Abstract
Replication forks in both prokaryotic and eukaryotic systems pause at random sites due to depletion of dNTP pools, DNA damage, tight binding nonhistone proteins or unusual DNA sequences and/or structures, in a mostly non-polar fashion. However there is also physiologically programmed replication termination at sequence-specific authentic replication termini. Here, the structure and functions of programmed replication termini, their mechanism of action and their diverse physiological functions in prokaryotes and eukaryotes have been reviewed.
Keywords: Replication termination, Terminator protein, Crystal structure, Recombination, Replicative aging, Gene silencing
The replicon hypothesis introduced the concepts of a unit of replication consisting of a cis-acting replication origin (ori) and a trans-acting initiator protein which interacted with the ori to control replication initiation. However, the model did not take into consideration whether there might also be defined replication termini (Ter) at which replication was completed [1].The prevailing notion at the time was that replication stopped wherever the two divergent forks, initiated from a fixed ori, met each other on the circular chromosome. While bacterial and certain plasmid chromosomes (e.g. of R6K) contain sequence-specific replication termini, some other plasmids (e.g.,ColE1) and all phage chromosomes, that have been studied so far, do not seem to have specific Ter sites [see reviews in [2-7]].
DNA replication forks can stall randomly because of various reasons such as depletion of the dNTP pool, DNA damage, at barriers formed by certain DNA sequences or strong DNA binding nonhistone proteins etc. [8,9]. Forks are also arrested at physiologically programmed authentic replication termini, usually in a polar mode, to facilitate certain DNA transactions. This latter class is the subject of this review. The first part of this review is focused on prokaryotic replication termination (of E. coli, B. subtilis and the plasmid R6K). It discusses the Ter sequences, crystal structures of the corresponding terminator proteins, their structure-function analysis, a critical evaluation of the models of polar fork arrest and its physiological significance. The second part discusses similar points mostly from two model eukaryotic systems namely S. cerevisiae and S. pombe, along with brief references to fork arrest in mammalian and other systems.
Replication termination in prokaryotes
E. coli and plasmid Ter systems
The first clear evidence for a site specific terminator site(s) was uncovered in the plasmid R6K. The plasmid has 3 ori sequences called α, β and γ that are located within a kb of each other. The origins initiate unidirectional replication with one ori firing per molecule. From the ori(s) located at 12 o'clock in the circular DNA, fork travel unidirectionally until reaching the 2 Ter sites (present as inverted repeats) at 2 o'clock. Then the second fork at the ori fires unidirectionally in the opposite direction until meeting the first one stalled at the Ter sites. Thus, the topology of replication is sequentially bidirectional due to an asymmetrically located Ter region with respect to the origins [10,11]. The host replication termini were discovered shortly thereafter and were found to be located at the antipode (Fig.1A) with respect to the ori, in the circular chromosomes [12,13]. The replication terminus regions of R6K and of the host E. coli were cloned and sequenced [14,15]. The activity of the replication termini in vitro was first investigated using partially fractionated cell extracts [16] and then reconstituted with a system of 26 purified proteins [17].
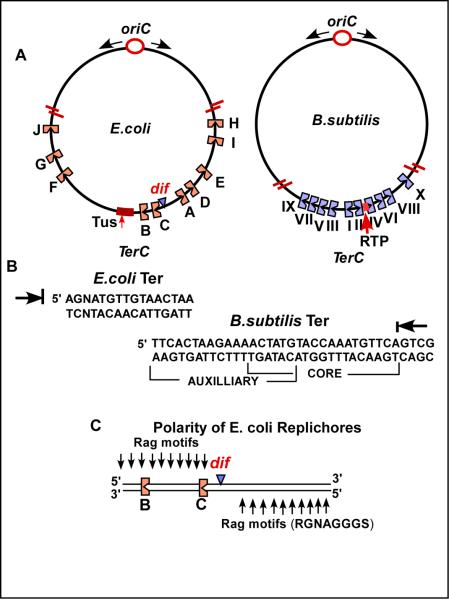
Fig.1.
The TerC regions of E. coli and B. subtilis and the terminator sequences. A, the relative locations of the Ter sites in the two TerC regions are shown including the regions encoding the terminator proteins Tus and RTP; the TerC regions are expanded and not drawn to scale; B, the rag sequence motifs and the polarized replicores in the TerC region of E. coli; C, the sequences of the Ter sites of E. coli and B. subtilis with the arrows showing the ends that arrest forks; the Ter site of B. subtilis shows the overlapping core and auxiliary sites.
Hill cloned the Tus gene of E. coli that encodes the host terminator protein [18-20]. The Tus protein was independently identified and purified by other groups [21,22]. The protein was shown to bind specifically to Ter sites [18,22,23]. E. coli chromosome contains two groups of 5 Ter sites of opposite polarity that are arranged in such a way that they form a replication trap that restricts fork arrest to the antipodal TerC region. Each cluster of 5 Ter sites of the same polarity located on one replichore (also called chirochore). Replichore refers to each arm of the chromosome from ori to Ter traced clockwise and anticlockwise that have sequence polarity [see Fig.1A and [24]]. Each replichore is marked by G rich sequences called Rag motifs that switch to polarity at the antipode (Fig.1C).
Crystal structure of the Tus-Ter system and structure- guided mutagenesis
How does a Tus-Ter complex arrest a replication fork in a polar mode? At least a partial understanding of the mechanism of polar fork arrest came from in vitro replication work using purified Tus supplementing a partially fractionated cell extract and a plasmid DNA substrate containing a unidirectionally replicating ori and a Ter placed in opposite orientations with respect to the origin. The reasoning was that since the DnaB helicase is the main biological motor that drives the fork, in coordination with DNA polymerase, it could be a target of the Tus-Ter complex which might have a polar contra-helicase (or antihelicase) activity. We and others investigated this hypothesis and confirmed that Tus-Ter complex impedes both fork movement and DnaB catalyzed duplex unwinding with the same polarity [25,26]. DNA polymerase I was also impeded in a non-polar fashion by Tus-Ter and this is believed to be of a non-physiological nature [27].
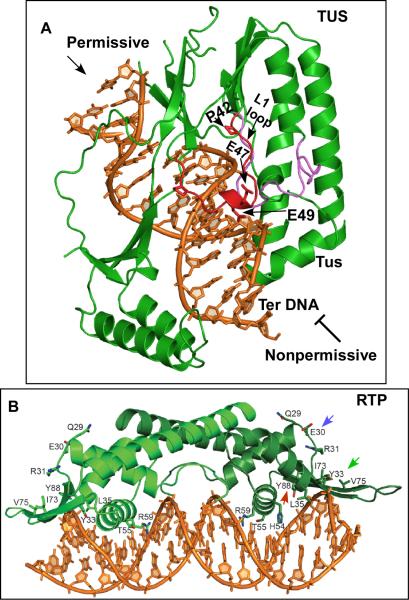
In order to further uncover the mechanism of polar fork arrest the crystal structure of the Tus-Ter complex needed to be solved. Historically, the first replication terminator protein structure to be solved was that of the RTP apoprotein (Replication Terminator Protein) of B. subtilis [28] followed by the crystal structure of a Tus-Ter complex of E. coli [29]. The termination system of B. subtlis will be discussed in a following section. Tus has a bilobed structure with its beta strands invading the major groove of the DNA that generates some helix distortions. It looks like a saddle sitting on the longitudinal axis of Ter DNA. The blocking and nonblocking ends of the structure are different. The blocking end has a loop called the L1 loop that includes the residues 42 47 and 49 (Fig.2A, shown in red)).
