Abstract
What if there was a rapid, inexpensive, and accurate blood diagnostic that could determine which patients were infected, identify the organism(s) responsible, and identify patients who were not responding to therapy? We hypothesized that systems analysis of the transcriptional activity of circulating immune effector cells could be used to identify conserved elements in the host response to systemic inflammation, and furthermore, to discriminate between sterile and infectious etiologies. We review herein a validated, systems biology approach demonstrating that 1) abdominal and pulmonary sepsis diagnoses can be made in mouse models using microarray (RNA) data from circulating blood, 2) blood microarray data can be used to differentiate between the host response to Gram-negative and Gram-positive pneumonia, 3) the endotoxin response of normal human volunteers can be mapped at the level of gene expression, and 4) a similar strategy can be used in the critically ill to follow septic patients and quantitatively determine immune recovery. These findings provide the foundation of immune cartography and demonstrate the potential of this approach for rapidly diagnosing sepsis and identifying pathogens. Further, our data suggest a new approach to determine how specific pathogens perturb the physiology of circulating leukocytes in a cell-specific manner. Large, prospective clinical trails are needed to validate the clinical utility of leukocyte RNA diagnostics (e.g., the riboleukogram).
Keywords: microarray, genomics, diagnostics, critical care, interactome
As described elsewhere in this issue, a major hurdle for clinical trials in the critically ill or injured is the difficulty of accurate and timely diagnosis, coupled with the difficulties of predicting and monitoring the response to therapy. There are a host of probabilistic tools available (e.g., Acute Physiology and Chronic Health Evaluation III) based on clinical examination and physiologic parameters. These tools perform well when defining the outcome of groups of patients at the time of admission, but as reported in numerous studies, are less reliable at defining or classifying individuals. What the current tools lack is resolution at the level of the individual, a measure of heritable predisposition to a given clinical trajectory, and an account of the individual’s response to therapy. Today, it is possible to both evaluate an individual’s genetic susceptibility to disease and measure their physiologic response to therapy using multiplexed molecular assays. The biological assumptions underlying these efforts are that 1) predisposition is determined to a greater or lesser degree by our genetics and 2) the host response can be measured and/or predicted and/or individualized at the molecular level. The promise of molecular monitoring is that it will be possible to get preliminary data as patients are admitted, and with a few repeated measures, answer the questions “what is going on?” and “how are they going to do?” (1–5).
In recent articles, we proposed an investigative strategy—functional genomics coupled with systems biology—for applying genome-wide technology to the study of critical illness and injury (4 – 6). This strategy proposes using information at the genome (DNA) level to discover predisposition to a given outcome, and uses data at the transcriptome (RNA) and proteome (protein) levels to make diagnoses and gauge the response to therapy (prognoses). These data in turn can be linked to physiologic data and clinical parameters using systems approaches to mathematically describe the host response to critical illness or injury. Associated with these high-throughput genomic technologies are a number of theoretical and technical challenges that have delayed their widespread implementation in the clinical setting. These include: 1) the requirement for investigators with diverse skill sets to develop effective communication and methodologic strategies, 2) the accumulation of sufficient technical expertise and experience to generate high-quality data, and 3) the development and application of data storage and analysis tools (4 – 6). Given the complexities of these interactions, the relatively meager resources available to individual investigators, and the need to share experiences and build collaborations, there exists a need for focused scientific discussions apart from mainstream critical care and trauma meetings. Two opportunities for intensivists to explore systems biology are the National Institutes of Health annual Functional Genomics of Critical Illness and Injury Symposium and the Society for Complexity in Acute Illness.
The ongoing challenge of accurately diagnosing infection in the intensive care unit— differentiating sterile from infectious causes of systemic inflammation—motivates a search for molecular diagnostics (7). Recent advances in genome sequencing, robotics, and miniaturization have significantly expanded the search for biomarkers; a number of strategies (both new and old) are being evaluated (7–9). The human tissue that is most easily accessible for longitudinal profiling is peripheral (circulating) blood. To this end, we have been testing a systems approach to develop a novel strategy for blood immunomonitoring (4, 5), what we now call immune cartography. An early example, the riboleukogram (10), is described herein. The hypothesis is that changes in circulating leukocyte RNA can be used to quantitatively determine the inflammatory response and thereby improve sepsis diagnostics and prognostics. We also expect that these data will inform dynamic models of the host response and identify functional modules and gene targets for further study.
