Abstract
Tsetse flies are the sole vectors of human African trypanosomiasis throughout sub-Saharan Africa. Both sexes of adult tsetse feed exclusively on blood and contribute to disease transmission. Notable differences between tsetse and other disease vectors include obligate microbial symbioses, viviparous reproduction, and lactation. Here, we describe the sequence and annotation of the 366-megabase Glossina morsitans morsitans genome. Analysis of the genome and the 12,308 predicted protein–encoding genes led to multiple discoveries, including chromosomal integrations of bacterial (Wolbachia) genome sequences, a family of lactation-specific proteins, reduced complement of host pathogen recognition proteins, and reduced olfaction/chemosensory associated genes. These genome data provide a foundation for research into trypanosomiasis prevention and yield important insights with broad implications for multiple aspects of tsetse biology.
African trypanosomiasis is transmitted by the tsetse fly to humans (sleeping sickness) and livestock (nagana) throughout sub-Saharan Africa, with an estimated 70 million people at risk of infection. Rearing livestock in endemic areas is difficult to impossible and results in an economic loss in agricultural output of several billion U.S. dollars per year. Human infections are fatal if untreated, but tools for disease control are limited because it has not been possible to develop vaccines and current trypanocidal drug treatments result in undesirable side effects with growing reports of drug resistance. The reduction or elimination of tsetse populations is an effective method for disease control that could be improved with greater knowledge of their biology and genetics (1).
Tsetse flies are key representatives of the dipteran clade Calyptratae, which represents 12% of the known diversity within the dipteran order. Many of the calyptrate species are blood feeders of biomedical importance (2). In addition, members of the calyptrate family of Glossinidae and superfamily Hippoboscoidea, to which tsetse belong (fig. S1) (3), are defined by the ability to nourish intrauterine offspring from glandular secretions and give birth to fully developed larvae (obligate adenotrophic viviparity). Tsetse flies live considerably longer than other vector insects, which somewhat compensates for their slow rate of reproduction. Trypanosome infections in tsetse are acquired by blood feeding from an infected vertebrate host, and trypanosomes have to overcome multiple immune barriers to establish an infection within the fly. As a result, trypanosome infection prevalence is low in field populations and in experimentally infected tsetse (4). Tsetse have symbionts that compensate for their nutritionally restricted diet by the production of specific metabolites and influence multiple other aspects of the fly’s immune and reproductive physiology (5).
In 2004, the International Glossina Genome Initiative (IGGI) was formed (6) to expand research capacity for Glossina, particularly in sub-Saharan Africa, through the generation and distribution of molecular resources, including bio-informatics training. An outcome of the effort undertaken by IGGI is the annotated Glossina morsitans genome presented here and further developed in satellite papers on genomic and functional biology findings that reflect the unique physiology of this disease vector (7–14).
Characteristics of the Glossina Genome
A combination of sequencing methods were used to obtain the Glossina morsitans morsitans (Gmm) genome, including Sanger sequencing of bacterial artificial chromosomes (BACs), small-insert plasmid and large-insert fosmid libraries, and 454 and Illumina sequencing (tables S1 and S2). The sequences were assembled into 13,807 scaffolds of up to 25.4 Mb, with a mean size of 27 kb and half the genome present in scaffolds of at least 120 kb. The 366-Mb genome is more than twice the size of the Drosophila melanogaster genome (fig. S2A and table S3). Clear conservation of synteny was detected between Glossina and Drosophila, but with the blocks of synteny tending to be twice as large in Glossina due to larger introns and an increase in the size of intergenic sequences, possibly as a result of transposon activity and/or repetitive sequence expansions. Sequences from most of the major groups of retrotransposons and DNA transposons are found in the Glossina genome (table S4). These sequences comprise ~14% of the assembled genome, in contrast to 3.8% of the Drosophila euchromatic genome (15). The Glossina genome is estimated to contain 12,308 protein-encoding genes based on automated and manual annotations. Although this number is fewer than Drosophila, the average gene size in Glossina is almost double that of Drosophila (fig. S2B). The number of exons and their average size is roughly equivalent in both fly species (fig. S2C), but the average intron size in Glossina appears to be roughly twice that of Drosophila (fig. S2D).
Orthologous clusters of proteins were generated by comparing the predicted Glossina protein sequences to five other complete Dipteran genomes (Drosophila melanogaster, Aedes aegypti, Anopheles gambiae, Culex quinquefasciatus, and Phlebotomus papatasi). Each cluster contained members from at least two taxa; groups from a single taxon were considered species-specific paralogs.
In total, 9172 (74%) of Glossina genes (from 8374 orthologous clusters) had a Dipteran ortholog, 2803 genes (23%) had no ortholog/paralog, and 482 (4%) had a unique duplication/paralog in Glossina. The ortholog analysis across the Diptera (fig. S3A) shows that 94% (7867 of 8374) of clusters that contain a Glossina gene also contain an ortholog with Drosophila (fig. S3B).
Blood Feeding and Nutrition
Blood feeding has originated at least 12 times in Diptera, and this genome facilitates a perspective for the comparative evolutionary biology of hematophagy (2). Unlike its distantly related blood-feeding relatives in the suborder Nematocera (such as mosquitoes and sandflies), which supplement their diet with plant nectar, both male and female Glossina use blood as their sole source of nutrients and energy.
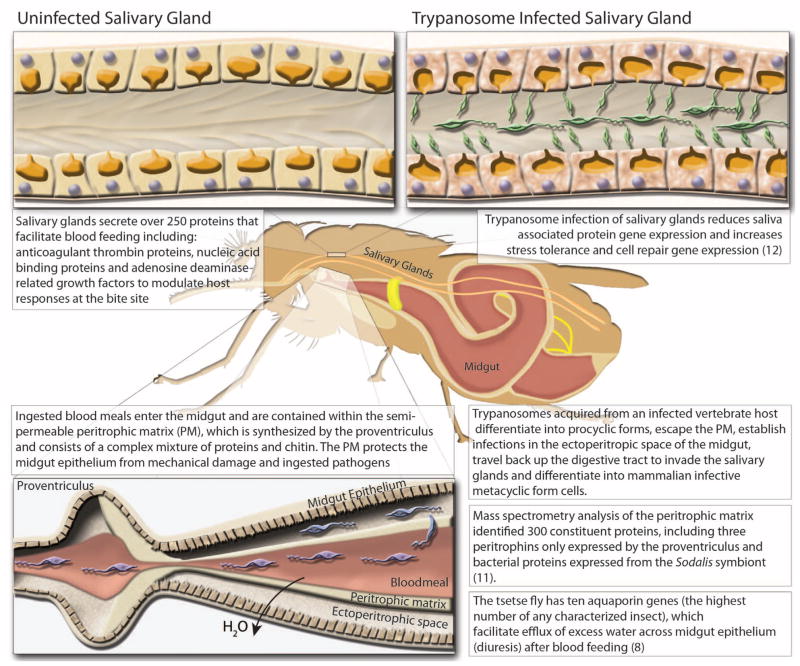
Adult tsetse have several salivary molecules that are essential for efficient blood feeding and digestion because they counteract the complex physiological responses of the host that impede blood feeding, including coagulation, blood platelet aggregation, and vasoconstriction (table S5 and Fig. 1) (16). One gene family, tsal, is the most abundant in the Glossina sialome (16) and encodes high-affinity nucleic acid–binding proteins that lack strong endonuclease activity (17). Orthologs to tsal are not found in Drosophila, but they are present in sandflies (Phlebotomus genus) and mosquitoes (Culex species only). In mosquitoes and sandflies, a single gene is responsible for the production of salivary endonucleases with hydrolysis activity (18). Glossina carries three distinct tsal genes (GMOY012071, GMOY012361, and GMOY012360) that colocalize to a 10-kb locus.