Fig.2. The crystal structures of the terminator protein-Ter complexes.
A, crystal structure of the Tus-Ter complex and the residues in the L1 loop at the blocking end that contribute to interaction with DnaB and fork arrest; B, structure of the RTP-Ter complex showing the winged helix structure; the residues believed to interact with the helicase namely E30 and Y33 and Y88 that contributes to dimer-dimer interaction are shown.
Using yeast forward (Y2H) and reverse 2-hybrid (YR2H) analysis it was shown that point mutations at the residues 47 and 49 caused loss of fork arrest without loss of DNA binding. The P42L mutation, although defective in interaction with DnaB, caused some loss in Ter-DNA binding. In contrast, the mutations at 47 and 49 were defective in both helicase and fork arrest in vitro without suffering any detectable loss of DNA binding. Biochemical experiments also confirmed loss of physical interactions between the Tus mutants and DnaB [30]. Taken together, the results supported the general conclusion that there was physical interaction between Tus and DnaB and that mutational alterations of the interacting residue in Tus led to the loss of ability to arrest both forks and DnaB in vitro. Mutational analysis of Tus carried out by another laboratory had originally shown that the mutations at the residue 47 and 49 showed no loss of DNA binding in vitro but impairment of fork arrest in vivo [31,32]. These authors also concluded that Tus-Ter binding by itself could not account for polar fork arrest.
Alternative mechanisms of polar fork arrest
At least two alternative explanations for Tus-Ter mediated polar fork arrest have been proposed. Despite the multifaceted evidence supporting Tus-DnaB interactions discussed above, it was persistently proposed that tight binding of Tus to Ter and not Tus-DnaB interaction, was a critical factor in polar fork arrest [33]. A second model proposed that in addition to Tus-Ter protein-DNA interaction, the binding of Tus with DnaB (and possibly other components of the replisome) contributed to polar termination of forks [30]. On the basis of a crystal structure of a forked DNA bound to Tus, it was reported that the C residue at position 6 (model II, Fig.3A, B) flipped out and was captured by Tus. The authors claimed that a Ter sequence, when partially unwound, up to but not including C6, and having a flipped out C6 had a very high affinity for Tus at relatively high salt (0.2M-0.25M) concentration as measured by surface plasmon resonance (SPR). It was further suggested that this flipped out C6 and the partially melted DNA caused polarity of fork arrest because helicase approaching the opposite face of Tus might not make contact with C6 until the Ter site got completely melted, thereby dislodging Tus [33]. The authors concluded that the physical protein-protein interaction between Tus and the replisome was not necessary fork arrest.
Fig.3.
The stratagem to show that a sliding DnaB helicase that does not unwind DNA is still arrested at Tus-Ter when present in the blocking orientation. See the text for further details.
This model was critically reexamined because it had at least three significant inconsistencies with the published evidence: (i) there is ample evidence, as discussed above, for biologically relevant Tus-DnaB as a factor in polar fork arrest; (ii) it seemed strange that the high DNA binding affinity of the synthetic forked DNA with a base flip out was observed only at high salt concentrations that severely inhibit (by 75% - 85%) both helicase and DNA replication activities in vitro, and (iii) it did not explain how Tus-Ter would arrest helicases that progress in a 3’ to 5’ direction (e.g. priA helicase [34].
The melting-base flip out model was critically tested as shown in Fig.3C. A helicase substrate was prepared that had a 5’-overhang and a complementary strand starting with a 3’-blunt end. The complementary strand was in 2 pieces in which a terminal 3’tailed, labeled strand served as a reporter for measuring DnaB catalyzed melting (shown in red). The rationale was as follows. DnaB would enter the substrate at the 5’ SS DNA end, undergo ATP-dependent sliding from left to right so that both strands of the DNA would pass through the central channel [35,36]. The sliding was expected to be arrested when it reached the Tus-Ter complex present in the blocking orientation (Fig. 3C, b). However, when the Tus-Ter was in the nonblocking orientation, the helicase was expected to slide thorough, displace Tus from Ter and melt the downstream reporter strand (Fig. 2C; c,d). Fig.2D provides the data that support the expected polar arrest of the sliding DnaB. Since model II specifically requires both DNA melting and base flipping, the data in Fig.2D do not support model II. The result was further confirmed by introducing two site-directed crosslinks just before C6 so that no unwinding was possible beyond the X-links. The helicase was still able to slide over the X-links and was arrested in a polar fashion by Tus-Ter. A mutant form of Tus E49K (Fig. 2) that was defective in helicase arrest but bound to Ter with normal affinity was able to bypass Ter in the blocking but not in the non-blocking orientation, thereby supporting model I [36]. In summary, polar fork arrest is probably the result of not only Tus-Ter interaction but also that between Tus-DnaB.
Physiological significance of programmed fork arrest at the antipode of a bacterial chromosome - dimer resolution
When two replication forks meet each other at the Ter C region generating two daughter molecules, odd numbers of crossovers between the daughter chromosomes would generate dimers that would not be able to segregate a single chromosome into to each daughter cell. The dimers therefore need to be resolved into monomers. This requires a specialized recombination system in which two recombinases called XerC and XerD act at the dif site to convert dimers back to monomers. This reaction also needs a membrane anchored protein called FtzK that has an N-terminal membrane spanning domain and a C-terminal AAA ATPase domain. The protein acts as a translocase, and works together with Xer C and XerD in resolving dimers. It reverses the order of the reaction so that XerD first generates Holiday junctions at dif that are resolved by XerC and FtzK following which the a monomeric daughter chromosome is pushed past the septum into the daughter cell [37]. The dif site does not work unless it has two replichores of opposite polarity on either side (Fig.1C) [24,38]. Therefore, it is understandable as to why the dif site needs to be located at or near the antipode. The localization of FtzK at the septum at or near the dif site, dimer resolution and efficient segregation of the daughter chromosomes requires that the Ter sites be located close to and around the dif locus.
What are the additional reactions necessary for the segregation of daughter molecules? It has been suggested that the last catenated segment holding the two daughter circular chromosomes at TerC is removed by the combined action of helicase and DNA polymerase, resulting in two catenated, covalently closed daughters. The catenanes in E. coli and possibly other bacteria are resolved by Topo IV rather than DNA gyrase. Although the latter can put in negative superhelical turns into DNA, it is a very inefficient decatenator. However, its afore-mentioned biochemical activities make it suitable for generating substrates for topo IV [39-41] (Fig. 4A). What functions do Ter sites perform in a plasmid replicon? It has been suggested that without the terminator site and Tus, the 3’ end a growing DNA strand would displace the 5’ end of the same strand thereby initiating rolling circle replication that would be detrimental to segregation of daughter molecules, as has been reported in plasmid R1 [42].
