Abstract
Urea-based inhibitors of human glutamate carboxypeptidase II (GCPII) have advanced into clinical trials for imaging metastatic prostate cancer. In parallel efforts, agents with increased lipophilicity have been designed and evaluated for targeting GCPII residing within the neuraxis. Here we report the structural and computational characterization of six complexes between GCPII and P1′-diversified urea-based inhibitors that have the C-terminal glutamate replaced by more hydrophobic moieties. The X-ray structures are complemented by quantum mechanics calculations that provide a quantitative insight into the GCPII/inhibitor interactions. These data can be used for the rational design of novel glutamate-free GCPII inhibitors with tailored physicochemical properties.
Keywords: GCPII, prostate-specific membrane antigen, PSMA, metallopeptidase, X-ray crystallography, structure-based drug design, urea-based inhibitor
Urea-based small-molecule inhibitors targeting human glutamate carboxypeptidase II (GCPII) were originally developed for application within the central nervous system (CNS), however, they were first used in vivo to image a peripheral version of GCPII known as the prostate-specific membrane antigen (PSMA) and prostate cancer (PCa).1–3 GCPII/PSMA (referred to throughout as GCPII) is now a well-established biomarker for imaging PCa, as this membrane-tethered metallopeptidase is over-expressed on the surface of castrate-resistant prostate tumors with its active site facing the extracellular milieu. Additionally, GCPII expression in the neovasculature of most solid tumors, but not normal vasculature, expands the utility of the enzyme for imaging/therapy of tumors other than prostate.4 In the nervous system, GCPII cleaves N-acetylaspartylglutamate (NAAG), releasing N-acetylaspartate and glutamate. Excessive glutamate production and release may overstimulate several glutamate receptor subtypes and lead to glutamate-associated neurotoxicity. Furthermore, a glutamate imbalance is linked to the pathophysiology of certain neurological diseases including schizophrenia, amyotrophic lateral sclerosis, neuropathic/diabetic pain and ischemia. Given the involvement of GCPII in a glutamate metabolism, inhibition of GCPII can be used as a therapeutic option for the prevention and treatment of neurological disorders as documented in several animal models of aforementioned diseases.5
Imaging agents targeting GCPII can be divided into three categories that include antibodies, aptamers and small-molecule ligands. Currently, only ProstaScint®, an 111In-labeled monoclonal antibody, has been approved by the FDA and is used clinically for imaging PCa, although with variable degrees of success.6 Because of the long biological half-life of antibodies, leading to excessive non-specific binding, several small-molecule ligands have recently entered clinical trials as viable alternatives for imaging PCa.7, 8 Typically, GCPII inhibitors for imaging applications are derivatives of NAAG with the principal C-terminal (P1′) docking glutamate moiety and the distal (P1) moiety that fine-tunes inhibitor affinity towards GCPII and harbors a radioactive or fluorescent tracer. The P1 and P1′ functionalities are connected via a zincbinding group (ZBG), resistant to hydrolysis. The most common ZBGs are phosphonates, phosphinates, phosporamidates, and ureas, with the latter being closest to become human medicines.9–13
Inhibitors of GCPII that target the CNS or peripheral nervous system, show promise in various animal models of neurological disorders.14, 15 For example, 2-phosphonomethylpropanedioic acid (2-PMPA), a phosphonate-based picomolar GCPII inhibitor, was used successfully to provide neuroprotection following middle cerebral artery occlusion, attenuate cocaine/ethanol-induced drug-seeking behavior, and alleviate hyperalgesia/allodynia in rat pain models.16–18 The urea-based GCPII inhibitor, ZJ-43, was effective in several pain as well as brain and spinal cord injury models.18, 19 The main pitfall associated with a use of NAAG-based or glutamate-based inhibitors is their high polarity, which stems from the presence of the C-terminal glutamate moiety. In consequence, such inhibitors poorly penetrate the blood-brain barrier (BBB) and their efficacy is limited. Several strategies are being developed to address that problem, including a use of lipophilic prodrugs and the substitution of the P1′ glutamate with a less polar functionality.20–22
Recently, we have published a comprehensive study of structure-activity relationship (SAR), in which we described modifications of the urea-based inhibitor, DCIBzL [compound (7)], at the P1′ glutamate.23 The aim of that study was to map the specificity of the S1′ pocket in GCPII towards non-glutamate moieties. We have showed that a variety of isosteres in the P1′ position is tolerated by the enzyme, however, substitution of the C-terminal glutamate inevitably leads to a decrease in inhibitor affinity by several orders of magnitude. Despite that drop in potency, the most potent isosteres still displayed low-nanomolar inhibition constants and were suitable for imaging GCPII-positive peripheral tumors in mice. Retention of high affinity combined with a significant increase in lipophilicity of the new isosteres suggest that further optimization of a functionality placed at the P1′ position might provide BBB-penetrable compounds.