Fig. 1. Diagrammatic presentation of major findings regarding the effects of trypanosome infection of the salivary glands and midgut, proteomic analysis of the peritrophic matrix, and the role of aquaporin proteins in blood meal digestion and diuresis.
(Top) comparison of trypanosome-uninfected and -infected states of Glossina salivary glands. (Left) Representative protein components of Glossina salivary secretions. (Right) Pathogenic effects of trypanosome infection on salivary gland function. (Bottom) Glossina digestive physiology and the infection process by trypanosomes. Associated satellite references for these findings are listed within the figure as numbers in parentheses and correspond to the reference list (8, 11, 12).
Another family of abundant salivary proteins, related to adenosine deaminases and insect growth factors (ADGFs) are thought to reduce the inflammation and irritation resulting from adenosine and inosine-induced mast cell activation. In tsetse, the ADGF genes are uniquely organized as a cluster of four genes in a 20-kb genomic locus (GMOY002973, GMOY012372, GMOY012373, and GMOY012374). An adenosine deaminase (ada) gene (GMOY008741) without the putative growth factor domain is encoded elsewhere in the genome. In Drosophila, five ADGF genes can be found in various loci and are associated with developmental regulation (19). Nematoceran Diptera, including sandflies and mosquitoes, have a maximum of three ADGF genes. Other arthropods, such as Ixodes scapularis, Rhodnius prolixus, and Pediculus humanus, have only bona fide adenosine deaminases.
Recent studies show that specific genes and proteins are down-regulated in salivary glands during parasite infection, which promotes trypanosome transmission because feeding efficiency is reduced and feeding time is extended (20). RNA-seq analysis of salivary gland gene expression during parasite infection confirmed the reduction of transcript abundance for previously identified genes, such as ada, tsal1, tsal2, and 5′ nucleotidase, as well as of many other putative secreted salivary protein genes (12). Additionally, genes involved in stress tolerance and cell repair showed increased expression, indicating that considerable salivary gland tissue damage is caused by trypanosome infections.
Upon blood-meal ingestion, the peritrophic matrix (PM), which separates the midgut epithelium from the blood bolus, protects gut cells from damaging or toxic dietary elements, allows for compartmentalized digestion and metabolism of the blood meal, and is a barrier against infection (5). Glossina produces a type-II PM, which is secreted continuously as concentric sleeves by the proventriculus and separates the lumen of the midgut (endoperitrophic space) from the monolayer of epithelial cells (21). Type-II PMs are generally composed of chitin, peritrophin proteins, glycosaminoglycans (GAGs), and mucin-like molecules. Analysis of isolated PMs of male flies by mass spectrometry identified ~300 proteins, including multiple uncharacterized peritrophins and peritrophin-like glycoproteins. This proteomic data identified the corresponding genes in the Glossina genome. Three of these genes are exclusively expressed by the proventriculus (table S6) (11).
Glossina takes a blood meal that is almost equivalent to its own weight, and excess water is rapidly excreted by means of the Aquaporin family of transport proteins (22). Ten aquaporin genes (aqps) were identified in Glossina, compared with six and eight in mosquitoes and Drosophila, respectively (table S7). In Glossina, two aqp genes are duplicates: the orthologs of the aqp2 and the Drosophila integral protein (drip) genes. Knockdown of aquaporins inhibited post–blood meal diuresis, increased dehydration tolerance, reduced heat tolerance, and extended the duration of lactation and pregnancy in females. The drip orthologs are particularly abundant in the female accessory gland (milk glands), suggesting a role in hydration of glandular secretions (8).
In comparison with mosquitoes and sand-flies, Glossina has a marked reduction in genes associated with carbohydrate metabolism, instead using a proline-alanine shuttle system for energy distribution and triglycerides and diglycerides for storage in the fat body and milk secretions. Little to no sugar nor glycogen is detectable in these flies (23). Genes involved in lipid metabolism are generally conserved, with gene expansions associated with fatty acid synthase, fatty acyl-CoA reductase, and 3-keto acyl-CoA synthase functions (table S8). In addition, three multi-vitamin transporters from the solute:sodium symporter (SSS) family are found in Glossina and mosquitoes, but not in Drosophila, suggesting an association with blood-meal metabolism (table S9).
