Abstract
Gene expression within human glioblastomas were analyzed from data on 20,083 genes entered into the on-line Human Protein Atlas. In selecting genes that are strongly expressed within normal human brain tissue, 58 genes were identified from a search of the 20,083 entries that were rated as showing 90% or greater intensity of expression within normal brain tissues. Of these 58, a subset of 48 genes was identified that not only had expression data for human glioblastomas but also for the human glioblastoma cell line U-251. Four of these 48 selected genes were found to be strongly expressed within the cytoplasm when assessed by both histologic sampling of high grade glioma patient cases as well as U-251 glioblastoma cell line immunofluoresence analysis. These four human genes are: AGBL2 (ATP/GTP binding protein-like 2), BLOC1S6 (biogenesis of lysosomal organelles complex-1, subunit 6), MAP1A (microtubule-associated protein 1A) and ZSWIM5 (zinc finger, SWIM-type containing 5, also known as KIAA1511). Further research is advocated to investigate the role of ZSWIM5 and AGBL2 in glioma cell biology.
Key words: brain tumor, glioma, gene, atlas
Introduction
As noted by Pertea and Salzberg,1 a grand total of 22,333 human protein-coding genes are known to exist within the human genome, and are listed by the NCBI’s RefSeq database. A significantly high fraction (90%) of theses genes have been categorized within an on-line expression atlas of these genes within normal human tissue as well different human neoplasms including high grade gliomas.2 For gliomas, immunohistochemical data is available not only for tissue samples from patient cases, but from also human glioma cell lines including the well studied U-251 glioblastoma cell line.
Gene expression within glioblastoma was analyzed in this research study on 20,083 genes entered into the on-line Human Protein Atlas. In order to identify those genes which are the most highly expressed within human gliomas, this study employed the operational strategic hypothesis that those genes which are strongly expressed by normal glial cells but not at all by neurons will be more likely to show higher levels of glioma expression. Although other similar large scale human genome investigations of glioma expression have been published in the past,3 they have largely been conducted by PCR micro-array analysis of very small samples of tissue; the strength of this study comes from the use of immunohistochemistry and immunofluoresence as the basis for evaluation from diverse clinical cases as well as the established U-251 glioblastoma cell line. Finally, this study was also conducted with the long term goal of identifying potential targets for anti-sense oligonucleotide gene therapy to interfere with malignant cytoplasmic over-expression of key proteins essential to glioma cell survival.
Materials and Methods
Tissue samples examined within The Human Protein Atlas include those from 46 different normal tissue types from 138 individuals and 20 different types of cancer from 216 patients, each sample represented by a 1 mm circular core sample;2 this study focused on normal brain expression within the cerebral cortex and hippocampus whereas neoplastic tissue included cases archived from surgical resections as well the U-251 glioblastoma cell line. The protein atlas also contains high resolution, multicolor images of immunofluorescently stained cells and provides spatial information on protein expression patterns on a fine cellular and subcellular level. For further details on atlas data methods, please see: http://www.proteinatlas.org/about/assays+annotation.
By manually sorting through 20,083 entries into the Human Protein Atlas, a subset of 58 genes were identified that were rated by histologic criteria as being strongly expressed within brain tissue at a 90% or greater levels. Attention was subsequently focused on identifying those genes from this selected group of 58 which have strong cytoplasmic expression both within patient tissue samples of high grade gliomas as well as the well the U-251 glioma cell line.
Results
After looking for genes that are strongly expressed within normal human brain tissue, 58 genes were identified from a search of the 20,083 entries that were rated as showing 90% or greater intensity of expression within normal brain tissues. Of these 58, research then focused on a subset of 48 genes strongly expressed within normal brain tissue that not only had immunohistochemical expression data available for human high grade glioma tissue samples from individual cases, but also had data for the human glioblastoma cell line U-251. A photo-montage of the 48 genes is illustrated in Figure 1 that compares expression within normal brain tissue (left column), high grade glioma patient cases (center column) and the U-251 cell line (right column).
Figure 1.
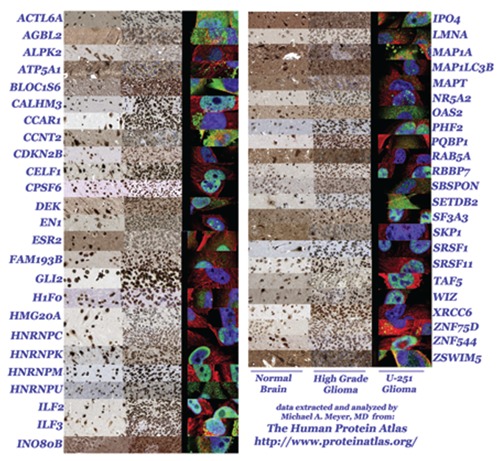
Montage of 48 genes selected from the Human Protein Atlas that display strong staining of normal brain tissue (left column) and also have both immunohistochemical data available for surgical resections of high grade gliomas from individual patient cases (middle column) and immunofluoresence data from the U-251 glioma cell line (right column).
General observations of the results shown in Figure 1 one include noting that 30 of the 48 genes were strongly positive for both high grade glioma patient cases as well the U-251 cell line, regardless of whether the pattern was cytoplasmic and/or nuclear in location.
Eleven of 48 genes were found to be cytoplasmically positive for strong expression by the U-251 human glioma cell line when assessed by immunofluoresence. These genes included: ACTL6A, AGBL2, BLOC1S6, CCNT2, HNRNPU, ILF3, IPO4, MAP1A, OAS2, SETDB2, ZSWIM5.