In Vitro and Animal Studies
There is substantial preclinical data supporting RNA diagnostics in critical illness and the use of systems approaches to identify new gene targets. In 2001, studies using cultured human cells suggested that instead of a single marker (e.g., interleukin-6), a suite of molecular markers could be used to better describe the cellular response to inflammatory stimuli. The authors concluded that human leukocytes in vitro alter RNA transcriptional profiles in response to diverse types of pathogens, including bacteria, fungi, and yeast. Importantly, the leukocyte responses observed exhibited both generic and pathogenic-specific responses to these agents (11–13). Thus, depending on the question to be answered, leukocyte expression profiles can be queried to search for generic changes in response to diverse agents, pathogen-specific responses, or both (14).
The in vitro reports described above suggested that genome-wide profiling of transcription holds promise as a molecular diagnostic tool, capable of generating profiles from leukocytes that are sensitive, specific, and timely for pathogen detection (6). We hypothesized that leukocyte gene expression profiles obtained using DNA microarrays could be used to predict septic states; in particular, distinguishing between sterile and infectious sources of systemic inflammation, a common conundrum in caring for the critically ill or injured (15). We tested this hypothesis in an in vivo model, subjecting C57BL/6 male mice to cecal ligation and puncture or to intraperitoneal lipopolysaccharide. Control mice had sham laparotomy or injection of intraperitoneal saline, respectively. A classification model was developed and tested on blood samples from septic mice (15, 16). Classifiers were constructed using data from a training data set of 26 Affymetrix GeneChip microarrays (Santa Clara, CA). The error rate of the classifiers was estimated on seven deidentified microarrays, and then on a subsequent cross-validation for all 33 blood microarrays. All seven of the de-identified microarrays (100%) were correctly classified. Considering all 33 microarrays, nested cross-validation estimates of classification accuracy of diagnosing sepsis from mouse blood was 94.4%. We concluded that sepsis induces changes in mouse blood gene expression that can be used to diagnose sepsis apart from noninfectious causes of systemic inflammation (15). Lists of genes with significant changes in expression between study and control groups were used to identify nine mouse common response genes for peritonitis, six of which were mapped into a single network using contemporary pathway analysis tools. Given that this list of nine genes was based on changes in relative RNA abundance across a number of cell types, the network analysis performed served as an exploratory tool, validating in silico the role of six of the nine genes in canonical pathways for inflammation, apoptosis, and signal transduction: inhibitor of DNA binding 2, calgranulin A and B (S100A8 and S100A9), interferon regulatory factor 7, lipocalin 2, and formyl peptide receptor-like 1.
In the follow-up study, we hypothesized that the circulating leukocyte response to infection could not only differentiate between infected and noninfected states but could also be used to differentiate between the host response to infectious agents, and to mathematically represent the host response to infectious perturbations (10). We used a translational research paradigm wherein mouse data indicated a novel clinical strategy to apply at the bedside. Murine peripheral blood leukocyte transcriptional responses at 24 hrs were examined to identify genes that could distinguish between different, clinically relevant insults: pneumococcal pneumonia, Pseudomonas pneumonia, and Pseudomonas lipopolysaccharide pneumonitis. We then used those genes to test whether there was a conserved transcriptional response to pulmonary infectious challenge. Lastly, we determined whether the human orthologs to these murine genes were informative with regard to the onset of infection in critically ill patients (see “Clinical Studies” section). Our results from this single time point mouse model demonstrated that 219 probe sets from mouse buffy coat reliably differentiated between the host response to these prototypical Gram-negative and Gram-positive insults. Leaving out one cross-validation resulted in an estimated classification accuracy of 93%. This result suggested that the transcriptional activity of buffy coat may be diagnostic not only for the onset of infection but also for the type of bacterial pathogen.
To complement the system classification tools described above, we also explored the use of network analysis to identify regulatory nodes and new gene targets of interest to sepsis investigators (“molecular cartography”) (17). For example, mouse gene expression profiles were used to examine the mechanisms responsible for the beneficial effect of bcl-2 overexpression on outcome from sepsis. We reported that splenocyte gene interaction network analysis implicated bim as a key player (node) in the apoptosis module, responsible in part for the beneficial effect of bcl-2 on survival in both pneumonia and cecal ligation and puncture models of sepsis (18). Subsequent study of targeted gene deletion of bim by our colleague Dr. Richard Hotchkiss confirmed its central role in the mouse sepsis model, as bim knock-outs experienced significantly less apoptosis and improved sepsis survival (19).
Collectively, these blood and spleen gene expression data from mouse models of sepsis corroborate early reports from in vitro leukocyte studies, demonstrating that microarray gene expression profiles are exquisitely sensitive tools that can be used to identify both generic and pathogen-specific host responses. Furthermore, these reports provide proof-of-feasibility that systematic analysis of changes in leukocyte RNA abundance can classify systemic inflammatory states and identify new gene targets. A major limitation of these single time point studies, however, is their failure to provide a context within which to identify the (mal)adaptive transitions in the host response that occur during the onset, progression, and resolution of disease. The human studies described below address this limitation and explore the potential of genomics and systems biology for clinical diagnostics and prognostics in the intensive care unit.