Microbiome
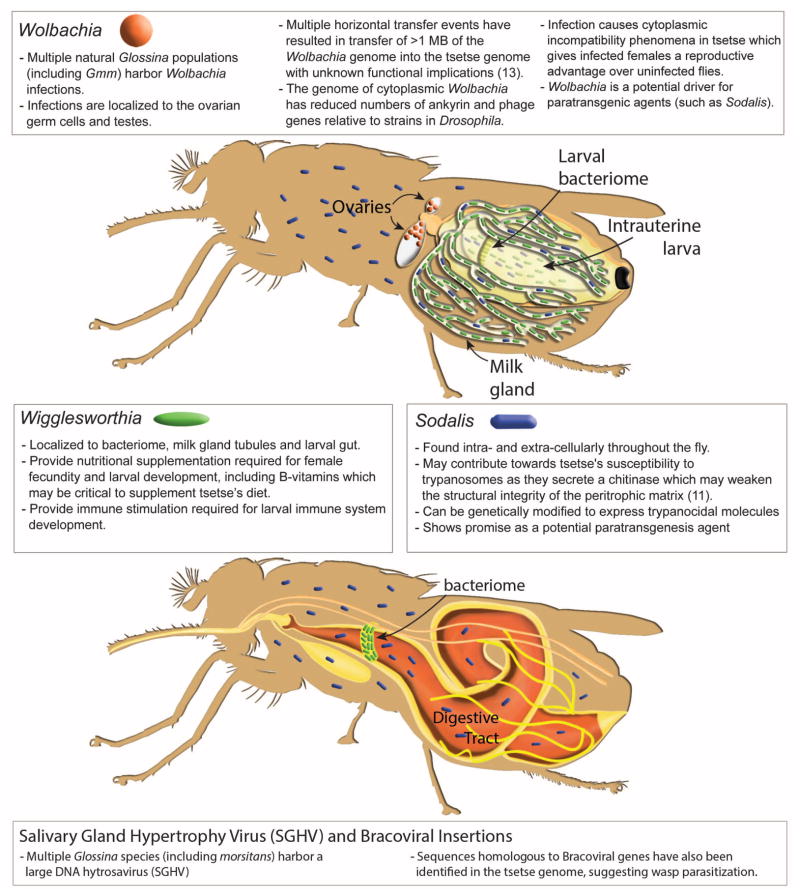
Glossina harbor multiple maternally transmitted mutualistic and parasitic microorganisms, including the obligate Wigglesworthia glossinidia, which reside intracellularly in cells that compromise the midgut-associated bacteriome organ as well as extracellularly in the milk gland lumen (Fig. 2). In the absence of Wigglesworthia, female flies tend to prematurely abort their larval offspring unless they receive dietary supplements (18). However, the larvae that have undergone intrauterine development in the absence of Wigglesworthia metamorphose into immune-compromised adults (24).
Fig. 2. Diagrammatic presentation of the Glossina microbiome, tissue localization of bacterial symbionts, physiological importance, and summary of genomic interactions.
(Top) Glossina reproductive physiology and associated symbiont localizations. (Bottom) Glossina digestive physiology and the associated symbiont localizations. Associated text describes significant findings regarding the Glossina microbiome and the associated impacts in terms of Glossina immunity, nutrition, vectorial capacity, and vector control. Associated satellite references are listed within the figure as numbers in parentheses and correspond to the reference list (11, 13).
The predicted proteome of Wigglesworthia indicates a capacity for B vitamin biosynthesis (25) and synthesis of thiamine monophosphate (TMP). Glossina lacks this capacity; however, it carries genes for thiamine transporters, a member of the reduced folate carrier family (GMOY009200), and a folate transporter (GMOY005445).
Wolbachia is another symbiont present in some wild Glossina populations (and in the strain sequenced here), which resides in gonadal tissues. Laboratory studies have shown that this associated Wolbachia strain induces cytoplasmic incompatibility (CI) in Glossina morsitans (26). Furthermore, at least three horizontal transfer events (HTEs) from Wolbachia were detected in Glossina chromosomes. The two largest insertions carry a total of 159 and 197 putative functional protein-coding genes, whereas the third lacks any protein coding genes. In situ staining of Glossina mitotic chromosomes with Wolbachia-specific DNA probes identified multiple insertions on the X, Y, and B chromosomes (table S10) (13). Although no Wolbachia-specific transcripts were detected arising from chromosomal insertions, the functional and evolutionary implications of these insertions require study.
Many Glossina species, including the strain sequenced here, harbor a large DNA hytrosavirus, the Glossina pallidipes salivary gland hypertrophy virus (GpSGHV) (27). The virus can reduce fecundity and life span in Glossina and cause salivary gland pathology and swelling at high densities. Also, the analysis of a group of genes lacking dipteran orthologs revealed many putative bracoviral genes [Basic Local Alignment Search Tool (BLAST) E values of <1 × 10−50)] spread over 151 genomic scaffolds (tables S11 and S12). The putative bracoviral sequences bear highest homology to those identified from the parasitic braconid wasps Glyptapanteles flavicoxis and Cotesia congregata. This suggests that Glossina was or is parasitized by an as-yet-unidentified braconid wasp.
Immunity
Multiple factors, including age, sex, nutritional status, and symbionts, influence tsetse’s competence as a vector for trypanosomes. Peptidoglycan (PGN) recognition proteins (PGRPs), antimicrobial effector peptides (AMPs) produced by immune deficiency (IMD) pathway, midgut lectins, antioxidants, EP protein (defined by its glutamic acid and proline repeats), and the gut-associated peritrophic matrix structure are all components that regulate the nature of the interactions between the fly and its symbionts (28).
Microbial detection is a multistep process that requires direct contact between host pattern recognition receptors (PRRs) and pathogen-associated molecular patterns (PAMPs). Drosophila has 13 PGRPs that play a role in the recognition of PGN, an essential component of the cell wall of virtually all bacteria. In Glossina, only six pgrp genes were identified, four in the long subfamily (pgrp-la, -lb, -lc, and -ld) and two in the short subfamily (pgrp-sa and -sb), whereas Drosophila has a gene duplication resulting in two related forms of pgrp-sb. Based on both genome annotation and transcriptome data, Glossina lacks orthologs of the PGN receptors, -le, -lf, -sc, and -sd, found in Drosophila. The reduced pgrp repertoire of Glossina may reflect its blood-specific diet, which likely exposes its gut to fewer microbes than Drosophila. In the Drosophila gut, PGRP-LE functions as the master bacterial sensor, which induces both responses to infectious bacteria and tolerance to microbiota by up-regulating suppressors of the IMD pathway, including PGRP-LB (29). In the case of Glossina, loss of amidase -SC1 along with PGRP-LE may indicate a host immune response that has evolved to protect the symbiosis with Wigglesworthia. Reduced immune capacity is also observed in aphids that also harbor obligate symbionts (30). A complete listing of orthologs to Drosophila immune genes is presented in table S13.
Reproduction and Developmental Biology
The reproductive biology of tsetse is unique to the Hippoboscoidea superfamily. The evolution of adenotrophic viviparity (intrauterine larval development and nourishment by glandular secretions) has required ovarian follicle reduction (two follicles per ovary compared with 30 to 40 in Drosophila), expansion and adaptation of the uterus to accommodate developing larvae, and adaptation of the female accessory gland to function as a nutrient synthesis and delivery system.
