Abstract
Incorporation of degradable moieties into synthetic hydrogels has greatly increased the utility of these three-dimensional matrices for in vitro cell culture as well as tissue engineering applications. A common method for introducing degradability is the inclusion of oligopeptides sensitive to cleavage by matrix metalloproteinases (MMPs), enabling cell-mediated remodeling and migration within the material. While this strategy has been effective, characterization and measurement of cell-mediated degradation in these materials has remained challenging. There are 20+ MMP family members whose activity is regulated in space and time by a number of biochemical and biophysical cues. Thus, the typical approach of characterizing cleavage of degradable moieties in solution with recombinant enzymes does not easily translate to three dimensional cell-mediated matrix remodeling. To address this challenge, we report here the synthesis of a cell-laden hydrogel matrix functionalized with a fluorogenic peptide substrate to provide real-time, quantitative monitoring of global MMP activity. Using this system, stimulation of MMP activity was observed with growth factor treatment in mammary epithelial cells and compared to classical zymography results. Further, the effect of biophysical cues on MMP activity of human mesenchymal stem cells was also investigated where more rigid hydrogels were observed to increase MMP activity. The regulation of MMP activity by these biochemical and biophysical cues highlights the need for in situ, real time measurement of hydrogel degradation, and use of these functionalized hydrogels will aid in future rational design of degradable synthetic hydrogels for in vitro cell studies and tissue engineering applications.
Keywords: biosensor, degradation, hydrogel, matrix metalloproteinases, mechanical properties, peptide
Introduction
Cellular encapsulation in synthetic hydrogels is a powerful methodology that has played a significant role in the field of tissue engineering from the design of cell delivery systems to more physiologically relevant three-dimensional culture systems [1,2]. Importantly, synthetic hydrogel scaffolds can be engineered to provide cells with precisely defined chemical and mechanical microenvironments that recapitulate critical aspects of the native extracellular matrix (ECM). For example, adhesive ligands such as RGD are routinely incorporated into synthetic hydrogels to promote integrin-mediated cell attachment and survival (reviewed in [3]). Cell-mediated degradation is another key characteristic of the native ECM that has been incorporated into synthetic hydrogel systems by the inclusion of enzyme sensitive peptides in the polymer networks [4–6]. Susceptibility to enzymatic degradation allows for cell-mediated remodeling and migration within cell-laden materials, which are important for applications focused on delivery or recruitment of endogenous cells for tissue repair. As these processes play critical roles in pathological disease as well as in normal tissue morphogenesis and homeostasis, enzymatically degradable hydrogels are now widely used for in vitro studies related to fundamental cell biology (e.g., cancer biology [7], vasculogenesis [8,9], differentiation [10–12]) to provide a more physiologically relevant three-dimensional microenvironment and also as a degradable scaffolds for in vivo tissue repair [6,13–16].
In developing these biomaterials systems, a common approach for introducing degradability has been the inclusion of oligopeptides with specific sequences sensitive to cleavage by matrix metalloproteinases (MMPs), as MMPs play an integral role in ECM remodeling in vivo. However, when deciding on the oligopeptide sequence, characterization of the degradation kinetics is typically performed with the peptide in solution rather than covalently tethered in a hydrogel matrix. In addition, the peptides are typically degraded with collagenase, a mixture of MMPs secreted by bacteria, or individual recombinant MMPs. Consequently, translating enzymatically driven degradation properties quantified in solution to those occurring in materials designed for in vitro three dimensional (3D) culture of cells or in vivo implantation applications is not necessarily straightforward. For example, there are 20+ members of the MMP family whose activity is regulated at multiple levels in space and time. Thus, even with a complete understanding of degradation kinetics of peptides in solution, it remains difficult to ascertain how cellular activity within cell-laden hydrogels changes and ultimately modulates matrix properties over time.