Normal Volunteer Studies
The systemic response to critical illness or injury has been characterized by an early inflammatory phase followed by a compensatory anti-inflammatory phase, based on reports from both patients and animal models (20, 21). This suggests that time series data are required to best model (and thereby understand) critically ill states. It has been postulated that the relative magnitude and duration of these phases (whether secondary to trauma, sepsis, cardiogenic shock, etc.) will determine whether the patient develops organ dysfunction, influencing the likelihood of subsequent complications and recovery (4, 22, 23). “Mixed” inflammatory states (components of both proinflammatory and anti-inflammatory phases) have also been recognized, adding to the difficulty of classifying the host response (4). Thus, the ability to accurately and rapidly monitor the dynamics of the host immunoinflammatory response has been an explicit goal of shock researchers. The clinical relevance of these efforts is indicated by recent reports demonstrating that more accurate immune classification, including an estimate of the risk of death, would likely improve the efficacy of anti-inflammatory therapies (24, 25).
The Inflammation and Host Response to Injury Program is a large-scale collaborative research grant funded by the NIH to explain differences in the host response to blunt trauma and burn injury. Before embarking on patient studies, normal human volunteers were used to develop blood sampling and processing protocols for microarray analysis. In a collaborative project, we discovered that gene expression profiling results can be confounded by differences in blood processing protocols, but, when standardized, can provide highly reproducible data that are informative with regard to human health and disease (6, 26). We hypothesized that circulating leukocyte gene expression profiles over time could be used first to mathematically quantify the human response to a prototypical inflammatory stimulus, and then to apply such an approach to the clinical setting. Intravenous endotoxin was used to validate this approach in a preclinical model and to study the dynamics of the systemic inflammatory response (27). As reported previously by Calvano et al, endotoxemia in normal volunteers produced a mild, reproducible, self-limited inflammatory state with flu-like symptoms (28, 29). Four normal subjects were treated with a reference dose of endotoxin; four additional normal subjects were studied similarly after injection of intravenous normal saline (placebo control). Using the protocols described above, whole genome expression profiling was performed on circulating leukocytes at 0, 2, 4, 6, 9, and 24 hrs after intravenous challenge. Two previous reports described the data mining methods applied to this dataset, including a network-based analysis of informational gene lists (27, 30). The results provided a global view of innate immune system tolerance and new insight into the dysregulation of leukocyte energy functional modules and translational machinery.
In a complementary analysis, we tested the ability of these microarray data (27) to classify the responses of the volunteers, providing information that potentially would be useful in a clinical setting (Fig. 1). In doing so, a number of computational challenges were encountered typical of applying genomics to clinical studies (6), including noisy measurements, stochastic data, low time resolution, few replicates, and large number of data elements (genes). The computational methods used were reported recently (10). To better visualize temporal changes in circulating leukocyte RNA abundance, the data were projected onto a smaller dimensional space using a series expansion method, Karhunen-Loeve Decomposition, a variant of principal components analysis. A successive decomposition was performed that easily distinguished between temporal profiles of the host response to the two treatments, providing a novel strategy to classify systemic inflammatory states.
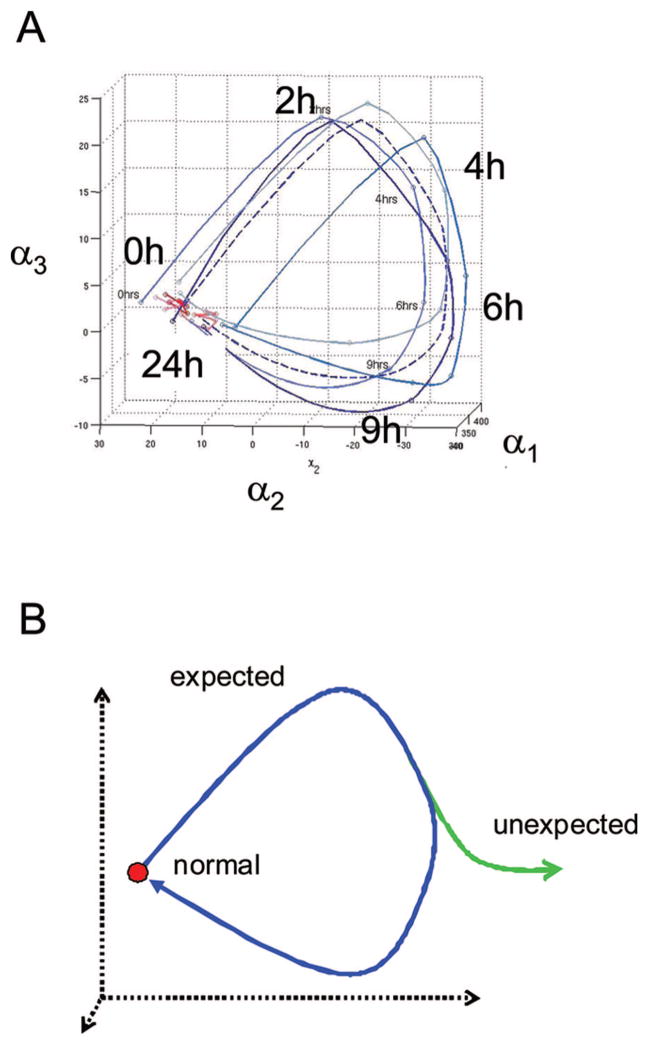
Figure 1.