Glossina, Drosophila, and other Brachyceran flies use lipase-derived yolk proteins for vitellogenesis, unlike non-Brachyceran flies that use the vitellogenin family of yolk proteins (31). Drosophila and Brachyceran flies outside of the Hippoboscidae superfamily produce multiple oocytes per gonotrophic cycle. However, Glossina only develops a single oocyte each cycle. Unlike Drosophila, which has three yolk protein genes (yp1, yp2, and yp3) localized on the X chromosome, Glossina has a single yolk protein gene, which is orthologous to Drosophila yp2 (GMOY002338), expressed only in the ovaries, and lacks fat body–associated expression. Multiple yolk proteins have been identified in other Brachyceran flies, indicating that Glossina may have lost these genes in association with its reduction in reproductive capacity (31).
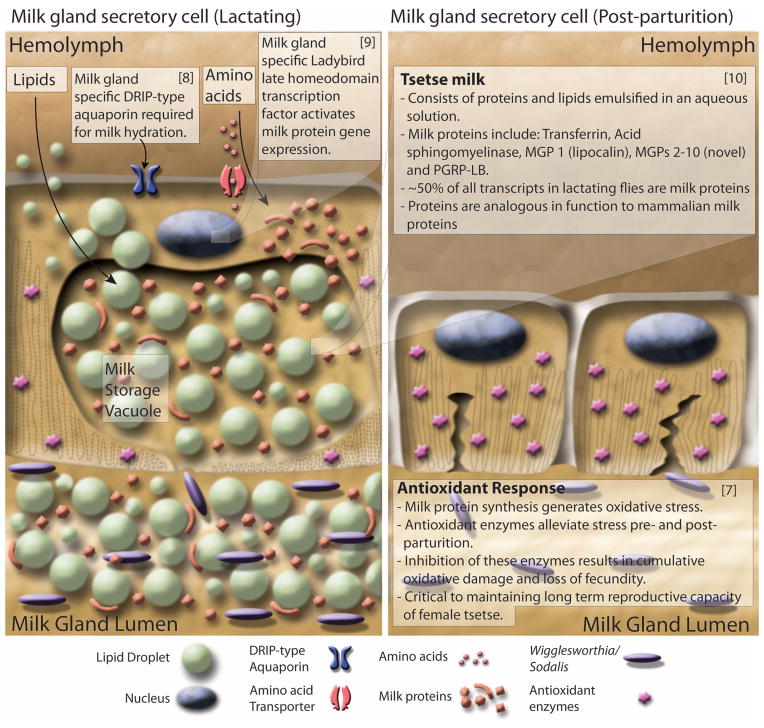
Glossina larvae are dependent on their mother’s milk gland secretions for nutrition, as well as for transfer of symbiotic fauna (Fig. 3). This gland is highly specialized and secretes a complex mixture of stored lipids and milk proteins. Analysis of differential gene expression in lactating versus nonlactating females confirmed the presence of previously characterized milk protein genes—including a lipocalin (mgp1), a transferrin (trf), an acid sphingomyelinase (asmase), milk proteins 2 + 3 (mgp2 and -3), and pgn recognition protein lb (pgrp-lb) (28)—but also revealed a previously undiscovered suite of eight paralogs to the mgp2 and -3 genes. Annotation of the 40-kb genomic loci encompassing mgp2 and mgp3 revealed that these genes have arisen via gene duplication events. These genes have similar exon/intron structures and are expressed in the same stage- and tissue-specific manner as mgp2 and mgp3 (10). The newly identified milk proteins may function as lipid emulsification agents, sources of amino acids, and phosphate carriers. The 12 genes associated with milk synthesis make up almost half of all maternal transcriptional activity during lactation (table S14) (10). The combined suite of Glossina milk proteins bear remarkable functional similarities to those of placental mammals and marsupials (Fig. 3).
Fig. 3. Diagrammatic presentation of milk gland secretory cell physiology and milk production during lactation and after parturition.
(Left) Lactation. Nutrients including lipids, amino acids, and water are taken up by the cell through various transporters. Lipids are aggregated into droplets while amino acids are incorporated into the synthesis of a battery of milk proteins. During pregnancy, milk protein genes are up-regulated by the Ladybird Late homeodomain protein. Lipids, proteins, and water are combined to form the milk constituents, which are stored in a large extracellular secretory reservoir. Stored milk is secreted into the lumen, which also houses the extracellular obligate bacterial symbiont Wigglesworthia. The milk and symbionts are transported through the gland to the uterus, where the developing larvae feeds upon these secretions. (Right) Involution and recovery. After parturition, milk gland cells shrink, undergo autophagy, and express antioxidant enzymes to inhibit oxidative damage. The recovered cells prepare for the next round of lactation by regeneration of protein synthesis and structural components. Associated satellite references for these findings are listed within the figure as numbers in parentheses and correspond to the reference list (7–10).
The massive level of protein synthesis during lactation generates substantial oxidative stress in tsetse females, but females can undergo this process 8 to 10 times during their life spans without evidence of reproductive senescence. Transcriptional analysis of antioxidant enzyme (AOE) gene expression revealed an increase in abundance of these genes during lactation and after birth (7) (table S15), such that knockdown of these enzymes decreases fecundity in subsequent reproductive cycles. The mediation of oxidative stress by AOEs at key points in tsetse reproduction appears critical to preservation of fecundity late into Glossina’s life span (7) (Fig. 3).
The milk proteins produced by tsetse are under tight transcriptional regulation and are only expressed in the female accessory gland. The expression level of these genes is coordinated with the stage of pregnancy and increases with larval development. The system regulating these genes appears conserved as transgenic Drosophila carrying the mgp1 gene promoter sequence drive the expression of a green fluorescent protein reporter gene exclusively in the female accessory glands in coordination with oogenesis/ovulation. Comparative analysis of the promoter sequences from multiple milk protein genes revealed the presence of conserved binding sites for homeo-domain transcription factors. Analysis of the homeodomain transcription factors in Glossina (table S16) identified a gene, ladybird late (lbl), which is expressed exclusively in the milk gland of adult female flies and the female accessory glands of Drosophila. Knockdown of lbl results in a global reduction of milk gland protein expression in tsetse and causes loss of fecundity (9) (Fig. 3).
Sensory Genes as Targets for Glossina Control Strategies
Glossina species differ in host preferences and vary in their response to chemical and visual cues from different mammalian hosts or for mate finding. Sensory proteins range from odorant binding proteins (OBP), chemosensory proteins (CSP), odorant receptors (OR), gustatory receptors (GR), and ligand-gated ionotropic receptors (IR) to sensory neuron membrane proteins (SNMP) (32).
Detailed annotation of Glossina sensory receptors reveals that they have fewer olfactory proteins relative to Drosophila, An. gambiae, and Apis mellifera (table S17) (14). Of note, six ORs are homologous to a single Drosophila OR, which is associated with female mating deterrence. In addition, GR genes associated with sweet tastes, present in all other Diptera, are missing in tsetse. These genetic differences are consistent with the combination of a restricted diet of vertebrate blood and their narrow host range.
