Abstract
We previously reported that the ESR1 XbaI, analyzed by PCR followed by restriction enzyme digestion, was associated with baseline and tamoxifen-induced serum lipid profiles. After re-analysis using more robust Taqman® assays, ~10% of the genotypes for the ESR1 XbaI SNP were revised. Other genotypes (ESR1 PvuII, ESR2, and CYP2D6) were nearly identical. When re-analyzed, previously reported associations between ESR1-Xba1 genotypes and baseline triglyceride levels or LDL cholesterol, or between ESR1 XbaI genotype and tamoxifen-induced changes in total cholesterol, triglycerides or HDL-cholesterol were no longer observed. However, the following observations from the original report have not changed: 1) decreased circulating lipids in women taking tamoxifen 2) an association of changes in triglycerides with ESR2-02 genotype, and 3) no association between either ESR1 PvuII or CYP2D6 and changes in serum lipid concentrations after tamoxifen treatment.
Keywords: Breast Cancer, tamoxifen, lipids, estrogen receptor, pharmacogenetics
Introduction
Tamoxifen is a selective estrogen receptor modulator that has tissue-specific estrogen agonistic or antagonistic properties(1). We and others have previously reported that tamoxifen induces favorable lipid profiles, presumably due to an estrogenic effect in liver, mediated through the estrogen receptors, alpha and beta (ERα, β)(2). The genes that encode for these receptors (ESR1 and 2) are highly polymorphic(3). Tamoxifen is a relatively inactive pro-drug, but is converted to active metabolites, including 4-hydroxy tamoxifen and 4-hydroxy-N-desmethyl tamoxifen (designated endoxifen)(4). Conversion of tamoxifen to endoxifen is highly dependent on the activity of the enzymatic product of CYP2D6, which is also highly polymorphic(5). Therefore, we hypothesized that the estrogenic effects of tamoxifen on lipid profile might be associated with inherited single nucleotide polymorphisms in the genes that encode for the targets or the metabolic enzymes that mediate its activity.
We previously reported(2) that tamoxifen significantly lowered LDL cholesterol (p < 0.0001) and increased triglycerides (p=0.006). In postmenopausal women, the ESR1 XbaI GG genotype was associated with relatively greater tamoxifen-induced reductions in total cholesterol compared to the GA or AA genotypes (p=0.03). The ESR1 XbaI refers to the SNP rs#9340799 and the ESR1 PvuII refers to the SNP rs#2234693. In premenopausal women, the ESR1 XbaI GG genotype was associated with a large tamoxifen-induced elevation in triglycerides, while tamoxifen-treated women with the AA and AG genotypes had little or no change in triglyceride levels (p=0.002; gene-dose effect). In contrast, premenopausal women with the ESR1 XbaI AA genotype experienced elevated HDL-cholesterol levels after four months of tamoxifen, while those with the AG or GG genotypes had no change or a reduction in HDL-cholesterol levels, respectively (p=0.004; gene dose effect). In postmenopausal women, the ESR2-02 GG genotype was significantly associated with tamoxifen-induced elevations in serum triglycerides compared to no discernable change or a reduction in women with AG or AA genotypes, resepectively (p=0.01; gene dose effect). We concluded that estrogen receptor genotyping might be useful in predicting which women would benefit more from tamoxifen.
The genotyping data for ESR1 on which our prior report was based were generated by PCR followed by restriction enzyme digestion (restriction fragment length polymorphism, RFLP), methods previously described by other investigators(6). We subsequently re-determined the genotypes using Taqman® assays and found that approximately 10% of the genotypes for the ESR1-XbaI SNP determined by RFLP were discordant. We report re-analysis of our data using the more accurate Taqman® assay.
Methods
Subjects, Study Design, Sample Collection, and Lipid Analysis
The subjects, study design, sample collection, and lipid analysis details have been previously described, including exclusion of perimenopausal women and patients taking lipid-lowering medications (ClinicalTrials.gov Identifier NCT00228930)(2). Archived genomic DNA specimens were selected for re-analysis using the Taqman® assay. Of the 176 cases included in the original report by Ntudikem et al, nine had been exhausted by previous testing, and therefore 167 (95%) of the original specimens were available for this re-analysis (Supplementary Figure-CONSORT Diagram).