Immune cartography of systemic inflammation. A, Previously reported circulating leukocyte gene expression values for 5150 probe sets were used to map immune trajectories in three dimensions for normal volunteers challenged with either intravenous endotoxin or normal saline (27). An interpolation scheme was used to accommodate the uneven and sparse sampling of leukocyte expression values over time; the methods are described elsewhere (10). Evident in endotoxin-treated subjects (blue curves) is a cyclic response, coincident with the transition in physiologic states from normalcy (t = 0 hrs), to symptomatic systemic inflammation (t = 2–9 hrs) and recovery (t = 24 hrs). These paths are easily distinguished from those of the subjects treated with saline (red curves). Similarly, the variance of trajectories over time reflects differences between the endotoxin- and saline-treated subjects. Intersubject differences likely contribute to the group variances observed. B, A hypothetical aggregate response is plotted to describe the “expected” systemic inflammatory response (blue curve), which begins at and returns to a normal (homeostatic) state. “Unexpected” or abnormal responses are anticipated to deviate from the expected path at bifurcation points (green curve). A return to the expected path (healing) would be achieved with successful therapy. We submit that immune cartography is thereby well suited for use as a monitoring tool.
Clinical Studies
Given the goal of creating dynamic models of systemic inflammation for clinical use (4, 5), we began by applying this methodology to data collected from mechanically ventilated critically ill patients, as reported recently (10). Specifically, we studied the temporal behavior of time series DNA microarray data from circulating leukocytes in 27 patients before, during, and after ventilator-associated pneumonia (VAP). This study was motivated, in part, by our interest in exploring the translational research paradigm, bringing to the bedside the mouse pneumonia data described above. Initially, we examined the behavior of the human orthologs to the 219 genes identified in the mouse pneumonia study (10). Similar to the observations in Figure 1, we found that the onset of systemic inflammation (in this case due to bacterial pneumonia in patients) coincided with translation along the axis of a principal component. Importantly, this translation ceased after 5– 6 days, coincident with a clinical response to appropriate antibiotic therapy. Of interest, the abundance of 20 plasma cytokines (including procalcitonin) measured in the same patient samples generated a comparable trajectory using principal components analysis. However, in line with previous reports, the variance (“noise”) in plasma cytokine abundance either examined individually or collectively was large enough to prevent its use as a VAP diagnostic (10).
In contrast, when the leukocyte genes were selected in patients by explicitly accounting for time, a set of 85 genes were identified whose microarray expression levels changed consistently across all patients around the time of VAP diagnosis (10). In addition, two other aspects of this analysis were noteworthy. Testing for the effect of covariates on gene expression (including age, gender, and type of bacteria) identified ethnic background as having the largest impact, as measured by the number of genes altered in response to VAP (~2700 genes or 32%). These findings are strikingly similar to those reported based on variance in gene expression in cultured cells derived from different ethnic groups (25% of the genes studied in a recent report) (31). As the sepsis mortality rate is highest for African American males, and the reasons for this difference remain obscure (32), further study is indicated to gain molecular insight into health disparities. These studies may also provide important new insight into genetic predisposition for sepsis. Finally, we discovered that the variance in leukocyte gene expression as visualized using principal components analysis decreased significantly as the patients recovered from VAP and critical illness. In phase space analysis, the patient-specific trajectories seemed to converge, consistent with return to homeostasis (what physicists would call an attractor state, Fig. 2). This property also can be observed in the data from normal volunteers treated with endotoxin (Fig. 1), as those trajectories reflect health (t = 0 hrs) then systemic inflammation (t = 4 –9 hrs) then a return back to baseline (t = 24 hrs). In summary, circulating leukocyte gene expression profiles can map the dynamics of the human systemic inflammatory response generated by both infectious (VAP) and noninfectious (lipopolysaccharide) insults.