The visual system of Glossina is very similar to that of other calyptrate Diptera, which are generally fast flying, such as the house fly Musca domestica and the blow fly Calliphora vicina (33). In tsetse, both sexes employ vision for rapid host identification and pursuit (34); males, however, also depend on vision for long-distance spotting and tracking of female mating partners (35). Morphology and function of the compound eye retina is highly conserved throughout the Brachycera, allowing for direct comparisons with Drosophila (36). The search for vision-associated genes revealed that all of the core components of the highly efficient Drosophila phototransduction cascade are conserved in Glossina (table S18). This is also the case for four of the five opsin transmembrane receptor genes that are differentially expressed in the photoreceptors of the Drosophila compound eye: Rh1, Rh3, Rh5, and Rh6. Most important, the recovery of opsin Rh5 indicates the likely presence of blue-sensitive R8p photoreceptors in Glossina that have been missed in experimental studies (33). This finding is consistent with tsetse’s attraction to blue/black, which has been widely exploited for the development of traps to reduce vector populations (37). It is further notable that the study of opsin conservation and expression in the blow fly retina recovered the same four opsin paralogs (38), suggesting that the deployment of a single ultraviolet (UV)–sensitive opsin (Rh3) represents the ground state for calyptrate Diptera, in contrast to the expression of two UV-sensitive opsins (Rh3 and Rh4) in the eyes of Drosophila. The Glossina genome also contains the ortholog of the Drosophila Rh7 opsin gene, which is still of unknown function in Drosophila.
Future Directions
The assembly and annotation of the Glossina genome highlights its unique biology and facilitates the application of powerful high-throughput technologies in a way that was previously impossible. In addition, genomic and transcriptomic data on five Glossina species (G. fuscipes fuscipes, G. palpalis gambiensis, G. brevipalpis, G. austeni, and G. pallidipes) are being generated to produce additional genome assemblies for evolutionary and developmental analyses to study genomic differences associated with host specificity, vectorial capacity, and evolutionary relationships.
Acknowledgments
The public release and future updates of the genome sequence and associated information are hosted at VectorBase (www.vectorbase.org). The genome sequence can also be found at GenBank under the accession no. CCAG010000000. The Glossina morsitans genome project was funded by the Wellcome Trust (grants 085775/Z/08/Z and 098051) and World Health Organization (WHO) Special Programme for Research and Training in Tropical Diseases (TDR) (project no. A90088) and the Ambrose Monell Foundation. Authors also acknowledge support by the Food and Agriculture Organization/International Atomic Energy Agency Coordinated Research Program “Improving SIT for Tsetse Flies through Research on their Symbionts,” the European Union Cooperation in Science and Technology (COST) Action FA0701 “Arthropod Symbiosis: From Fundamental Studies to Pest and Disease Management,” and a grant-in-aid for Scientific Research on Priority Areas “Comprehensive Genomics” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to M.H.). BAC libraries were generated through National Institute of Allergy and Infectious Diseases resources, and sequencing was supported by RIKEN Japan. We thank the staff in the library construction, sequence production, and informatics support groups at the Wellcome Trust Sanger Institute.
Members of the International Glossina Genome Initiative (IGGI)
Project leadership and conception: Junichi Watanabe,76 Masahira Hattori,6 Matthew Berriman,64 Michael J. Lehane,43 Neil Hall,53,79 Philippe Solano,49 Serap Aksoy,36 Winston Hide,67,80 Yeya Touré68. Manual annotation coordinator, editor, and illustrator: Geoffrey M. Attardo36. Sequence production, assembly and global analysis: Alistair C. Darby,53 Atsushi Toyoda,7 Christiane Hertz-Fowler,64 Denis M. Larkin,51 James A. Cotton,64 Junichi Watanabe,76 Mandy J. Sanders,64 Martin T. Swain,51 Masahira Hattori,6 Matthew Berriman,64 Michael A. Quail,64 Noboru Inoue,63 Sophie Ravel,50 Todd D. Taylor,66 Tulika P. Srivastava,66,74 Vineet Sharma,78,66 Wesley Warren,69 Richard K. Wilson,69 Yutaka Suzuki6. Annotation automatic, manual annotation capture, and public release: Daniel Lawson,47 Daniel S. T. Hughes,47 Karyn Megy47. Olfaction group leaders: Daniel K. Masiga,61 Paul O. Mireji10. Reproduction and development group leader: Geoffrey M. Attardo36. Signaling group leader: Immo A. Hansen21. Salivary group leader: Jan Van Den Abbeele24. Metabolism and stress response group leader: Joshua B. Benoit14,36. Horizontal transfer group leader: Kostas Bourtzis34,3,35. Digestion group leader: Michael J. Lehane43. Immunity group leader: Serap Aksoy36. Sensory annotations: Daniel K. Masiga,61 George F. O. Obiero,61,67 Hugh M. Robertson,33 Jeffery W. Jones,17 Jing-Jiang Zhou,13 Linda M. Field,13 Markus Friedrich,17 Paul O. Mireji,10 Steven R. G. Nyanjom11. Salivary annotations: Erich L. Telleria,36 Guy Caljon,24 Jan Van Den Abbeele,24 José M. C. Ribeiro57. Midgut and digestion annotations: Alvaro Acosta-Serrano,42,43 Joshua B. Benoit,14,36 Cher-Pheng Ooi,43 Clair Rose,42 David P. Price,21 Lee R. Haines,43 Michael J. Lehane43. Metabolism annotations: Alan Christoffels,67 Cheolho Sim,19 Daphne Q. D. Pham,16 David L. Denlinger,31 Dawn L. Geiser,40 Irene A. Omedo,26 Joshua B. Benoit,14,36 Joy J. Winzerling,39 Justin T. Peyton,37 Kevin K. Marucha,10 