Fig.4.
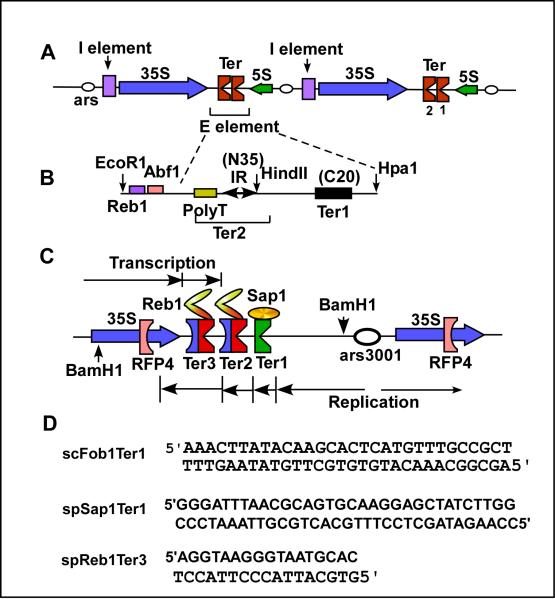
Replication termini of S. cerevisiae and S. pombe located in the nontranscribed spacer regions of the respective rDNA. A, structure of the rDNA of S. cerevisiae; the I and E elements present in cis promote recombination at ectopic sites; the twin Ter sites bind to the terminator protein Fob1; expanded view of Ter1 and Ter2, the latter contains an inverted repeat that binds to Fob1, the preceding AT-rich region and the succeeding ~20 bp are also needed for fork arrest; C, Ter sites of the spacer region of the rDNA S.pombe; Ter1 arrests replication moving from right to left by forming a complex with Sap1, both Ter2 and Ter3 bind to Reb1 that arrests replicati9on proceeding from right to left and pol I catalyzed transcription proceeding from the left to the right, both in a polar mode; fork arrest at RFP 4 is believed to be caused by collision between transcription and replication; D, sequences of Ter sites recognized by Fob1, Reb1 and Sap1.
Termination system of B. subtilis
The replication termini of B. subtilis and the terminator protein called RTP (Replication Terminator Protein) were discovered by Wake and his collaborators [43,44] The replication termination sequence has overlapping core (stronger RTP binding) and an auxiliary (weaker binding) sequence. Forks approaching the core end are arrested in a polar mode (Fig.1B). The B. subtilis chromosome contains 10 polar Ter sites present at the antipode to oriC (Fig.1A). As in E. coli, these 10 Ter sites constitute a replication trap with Ter1 being the most frequently used site in vivo[45]. Gel mobility shift experiments showed that each Ter site binds to two interacting dimers of RTP in a cooperative fashion [46]. The binding of RTP to Ter DNA causes a slight bending of the DNA and this bending might contribute to a better fit between the protein and the cognate DNA binding site. Whether the bending also contributes to any other biological function is not known [47].
Structure of RTP and the Replication termination mechanism
In order to address the structure-function questions and to perform structure guided mutagenesis, we determined the crystal structure of WT apoprotein and of two mutant forms [28,48,49]. The alpha carbon conformation of the WT and the mutant forms were compared to make sure that the mutations did not cause any visible distortions of the WT structure. The apoprotein structure at high resolution revealed that the protein is in a winged helix form [28]. The DNA binding surface was modeled by using a docking program and furthermore, by converting the protein to a chemical nuclease, its contact points in solution on the DNA were established. Subsequent work solved, first the structure of a single dimer bound to DNA (Fig. 2) and subsequently of two dimers binding to the overlapping core and auxiliary sequences [50]. The latter structure confirmed the prediction made from biochemical experiments that the two symmetrical dimers of RTP apoprotein should generate asymmetry upon DNA binding to the core and the auxiliary sites.
The residue Y88 was identified by biochemical and crystallographic analyses to be involved in dimer-dimer interaction and the mutant Y88F (red arrow, Fig. 2B) was unable to block DnaB helicase or a replication fork in an E. coli based in vitro system. The biochemical work was greatly aided by the discovery that RTP promotes polar arrest of DnaB helicase of E. coli and arrest forks at the cognate site when a plasmid of E. coli was replicated in vitro in the presence of the cognate protein [51]. Interestingly, a plasmid pT181 of S. aureus that requires the PcrA helicase and supports rolling circle replication was not arrested by RTP in vitro [51]. Further evidence of such helicase specificity and replisome specificity was found in vitro [34,52] and in vivo as revealed by extremely low level of fork arrest by E. coli Tus-Ter system in vivo in B. subtilis [53].
Additional evidence supporting the conclusion that RTP-Ter interaction is not sufficient for polar fork arrest and that interaction(s) between the protein and the helicase (or some other component of the replisome) was provided by the observation that a GFP peptide fused to the N-terminus of RTP retained undiminished DNA binding activity in but had very feeble fork arrest activity in comparison with the WT protein [54]. Keeping in mind that there is always a certain amount of fork arrest just due to DNA-protein interaction, even when the fork approaches the permissive face of the RTP-Ter complex, the fork bypass through Ter bound GFP-RTP fusion in the blocking orientation suggests that the GFP peptide seems to have functioned as a molecular shield preventing direct contact between the RTP-Ter complex and the replisome that is believed to be necessary for fork arrest.
Replication termination in eukaryotes
Since this topic has been reviewed recently at some length in another publication [5], it is presented briefly here, with the focus on physiological activities of eukaryotic replication termini and the cognate proteins. We will mostly discuss published results from both budding and fission yeasts taken as model systems. The nontranscribed spacer regions of rDNA of both budding and fission yeasts contain replication termini that arrest replication forks moving opposite to the direction of transcription of 35S RNA. Thus, replication emanating from the ars present in the spacer regions of rDNA to the right of the Ter sites, are forced to proceed in the same direction as the major transcription unit [55,56](Fig. 4A). It has been suggested that such restrictions on fork movement promotes genome stability by preventing the collision between transcription and replication [57,58]. In budding yeast, the twin Ter sites in the rDNA bind to a protein called Fob1, despite some sequence dissimilarities between Ter1 and Ter2 [59,60].