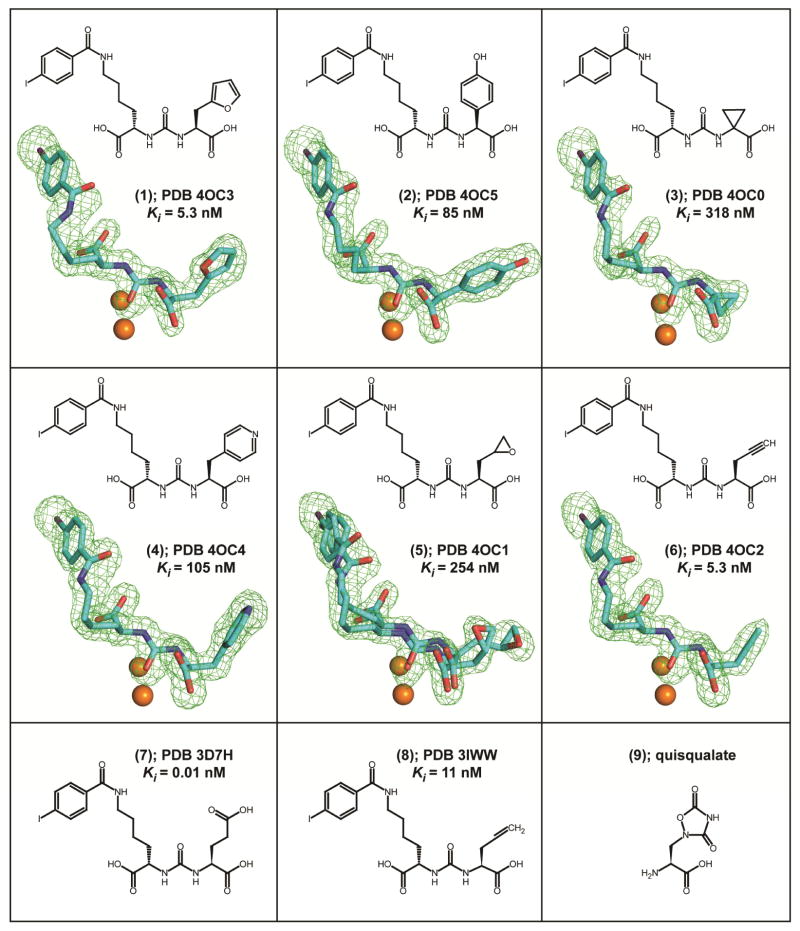
Here, we present the follow-up report, detailing interactions between GCPII and a series of six selected isosteres of (7) in the P1′ position. By the combination of X-ray crystallography and quantum mechanics (QM) calculations, we aim to increase an understanding of interactions between non-glutamate moieties in the P1′ position of an inhibitor and the S1′ pocket of the enzyme. We selected six compounds to encompass a diversity of isosteres that span affinities for GCPII across two orders of magnitude (Figure 1). Included are the most potent isosteres (6), Ki= 5.3 nM, and (1), Ki = 5.3 nM, representing an unsaturated linear chain and a five-membered heteroaromatic ring, respectively, less active (4), Ki = 105 nM, and (2), Ki = 85 nM, featuring six-membered aromatic rings, and finally, high nanomolar (3), Ki = 318 nM, and (5), Ki = 254 nM, three-membered ring compounds. Additionally, we included the parent molecule (7), featuring a glutarate moiety at P1′ with Ki = 10 pM. Based on previous SAR and structural reports, all compounds have a P1′ configuration corresponding to L-glutamate, with the exception of (3), which has no stereogenic center at the P1′ position. The L-stereoisomers typically bind to GCPII with affinities that are several orders of magnitude higher than their D-counterparts, which are unlikely to generate lead compounds. Additionally, excluded were inhibitors lacking the P1′ side chain altogether (i.e. glycine in the P1′) or missing the α-carboxylate functionality of the P1′ moiety. Interactions between the latter and the guanidinium group of Arg210 from GCPII were shown to be crucial to retain affinity to GCPII in both SAR and mutagenesis studies.23, 24
Figure 1.
Chemical formulas, PDB codes, and inhibition constants of inhibitors used in this study. Fo-Fc maps (green) for individual inhibitors are contoured at 3.0 σ and modeled inhibitors are shown in stick representation with atoms colored cyan (carbon), red (oxygen), blue (nitrogen), and violet (iodine). The active-site zinc ions are shown as orange spheres.