Matrix degradation and regulation of MMP activity are highly dependent on the cellular microenvironment, and one might anticipate significant differences between 2D and 3D environments. For example, in 3D culture, MMP activity has been shown to be necessary for cell proliferation and migration in 3D [17–19], which is not necessarily true for many 2D cultures. While a number of techniques have been developed to characterize MMP activity, such as gene expression analysis, zymography, and immunochemical assays, these assays require extensive sample processing to obtain results, particularly for 3D cell culture, and do not easily provide insight into temporal aspects of the regulation of MMP activity. To provide a complementary approach and circumvent some of these limitations, fluorogenic substrates that allow one to visualize MMP activity more easily have been developed. Specifically, native proteins, such as collagen and fibronectin, have been heavily labeled with fluorophores, rendering the intact protein non-fluorescent due to self-quenching. Upon cleavage of the labeled protein by cellular secreted MMPs, the fluorophores are separated and the fluorescence signal increases. Use of these heavily labeled proteins in cell culture materials has enabled visualization of MMP activity and matrix degradation in space and time without further sample processing [20–22]. However, the sensitivity of labeled native proteins is often limited due to high background signal, and the cleavage sites are ill-defined as the proteins are labeled randomly. To overcome these limitations, quenched fluorescent peptide substrates have been developed, whereby close proximity of two fluorophores on the intact peptide results in quenching, and cleavage by MMPs separates the fluorophores. Incorporation of these fluorogenic peptide substrates into 3D hydrogel matrices has enabled visualization of MMP activity and matrix degradation by migrating fibroblasts and tumor cells [23–25]. The increased resolution of these probes have allowed researchers to observe the localization of MMP activity at the leading edge of migrating cells [24]. Further, these substrates have also been used to visualize MMP-2 activity in vivo in mouse tumor models [26].
Motivated by the ability of these fluorogenic substrates to capture the spatial distribution of cellular MMP activity, we sought to synthesize functional hydrogel systems that would allow real time measurements in global MMP activity and matrix degradation in a quantitative manner. The ability to measure changes in global MMP activity is a critical step towards better understanding how cells locally remodel and degrade matrices over time, thereby affecting the physical and chemical properties of their microenvironment. Such knowledge is important to improving the field’s understanding of how cells exchange and receive information from the extracellular environment and will aid in the engineering of biomaterials that promote cell motility and tissue regeneration.
Here, we report the synthesis of a cell-laden hydrogel matrix functionalized with fluorogenic peptides to provide real-time, quantitative monitoring of global MMP activity. This approach is based on a quenched fluorogenic MMP degradable peptide substrate (Dabcyl-GGPQG↓IWGQK-Fluorescein-AhxC) that is covalently tethered to the matrix as a pendant functionality; the fluorescence of this reporter probe can be monitored over time in a multi-well format by a standard plate reader for facile measurement of MMP activity in 3D cell cultures. The design and characterization of the quenched fluorogenic MMP sensitive peptide in solution and coupled to a hydrogel are detailed. Additionally, the utility of the fluorogenic substrate functionalized hydrogels is demonstrated by investigating the effects of both biochemical and biophysical cues on MMP activity in 3D cell culture.
