2008;280(4):275–285. doi: 10.1007/s00438-008-0363-0. [DOI] [PubMed] [Google Scholar]