Crystallization experiments were carried out using an extracellular part of human GCPII (amino acids 44 – 750) that was heterologously expressed in Drosophila S2 cells and purified to homogeneity according to established protocols.25 Following the final size-exclusion purification step, the protein was concentrated to 10 mg/mL (in 20 mM Tris-HCl, 150 mM NaCl, pH 8.0), flash-frozen in liquid nitrogen and kept at −80°C until further use.
Diffracting crystals of GCPII/inhibitor complexes were obtained by first preparing a mixture of GCPII and a given inhibitor [mixing stock solution of GCPII (10 mg/mL) and inhibitor (20 mM) at 9:1 (v/v) ratio] and then mixing the GCPII/inhibitor solution with the same volume of the reservoir solution [33% pentaerythritol propoxylate (Sigma), 1.5% polyethylene glycol 3350 (Sigma), and 100 mM Tris–HCl, pH 8.0]. Crystals were grown using the hanging-drop vapor-diffusion setup at 293 K and diffraction intensities for each complex were collected from a single crystal at 100 K using synchrotron radiation at the SER-CAT beamlines 22-ID and 22-BM at the Advanced Photon Source (Argonne, USA). For each complex, a complete dataset was collected from a single crystal and data were processed using the HKL2000 software package.26 Difference Fourier methods were used to determine structures of GCPII/inhibitor complexes with the ligand-free GCPII structure (PDB code 2OOT)27 used as a starting model. Calculations were performed using the program Refmac 5.5.28 and the structural refinement was interspersed by manual corrections to the model with aid of the program Coot 0.6.29 The PRODRG server was used to generate the restrains library and the coordinate files for individual inhibitors.30 Detailed procedures, the data collection and refinement statistics are summarized in Supplementary Table S1. Atomic coordinates of the present structures together with the experimental structure factor amplitudes were deposited at the RCSB Protein Data Bank under accession numbers shown in Figure 1.
Structures of GCPII/inhibitor complexes were refined at the resolution range between 1.65 and 1.85 Å, with suitable crystallographic parameters (Table S1). The overall fold of the protein component is nearly invariant as reflected by the maximum root mean square deviation of 0.16 Å for the 683 equivalent Cα pairs between GCPII/(1) and GCPII/(2) complexes. During the final stages of refinement, inhibitors were modeled into the well-defined Fo-Fc positive density peaks with high confidence for all six inhibitors (Figure 1).
The 4-iodobenzoyl-ε-lysine, which is derived from the parent DCIBzL (7) molecule, is a structural motif common to all inhibitors presented here. This motif was included in the inhibitor design as its addition increases the affinity of a given compound nearly ten-fold compared to urea-based scaffolds lacking this functionality.31 That effect results primarily from the engagement of the terminal 4-iodobenzoyl group with an S1 hydrophobic ‘accessory pocket’ of GCPII shaped by the side chains of Glu457, Arg463, Asp465, Arg534, and Arg536. As expected, all structures feature the 4-iodobenzoyl group inserted into the pocket. The benzene ring of the inhibitor is parallel to guanidinium groups of Arg463 and Arg534, highlighting the importance of π-cation interaction for the inhibitor binding.
The lysine linker connecting the distal 4-iodobenzoyl functionality to the urea isostere is somewhat flexible, yet its conformation is constrained by the invariant positioning of the 4-iodobenzoyl group at one end and the urea at the other. Moreover, the P1 carboxylate group forms hydrogen bonds with side-chains of Asn519 (2.9 Å; in GCPII/(6) complex), Arg534 (2.9 Å), and Arg536 (2.9 Å and 3.0 Å), adding additional constraints. The P1 carboxylate has been reported to be an important signature of the NAAG-based inhibitors as its absence or substitution results in a weaker binding to GCPII.23, 32
Similar to structures of urea-based inhibitors described previously,31, 33 the ureido group of the six inhibitors studied here mimics a planar peptide bond of a GCPII substrate and interacts with several residues in the active site. The ureido carbonyl oxygen is polarized by the catalytic Zn ion (2.7 Å) and also interacts with side chains of Tyr552 (OH, 2.6 Å) and His553 (Nε2, 3.2 Å), and with the hydroxide anion (2.9 Å). The N1 and N2 ureido nitrogen atoms form pairs of hydrogen bonds with the Gly518 main-chain carbonyl (3.0 Å) and the Glu424 γ-carboxylate (Oε2, 3.1 Å), and with the Gly518 carbonyl oxygen (2.9 Å) and the hydroxide anion (3.2 Å), respectively.