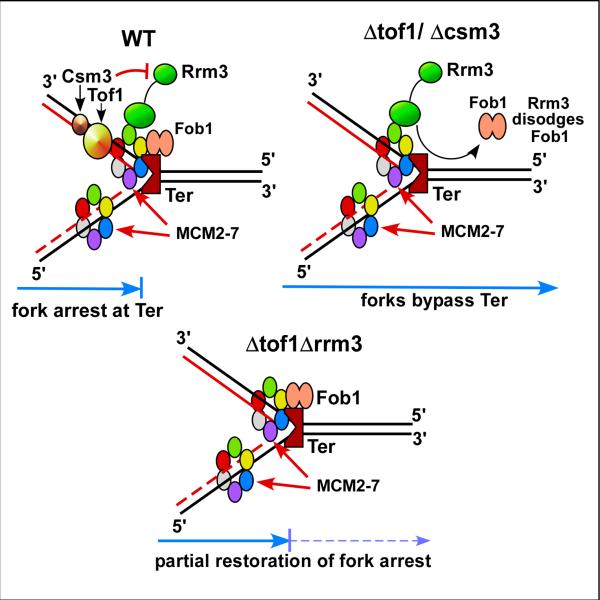
Stable fork arrest at a Fob1-Ter complex (in fact at almost any strongly bound nonhistone protein barrier), requires the activities of Tof1 (yeast equivalent of mammalian Timeless) and interacting Csm3 (its mammalian equivalent is called TIPIN) [61-63]. How does the Tof1-Csm3 complex ensure stable fork arrest at Ter ? We have reported that Tof1 and Csm3 counteract the activity of the Rrm3 helicase that tends to remove nonhistone proteins from in front of forks [64](Fig.5). Consistent with this observation, double mutants tof1rrm3 or csm3rrm3 restored at least partially fork arrest at Ter [61]. That the restoration was not complete may b due to the ability of the replicative helicase MCM2-7 to cause some protein displacement at Ter. This result, first obtained by the analysis of 2D gels has since been confirmed by independent methods [65,66]. Although Tof1 and Csm3 form a trimeric complex with Mrc1 [67], the latter is not required for stable fork arrest [61].
Fig.5.
Diagrams showing the mode of action of Tof1 and Csm3, that antagonize the action of the helicase Rrm3 to promote stable fork arrest in S. cerevisiae. The diagrams explain how forks are arrested in the WT, allowed to pass through by the unfettered action of Rrm3 when either tof1 or csm3 have been inactivated and partial restoration of fork arrest when both rrm3 and tof1 (or csm3) have been inactivated. See text for details.
Other functions of Ter and Fob1 in S. cerevisiae
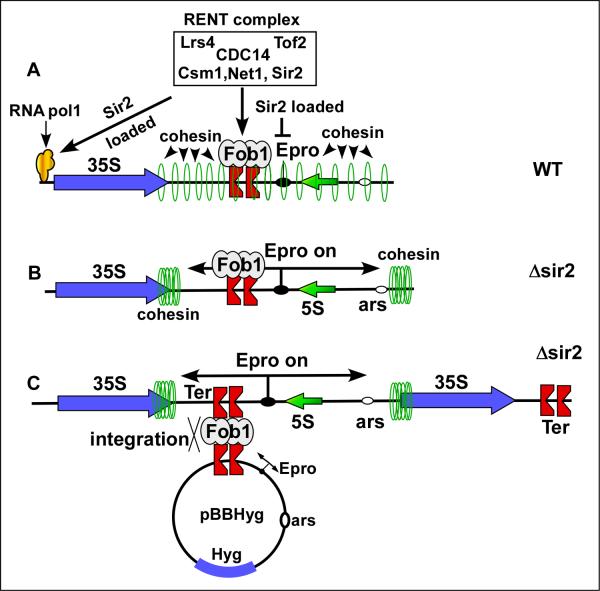
The existence of ~200 tandem copies of rDNA in chromosome XII of S. cerevisiae would tend to make it particularly susceptible to recombination leading to nonphysiological increase and decrease in rDNA repeats. To control such unwarranted and toxic recombination, yeast cells have developed a mechanism called rDNA silencing. A protein complex consisting of Net1, Sir2, CDC14, Lrs4, Csm1 and Tof2 called RENT (Regulator of Nucleolar Silencing and Telophase exit) delivers the NAD-dependent histone deacetylase Sir2 to the Ter site and to the promoter region of 35S rRNA encoding region. Net1 physically interacts with Ter bound Fob1and delivers Sir2 on to DNA near Ter. It also delivers Sir2 to the promoter region of 35S DNA by interaction of Net1 with a subunit of RNA pol I (Fig. 6) [68-70]. Sir2 causes silencing of RNA pol II but not of pol I or pol III in the rDNA array. A bidirectional promoter called Epro is repressed by the rDNA silencing. In the absence of Sir2 two transcripts are initiated from Epro (catalyzed by RNA pol II) that sweep cohesin molecules present all over the nontranscribed spacer region of the rDNA repeats to either ends (Fig. 6) [71]. In the absence of the restraining chromatid bundling activity of the ring shaped cohesin, interrepeat recombination occurs causing gain or loss of rDNA repeats from the array. Sir2 antagonizes this recombination without impairing intrarepeat recombination [72]. The intrarepeat recombination probably corrects errors caused by mutations whereas the interrepeat one causes reduction of rDNA repeat number in slowly growing cells and its expansion in fast growing cells. The interrepeat recombination that requires fork arrest by Fob1 and can be conveniently investigated by introducing a plasmid containing the Ter site and regions including the bipolar Epro promoter. In a Δsir2 strain, the plasmid integrates into the rDNA with high frequency and can be visualized and measured in quantitative Southern blots (Fig. 6C)[65,73].
Fig.6.
Recombination between repeats of S. cerevisiae rDNA is controlled by silencing of the rDNA region by loading of the RENT complex at two sites-at Ter by interaction of RNT1 with Fob1 and at the promoter region by interaction wiyth RNA pol I. A, loading of the Sir2 histone deacetylase by the RENT complex, B, the bidirectional E pro promoter is turned on when Sir2 is missing or is present at a low concentration and transcription displaces cohesin from the rDNA permitting subsequent interrepeat recombination, C, interrepeat recombination is easily measured by the integration frequencies (and excision) of the circular DNA with the ectopic Ter sites into (or out of) the rDNA array.
An important biological consequence of interrepeat repeat recombination is excision of rDNA circles from the rDNA array in senescent and aging cells presumably because of a deficit in the concentration of Sir2 in the nucleolus. The circles accumulate in the mother cell leading to its replicative aging. Presumably the episomal DNA acts as parasites that compete with the chromosomes resources needed for DNA transaction [74,75]. The recombination also requires fork arrest by Fob1 that leads to recombinogenic ds DNA breaks at Ter. It has been suggested that the formation of rDNA circles is a yeast-specific aging mechanism and that there are other pathways of aging that involve the kinases called Tor and Sch9 that control aging by controlling caloric uptake – an aging mechanism that yeast shares with other systems [76].
Fob1 also acts as a repressor of the FEAR (CDC Fourteen Early Anaphase Release network that controls the first release of the CDC14 phosphatase, sequestered in the RENT complex, into an active form that promotes escape from anaphase. Fob1 forms a complex with Spo12 and keeps it sequestered in an inactive form as a part of the RENT complex. Cell cycle dependent phosphorylation causes Spo12 that is needed for escape from mitosis, causes it to dissociate from Fob1. Phosphorylation of the Net1 protein also causes release of CDC14 that eventually leads to activation of the MEN (Mitotic Escape Network) pathway and escape from mitosis [77]. Therefore, Fob1 is a negative regulator of the FEAR pathway.
Replication termination in fission and other systems: Replication termination in fission yeast and in other systems
Unlike S. cerevisiae that has only one known terminator protein called Fob1, S. pombe is known to have 3 such proteins. One called Reb1 binds to the cognate sites located in the spacer regions of rDNA and elsewhere on the 3 chromosome, Sap1 that binds to Ter1 in the rDNA spacers [78-80] and Rtf1 that binds to the pause element MPS1 at or near the imprinted region that triggers mating type switching and also to an upstream Ter site (RTS1) that prevent replication forks from entering the switching site from the wrong direction (Fig. 7).