Genotyping
In our prior report(2), genotyping for ESR1 was conducted using the RFLP method that had been previously described in regards to estrogen effects on lipids in postmenopausal women(6). In our prior report, ESR2 genotyping was performed using the Taqman® method(2). To ensure quality control and analysis, in the current report, all specimens were re-genotyped for ESR1 and ESR2 using Taqman® assays according to manufacturer’s instructions (Applied Biosystems, Inc., Foster City, CA) as previously described and validated (7) (http://snp500cancer.nci.nih.gov; ESR2-01: rs#1256049; ESR2-02: rs#4986938). ESR1 genotypes for rs2234693 (ESR1 PvuII) and rs9340799 (ESR1 XbaI) were determined with the following TaqMan assays: C_3163590_10 for the rs#2234693 and C_3163591_10 for the rs#9340799). In a previous publication by COBRA investigators, the concordance rate between these two assays and manual DNA sequencing was >99% in 220 subjects (7). As a further confirmation, each specimen was genotyped twice with Taqman® assays and any samples that were discordant with the previous RFLP assay were confirmed by DNA sequencing. All samples were also regenotyped for ESR2 using the Taqman® assay previously described (2). In our previous report, CYP2D6 genotyping had already been performed using at least two different methodologies with 100% concordance, and therefore we did not repeat CYP2D6 genotyping in the current analysis.
Statistical Analysis
Detailed methods for statistical analytical correlations among clinical factors, lipids, and genotypes have been previously described(2) and the re-analyses of these correlations in the current report were performed identically. In addition, a similar re-analysis was performed by a completely separate and independent statistical team with identical results (see Supplemental Material and Acknowledgements).
Results
Comparison of Genotype Results Obtained by Taqman® vs. RFLP
Each of the remaining 167 specimens was re-genotyped using Taqman® methodology for each of the genotypes (ESR1 XbaI and PvuII; ESR2-01 and -02) reported by Ntudikem et al(2). Each Taqman® genotype was performed on two separate occasions. All duplicate Taqman® results for each genotype were identical.
Concordance between Taqman® and RFLP genotyping for ESR1 PvuII, ESR2-01 and -02 were all 98% or higher. Taqman® genotyping could be successfully determined for ESR1 XbaI in 157 of the 167 available DNA specimens (Supplementary Figure CONSORT Diagram). Of these, 17 (10.8%) were discordant between the two assays.
Correlation Between Genotype and Phenotype
Associations between lipid levels and clinical features and ESR1 PvuII, ESR2, and CYP2D6 genotypes
In the re-analysis, we observed that the previously reported associations between menopausal status and serum lipid concentrations at baseline, and between tamoxifen use and tamoxifen-associated change in lipid levels, remained unchanged. Likewise, as in the original report, changes in triglycerides were significantly associated with ESR2-02 genotype while there was no association between either ESR1 PvuII or CYP2D6 genotypes and changes in serum lipid concentrations after tamoxifen treatment (Supplemental Material).
Associations between ESR1 XbaI and lipids
We previously reported (2) a statistically significant association between total cholesterol at baseline before tamoxifen treatment in all women and, in subgroup analysis, in premenopausal women with ESR1 Xba1 genotype. In the re-analysis with Taqman® genotyping, these associations were not observed (p=0.23 and 0.94, respectively; Table 1). Likewise, previously reported associations between LDL cholesterol in the entire dataset and in the premenopausal subgroup according to ESR1 XbaI genotypes were no longer detected (p=0.15 and 0.93, respectively; Table 1).
Table 1.
Mean baseline lipid concentrations according to ER-α Xbal genotype and menopausal status.
| ER-α Xbal genotype | Lipid Particle | AA (N=48) | AG (N=58) | GG (N=16) | P-Valuea |
|---|---|---|---|---|---|
| All women (N=122)b | Total Cholesterol | 227 (213, 241) | 215 (204, 225) | 234 (212, 256) | 0.23 |
| Triglycerides | 141 (120, 161) | 135 (106, 164) | 131 (101, 160) | 0.69 | |
| HDL-cholesterol | 61 (57, 65) | 64 (60, 69) | 64 (56, 72) | 0.78 | |
| LDL-cholesterol | 137 (125, 149) | 124 (115, 133) | 144 (126, 162) | 0.15 | |
| Premenopausal (N=48) | (N=17) | (N=28) | (N=3) | ||