Glutarate functionality at the P1′ (C-terminal) position is a hallmark of virtually all GCPII inhibitors.10 As the S1′ pocket of the enzyme is fine-tuned for glutamate binding, the presence of P1′ glutamate ensures high affinity of the inhibitor and its specificity towards GCPII (and orthologous GCP334). Yet, isostere replacements of the P1′ glutamate are possible, primarily to increase the lipophilicity of an inhibitor with the aim to facilitate passive diffusion across biological membranes, including the BBB.22, 23
Both structure-activity relationships (SAR) and site-directed mutagenesis studies indicate the P1′ α-carboxylate function of an inhibitor as an element prominently contributing to affinity towards GCPII. That contribution is realized via interactions with Arg210 of the enzyme. A role of that interaction is well-illustrated by a decrease of approximately five orders of magnitude in IC50 of 2-PMPA in the case of an Arg210Ala mutant as compared to the wild-type enzyme,24, 35 as well as in a setup pairing the wild-type GCPII with an inhibitor missing the α-carboxylate.23 Consequently, all inhibitors in this study feature the α-carboxylate function that predictably engages the guanidinium group of Arg210 and forms additional hydrogen bonds with an invariant water molecule (2.5 Å) and the hydroxyl groups of Tyr552 (3.4 Å) and Tyr700 (2.6 Å).
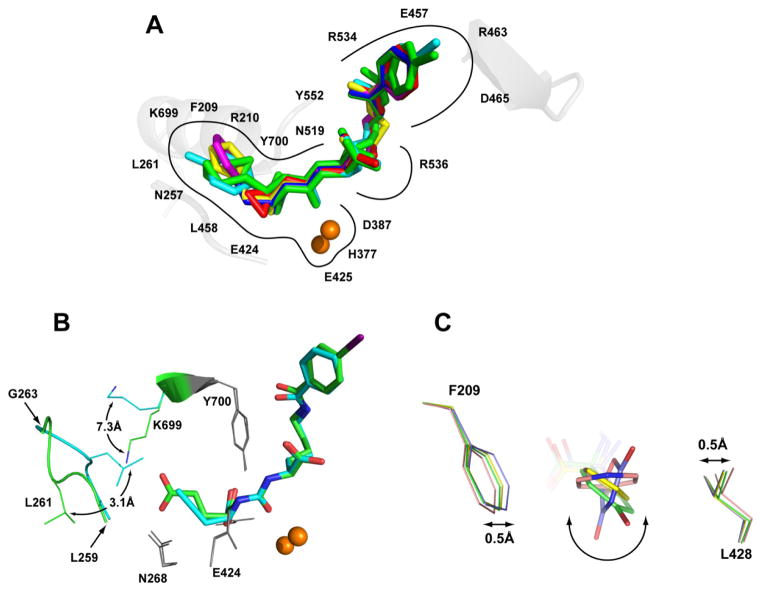
Although the S1′ pocket of GCPII is evolutionarily optimized for the binding of the glutamate moiety and structurally can be viewed as more rigid compared to the S1 pocket, we have recently observed limited plasticity of the S1′ pocket that allows for non-glutamate functionalities. Those limited variations in the shape of the S1′ pocket are permitted by the flexibility of the Asn257 side chain, 35 the swinging motion of Lys699,22 and repositioning of the Leu259 – Gly263 amino acid stretch closing the bottom of the S1′ pocket.23 Concurrently, fixed conformations of side chains in Phe209 and Leu428 define the width of the S1′ pocket, represented by an approximate distance of 8.2 Å between the Leu428 Cβ and Phe209 Cζ atoms. The rigidity of the S1′ pocket limits the orientation and size of side chains that can be placed on the P1′ portion of the inhibitor (but see below).
In structures presented here the major re-arrangement of the S1′ pocket is observed in the GCPII/(6) complex. In that complex, the Leu259 – Gly263 amino acid stretch is repositioned from the “canonical” conformation by 3.1 Å (as measured for the Cα of Leu261), resulting in a smaller S1′ pocket (Figure 2B). That change is accompanied by the relocation of the Lys699 side chain, represented by the 7.3 Å shift of the Nζ atom, to avoid clash with the Leu261 side chain. It is noteworthy that a similar rearrangement was observed by us previously for GCPII/(8).23 Therefore, those conformational adjustments reflect the inherent plasticity of the S1′ pocket of GCPII and not a crystallization artifact. On the other hand, small molecules featuring longer aliphatic side-chains of the terminal moiety, as opposed to the short alkenes/alkynes reported here, can be accommodated by the S1′ pocket with conformational changes limited only to the side chain of Lys699.22 Furthermore, flexibility of the Leu259 – Gly263 segment is observed in nearly all structures of GCPII and is reflected by elevated B-factors and inferior electron density as compared to other regions of the enzyme.