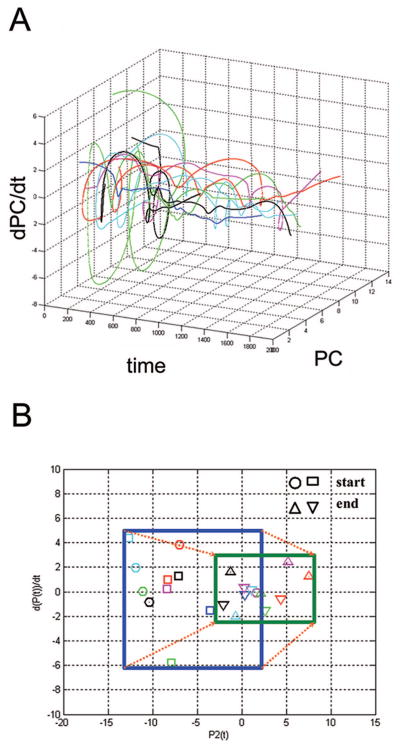
Figure 2.
Riboleukograms of critically ill and injured patients. As reported recently, application of immune cartography in the clinical setting provides dynamic, RNA-based graphs of the leukocyte response, which we term riboleukograms (10). A, Phase space analysis of circulating leukocyte expression profiles for human genes informational for ventilator-associated pneumonia complicating recovery. All 11 patients were treated with the appropriate antibiotic(s) for their infecting pathogen and all patients were discharged from the intensive care unit. Individual patient trajectories start at various points in space and marked heterogeneity is observed, but all of the riboleukograms converge as they move to the right to a common region of the graph. Furthermore, these data suggest that a trajectory specific for each patient (riboleukogram) could be plotted daily based upon leukocyte RNA data, and the distance from the attractor could be measured indicating whether the patient was healing or not (“genomic vital signs”). B, Starting points (critically ill, blue box) and ending points (recovery, green box) of riboleukograms for the same 11 patients. All paths move to the right from a larger (blue) to a smaller (green) box. This behavior mimics that of an attractor in the physical sciences, consistent with critical illness complicated by ventilator-associated pneumonia perturbing a stable immune state that returns to homeostasis. We submit that this general method can be used to characterize, monitor, and ultimately recognize infection in patients in whom the diagnosis of sepsis is particularly difficult to make (10).
We coined the term riboleukogram to refer to these dynamic maps of leukocyte gene expression, in essence, an electrocardiogram for the immune system (10). Riboleukograms reflect the plasticity of immune responsiveness and suggest the existence of an immune attractor state. They may also provide a better understanding of immune health disparities based on genotype (10, 32).
Confirmatory findings in patients have been reported recently by others. For example, Ramilo et al (33) used blood gene expression analysis to develop discriminative transcriptional signatures as novel RNA diagnostics for acute infections in 138 children. Microarray patterns were observed to differentiate between four common pathogens—influenza A virus, Escherichia. coli, Staphylococcus aureus, and Streptococcus pneumoniae—with a diagnostic accuracy of 85%–95%. Similarly, in 90 trauma patients with systemic inflammatory response syndrome, Johnson et al (34) were able to discriminate between those who recovered with or without sepsis. Gene annotation and pathway analysis tools were used in that study to identify overrepresented modules associated with leukocyte RNA abundance in septic trauma patients: innate immunity, cytokine receptors, T-helper cell differentiation, and protein synthesis. Finally, Tang et al (35) tested whether circulating neutrophil-specific gene expression profiles could identify candidate genes in sepsis, and, in the process, found a profile capable of diagnosing sepsis in 94 critically ill patients. They described an expression signature of 50 genes that identified sepsis with an adjusted prediction accuracy of 82% in the validation cohort. This molecular signature of sepsis was associated with functional modules of inflammation, immune regulation, and mitochondrial function, a conclusion similar to that reported based on data from endotoxin-treated normal volunteers (27).
All four of these clinical studies are limited by small sample size and they differ with regard to sampling protocols, technology platforms, statistical methods, and the lists of informational genes identified. Nevertheless, these reports provide convincing evidence supporting the development of circulating leukocyte RNA profiles as novel sepsis diagnostics in both children and adults. These transcriptional signatures also provide important insight into the host response in a pathogen-specific and leukocyte-specific manner.