Materials and Methods
Synthesis of fluorescently labeled peptide substrates
Peptides (GGPQG↓IWGQK(Dde)AhxC, GGIQQWGGPK(Dde)AhxC) were synthesized using solid phase peptide synthesis on a Tribute Protein Synthesizer (Protein Technologies) with a Rink Amide MBHA resin (Novabiochem). Fmoc protected amino acids were purchased from Chem-Impex. A dabcyl succinimidyl ester (Anaspec) was coupled on resin to the amino terminus in dimethyl formimide (DMF) with 6 equivalents N,N’-diisopropylethylamine (DIPEA) and reacted overnight. Reaction completion was confirmed by a negative ninhydrin test. An orthogonally protected lysine (Lysine-Dde) (Anaspec) was deprotected two times using 2% hydrazine monohydrate in DMF for 10 minutes, and exposure of the free amine was confirmed by a positive ninhydrin test. Fluorescein NHS ester (Life Technologies) was coupled to the free amine as described for the dabcyl NHS ester. Peptides were cleaved from the resin using trifluoroacetic acid (TFA), phenol, triisopropyl silane, water (95/2.5/1.25/1.25 v/v), allowed to react at room temperature for 4 hrs, and precipitated three times in chilled diethyl ether. Peptides were purified by semi-preparative reverse phase high pressure liquid chromatography (AutoPurification HPLC, Waters) using a 15 min (30–60%) gradient of acetonitrile in water and expected mass was confirmed by matrix assisted laser desorption ionization time-of-flight mass spectrometry (Applied Biosystems DE Voyager).
Hydrogel precursor chemistry and characterization
PEG-norbornene (PEG-NB) was synthesized from a 4-arm PEG hydroxyl (Mn~20,000) (JenKem Technology USA) as described elsewhere [27]. Briefly, 5-norbornene-2-carboxylic acid (predominantly endo isomer; Alfa Aesar) was first converted to a dinorbornene anhydride using N,N’-dicyclohexylcarbodiimide (0.5 eq. to norbornene; Sigma Aldrich) in dichloromethane. The 4-arm PEG monomer was then reacted overnight with the norbornene anhydride (5 eq. to PEG hydroxyls) in dichloromethane. Pyridine (5 eq. to PEG hydroxyls) and 4-dimethylaminopyridine (0.05 eq. to PEG hydroxyls) were also included. The reaction was conducted at room temperature under argon. End group functionalization was verified by 1H NMR. The photoinitiator lithium phenyl-2,4,6-trimethylbenzoylphosphinate (i.e., lithium acylphosphinate; LAP) was synthesized as described [28]. The peptide crosslinker, KCGPQG↑IWGQCK, and cell adhesion peptide, CRGDS, were purchased from American Peptide Company.
Cell culture
hMSCs were isolated from bone marrow (Lonza) and cultured in low glucose DMEM (LG DMEM) supplemented with 10% fetal bovine serum (FBS), 1 ng/mL human fibroblast growth factor (PeproTech), 50 U/mL penicillin and 50 U/mL streptomyosin (Life Technologies). Cells were passaged at 80% confluency, and used for experiments up to passage 6. MCF10A cells (ATCC) were cultured as described [29] and treated with BSA vehicle control or 1 ng/ml TGF-β1 (PeproTech). For MMP inhibitor experiments, hMSCs were treated with DMSO control or 100 μM GM6001 (EMD Millipore). For studies comparing substrates coupled to the hydrogel or in solution, 25 nmoles of the fluorogenic MMP degradable peptide or a commercial fluorogenic peptide substrate, Mca-PLGL-Dpa-AR-NH2 (R&D Systems), were added to 1 mL of 1% FBS LG DMEM for 24 hrs.
Hydrogel preparation and cell encapsulation
3 mM (6 wt%) PEG-NB, MMP degradable crosslinker (KCGPQG↓IWGQCK), 0.5 mM CRGDS, 2 mM photoinitiator LAP, and 0.5 mM fluorogenic peptide substrate were vortexed briefly. For low, medium, and high crosslinked gels, 3.9, 4.5, and 5.5 mM MMP degradable peptide were used, respectively. Cells were encapsulated by adding a concentrated cell suspension (10×106 cells/mL) in phosphate buffered saline (PBS) to the PEG precursor solution to achieve the desired seeding density. Cells were encapsulated at a density of 1×106 cells/mL unless otherwise noted. 50 μL of PEG precursor-cell solution were pipetted into a rubber gasket with a 6 mm (inner diameter) on a thiolated coverslip to covalently link the hydrogel to the coverslip, and exposed to 365 nm light at an intensity of 4 mW cm−2 for 3 minutes. Coverslips (12 mm, No. 1.5, Fisher Scientific) were thiolated as previously described [30] via liquid deposition (0.5% (3-mercaptopropyl)trimethoxysilane (Sigma Aldrich)) in 95% ethanol/water solution for 3 min, rinsed in 95% ethanol/water, and dried at 80°C. Post-encapsulation, gels were incubated in 10% FBS LG DMEM for 2 hrs, then 1% FBS LG DMEM for the duration of the experiment.