Mario Jonas,67 Megan E. Meuti,31 Neil D. Rawlings,60 Paul O. Mireji,10 Qirui Zhang,31 Rosaline W. Macharia,5,67 Veronika Michalkova,36,54 Zahra Jalali Sefid Dashti67. Signaling annotations: Aaron A. Baumann,65 Gerd Gäde,15 Heather G. Marco,15 Immo A. Hansen,21 Jelle Caers,20 Liliane Schoofs,20 Michael A Riehle,32 Wanqi Hu,12 Zhijian Tu12. Reproduction and development annotations: Aaron M Tarone,30 Anna R. Malacrida,18 Caleb K. Kibet,61 Joshua B. Benoit,14,36 Francesca Scolari,18 Geoffrey M. Attardo,36 Jacobus J. O. Koekemoer,44,46 Judith Willis,25 Ludvik M. Gomulski,18 Marco Falchetto,18 Maxwell J. Scott,29 Shuhua Fu,9 Sing-Hoi Sze,28 Thiago Luiz36. Immunity annotations: Brian Weiss,36 Deirdre P. Walshe,43 Jingwen Wang,36 Joshua B. Benoit,14,36 Geoffrey M. Attardo,36 Mark Wamalwa,67,77 Sarah Mwangi,67 Serap Aksoy,36 Urvashi N. Ramphul43. Horizontal transfer and microbiome annotations: Anna K. Snyder,23 Corey L. Brelsfoard,38 Gavin H. Thomas,22 George Tsiamis,35 Kostas Bourtzis,34,3,35 Peter Arensburger,2 Rita V. M. Rio,23 Sandy J Macdonald,22 Sumir Panji67,8. IGGI annotation workshop contributors: Adele Kruger,67 Alan Christoffels,67 Alia Benkahla,48 Apollo S. P. Balyeidhusa,59 Atway Msangi,70 Cher-Pheng Ooi,43 Chinyere K. Okoro,72 Daniel K. Masiga,61 Dawn Stephens,58 Deirdre P. Walshe,43 Eleanor J. Stanley,47 Feziwe Mpondo,67 Florence Wamwiri,56 Furaha Mramba,70 Geoffrey M. Attardo,36 Geoffrey Siwo,45 George F. O. Obiero,61,67 George Githinji,75 Gordon Harkins,67 Grace Murilla,56 Heikki Lehväslaiho,1 Imna Malele,70 Jacobus J. O. Koekemoer,44,46 Joanna E. Auma,56 Johnson K. Kinyua,11 Johnson Ouma,56,71 Junichi Watanabe,76 Karyn Megy,47 Loyce Okedi,62 Lucien Manga,73 Mario Jonas,67 Mark Wamalwa,67,77 Martin Aslett,64 Mathurin Koffi,49 Matthew Berriman,64 Michael J. Lehane,43 Michael W. Gaunt,41 Mmule Makgamathe,58 Neil Hall,53,79 Nicola Mulder,8 Oliver Manangwa,70 Patrick P. Abila,62 Patrick Wincker,4 Paul O. Mireji,10 Richard Gregory,53 Rita V. M. Rio,23 Rosemary Bateta,10 Ryuichi Sakate,55 Serap Aksoy,36 Sheila Ommeh,52 Stella Lehane,43 Steven R. G. Nyanjom,11 Tadashi Imanishi,55 Todd D. Taylor,66 Victor C. Osamor,27 Vineet Sharma,66,78 Winston Hide,67,80 Yoshihiro Kawahara55,81 Joshua B. Benoit14,36
Footnotes
Biological and Environmental Sciences and Engineering, King Abdullah University of Science and Technology (KAUST), 4700 King Abdullah University of Science and Technology, Thuwal, 23955-6900, Kingdom of Saudi Arabia.
Biological Sciences Department, California State Polytechnic University Pomona, 3801 West Temple Avenue, Pomona, CA 91768, USA.
Biomedical Sciences Research Center, Alexander Fleming Biomedical Sciences Research Center, 34 Fleming Street, Vari, 16672, Greece.
CEA, Genoscope, 2 Rue Gaston Crémieux, CP5706, Evry Cedex, 91507, France.
Center for Biotechnology and Bioinformatics, University of Nairobi, Post Office Box 30197-00100, Nairobi, Kenya, Nairobi, Kenya.
Center for Omics and Bioinformatics, Department of Computational Biology, Graduate School of Frontier Sciences, The University of Tokyo, 5-1-5 Kashiwanoha, Kashiwa, Chiba, 277-8561, Japan.
Comparative Genomics Laboratory, National Institute of Genetics, 411-8540 Yata 1111, Mishima, Shizuoka, 411-8540, Japan.
Computational Biology Group, IIDMM, University of Cape Town Faculty of Health Sciences, Cape Town, 7925, South Africa.
Department of Biochemistry and Biophysics, Texas A&M University, 328B TAMU, College Station, TX 77843, USA.
Department of Biochemistry and Molecular Biology, Egerton University, Post Office Box 536, Njoro, Kenya.
Department of Biochemistry, Jomo Kenyatta University of Agriculture and Technology (JKUAT), Post Office Box 62000-00200, Nairobi, Kenya.
Department of Biochemistry, Virginia Polytechnic Institute and State University, 309 Fralin Hall, Blacksburg, VA 24061, USA.
Department of Biological Chemistry and Crop Protection, Rothamsted Research, West Common, Harpenden, Herts, AL5 2JQ, UK.
Department of Biological Sciences, McMicken College of Arts and Sciences, University of Cincinnati, Cincinnati, OH 45221, USA.
Department of Biological Sciences, University of Cape Town, Private Bag, Rondebosch, ZA-7700, South Africa.
Department of Biological Sciences, University of Wisconsin–Parkside, 900 Wood Road, Kenosha, WI 53144, USA.
Department of Biological Sciences, Wayne State University, 5047 Gullen Mall, Detroit, MI 48202, USA.
Department of Biology and Biotechnology, University of Pavia, Via Ferrata 9, Pavia, 27100, Italy.
Department of Biology, Baylor University, Waco, TX 76798, USA.
Department of Biology, KU Leuven, Naamsestraat 59, Leuven, B-3000, Belgium.
Department of Biology, New Mexico State University, Foster Hall 263, Las Cruces, NM 88003, USA.
Department of Biology, University of York, Wentworth Way, York, Y010 5DD, UK.
Department of Biology, West Virginia University, 53 Campus Drive, 5106 LSB, Morgantown, WV, USA.
Department of Biomedical Sciences, Institute of Tropical Medicine Antwerp, Nationalestraat 155, Antwerp, B-2000, Belgium.
Department of Cellular Biology, University of Georgia, 302 Biological Sciences Building, Athens, GA 30602, USA.
Department of Clinical Research, KEMRI-Wellcome Trust Programme, CGMRC, Post Office Box 230-80108, Kilifi, Kenya.
Depart-ment of Computer and Information Sciences, College of Science and Technology, Covenant University, P.M.B. 1023, Ota, Ogun State, Nigeria.
Department of Computer Science and Engineering, Department of Biochemistry and Biophysics, Texas A&M University, HRBB 328B TAMU, College Station, TX 77843, USA.
Department of Entomology, North Carolina State University, Campus Box 7613, Raleigh, NC 27695–7613, USA.