Figure 2.
Panel A: The superposition of the active-site bound inhibitors. Complexes of GCPII/inhibitor were superimposed on corresponding Cα atoms of the enzyme. Individual inhibitors are colored yellow, cyan, red, magenta, green, and blue for (1), (2), (3), (4), (5), and (6), respectively. The zinc ions are shown as orange spheres. Panel B: Re-arrangement within the S1 pocket of GCPII induced by the inhibitor binding. Structures of GCPII/(6) (cyan) and GCPII/(7)(PDB code 3D7H; green) were superimposed on corresponding Cα atoms of the enzyme. The active site-bound inhibitors are shown in stick representation, while selected residues forming the S1′ site of the enzyme are shown as lines, the active-site zinc ions as spheres, and the Leu259 – Gly263 loop in the cartoon representation. Notice the repositioning of the Leu261 side chain (3.1 Å for Cα’s) accompanied by the swing motion of the Lys699 (7.3 Å for the Nζ). Positions of remaining residues forming the S1′ pocket are unchanged (grey). Panel C: Conformations of ring functionalities of inhibitors in the S1′ pocket. Complexes of GCPII/(2) (green), GCPII/(4) (pink), GCPII/(1)(yellow), and GCPII/(9) (blue) were superimposed on corresponding Cα atoms of the enzyme. The active site-bound inhibitors are shown in stick representation, while Phe209 and Leu428, the two residues defining the side-walls of the S1′ pocket, are shown as lines. The quisqualate oxadiazolidine ring is placed virtually parallel to the walls of the S1 pocket, while the plane of the (4) pyridine ring is rotated by 70°. This orientation brings about the increase in the Phe209-Leu428 side chain spacing by approximately 1.0 Å.
Several GCPII inhibitors harbor five- or six- membered rings at the P1′ position. Those include (1), (2) and (4), reported here as well as quisqualate (9) a natural glutamate isostere, reported previously (PDB code 2OR4).35 Unexpectedly and somewhat counter-intuitively, the planar rings of the different inhibitors assume orientations that differ by up to 70° from each other (Figure 2C). Apparently, the positioning of the ring planes is determined by (i) chemical composition of the ring, (ii) interactions of the ring with the reminder of the inhibitor molecule, and (iii) interactions of the ring and its substituents with the enzyme. The plane of the quisqualate oxadiazolidine ring (PDB code 2OR4) is placed parallel to side-walls of the S1′ pocket, the latter defined by the side chains of Phe209 and Leu428. Previously, we proposed that more extensive van der Waals interactions between quisqualate and side chains of Phe209 and Leu428, as compared to the glutamate isostere, may be associated with higher affinity of the former towards GCPII.35 Interestingly, the plane of the pyridine ring in (4) is oriented at a 70° angle compared to the oxadiazolidine ring, positioning it nearly perpendicular to the “side walls” of the S1′ pocket. Such an orientation of the pyridine ring leads to an expansion of the S1′ pocket, realized by the increase of the distance between side chains of Phe209 and Leu428 by approximately 1.0 Å. The ring orientations in the two remaining complexes, (1) and (2), are between the two extremes, and side chains of Phe209 and Leu428 have conformations as in the GCPII/(9) (quisqualate) complex, i.e., corresponding to a compact S1′ pocket.
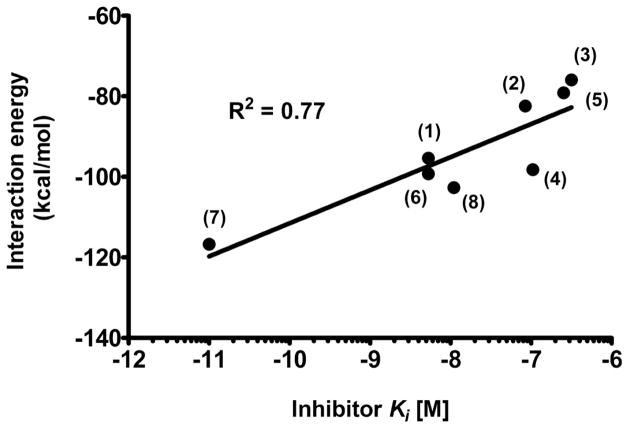
To gain a quantitative description of contributions by individual parts of inhibitors to GCPII binding and to elucidate the role of (de)solvation in complex formation, we performed detailed QM calculations for individual structures and compared results with those obtained for the parent (7) (DCIBzL) complex. Initial geometries of the GCPII/ligand complexes were taken from individual crystal structures. The protein residues, water molecules, and ions located within 10 Å from the ligand atoms were selected using the VMD program.36 The radius of 10 Å was necessary to obtain a satisfactory correlation between experimental inhibition values and calculated interaction energies (Figure 3). In the initial calculations, we included only the elements of the enzyme located within a distance of 4 Å from the ligand, which allowed for the description of the system at a higher level of theory. However, for the 4 Å radius, energies calculated by PM7 and DFT-D,37 equivalent to TPSS38/TZVP39 with corrections for the dispersion and higher order correlation effects, did not correlate well with the experimental data.