Future Directions
The clinical findings above extend our earlier mouse and normal volunteer studies, demonstrating that systems biologists can use blood leukocyte transcriptional profiles to map the dynamics of the host response and differentiate septic from sterile sources of systemic inflammation (15). The overwhelming complexity of thousands of gene interactions among thousands of leukocyte contacts is thereby reduced to clinical trajectories that can be quantified and mapped. We suspect that immune cartography will provide important new biological insight into how the host balances “proinflammatory” and “anti-inflammatory” influences during injury and recovery, and how the concept of “immune paralysis” might be quantitatively defined (4, 10). Furthermore, we anticipate this approach will provide for a number of important innovations as “genomic vital signs” (5), including: 1) dynamic RNA profiles that can help distinguish between the host responses to various types of infecting organism, 2) robust, patient-specific computational models of the response to systemic inflammation and sepsis, and 3) phase space models of recovery to an immunologic attractor. Figures 1 and 2 provide examples of early prototypes (e.g., riboleukograms).
Before we get to that point, however, there are a number of technical, computational, and experimental hurdles to clear (6, 10, 36 –38), the most important of which is lack of standardization. This is as big an issue for the clinicians as it is for the experimentalists and theorists. How can one verify the clinical utility of new technology if systemic inflammatory states cannot be diagnosed with certainty? A few false-positive and false-negative findings in a small study will ensure a negative result when testing clinical utility. The syndromic nature of systemic inflammatory states, thus, is the motivation for and the challenge of validating new sepsis diagnostics. Demographic differences in patient cohorts across studies contribute an additional level of complexity, consistent with our data and those of others indicating that ethnic background, age, and gender substantially increase the variance in the gene expression signal observed (10, 31). This variance helps explain, in part, why there is so little overlap among the lists of genes and transcriptional patterns described above as diagnostic for sepsis. Although this may at first seem alarming, a similar lack of concordance was observed in early cancer studies touting transcriptional signatures as novel diagnostics and prognostics (39). Nevertheless, with careful attention to sound clinical trial design, biomarker verification, and rigorous validation, novel gene expression diagnostics can be developed (37, 38). Two of the first to receive regulatory (Food and Drug Administration) approval are aimed at the prognosis of breast cancer and the diagnosis of heart transplant rejection (40, 41).
The experiences above in the field of cancer emphasize the importance of multidisciplinary teams that integrate knowledge of rapidly evolving technology, new computational approaches, advances in the basic sciences, and improvements in patient care and trial design. For instance, the challenging nature of modeling patient responses requires that computational strategies be optimized for noisy, under-determined data. In addition, recent reports [including that by Tang et al (35)] suggest that additional molecular insight and diagnostic information will be provided by sampling protocols and technological advances that provide rapid, leukocyte-specific, gene expression profiles (35, 42– 44). How will this information be integrated into the development of current diagnostic technologies in the pipeline, many of which are based on reports of gene expression profiles from whole blood or buffy coat? (26). We also might find (again mirroring the experience of oncologists) that leukocyte gene expression profiles lead to the discovery of patterns that are clinically obscure but nevertheless have important treatment implications (45). Although these class discovery efforts would be expected to provide vital new information of use for sepsis clinical trials and drug testing, the large patient sample sizes necessary would likely take years to accrue. Scientific reviewers of clinical grants and manuscripts, therefore, might best prepare for a decade of “descriptive science” on how best to apply molecular profiling and immune cartography at the bedside. Progress in this new era of translational research and personalized medicine will depend on it.
Acknowledgments
This work was supported, in part, by NIH R21GM075023, NIH U54GM62119, NIH T32GM08795, and the Barnes-Jewish Hospital Foundation Research Award. Dr. McDunn has received an honoraria from Pfizer, Inc.
We thank our colleagues in the Washington University Center for Critical Illness and Health Engineering and the National Institute of General Medical Sciences Inflammation and Host Response to Injury Program for collaborative support.
Footnotes
The remaining authors have not disclosed any potential conflicts of interest.
Some of these data were reported in abstract form at the 2005 Critical Care Congress of the Society of Critical Care Medicine (Phoenix, AZ). For that presentation, Dr. Polpitiya received one of the Society’s Annual Scientific Awards (2005).