Department of Entomology, Texas A&M University, 2475 TAMU, College Station, TX 77843, USA.
Department of Entomology, The Ohio State University, 400 Aronoff Laboratory, 318 West 12th Avenue, Columbus, OH 43210, USA.
Department of Entomology, University of Arizona, 1140 East South Campus Drive, Forbes 410, Tucson, AZ 85721, USA.
Department of Entomology, University of Illinois at Urbana-Champaign, 505 South Goodwin Avenue, Urbana, IL 61801, USA.
Insect Pest Control Laboratory, Joint FAO/IAEA Programme of Nuclear Techniques in Food and Agriculture, Vienna, 1220, Austria.
Department of Environmental and Natural Resources Management, University of Patras, 2 Seferi Street, Agrinio, 30100, Greece.
Department of Epidemiology of Microbial Diseases, Yale School of Public Health, 60 College Street, New Haven, CT 06520, USA.
Department of Evolution, Ecology, and Organismal Biology, The Ohio State University, 300 Aronoff Laboratory, 318 West 12th Avenue, Columbus, OH 43210, USA.
Department of Natural Sciences, St. Catharine College, 2735 Bardstown Road., St. Catharine, KY 40061, USA.
Department of Nutritional Sciences, University of Arizona, Career and Academic Services, College of Agriculture and Life Sciences, Forbes Building, Room 201, Post Office Box 210036, Tucson, AZ 85721–0036, USA.
Department of Nutritional Sciences, University of Arizona, Shantz 405, 1177 East 4th Street, Tucson, AZ 85721–0038, USA.
Department of Pathogen Molecular Biology, London School of Hygiene and Tropical Medicine, Keppel Street, London, WC1E 7HT, UK.
Department of Parasitology, Liverpool School of Tropical Medicine, Pembroke Place, Liverpool, L3 5QA, UK.
Department of Vector Biology, Liverpool School of Tropical Medicine, Pembroke Place, Liverpool, L3 5QA, UK.
Entomology Section, Onderstepoort Veterinary Institute, Private Bag X5, Onderstepoort, 110, South Africa.
Eck Institute for Global Health, Department of Biological Sciences, University of Notre Dame, Notre Dame, IN 46556, USA.
Department of Veterinary Tropical Diseases, University of Pretoria, Private Bag X04, Onderstepoort, 110, South Africa.
European Molecular Biology Laboratories, European Bio-informatics Institute (EMBL-EBI), Wellcome Trust Genome Campus, Hinxton, Cambridge, Cambridgeshire, CB10 1SA, UK.
Group of Bioinformatics and Modeling, Laboratory of Medical Parasitology, Biotechnology, and Biomolecules, Institut Pasteur de Tunis, 13, Place Pasteur, BP74, Belvédère, Tunis, 1002, Tunisia.
Institut de Recherche pour le Développement (IRD), UMR 177 IRD-CIRAD INTERTRYP, CIRDES Bobo-Dioulasso, Burkina Faso.
Institut de Recherche pour le Développement (IRD), UMR 177 IRD-CIRAD INTERTRYP, LRCT Campus International de Baillarguet, Montpellier, France.
Institute of Biological, Environmental, and Rural Sciences, University of Aberystwyth, Old College, King Street, Aberystwyth, Ceredigion, SY23 3FG, UK.
Institute of Biotechnology Research, Jomo Kenyatta University of Agriculture and Technology (JKUAT), Post Office Box 62000-00200, Nairobi, Kenya.
Institute of Integrative Biology, The University of Liverpool, Crown Street, Liverpool, L69 7ZB, UK.
Institute of Zoology, Slovak Academy of Sciences, Dúbravská cesta 9, Bratislava, 845 06, Slovakia.
Integrated Database Team, Biological Information Research Center, National Institute of Advanced Industrial Science and Technology, Aomi 2-4-7, Kotoku, Tokyo, 135-0064, Japan.
Kenya Agricultural Research Institute Trypanosomiasis Research Centre, Post Office Box 362, Kikuyu, 902, Kenya.
Laboratory of Malaria and Vector Research, National Institute of Allergy and Infectious Diseases, 12735 Twinbrook Parkway, Room 2E-32D, Rockville, MD 20852, USA.
Technology Innovation Agency, National Genomics Platform, Post Office Box 30603, Mayville, Durban, 4058, South Africa.
Department of Biochemistry and Sports Science, Makerere University, Post Office Box 7062, Kampala, Uganda.
Bateman Group, Wellcome Trust Sanger Institute, EMBL European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, Cambridgeshire, CB10 1SA, UK.
Molecular Biology and Bioinformatics Unit, International Center of Insect Physiology and Ecology, Duduville Campus, Kasarani, Post Office Box 30772-00100, Nairobi, Kenya.
National Livestock Resources Research Institute (NaLIRRI), Post Office Box 96, Tororo, Uganda.
National Research Center for Protozoan Diseases, Obihiro University of Agriculture and Veterinary Medicine, Inada-cho, Obihiro, Hokkaido, 080-8555, Japan.
Parasite Genomics Group, Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, Cambridgeshire, CB10 1SA, UK.
Riddiford Laboratory, Janelia Farm Research Campus, Howard Hughes Medical Institute, 19700 Helix Drive, Ashburn, VA 20147, USA.
RIKEN Center for Integrative Medical Sciences, 1-7-22 Suehiro-cho, Tsurumiku, Yokohama, Kanagawa, 230-0045, Japan.
South African National Bioinformatics Institute, South African MRC Bio-informatics Unit, University of the Western Cape, 5th Floor Life Sciences Building, Modderdam Road, Bellville 7530, South Africa.
Special Programme for Research and Training in Tropical Diseases (TDR), WHO, Avenue Appia 20, 1211 Geneva 27, Switzerland.
The Genome Institute, Washington University School of Medicine, St. Louis, MO 63110, USA.
Tsetse and Trypanosomiasis Research Institute (TTRI), Majani Mapana, Off Korogwe Road, Post Office Box 1026, Tanga, Tanzania.
Vector Health International, Post Office Box 15500, Arusha, Tanzania.
Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, Cambridgeshire, CB10 1SA, UK.
WHO Regional Office for Africa, WHO, Cité du Djoué, Post Office Box 06, Brazzaville, Congo.
School of Basic Sciences, Indian Institute of Technology, Mandi 175001, Himachal Pradesh, India.
Department of Parasite, Vector, and Human Biology, KEMRI-Wellcome Trust Programme, CGMRC, Post Office Box 230-80108, Kilifi, Kenya.