Figure 3.
The correlation between the calculated interaction energies and experimental inhibition constants. The PM7 interaction energy was calculated using implicit solvation in MOPAC2012 with the initial geometries of GCPII/ligand complexes taken from individual crystal structures. Calculated interaction energy and experimental inhibition constants correlate with the R2 value for the least square linear fit equal to 0.77. Corresponding compound descriptors are attached to each individual experimental data point.
Hydrogen atoms were added to experimental structures and their positions were optimized using the semi-empirical method PM740 as implemented in the MOPAC2012 package.41 The geometry of the entire model was partially relaxed using the same method combined with the ‘geo_ref’ keyword and the proportionality constant equal to 10. Water molecules, with the exception of the hydroxide anion shared by the two active-site zinc ions, were removed and the PM7 interaction energy was calculated using implicit solvation in MOPAC2012. The implicit solvation is preferable as different resolutions of individual structures resulted in different numbers of experimental water molecules, from 29 (GCPII/(8)complex) to 56 (GCPII/(7)), found inside the region defined by the 10-Å selection radius.
Finally, the calculated interaction energies were correlated with log(Ki) and the result is shown in Figure 3. Calculated and experimental energies correlated reasonably well with the R2 value for least square linear fit equal to 0.77. In an attempt to improve further the correlation coefficient, we included the effects of the solvation free energy of ligands using COSMO calculations. We also accounted for the deformation energy, resulting from the geometry change induced by the binding of ligands to the binding pocket of GCPII. However, the inclusion of any of those contributions did not lead to an improvement in correlation. Although we succeeded in correlating the overall interaction energy with the experimental data, attempts to partition the energy and identify (de)stabilizing contributions of groups of residues showed poor additivity of energy, making interpretation unreliable, especially at the level of individual amino acid residues. Only a weak correlation (R2 = 0.54) was found for the interaction of the ligands with the active site zinc alone (2xZn2++ OH−). Our findings reflect the generally accepted notion that QM calculations for zinc-containing systems are very challenging, even when extensive experimental information is available.42
In conclusion, here we extend a structural insight into the S1′ pocket of GCPII. Our data reveal previously unreported plasticity of the S1′ site, a property required to accommodate bulkier functionalities at the P1′ position of an inhibitor. These findings will be valuable for the rational design of novel GCPII inhibitors with non-glutamate moieties at the P1′ position, eventually enabling compounds that cross the BBB.
Supplementary Material
Data Collection and Refinement Statistics.
Acknowledgments
The authors acknowledge the use of beamline 22-ID of the Southeast Regional Collaborative Access Team (SER-CAT), located at the Advanced Photon Source, Argonne National Laboratory. Use of the APS was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. C.B. acknowledges the support from the EMBO (Installation grant 1978) and IRG (project number 249220). This publication is supported by the project „BIOCEV – Biotechnology and Biomedicine Centre of the Academy of Sciences and Charles University“ (CZ.1.05/1.1.00/02.0109), from the European Regional Development Fund, by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research (to J.L.), by NIH R01 CA134675 (to M.G.P.), and by the National Research Foundation of Korea (NRF) grant funded by the Korea Government (MEST) (No.2012R1A1A1010000; to Y.B.).