Fig.7.

Diagram showing the mating type switch region of S. pombe. The terminator protein Rtf1 binds to both the pause site (MPS1) that is necessary for imprinting of the lagging strand of the DNA and also to RTS1 where fork arrest of the fork moving from left to right is arrested to prevent erasure of the imprint. SAS1 is the binding site for Sap1 that promotes the switching but does not arrest forks; H1 and H2 are boundary elements.
Reb1 binds to cognate Ter sites 2 and 3 in the spacer region of rDNA [81-83] and to similar sites present out side the rDNA, in all of the 3 chromosomes. The protein has an N-terminal dimerization domain and internal twin myb-associated followed by twin myb domains (Fig. 4C). Recently, we have solved the crystal structure of the protein bound to its cognate DNA (publication in preparation). Unlike Sap1 that arrests forks in a polar fashion but does not arrest transcription, Reb1 not only arrests replication from one direction but also transcription catalyzed by RNA pol I from the opposite direction [81]. The protein also activates transcription by RNA pol II [84,85].
The Sap1 protein binds to the SAS1 site [86] but does not cause fork arrest [80]. Recently, it was reported that in the absence of CenBP, replication forks stall at LTR elements and that the stalling is caused by Sap1[87].
Using a mutated Ter site that has greatly reduced affinity for Reb1, it was shown that when the mutated site was placed 800-100bp away from a wt site in a plasmid with an ars of S. pombe, the wt sites looped to the mutant site and was able to rescue its function,i.e., fork arrest. Furthermore, using the same mutant, it was shown that a Ter site on chromosome 2 was able to interact with a major Ter site and minor one located also on chromosome 1 by “chromosome kissing”. The major and minor refer to the magnitude of interactions of the sites with the one located in chromosome II (chr). No such interaction was detected with the array of Ter sites located on rDNA present at both ends of chromosome III. That the chromosome kissing between the site on chr II and chr I was physiologically relevant was shown by mutating one of pair of sites that abolished kissing and fork arrest [88,89]. Because Reb1 is also a transcription terminator and a transcription initiator, it seems likely that kissing interaction probably also control these additional 2 transactions.
In their very elegant work, Dalgaard and collaborators showed that the protein Rtf1 binds to RFP1 to cause pausing of replication forks necessary for imprinting of a particular DNA strand by incorporation of short piece of RNA. It subsequently converted to ds breaks that induce mating type switching (Fig.6). Furthermore, Rtf1 binds to the upstream RTS1 to arrest forks that otherwise would have invaded the imprinted site from the wrong direction, thereby erasing the imprint. Further work by Dalgaard and coworkers revealed that fork arrest also required three other proteins namely Swi1, Swi3 (the equivalent of Tim and TIPIN in mammalian cells) and Rtf2 [90].[91]. That Swi1 and Swi3 also are necessary at the 3 Ter sites in each rDNA repeat but with some difference. A mutant form of Swi1 that failed to promote pausing at RPS1 was functional at the Ter sites of rDNA [83]. The Rtf2 is a PCNA binding protein that promotes efficient fork arrest by preventing replication restart [90].
Recently another pair of proteins called Lsd1 (lysine methylase) and Lsd2 (amine oxidase) have been shown to be necessary not only for fork arrest at Ter sites but also at the pause element at the switch locus [92].
In mammalian cells, there is a protein equivalent to Reb1 of fission yeast that also causes fork arrest [93]. Similar protein(s) have been discovered in Xenopus that seem to be developmentally regulated. In Xenopus oocytes no fork arrest at the rDNA Ter sites was observed until after the midblastula transition [94].
How are the two daughter chromosomes separated from a catenated structure after termination? In S. cerevisiae and probably also in S. pombe probably topoisomerase II is involved in the process [95,96]. However, the helicase Pfh1 that is a Pif1 family of helicase is necessary for daughter molecule resolution after fork arrest at a Ter site of S. pombe [97].
Future work should reveal not only additional functions of eukaryotic Ter proteins but by utilizing the crystal structures (yet to be determined excepting for Reb1) additional insights in to the mechanism of both replication and transcription termination is likely to be uncovered.
Highlights.
Bullet Points
Replication Terminus is a specific sequence (s) called Ter at which replication forks are arrested in a polar mode. Terminator proteins bind to the Ter sequences and arrest the replicative helicase in a polar fashion. Fork arrest inn several eukaryotes are turned on as a function of development. The terminator proteins are polar contrahelicases (antihelicases) that anatagonize DNA unwinding in a directional mode. In the cases where in vitro biochemical analysis has been possible to do, the terminator proteins physically interact with the helicase and the interaction seems to play a role in fork arrest.
Replication terminators require additional proteins for stable and efficient fork arrest. The Timeless and TIPIN class of fork protection proteins are required for polar fork arrest in both budding and fission yeasts. The protein complex directly interacts with the MCM helicase and DNA polymerase and enhances the catalytic activities of both. In budding yeast, it has been shown that the corresponding proteins called Tof1 and Csm3 antagonize the activity of the Rrm3 helicase that sweeps nonhistone proteins from the path of replication forks to facilitate fork movement. Additional proteins called Rts2 and Lsd1 and Lsd2 are also required fork arrest,
Besides replication termination, the terminator complex and in some cases the cognate terminator proteins promote other critical functions such as preservation of genome stability by preventing replication-transcription collision, promotion of recombination to preserve rDNA copy number homeostasis, escape from mitosis and cellular aging. In fission yeast, it controls cellular development by initiation and maintenance of mating type switching.