For information regarding this article, cobb@wustl.edu
References
- 1.Cobb JP, Brownstein BH, Watson MA, et al. Injury in the era of genomics. Shock. 2001;15:165–170. doi: 10.1097/00024382-200115030-00001. [DOI] [PubMed] [Google Scholar]
- 2.Chung TP, Laramie JM, Province M, et al. Functional genomics of critical illness and injury. Crit Care Med. 2002;30(Suppl 1):S51–S57. [PubMed] [Google Scholar]
- 3.Hopf HW. Molecular diagnostics of injury and repair responses in critical illness: What is the future of “monitoring” in the intensive care unit? Crit Care Med. 2003;31:S518–S523. doi: 10.1097/01.CCM.0000081433.98328.4B. [DOI] [PubMed] [Google Scholar]
- 4.Cobb JP, O’Keefe GE. Injury research in the genomic era. Lancet. 2004;363:2076–2083. doi: 10.1016/S0140-6736(04)16460-X. [DOI] [PubMed] [Google Scholar]
- 5.McDunn JE, Chung TP, Laramie JM, et al. Physiological genomics. Surgery. 2006;139:133–139. doi: 10.1016/j.surg.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Cobb JP, Mindrinos M, Miller-Graziano C, et al. Application of genome-wide expression analysis to human health and disease. Proc Natl Acad Sci U S A. 2005;102:4801–4806. doi: 10.1073/pnas.0409768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy MM, Fink MP, Marshall JC, et al. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit Care Med. 2003;31:1250–1256. doi: 10.1097/01.CCM.0000050454.01978.3B. [DOI] [PubMed] [Google Scholar]
- 8.Gibot S, Cravoisy A, Levy B, et al. Soluble triggering receptor expressed on myeloid cells and the diagnosis of pneumonia. N Engl J Med. 2004;350:451–458. doi: 10.1056/NEJMoa031544. [DOI] [PubMed] [Google Scholar]
- 9.Christ-Crain M, Jaccard-Stolz D, Bingisser R, et al. Effect of procalcitonin-guided treatment on antibiotic use and outcome in lower respiratory tract infections: Cluster-randomised, single-blinded intervention trial. Lancet. 2004;363:600–607. doi: 10.1016/S0140-6736(04)15591-8. [DOI] [PubMed] [Google Scholar]
- 10.McDunn JE, Husain KD, Polpitiya AD, et al. Plasticity of the systemic inflammatory response to acute infection during critical illness: Development of the riboleukogram. PLoS ONE. 2008;3:e1564. doi: 10.1371/journal.pone.0001564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Q, Liu D, Majewski P, et al. The plasticity of dendritic cell responses to pathogens and their components. Science. 2001;294:870–875. doi: 10.1126/science.294.5543.870. [DOI] [PubMed] [Google Scholar]
- 12.Nau GJ, Richmond JF, Schlesinger A, et al. Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci U S A. 2002;99:1503–1508. doi: 10.1073/pnas.022649799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feezor RJ, Oberholzer C, Baker HV, et al. Molecular characterization of the acute inflammatory response to infections with gram-negative versus gram-positive bacteria. Infect Immun. 2003;71:5803–5813. doi: 10.1128/IAI.71.10.5803-5813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- 15.Chung TP, Laramie JM, Meyer DJ, et al. Molecular diagnostics in sepsis: From bedside to bench. J Am Coll Surg. 2006;203:585–598. doi: 10.1016/j.jamcollsurg.2006.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao R, Wang X, Spitznagel EL, et al. Primary and secondary transcriptional effects in the developing human Down Syndrome brain and heart. Genome Biol. 2005;6:R107. doi: 10.1186/gb-2005-6-13-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guimera R, Mossa S, Turtschi A, et al. The worldwide air transportation network: Anomalous centrality, community structure, and cities’ global roles. Proc Natl Acad Sci U S A. 2005;102:7794–7799. doi: 10.1073/pnas.0407994102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner TH, Drewry AM, MacMillan SK, et al. Surviving sepsis: bcl-2 overexpression modulates splenocyte transcriptional responses in vivo. Am J Physiol Regul Integr Comp Physiol. 2007;292:R1751–R1759. doi: 10.1152/ajpregu.00656.2006. [DOI] [PubMed] [Google Scholar]
- 19.Chang KC, Unsinger J, Davis CG, et al. Multiple triggers of cell death in sepsis: Death receptor and mitochondrial-mediated apoptosis. FASEB J. 2007;21:708–719. doi: 10.1096/fj.06-6805com. [DOI] [PubMed] [Google Scholar]
- 20.Bone RC, Balk RA, Cerra FB, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644–1655. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 21.Bone RC. Sir Isaac Newton, sepsis, SIRS, and CARS. Crit Care Med. 1996;24:1125–1128. doi: 10.1097/00003246-199607000-00010. [DOI] [PubMed] [Google Scholar]
- 22.Hochman JS. Cardiogenic shock complicating acute myocardial infarction: Expanding the paradigm. Circulation. 2003;107:2998–3002. doi: 10.1161/01.CIR.0000075927.67673.F2. [DOI] [PubMed] [Google Scholar]