Abbreviations Used
- BBB
blood-brain barrier
- GCPII
glutamate carboxypeptidase II
- NAAG
N-acetylaspartyl-glutamate
- QM
quantum mechanics
Footnotes
PDB ID Codes: Atomic coordinates of the present structures together with the experimental structure factor amplitudes were deposited at the RCSB Protein Data Bank under accession numbers 4OC3 (complex with (1)), 4OC5 (complex with (2)), 4OC0 (complex with (3)), 4OC4 (complex with (4)), 4OC1 (complex with (5)), and 4OC2 (complex with (6)).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Foss CA, Mease RC, Fan H, Wang Y, Ravert HT, Dannals RF, Olszewski RT, Heston WD, Kozikowski AP, Pomper MG. Clin Cancer Res. 2005;11:4022. doi: 10.1158/1078-0432.CCR-04-2690. [DOI] [PubMed] [Google Scholar]
- 2.Nan F, Bzdega T, Pshenichkin S, Wroblewski JT, Wroblewska B, Neale JH, Kozikowski AP. J Med Chem. 2000;43:772. doi: 10.1021/jm9905559. [DOI] [PubMed] [Google Scholar]
- 3.Pomper MG, Musachio JL, Zhang J, Scheffel U, Zhou Y, Hilton J, Maini A, Dannals RF, Wong DF, Kozikowski AP. Mol Imaging. 2002;1:96. doi: 10.1162/15353500200202109. [DOI] [PubMed] [Google Scholar]
- 4.Chang SS, Reuter VE, Heston WD, Bander NH, Grauer LS, Gaudin PB. Cancer Res. 1999;59:3192. [PubMed] [Google Scholar]
- 5.Barinka C, Rojas C, Slusher B, Pomper M. Curr Med Chem. 2012;19:856. doi: 10.2174/092986712799034888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taneja SS. Rev Urol. 2004;6(Suppl 10):S19. [PMC free article] [PubMed] [Google Scholar]
- 7.Mease RC, Foss CA, Pomper MG. Curr Top Med Chem. 2013;13:951. doi: 10.2174/1568026611313080008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Afshar-Oromieh A, Malcher A, Eder M, Eisenhut M, Linhart HG, Hadaschik BA, Holland-Letz T, Giesel FL, Kratochwil C, Haufe S, Haberkorn U, Zechmann CM. Eur J Nucl Med Mol Imaging. 2013;40:486. doi: 10.1007/s00259-012-2298-2. [DOI] [PubMed] [Google Scholar]
- 9.Banerjee SR, Pullambhatla M, Byun Y, Nimmagadda S, Green G, Fox JJ, Horti A, Mease RC, Pomper MG. J Med Chem. 2010;53:5333. doi: 10.1021/jm100623e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferraris DV, Shukla K, Tsukamoto T. Curr Med Chem. 2012;19:1282. doi: 10.2174/092986712799462658. [DOI] [PubMed] [Google Scholar]
- 11.Kozikowski AP, Nan F, Conti P, Zhang J, Ramadan E, Bzdega T, Wroblewska B, Neale JH, Pshenichkin S, Wroblewski JT. J Med Chem. 2001;44:298. doi: 10.1021/jm000406m. [DOI] [PubMed] [Google Scholar]
- 12.Liu T, Wu LY, Choi JK, Berkman CE. Prostate. 2009;69:585. doi: 10.1002/pros.20909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pavlicek J, Ptacek J, Barinka C. Curr Med Chem. 2012;19:1300. doi: 10.2174/092986712799462667. [DOI] [PubMed] [Google Scholar]
- 14.Rahn KA, Slusher BS, Kaplin AI. Curr Med Chem. 2012;19:1335. doi: 10.2174/092986712799462649. [DOI] [PubMed] [Google Scholar]
- 15.Wozniak KM, Rojas C, Wu Y, Slusher BS. Curr Med Chem. 2012;19:1323. doi: 10.2174/092986712799462630. [DOI] [PubMed] [Google Scholar]
- 16.Slusher BS, Thomas A, Paul M, Schad CA, Ashby CR., Jr Synapse. 2001;41:22. doi: 10.1002/syn.1056. [DOI] [PubMed] [Google Scholar]
- 17.Slusher BS, Vornov JJ, Thomas AG, Hurn PD, Harukuni I, Bhardwaj A, Traystman RJ, Robinson MB, Britton P, Lu XC, Tortella FC, Wozniak KM, Yudkoff M, Potter BM, Jackson PF. Nat Med. 1999;5:1396. doi: 10.1038/70971. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto T, Saito O, Aoe T, Bartolozzi A, Sarva J, Zhou J, Kozikowski A, Wroblewska B, Bzdega T, Neale JH. Eur J Neurosci. 2007;25:147. doi: 10.1111/j.1460-9568.2006.05272.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhong C, Zhao X, Sarva J, Kozikowski A, Neale JH, Lyeth BG. J Neurotrauma. 2005;22:266. doi: 10.1089/neu.2005.22.266. [DOI] [PubMed] [Google Scholar]