Acknowledgements
We thank Shaun Olsen for his generous help in preparing some of the illustrations. Our work was supported by 5R01GM098013-04.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jacob F, Brenner S, Cuzin F. On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp. Quant. Biol. 1964;28:329–348. [Google Scholar]
- 2.Bastia D, Mohanty BK. Mechanisms for completing DNA replication. In: DePamphilis M, editor. DNA Replication in Eukaryotic Cells. Cold Spring harbor Laboratory Press; Plainview, NY: 1996. pp. 177–215. [Google Scholar]
- 3.Kaplan DL, Bastia D. Mechanism of polar arest of a replication fork. Mol. Microbiol. 2009;72:279–285. doi: 10.1111/j.1365-2958.2009.06656.x. [DOI] [PubMed] [Google Scholar]
- 4.Hill TM. Arrest of bacterial DNA replication. Ann. Rev. Microbiol. 1992;46:603–636. doi: 10.1146/annurev.mi.46.100192.003131. [DOI] [PubMed] [Google Scholar]
- 5.Dalgaard JZ, Godfrey E, MacFarlane RJ. Selligman, editor. Eukaryotic replication barriers :Why, Where forks stall; in DNA Replication-current advances. InTech., InTech. 2011 [Google Scholar]
- 6.Bussiere DE, Bastia D. Termination of DNA replication of bacterial and plasmid chromosomes. Mol. Microbiol. 1999;31:1611–1618. doi: 10.1046/j.1365-2958.1999.01287.x. [DOI] [PubMed] [Google Scholar]
- 7.Bastia D, Mohanty BK. Termination of DNA Replication. In: DePamphilis M, editor. DNA replication and human disease. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2006. pp. 155–174. [Google Scholar]
- 8.Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voineagu I, Narayanan V, Lobachev KS, Mirkin SM. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc Natl Acad Sci U S A. 2008;105:9936–9941. doi: 10.1073/pnas.0804510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crosa JH, Luttrop LK, Falkow S. Mode of replication of the conjugative R-plasmid R6K in Escherichia coli. J. Bacteriol. 1976;126:454–456. doi: 10.1128/jb.126.1.454-466.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolter R, Helinski DR. Activity of the replication terminus of plasmid R6K in hybrid replicons in Escherichia coli. Journal of Molecular Biology. 1978;124:425–441. doi: 10.1016/0022-2836(78)90180-8. [DOI] [PubMed] [Google Scholar]
- 12.Kuempel PL, Henson JM, Dircks L, Tecklenberg M, Lim dif DF. a recA-independent recombination site in the terminus region of the chromosome of Escherichia coli. New Biol. 1991;3:799–811. [PubMed] [Google Scholar]
- 13.Kuempel P, Duerr SA, Seely NR. Terminus region of the chromosome of E. coli inhibits replication forks. Proc. Natl. Acad. Sci. USA. 1977;74:3927–3931. doi: 10.1073/pnas.74.9.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bastia D, Germino J, Crosa JH, Ram J. The nucleotide sequencesurrounding the replication terminus of R6K. Proc. Natl. Acad. Sci. USA. 1981;78:2095–2099. doi: 10.1073/pnas.78.4.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horiuchi T, Hidaka M. Core sequence of two separable terminus sites of R6K plasmid that exhibit polar inhibition of replication is a 20 bp inverted repeat. Cell. 1988;54:515–523. doi: 10.1016/0092-8674(88)90073-6. [DOI] [PubMed] [Google Scholar]
- 16.Germino J, Bastia D. Termination of DNA replication in vitro at a sequence specific replication terminus. Cell. 1981;23:681–687. doi: 10.1016/0092-8674(81)90431-1. [DOI] [PubMed] [Google Scholar]
- 17.Abhyankar MM, Zzaman S, Bastia D. Reconstitution of R6K DNA Replication in Vitro Using 22 Purified Proteins. J Biol Chem. 2003;278:45476–45484. doi: 10.1074/jbc.M308516200. [DOI] [PubMed] [Google Scholar]
- 18.Hill TM, Tecklenberg M, Pelletier AJ, Kuempel tus PL. the trans-acting gene required for termination of DNA replication in Escherichia coli, encodes a DNA binding protein. Proc. Natl. Acad. Sci. USA. 1989;86:1593–1597. doi: 10.1073/pnas.86.5.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill TM, Pelletier AJ, Tecklenberg ML, Kuempel PL. Identification of DNA sequences from the E. coli terminus region that halts replication forks. Cell. 1988;55:459–466. doi: 10.1016/0092-8674(88)90032-3. [DOI] [PubMed] [Google Scholar]
- 20.Coskun-Ari FF, Hill TM. Sequence-specific interactions in the Tus-Ter complex and the effect of base pair substitutions on arrest of DNA replication in Escherichia coli. J Biol Chem. 1997;272:26448–26456. doi: 10.1074/jbc.272.42.26448. [DOI] [PubMed] [Google Scholar]
- 21.Hidaka M, Kobayashi T, Takenaka S, Takeya H, Horiuchi T. Purification of a DNA replication terminus (ter) site-binding protein in Escherichia coli and identification of the structural gene. J Biol Chem. 1989;264:21031–21037. [PubMed] [Google Scholar]
- 22.Sista PR, Mukherjee S, Patel P, Khatri GS, Bastia D. A host-encoded DNA-binding protein promotes termination of plasmid replication at a sequence specific replication terminus. Proc. Natl. Acad. Sci. USA. 1989;86:3026–3030. doi: 10.1073/pnas.86.9.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sista PR, Hutchinson CA, 3rd, Bastia D. DNA-protein interaction at the replication termini of plasmid R6K. Genes & Development. 1991;5:74–82. doi: 10.1101/gad.5.1.74. [DOI] [PubMed] [Google Scholar]
- 24.Corre J, Louarn JM. Evidence from terminal recombination gradients that FtsK uses replichore polarity to control chromosome terminus positioning at division in Escherichia coli. J Bacteriol. 2002;184:3801–3807. doi: 10.1128/JB.184.14.3801-3807.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khatri GS, MacAllister T, Sista PR, Bastia D. The replication terminator protein of E. coli is a DNA sequence-specific contra-helicase. Cell. 1989;59:667–674. doi: 10.1016/0092-8674(89)90012-3. [DOI] [PubMed] [Google Scholar]
- 26.Lee EH, Kornberg A, Hidaka M, Kobayashi T, Horiuchi T. Escherichia coli replication termination protein impedes the action of helicases. Proc. Natl. Acad. Sci. U S A. 1989;86:9104–9108. doi: 10.1073/pnas.86.23.9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee EH, Kornberg A. Features of replication fork blockage by the E. coli terminus-binding protein. J. Biol. Chem. 1992;267:8778–8784. [PubMed] [Google Scholar]
- 28.Bussiere DE, Bastia D, White SW. Crystal structure of the replication terminator protein from B. subtilis at 2.6 A. Cell. 1995;80:651–660. doi: 10.1016/0092-8674(95)90519-7. [DOI] [PubMed] [Google Scholar]
- 29.Kamada K, Horiuchi T, Ohsumi K, Shimamoto N, Morikawa K. Structure of a replication-terminator protein complexed with DNA. Nature. 1996;383:598–603. doi: 10.1038/383598a0. [DOI] [PubMed] [Google Scholar]
- 30.Mulugu S, Potnis A, Shamsuzzaman, Taylor J, Alexander K, Bastia D. Mechanism of termination of DNA replication of Escherichia coli involves helicase-contrahelicase interaction. Proc Natl Acad Sci U S A. 2001;98:9569–9574. doi: 10.1073/pnas.171065898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henderson TA, Nilles AF, Valjavec-Gratian M, Hill TM. Site-directed mutagenesis and phylogenetic comparisons of the Escherichia coli Tus protein: DNA-protein interactions alone can not account for Tus activity. Mol Genet Genomics. 2001;265:941–953. doi: 10.1007/s004380100501. [DOI] [PubMed] [Google Scholar]
- 32.Skokotas A, Wrobleski M, Hill TM. Isolation and characterization of mutants of Tus, the replication arrest protein of Escherichia coli. J Biol Chem. 1994;269:20446–20455. [PubMed] [Google Scholar]