Institute of Medical Science, The University of Tokyo, 4-6-1 Shirokanedai, Minatoku, Tokyo, 108-8639, Japan.
Department of Biochemistry and Biotechnology, Kenyatta University, Post Office Box 43844-00100, Nairobi, Kenya.
Department of Biological Sciences, Indian Institute of Science Education and Research, Indore Bypass Road, Bhauri District, Bhopal, Madhya Pradesh, 462066, India.
Faculty of Science, King Abdulaziz University, Jeddah, 21589, SA.
Department of Biostatistics, Harvard School of Public Health, 655 Huntington Ave. Boston, MA 02461.
Bioinformatics Research Unit, Agrogenomics Research Center, National Institute of Agrobiological Sciences, 2-1-2, Kannondai, Tsukuba, Ibaraki 305-8602, Japan.
Supplementary Materials
www.sciencemag.org/content/344/6182/380/suppl/DC1
Materials and Methods
Supplementary Text Figs. S1 to S9
Tables S1 to S43
References (39–101)
References and Notes
- 1.Welburn SC, Maudlin I, Simarro PP. Parasitology. 2009;136:1943–1949. doi: 10.1017/S0031182009006416. [DOI] [PubMed] [Google Scholar]
- 2.Wiegmann BM, et al. Proc Natl Acad Sci USA. 2011;108:5690–5695. doi: 10.1073/pnas.1012675108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petersen FT, Meier R, Kutty SN, Wiegmann BM. Mol Phylogenet Evol. 2007;45:111–122. doi: 10.1016/j.ympev.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 4.Lehane MJ, Aksoy S, Levashina E. Trends Parasitol. 2004;20:433–439. doi: 10.1016/j.pt.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Weiss BL, Wang J, Maltz MA, Wu Y, Aksoy S. PLOS Pathog. 2013;9:e1003318. doi: 10.1371/journal.ppat.1003318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aksoy S, et al. Trends Parasitol. 2005;21:107–111. doi: 10.1016/j.pt.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Michalkova V, Benoit JB, Attardo GM, Medlock J, Aksoy S. PLOS ONE. 2014;9:e87554. doi: 10.1371/journal.pone.0087554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benoit JB, et al. PLOS Negl Trop Dis. 2014;8:e2517. doi: 10.1371/journal.pntd.0002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Attardo GM, et al. PLOS Negl Trop Dis. 2014;10:e2645. doi: 10.1371/journal.pntd.0002645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benoit JB, et al. PLOS Genet. 2014;10:e1003874. doi: 10.1371/journal.pgen.1003874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rose C, et al. PLOS Negl Trop Dis. 2014;8:e2691. doi: 10.1371/journal.pntd.0002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Telleria EL, et al. PLOS Negl Trop Dis. 2014;8:e2649. doi: 10.1371/journal.pntd.0002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brelsfoard C, et al. PLOS Negl Trop Dis. 2014;8:e2728. doi: 10.1371/journal.pntd.0002728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obiero GFO, et al. PLOS Negl Trop Dis. 2014;8:e2663. doi: 10.1371/journal.pntd.0002663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaminker JS, et al. Genome Biol. 2002;3:RESEARCH0084. doi: 10.1186/gb-2002-3-12-research0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alves-Silva J, et al. BMC Genomics. 2010;11:213. doi: 10.1186/1471-2164-11-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caljon G, et al. PLOS ONE. 2012;7:e47233. doi: 10.1371/journal.pone.0047233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribeiro JM, Mans BJ, Arcà B. Insect Biochem Mol Biol. 2010;40:767–784. doi: 10.1016/j.ibmb.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dolezal T, Dolezelova E, Zurovec M, Bryant PJ. PLOS Biol. 2005;3:e201. doi: 10.1371/journal.pbio.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Den Abbeele J, Caljon G, De Ridder K, De Baetselier P, Coosemans M. PLOS Pathog. 2010;6:e1000926. doi: 10.1371/journal.ppat.1000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehane MJ. Annu Rev Entomol. 1997;42:525–550. doi: 10.1146/annurev.ento.42.1.525. [DOI] [PubMed] [Google Scholar]
- 22.Campbell EM, Ball A, Hoppler S, Bowman AS. J Comp Physiol B. 2008;178:935–955. doi: 10.1007/s00360-008-0288-2. [DOI] [PubMed] [Google Scholar]
- 23.Norden DA, Paterson DJ. Comp Biochem Physiol. 1969;31:819–827. doi: 10.1016/0010-406x(69)92082-9. [DOI] [PubMed] [Google Scholar]
- 24.Weiss BL, Maltz M, Aksoy S. J Immunol. 2012;188:3395–3403. doi: 10.4049/jimmunol.1103691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rio RV, et al. MBio. 2012;3:e00240–11. [Google Scholar]
- 26.Alam U, et al. PLOS Pathog. 2011;7:e1002415. doi: 10.1371/journal.ppat.1002415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abd-Alla AM, et al. J Virol. 2008;82:4595–4611. doi: 10.1128/JVI.02588-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dyer NA, Rose C, Ejeh NO, Acosta-Serrano A. Trends Parasitol. 2013;29:188–196. doi: 10.1016/j.pt.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Bosco-Drayon V, et al. Cell Host Microbe. 2012;12:153–165. doi: 10.1016/j.chom.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Elsik CG. Genome Biol. 2010;11:106. doi: 10.1186/gb-2010-11-2-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hens K, Lemey P, Macours N, Francis C, Huybrechts R. Insect Mol Biol. 2004;13:615–623. doi: 10.1111/j.0962-1075.2004.00520.x. [DOI] [PubMed] [Google Scholar]
- 32.Liu R, et al. Insect Mol Biol. 2012;21:41–48. doi: 10.1111/j.1365-2583.2011.01114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardie R, Vogt K, Rudolph A. J Insect Physiol. 1989;35:423–431. [Google Scholar]
- 34.Gibson G, Torr SJ. Med Vet Entomol. 1999;13:2–23. doi: 10.1046/j.1365-2915.1999.00163.x. [DOI] [PubMed] [Google Scholar]
- 35.Brady J. Physiol Entomol. 1991;16:153–161. [Google Scholar]
- 36.Friedrich M. Encyclopedia of Life Sciences. Wiley; Chichester: 2010. [DOI] [Google Scholar]
- 37.Lindh JM, et al. PLOS Negl Trop Dis. 2012;6:e1661. doi: 10.1371/journal.pntd.0001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitt A, Vogt A, Friedmann K, Paulsen R, Huber A. J Exp Biol. 2005;208:1247–1256. doi: 10.1242/jeb.01527. [DOI] [PubMed] [Google Scholar]