- 20.Feng JF, Gurkoff GG, Van KC, Song M, Lowe DA, Zhou J, Lyeth BG. Brain Res. 2012;1469:144. doi: 10.1016/j.brainres.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng JF, Van KC, Gurkoff GG, Kopriva C, Olszewski RT, Song M, Sun S, Xu M, Neale JH, Yuen PW, Lowe DA, Zhou J, Lyeth BG. Brain Res. 2011;1395:62. doi: 10.1016/j.brainres.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plechanovova A, Byun Y, Alquicer G, Skultetyova L, Mlcochova P, Nemcova A, Kim HJ, Navratil M, Mease R, Lubkowski J, Pomper M, Konvalinka J, Rulisek L, Barinka C. J Med Chem. 2011;54:7535. doi: 10.1021/jm200807m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, Byun Y, Barinka C, Pullambhatla M, Bhang HE, Fox JJ, Lubkowski J, Mease RC, Pomper MG. Bioorg Med Chem Lett. 2010;20:392. doi: 10.1016/j.bmcl.2009.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mlcochova P, Plechanovova A, Barinka C, Mahadevan D, Saldanha JW, Rulisek L, Konvalinka J. FEBS J. 2007;274:4731. doi: 10.1111/j.1742-4658.2007.06021.x. [DOI] [PubMed] [Google Scholar]
- 25.Barinka C, Mlcochova P, Sacha P, Hilgert I, Majer P, Slusher BS, Horejsi V, Konvalinka J. Eur J Biochem. 2004;271:2782. doi: 10.1111/j.1432-1033.2004.04209.x. [DOI] [PubMed] [Google Scholar]
- 26.Minor W, Cymborowski M, Otwinowski Z. Acta Physica Polonica A. 2002;101:613. [Google Scholar]
- 27.Barinka C, Starkova J, Konvalinka J, Lubkowski J. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:150. doi: 10.1107/S174430910700379X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. Acta Crystallogr D Biol Crystallogr. 2011;67:355. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P, Lohkamp B, Scott WG, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2010;66:486. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuttelkopf AW, van Aalten DM. Acta Crystallogr D Biol Crystallogr. 2004;60:1355. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 31.Barinka C, Byun Y, Dusich CL, Banerjee SR, Chen Y, Castanares M, Kozikowski AP, Mease RC, Pomper MG, Lubkowski J. J Med Chem. 2008;51:7737. doi: 10.1021/jm800765e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barinka C, Hlouchova K, Rovenska M, Majer P, Dauter M, Hin N, Ko YS, Tsukamoto T, Slusher BS, Konvalinka J, Lubkowski J. J Mol Biol. 2008;376:1438. doi: 10.1016/j.jmb.2007.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klusak V, Barinka C, Plechanovova A, Mlcochova P, Konvalinka J, Rulisek L, Lubkowski J. Biochemistry. 2009;48:4126. doi: 10.1021/bi900220s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hlouchova K, Barinka C, Klusak V, Sacha P, Mlcochova P, Majer P, Rulisek L, Konvalinka J. J Neurochem. 2007;101:682. doi: 10.1111/j.1471-4159.2006.04341.x. [DOI] [PubMed] [Google Scholar]
- 35.Barinka C, Rovenska M, Mlcochova P, Hlouchova K, Plechanovova A, Majer P, Tsukamoto T, Slusher BS, Konvalinka J, Lubkowski J. J Med Chem. 2007;50:3267. doi: 10.1021/jm070133w. [DOI] [PubMed] [Google Scholar]
- 36.Humphrey W, Dalke A, Schulten K. J Mol Graphics Modell. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 37.Jurecka P, Cerny J, Hobza P, Salahub DR. J Comp Chem. 2007;28:555. doi: 10.1002/jcc.20570. [DOI] [PubMed] [Google Scholar]
- 38.Tao JM, Perdew JP, Staroverov VN, Scuseria GE. Phys Rev Lett. 2003;91:146401. doi: 10.1103/PhysRevLett.91.146401. [DOI] [PubMed] [Google Scholar]
- 39.Schafer A, Huber C, Ahlrichs R. J Chem Phys. 1994;100:5829. [Google Scholar]
- 40.Stewart JJP. J Mol Model. 2013;19:1. doi: 10.1007/s00894-012-1667-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart JJP. MOPAC2012. Stewart Computational Chemistry; Colorado Springs, CO, USA: 2012. [Google Scholar]
- 42.Raha K, Merz KM., Jr J Am Chem Soc. 2004;126:1020. doi: 10.1021/ja038496i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Collection and Refinement Statistics.