| Total Cholesterol | 211 (185, 238) | 210 (193, 227) | 208 (81, 335) | 0.94 | |
| Triglycerides | 141 (102, 180) | 115 (67, 163) | 92 (23, 161) | 0.19 | |
| HDL-cholesterol | 59 (54, 65) | 70 (63, 76) | 68 (6, 130) | 0.89 | |
| LDL-cholesterol | 123 (100, 146) | 118 (106, 130) | 122 (41, 202) | 0.93 | |
| Postmenopausal (N=74) | (N=31) | (N=30) | (N=13) | ||
| Total Cholesterol | 235 (218, 251) | 220 (206, 233) | 240 (217, 263) | 0.30 | |
| Triglycerides | 140 (115, 165) | 153 (119, 187) | 140 (106, 174) | 0.72 | |
| HDL-cholesterol | 62 (56, 68) | 60 (53, 67) | 63 (55, 71) | 0.58 | |
| LDL-cholesterol | 145 (131, 159) | 129 (116, 143) | 149 (129, 169) | 0.29 |
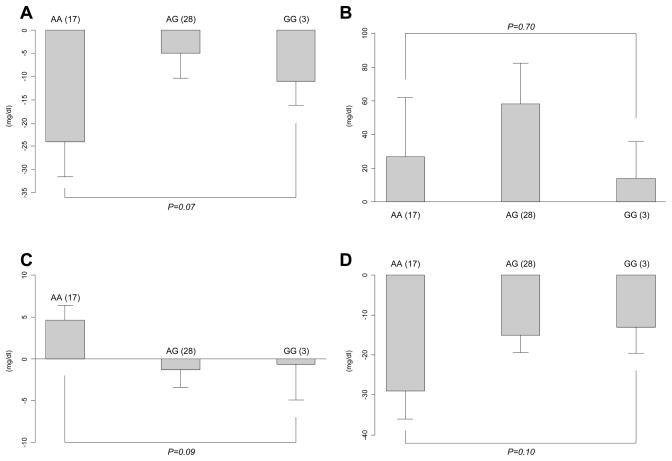
As was reported with RFLP genotyping, after four months of tamoxifen, there were no detectable associations between Taqman®-analyzed ESR1 XbaI genotypes and triglycerides, HDL-cholesterol or LDL-cholesterol (Table 2). Although the previously reported association between greater reduction in total cholesterol for the GG genotype of XbaI was again observed, it was not statistically significant in the re-analysis (reduction in total cholesterol for each genotype: AA (n=31) −28 mg/dl; AG (n=30) −13 mg/dl; and GG (n=13) −41 mg/dl; p=0.06)(Table 2). Likewise, the previously reported correlations between XbaI genotypes and triglyceride levels in premenopausal women were not observed in the current re-analysis (Figure 1B). Although associated changes for HDL-cholesterol by XbaI genotype were quantitatively similar to those previously observed in this patient group, they failed to reach statistical significance in the re-analysis (p=0.09)(Figure 1C). As before, no statistically significant associations were detected between XbaI genotypes and tamoxifen-induced changes in total cholesterol (p=0.07)(Figure 1A) or LDL-cholesterol levels (p=0.10)(Figure 1D).
Table 2.
Association of total cholesterol concentration change at 4 months with ER-α Xbal genotypes (postmenopausal group).
| ER-α Xbal genotype | ||||
|---|---|---|---|---|
| Lipid particle | AA (N=31) | AG (N=30) | GG (N=13) | P-valuea |
| Total cholesterol | −28 (−40, −16) | −13 (−22, −4) | −41 (−62, −20) | 0.06 |
| Triglycerides | 28 (4.5, 51) | 29 (−33, 90) | 27 (−37, 92) | 0.98 |
| HDL-cholesterol | −1 (−5.8, 3.6) | 1.9(−1.0, 4.8) | −1.5(−9.2, 6.1) | 0.61 |
| LDL-cholesterol | −32 (−43, −21) | −16 (−25, −8) | −39 (−60, −18) | 0.17 |
Figure 1. Response of serum lipid particle to tamoxifen in premenopausal women according to ESR1 XbaI genotype.
(A: Total cholesterol, B: triglycerides, C: HDL cholesterol and D: LDL cholesterol) The bars show the mean change in serum lipid concentration in mg/dl while the error bars are the standard error of the means. The Y-axis indicates the direction of the change in lipid concentration. Shown in brackets next to each genotype is the number of subjects in each genotype group. * P-value shown is for gene-dose effect
Discussion
Tamoxifen exerts its biological action through binding with its receptors, ERα and β, encoded by the genes ESR1 and 2, respectively. Using an older assay, we had reported selected associations between SNPs in these genes and the favorable effect of tamoxifen on lipid profiles. In the current report, we have identified that for one of these alleles (ESR1 XbaI), re-analysis with the more accurate assay, Taqman®, indicated that approximately 10% of the previously assigned genotypes were incorrect. In contrast, our previously reported genotypes for ESR1 PvuII, ESR2 and CYP2D6 were correct (98–100% concordance between assays).
In a re-analysis of the clinical associations of inherited germ line SNPs, using the genotypes derived from the Taqman® assays, we no longer detected the previously reported statistically significant association between ESR1 Xba1 genotypes and baseline triglyceride levels in all women, or between ESR1 Xba1 genotypes and baseline triglycerides or LDL cholesterol in the premenopausal subset. Likewise, in postmenopausal women, we no longer detected any statistically significant associations between ESR1 XbaI genotype and tamoxifen-induced changes in total cholesterol, nor in premenopausal women between ESR1 XbaI genotype and tamoxifen-induced changes in triglycerides or HDL-cholesterol. Most of these changes are due to genotype reassignment in the approximately 10% of subjects who were initially incorrectly assigned using RFLP genotyping.