- 23.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348:138–150. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 24.Eichacker PQ, Parent C, Kalil A, et al. Risk and the efficacy of antiinflammatory agents: Retrospective and confirmatory studies of sepsis. Am J Respir Crit Care Med. 2002;166:1197–1205. doi: 10.1164/rccm.200204-302OC. [DOI] [PubMed] [Google Scholar]
- 25.Minneci PC, Deans KJ, Banks SM, et al. Should we continue to target the platelet-activating factor pathway in septic patients? Crit Care Med. 2004;32:585–588. doi: 10.1097/01.CCM.0000110730.38696.9C. [DOI] [PubMed] [Google Scholar]
- 26.Feezor RJ, Baker HV, Mindrinos M, et al. Whole blood and leukocyte RNA Isolation for gene expression analyses. Physiol Genomics. 2004;19:247–254. doi: 10.1152/physiolgenomics.00020.2004. [DOI] [PubMed] [Google Scholar]
- 27.Calvano SE, Xiao W, Richards DR, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 28.Suffredini AF, Fromm RE, Parker MM, et al. The cardiovascular response of normal humans to the administration of endotoxin. N Engl J Med. 1989;321:280–287. doi: 10.1056/NEJM198908033210503. [DOI] [PubMed] [Google Scholar]
- 29.Richardson RP, Rhyne CD, Fong Y, et al. Peripheral blood leukocyte kinetics following in vivo lipopolysaccharide (LPS) administration to normal human subjects. Influence of elicited hormones and cytokines. Ann Surg. 1989;210:239–245. doi: 10.1097/00000658-198908000-00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Storey JD, Leek JT, Xiao W, et al. A significance method for time course microarray experiments applied to two human studies. Proc Natl Acad Sci U S A. 2005;102:12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spielman RS, Bastone LA, Burdick JT, et al. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet. 2007;39:226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin GS, Mannino DM, Eaton S, et al. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med. 2003;348:1546–1554. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- 33.Ramilo O, Allman W, Chung W, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson SB, Lissauer M, Bochicchio GV, et al. Gene expression profiles differentiate between sterile SIRS and early sepsis. Ann Surg. 2007;245:611–621. doi: 10.1097/01.sla.0000251619.10648.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang BM, McLean AS, Dawes IW, et al. The use of gene-expression profiling to identify candidate genes in human sepsis. Am J Respir Crit Care Med. 2007;176:676–684. doi: 10.1164/rccm.200612-1819OC. [DOI] [PubMed] [Google Scholar]
- 36.Yeang CH, Ramaswamy S, Tamayo P, et al. Molecular classification of multiple tumor types. Bioinformatics. 2001;17(Suppl 1):S316–S322. doi: 10.1093/bioinformatics/17.suppl_1.s316. [DOI] [PubMed] [Google Scholar]
- 37.Manolio T. Novel risk markers and clinical practice. N Engl J Med. 2003;349:1587–1589. doi: 10.1056/NEJMp038136. [DOI] [PubMed] [Google Scholar]
- 38.Ioannidis JP. Microarrays and molecular research: Noise discovery? Lancet. 2005;365:454–455. doi: 10.1016/S0140-6736(05)17878-7. [DOI] [PubMed] [Google Scholar]
- 39.Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays: A multiple random validation strategy. Lancet. 2005;365:488–492. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- 40.Couzin J. Amid debate, gene-based cancer test approved. Science. 2007;315:924. doi: 10.1126/science.315.5814.924. [DOI] [PubMed] [Google Scholar]
- 41.Salisbury MW. The heart of the matter. [Accessed November 17, 2008];Genome Technol. 2007 47 Available at: http://www.genome-technology.com/issues/2_2/intheclinic/138721-1.html. [Google Scholar]
- 42.Laudanski K, Miller-Graziano C, Xiao W, et al. Cell-specific expression and pathway analyses reveal novel alterations in trauma-related human T-cell and monocyte pathways. Proc Natl Acad Sci U S A. 2006;103:15564–15569. doi: 10.1073/pnas.0607028103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDunn JE, Turnbull IR, Polpitiya AD, et al. Splenic CD4+ T-cells have a distinct transcriptional response 6 hours after the onset of sepsis. J Am Coll Surg. 2006;203:365–375. doi: 10.1016/j.jamcollsurg.2006.05.304. [DOI] [PubMed] [Google Scholar]
- 44.Moldawer LL. Opening the window on genome-wide expression analyses in sepsis. Am J Respir Crit Care Med. 2007;176:631–632. doi: 10.1164/rccm.200707-972ED. [DOI] [PubMed] [Google Scholar]
- 45.Quackenbush J. Microarray analysis and tumor classification. N Engl J Med. 2006;354:2463–2472. doi: 10.1056/NEJMra042342. [DOI] [PubMed] [Google Scholar]