- 33.Mulcair MD, Schaeffer PM, Oakley AJ, Cross HF, Neylon C, Hill TM, Dixon NE. A Molecular Mousetrap Determines Polarity of Termination of DNA Replication in E. coli. Cell. 2006;125:1309–1319. doi: 10.1016/j.cell.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 34.Sahoo T, Mohanty BK, Lobert M, Manna AC, Bastia D. The contrahelicase activities of the replication terminator proteins of Escherichia coli and Bacillus subtilis are helicase-specific and impede both helicase translocation and authentic DNA unwinding. Journal of Biological Chemistry. 1995;270:29138–29144. doi: 10.1074/jbc.270.49.29138. [DOI] [PubMed] [Google Scholar]
- 35.Kaplan DL, O'Donnell M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol Cell. 2002;10:647–657. doi: 10.1016/s1097-2765(02)00642-1. [DOI] [PubMed] [Google Scholar]
- 36.Bastia D, Zzaman S, Krings G, Saxena M, Peng X, Greenberg MM. Mechanism of replication termination as revealed by Tus-mediated polar arrest of a sliding helicase. Proc.Natl.Acad.Sci.USA. 2008;105:12831–12836. doi: 10.1073/pnas.0805898105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blakely G, Colloms S, May G, Burke M, Sherratt D. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 1991;3:789–798. [PubMed] [Google Scholar]
- 38.Capiaux H, Lesterlin C, Perals K, Louarn JM, Cornet F. A dual role for the FtsK protein in Escherichia coli chromosome segregation. EMBO Rep. 2002;3:532–536. doi: 10.1093/embo-reports/kvf116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zechiedrich EL, Cozzarelli N. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes and Dev. 1995;9:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]
- 40.Adams DE, Shekhtman EM, Zechiedrich EL, Schmid MB, Cozzarelli NR. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- 41.Peng H, Marians KJ. The interaction of Escherichia coli topoisomerase IV with DNA. J Biol Chem. 1995;270:25286–25290. doi: 10.1074/jbc.270.42.25286. [DOI] [PubMed] [Google Scholar]
- 42.Krabbe M, Zabielski J, Bernander R, Nordstrom K. Inactivation of the replication-termination system affects the replication mode and causes unstable maintenance of plasmid R1. Mol Microbiol. 1997;24:723–735. doi: 10.1046/j.1365-2958.1997.3791747.x. [DOI] [PubMed] [Google Scholar]
- 43.Weiss AS, Wake RG. Impediment to replication fork movement at the terminus region of the Bacillus subtilis chromosome. J. Mol.Biol. 1984;179:745–750. doi: 10.1016/0022-2836(84)90165-7. [DOI] [PubMed] [Google Scholar]
- 44.Carrigan CM, Peck RA, Smith MT, Wake RG. Normal terC region of B. subtilis chromosome acts in a polar manner to arrest the clockwise replication fork. J. Mol. Biol. 1991;222:197–207. doi: 10.1016/0022-2836(91)90206-l. [DOI] [PubMed] [Google Scholar]
- 45.Wake RG, King G. A tale of two terminators: Crystal structure sharpens the debate on replication arrest mechanisms. S. Structure. 1997;5:1–5. doi: 10.1016/s0969-2126(97)00160-3. [DOI] [PubMed] [Google Scholar]
- 46.Langley DB, Smith MT, Lewis PJ, Wake RG. Protein-nucleoside contacts in the interaction between the replication terminator protein of B. subtilis and the DNA terminator. Mol. Microbiol. 1993;10:771–779. doi: 10.1111/j.1365-2958.1993.tb00947.x. [DOI] [PubMed] [Google Scholar]
- 47.Kralicek AV, Wilson PK, Ralston GB, Wake RG, King GF. Reorganization of terminus DNA upon binding replication terminator protein: implications for the functional replication fork arrest complex. Nucl. Acids Res. 1997;25:590–596. doi: 10.1093/nar/25.3.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manna AC, Pai KS, Bussiere DE, White S.W.a., Bastia D. The dimer-dimer interaction surface of the replication terminator protein of B. subtilis and termination of DNA replication. Proc. Natl. Acad. Sci. USA. 1996;93:3253–3258. doi: 10.1073/pnas.93.8.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manna AC, Pai KS, Bussiere DE, Davies C, White SW, Bastia D. Helicase-contrahelicase interaction and the mechanism of termination of DNA replication. Cell. 1996;87:881–891. doi: 10.1016/s0092-8674(00)81995-9. [DOI] [PubMed] [Google Scholar]
- 50.Vivian JP, Porter CJ, Wilce JA, Wilce MC. An asymmetric structure of the Bacillus subtilis replication terminator protein in complex with DNA. Journal of Molecular Biology. 2007;370:481–491. doi: 10.1016/j.jmb.2007.02.067. [DOI] [PubMed] [Google Scholar]
- 51.Kaul S, Mohanty BK, Sahoo T, Patel I, Khan SA, Bastia D. The replication terminator protein of the gram-positive bacterium Bacillus subtilis functions as a polar contrahelicase in gram-negative Escherichia coli. Proc Natl Acad Sci U S A. 1994;91:11143–11147. doi: 10.1073/pnas.91.23.11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sahoo T, Mohanty BK, Patel I, Bastia D. Termination of DNA replication in vitro: requirement for stereospecific interaction between two dimers of the replication terminator protein of Bacillus subtilis and with the terminator site to elicit polar contrahelicase and fork impedance. EMBO Journal. 1995;14:619–628. doi: 10.1002/j.1460-2075.1995.tb07038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andersen PA, Griffiths AA, Duggin IG, Wake RG. Functional specificity of the replication fork-arrest complexes of Bacillus subtilis and Escherichia coli: significant specificity for Tus-Ter functioning in E. coli. Mol Microbiol. 2000;36:1327–1335. doi: 10.1046/j.1365-2958.2000.01945.x. [DOI] [PubMed] [Google Scholar]
- 54.Duggin IG. DNA replication fork arrest by the Bacillus subtilis RTP-DNA complex involves a mechanism that is independent of the affinity of RTP-DNA binding. Journal of Molecular Biology. 2006;361:1–6. doi: 10.1016/j.jmb.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 55.Linskens MHK, Huberman JA. Organization of origin of replication of rDNA in S. cerevisiae. Mol. Cell. Biol. 1988;8:4927–4935. doi: 10.1128/mcb.8.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brewer BJ, Fangman WL. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 57.Brewer BJ. When polymerases collide: Replication and the transcriptional organization of the E. coli chromosome. Cell. 1988;53:679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- 58.Lin YL, Pasero P. Interference between DNA replication and transcription as a cause of genomic instability. Curr Genomics. 2012;13:65–73. doi: 10.2174/138920212799034767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kobayashi T. The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol. Cell. Biol. 2003;23:9178–9188. doi: 10.1128/MCB.23.24.9178-9188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mohanty BK, Bastia D. Binding of the replication terminator protein Fob1p to the Ter sites of yeast causes polar fork arrest. J Biol Chem. 2004;279:1932–1941. doi: 10.1074/jbc.M309078200. [DOI] [PubMed] [Google Scholar]