Eight of 48 genes were found to be cytoplasmically positive for strong immunohistochemical expression when assessed by histological sampling of high grade glioma patient cases. These genes included: AGBL2, ATP5A1, BLOC1S6, INO80B, MAP1A, MAP1LC3B, RAB5A, and ZSWIM5.
Four of 48 selected genes were strongly expressed within the cytoplasm when assessed by both histologic sampling of high grade glioma patient cases as well as U-251 gliobalstoma cell line immunofluoresence analysis. These four human genes are: AGBL2 (ATP/GTP binding protein-like 2), BLOC1S6 (biogenesis of lysosomal organelles complex-1, subunit 6, pallidin), MAP1A (microtubule-associated protein 1A) and ZSWIM5 (zinc finger, SWIM-type containing 5, also known as KIAA1511).
Significant discordance between intense gene expression within patient derived high grade gliomas despite no expression within the U-251 cell line was found for GLI2, RAB5A, and XRCC6.
Genes with selectively intense neuronal expression yet with weak to absent glial cell expression were found to be negative for U251 gliomas in 5 of 48 cases, with the contrast between the factors being most remarkable for three genes in particular: ALPK2, RAB5A, and SKP1; the strongest neuronal expression pattern amongst these three genes was ALPK2 (alpha-kinase 2) for antibody HPA028084 but weak with antibody HPA027976.
Discussion
This study brings to light the potential importance of a previously unstudied protein identified as ZSWIM5 (zinc finger, SWIM-type containing 5, also known as KIAA1511). No dedicated studies have been published on this unusual protein, which by the protein atlas data, displays intense expression within the brain with little expression elsewhere except for the seminiferous ductal cells of the testis and pancreatic Islets of Langerhans as well as placental decidual cells. Similarly, gliomas display intense staining for ZSWIM5 but weak to modest staining for most other neoplasms.
The detection of MAP1A (microtubule-associated protein 1A) as being one of four genes with strong cytoplasmic expression is of importance with relation to published data on antisense gene therapy.4 As found by Matsuno et al.,4 intra-tumoral delivery of antisense, sense and scramble phosphorothioate oligodeoxynucleotide for MAP 1A mRNA generated tumor diameters of 2.33, 4.625 and 5.25 mm respectively in animals, which showed statistically significant differences, suggesting the important role of MAP 1A in cell proliferation and its suppression may serve as a novel antitumour therapy for gliomas. However, review of data from the protein atlas also shows selectively intense normal neuronal expression, which may be a limiting factor on this approach with MAP1A.
Other genes found here to be of potential importance with strong cytoplasmic glioma expression include AGBL2 (ATP/GTP binding protein-like 2) as well as BLOC1S6 (biogenesis of lysosomal organelles complex-1, subunit 6, pallidin). With regards to AGBL2, there is data to show that AGBL2 is a RARRES1 interacting protein that regulates the tubulin tyrosination cycle.5 As explained by Sahab et al.,5 Retinoic acid receptor responder 1 (RARRES1) is lost in many cancer cells and normally blocks AGBL2 activity and thereby increases the proportion of tyrosinated tubulin and alters the interaction of kinesins with microtubules.
This study generated new and unanticipated data, as a prior cDNA microarray study did not list these selected four genes in their list of 54 upregulated genes showing expression increases varying from 3.01 to 19.91 fold change.3 Although Yokota et al. identified DNCH2 (5.27 fold change) as well as ARHGEF6 (3.74 fold change) as special genes with matching histologic over-expression within gliomas,3 this could not be verified by this study, which showed only modest expression as determined by the protein atlas. The reasons for this discrepancy remain unclear but might relate to antibody type and specificity; as shown above in this study for the case of ALPK2 (alpha-kinase 2), antibody HPA028084 generated strong neuronal staining but only weakly with antibody HPA027976.
Conclusions
In conclusion, AGBL2, BLOC1S6, MAP1A and ZSWIM5 have been identified here as genes of interest for human glioma with high cytoplasmic expression. Further research is clearly needed to investigate the relatively unknown protein ZSWIM5, which is selectively and strongly expressed within normal brain and malignant gliomas. In addition, the linkage of AGBL2 to tubulin dynamics may be of special importance for glioma cell cytoarchitecture and motility; further investigation into the physiological role that AGBL2 plays in glioma cell survival is therefore clearly needed.
Acknowledgements
The author wishes to thank Dr. Janet Bay for reviewing the initial version of this manuscript
References
- 1.Pertea M, Salzberg SL.Between a chicken and a grape: estimating the number of human genes. Genome Biol 2010;11:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 2010;28:1248-50 [DOI] [PubMed] [Google Scholar]
- 3.Yokota T, Kouno J, Adachi K, et al. Identification of histological markers for malignant glioma by genome-wide expression analysis: dynein, alpha-PIX and sorcin. Acta Neuropathol 2006;111:29-38 [DOI] [PubMed] [Google Scholar]
- 4.Matsuno A, Katayama H, Murakami M, Nagashima T.In vivo growth suppression of rat C6 glioma transplanted in rat brain using antisense oligonucleotide for microtubule-associated protein 1A messenger ribonucleic acid. Br J Neurosurg 2004; 18:343-6 [DOI] [PubMed] [Google Scholar]
- 5.Sahab ZJ, Hall MD, Me Sung Y, et al. Tumor suppressor RARRES1 interacts with cytoplasmic carboxypeptidase AGBL2 to regulate the -tubulin tyrosination cycle. Cancer Res 2011;71:1219-28 [DOI] [PMC free article] [PubMed] [Google Scholar]