These data may have relevance to many other studies conducted using RFLP assays in general, but specifically to those conducted with this assay for variants in ESR1, originally described by Herrington et al.(1) We are not certain of the cause of the original genotyping errors. We believe that incomplete digestion of the PCR product by the XbaI enzyme may have contributed, possibly due to a reagent problem, a tube heating block problem, or to an unknown factor in the DNA sample that partially blocked the enzyme. Alternatively, there may have been some peak suppression on the instrument that was used to analyze the digested PCR products. This circumstance could result from too little or too much DNA in the sample, a manufacturing problem with the chip or with the corresponding reagent. We repeated the RFLP assay on the DNA samples from these subjects and obtained the correct genotyping results, which indicates that there it is not a problem specific to the SNP that is affecting the assay. Since the frequencies of these errors are small, they were not observed in our original development and validation of the RFLP assay. In summary, we believe that these are random errors that may or may not be present in assays conducted by other investigators.
It is possible that some of the differences in our observations, such as the tamoxifen-induced changes total cholesterol in postmenopausal women (p=0.06; Table 2) and in HDL-cholesterol levels in premenopausal women (p=0.09, Figure 1C) may be due to loss of statistical power, since not all specimens were available for the re-analysis. However, power calculations for the results presented in Tables 1 and 2 (current version), and their corresponding tables (3A and 3B in the original version), respectively, do not support this hypothesis. It is clear that, if the number of samples assayed were decreased, but the size of the genetic effects were assumed to be the same, then the 5% reduction in sample size would have led to only a 3% to 4% reduction in power. Therefore, the difference between the original report and the reanalysis is unlikely to be due to decreased number of samples per se. Instead, it appears that the loss of statistical power is due to a decrease in the magnitude of the genetic effects observed due in turn to the reassignment of genotypes made necessary by the corrected genotyping. The following other observations from the original submission have not changed: 1) circulating lipids are decreased in women taking tamoxifen, which is consistent with other reports, 2) changes in triglycerides were statistically associated with ESR2-02 genotype, and 3) there was no association between either ESR1 PvuII or CYP2D6 genotype and changes in serum lipid concentrations after tamoxifen treatment.
Acknowledgments
Sources of Support. Supported in part by a Pharmacogenetics Research Network Grant # U-01 GM61373 (DAF) that supports the Consortium on Breast Cancer Pharmacogenomics (COBRA), a Clinical Pharmacology training grants 5T32-GM08425 (NN, MR) from the National Institute of General Medical sciences, National Institutes of Health, Bethesda, MD, Damon Runyon-Lilly Clinical Investigator Award CI-3 from the Damon Runyon Cancer Research Foundation (VS), Indiana University GCRC grant M01RR00750 from the National Institutes of Health, Bethesda, MD, University of Michigan GCRC grant M01-RR00042 from the National Institutes of Health, Bethesda, MD and by the Fashion Footwear Charitable Foundation of New York/QVC Presents Shoes on Sale™ (DFH).
We would like to thank Dr. Marylyn Ritchie, Associate Professor, Vanderbilt University Center for Human Genetics Research for her thorough re-analysis of the data included in this report (see Supplementary Material).
Footnotes
None of the authors reports any conflicts of interest regarding these results.
The description of the study design can be found on www.ClinicalTrials.gov (NCT00228930)
References
- 1.MacGregor JI, Jordan VC. Basic guide to the mechanisms of antiestrogen action. Pharmacol Rev. 1998;50:151–96. [PubMed] [Google Scholar]
- 2.Ntukidem NI, et al. Estrogen receptor genotypes, menopausal status, and the lipid effects of tamoxifen. Clin Pharmacol Ther. 2008;83:702–10. doi: 10.1038/sj.clpt.6100343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Massart F. Human races and pharmacogenomics of effective bone treatments. Gynecol Endocrinol. 2005;20:36–44. doi: 10.1080/09513590400019437. [DOI] [PubMed] [Google Scholar]
- 4.Stearns V, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95:1758–64. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 5.Jin Y, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–9. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 6.Herrington DM, et al. Estrogen-receptor polymorphisms and effects of estrogen replacement on high-density lipoprotein cholesterol in women with coronary disease. N Engl J Med. 2002;346:967–74. doi: 10.1056/NEJMoa012952. [DOI] [PubMed] [Google Scholar]
- 7.Onitilo AA, et al. Estrogen receptor genotype is associated with risk of venous thromboembolism during tamoxifen therapy. Breast Cancer Res Treat. 2008 doi: 10.1007/s10549-008-0264-2. [DOI] [PubMed] [Google Scholar]