- 61.Mohanty BK, Bairwa NK, Bastia The D. Tof1p-Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2006;103:897–902. doi: 10.1073/pnas.0506540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005;19:1905–1919. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tourriere H, Versini G, Cordon-Preciado V, Alabert C, Pasero P. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell. 2005;19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 64.Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 65.Mohanty BK, Bairwa NK, Bastia D. Contrasting roles of checkpoint proteins as recombination modulators at Fob1-Ter complexes with or without fork arrest. Eukaryot Cell. 2009;8:487–495. doi: 10.1128/EC.00382-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fernius J, Marston AL. Establishment of cohesion at the pericentromere by the Ctf19 kinetochore subcomplex and the replication fork-associated factor, Csm3. PLoS Genet. 2009;5:e1000629. doi: 10.1371/journal.pgen.1000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bando M, Katou Y, Komata M, Tanaka H, Itoh T, Sutani T, Shirahige K. Csm3, Tof1, and Mrc1 form a heterotrimeric mediator complex that associates with DNA replication forks. J Biol Chem. 2009;284:34355–34365. doi: 10.1074/jbc.M109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 69.Huang J, Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003;17:2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang J, Brito IL, Villen J, Gygi SP, Amon A, Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes & Development. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kobayashi T, Ganley AR. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science. 2005;309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- 72.Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell. 2004;117:441–453. doi: 10.1016/s0092-8674(04)00414-3. [DOI] [PubMed] [Google Scholar]
- 73.Benguria A, Hernandez P, Krimer DB, Schvartzman JB. Sir2p suppresses recombination of replication forks stalled at the replication fork barrier of ribosomal DNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:893–898. doi: 10.1093/nar/gkg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sinclair DA. Paradigms and pitfalls of yeast longevity research. Mech Ageing Dev. 2002;123:857–867. doi: 10.1016/s0047-6374(02)00023-4. [DOI] [PubMed] [Google Scholar]
- 75.Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 76.Rine Cell biology J. Twists in the tale of the aging yeast. Science. 2005;310:1124–1125. doi: 10.1126/science.1121310. [DOI] [PubMed] [Google Scholar]
- 77.Stegmeier F, Huang J, Rahal R, Zmolik J, Moazed D, Amon A. The replication fork block protein Fob1 functions as a negative regulator of the FEAR network. Curr Biol. 2004;14:467–480. doi: 10.1016/j.cub.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 78.Mejia-Ramirez E, Sanchez-Gorostiaga A, Krimer DB, Schvartzman JB, Hernandez P. The mating type switch-activating protein Sap1 Is required for replication fork arrest at the rRNA genes of fission yeast. Mol Cell Biol. 2005;25:8755–8761. doi: 10.1128/MCB.25.19.8755-8761.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krings G, Bastia D. Sap1p binds to Ter1 at the ribosomal DNA of Schizosaccharomyces pombe and causes polar replication fork arrest. J Biol Chem. 2005;280:39135–39142. doi: 10.1074/jbc.M508996200. [DOI] [PubMed] [Google Scholar]
- 80.Krings G, Bastia D. Molecular architecture of a eukaryotic DNA replication terminus-terminator protein complex. Mol Cell Biol. 2006;26:8061–8074. doi: 10.1128/MCB.01102-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Biswas S, Bastia D. Mechanistic insights into replication termination as revealed by investigations of the Reb1-Ter3 complex of Schizosaccharomyces pombe. Mol Cell Biol. 2008;28:6844–6857. doi: 10.1128/MCB.01235-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sanchez-Gorostiaga A, Lopez-Estrano C, Krimer DB, Schvartzman JB, Hernandez P. Transcription termination factor reb1p causes two replication fork barriers at its cognate sites in fission yeast ribosomal DNA in vivo. Mol. Cell. Biol. 2004;24:398–406. doi: 10.1128/MCB.24.1.398-406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krings G, Bastia D. swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 2004;101:14085–14090. doi: 10.1073/pnas.0406037101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao A, Guo A, Liu Z, Pape L. Molecular cloning and analysis of Schizosaccharomyces pombe Reb1p: sequence-specific recognition of two sites in the far upstream rDNA intergenic spacer. Nucleic Acids Res. 1997;25:904–910. doi: 10.1093/nar/25.4.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rodriguez-Sanchez L, Rodriguez-Lopez M, Garcia Z, Tenorio-Gomez M, Schvartzman JB, Krimer DB, Hernandez P. The fission yeast rDNA-binding protein Reb1 regulates G1 phase under nutritional stress. J Cell Sci. 2011;124:25–34. doi: 10.1242/jcs.070987. [DOI] [PubMed] [Google Scholar]
- 86.Arcangioli B, Klar AJ. A novel switch-activating site (SAS1) and its cognate binding factor (SAP1) required for efficient mat1 switching in Schizosaccharomyces pombe. Embo J. 1991;10:3025–3032. doi: 10.1002/j.1460-2075.1991.tb07853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zaratiegui M, Vaughn MW, Irvine DV, Goto D, Watt S, Bahler J, Arcangioli B, Martienssen RA. CENP-B preserves genome integrity at replication forks paused by retrotransposon LTR. Nature. 2011;469:112–115. doi: 10.1038/nature09608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Singh S, Sabatinos S, Forsburg SL, Bastia D. Regulation of replication termination by Reb1 protein-mediated action at a distance. Cell. 2010;142:868–878. doi: 10.1016/j.cell.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bastia D, Singh S. Regulation of replication termination by chromosome kissing. BioArchitect. 2011;1:24–28. doi: 10.4161/bioa.1.1.14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Inagawa T, Yamada-Inagawa T, Eydmann T, Mian IS, Wang TS, Dalgaard JZ. Schizosaccharomyces pombe Rtf2 mediates site-specific replication termination by inhibiting replication restart. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7927–7932. doi: 10.1073/pnas.0812323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dalgaard JZ, Klar AJ. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell. 2000;102:745–751. doi: 10.1016/s0092-8674(00)00063-5. [DOI] [PubMed] [Google Scholar]
- 92.Holmes A, Roseaulin L, Schurra C, Waxin H, Lambert S, Zaratiegui M, Martienssen RA, Arcangioli B. Lsd1 and lsd2 control programmed replication fork pauses and imprinting in fission yeast. Cell Rep. 2012;2:1513–1520. doi: 10.1016/j.celrep.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Putter V, Grummt F. Transcription termination factor TTF-I exhibits contrahelicase activity during DNA replication. EMBO Rep. 2002;3:147–152. doi: 10.1093/embo-reports/kvf027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maric C, Levacher B, Hyrien O. Developmental regulation of replication fork pausing in Xenopus laevis ribosomal RNA genes. J Mol Biol. 1999;291:775–788. doi: 10.1006/jmbi.1999.3017. [DOI] [PubMed] [Google Scholar]
- 95.Baxter J, Diffley JF. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol Cell. 2008;30:790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 96.Fachinetti D, Bermejo R, Cocito A, Minardi S, Katou Y, Kanoh Y, Shirahige K, Azvolinsky A, Zakian VA, Foiani M. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol Cell. 2012;39:595–605. doi: 10.1016/j.molcel.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Steinacher R, Osman F, Dalgaard JZ, Lorenz A, Whitby MC. The DNA helicase Pfh1 promotes fork merging at replication termination sites to ensure genome stability. Genes Dev. 2012;26:594–602. doi: 10.1101/gad.184663.111. [DOI] [PMC free article] [PubMed] [Google Scholar]