Fluorescence measurements
Fluorescence measurements were conducted with a Synergy H1 microplate reader (BioTek), at 494 nm excitation (ex)/ 521 nm emission (em) for the fluorogenic peptide and 560nm ex/ 590nm em for alamarBlue (Life Technologies). Solution studies of peptide cleavage used 10 μM peptide and 20 μg/ml of 200 units/mg collagenase (unless otherwise noted) (Sigma Aldrich) or 20 μg/ml proteinase K (Sigma Aldrich) in phosphate buffered saline (PBS pH 7.4). For studies with the fluorogenic peptides conjugated to PEG hydrogels and encapsulated cell experiments, an area scan was performed using a 24 well plate format with a 7×7 matrix, and the average was calculated for the entire matrix.
Enzyme Kinetics
Serial dilutions in PBS of the fluorogenic peptide substrate from 0.0625 to 250 μM were exposed to 1, 5, or 10 μg/ml collagenase and fluorescence measurements taken every 2 min for 60 min. Similar kinetic constants were observed for each concentration of collagenase, and data presented are from 5 μg/ml collagenase trials. IGOR Pro software (Wavemetrics, Inc.) was used to solve for k*.
Zymography
MCF10A cells were encapsulated (2×106 cells/ml), and gels incubated in growth media for 2 hr. Gels were rinsed 2x with PBS, and media replaced with DMEM-F12, 1% pen-strep. Conditioned media was collected after 24 hrs, frozen, and lyophilized. Samples were reconstituted in PBS and protein levels measured by absorbance at 280 nm with a spectrophotometer (Thermo Scientific, Nanodrop 1000). Samples were loaded into a 10% zymogram gel (Life Technologies) with Precison Plus Dual Color Protein Standard (Bio-Rad), and zymography assay performed as described [31]. Zymogram gels were scanned to digital images (Epson, Perfection 4490 Scanner), converted to gray scale, and levels adjusted with Adobe Photoshop. ImageJ (NIH) software was used for quantification.
Rheology
30 uL gels were polymerized in 1 mL syringes, swollen in PBS overnight, and loaded onto a rheometer (TA Instruments, Discovery HR-3). 8 mm parallel plate geometry were used to measure the swollen gel modulus at 25ºC using a frequency sweep between 0.5 and 100 rad/s at an oscillatory strain of 0.5% and a gap size of 1500–2000 μm. Measurements were taken in the linear viscoelastic response regime.
DNA content analysis
PEG-NB hydrogels were rinsed twice with PBS, and degraded in 1 mL of 2 mg/ml collagenase in PBS for 1 hr at 37ºC. The collagenase cell solution was transferred to an epitube and centrifuged at 1000 RPM for 5 min. 0.95 mL of collagenase supernatant was removed carefully to not disturb the cell pellet. The cell pellet was resuspended, washed in 0.95 mL of PBS, and centrifuged again. 0.95 mL of PBS were removed, and the cell pellet was resuspended in the remaining volume. Cells were frozen at −70ºC and thawed twice to lyse cells. DNA content was measured using Quant-It Picogreen DNA quantification kit (Life Technologies) according to manufacturer’s instructions.PEG-NB hydrogels without cells, but with the fluorogenic MMP peptide, were processed in a similar manner and used as a blank.
Data analysis
Each experiments was performed three independent times with two replicates per condition. Data was analyzed with Graphpad Prism 3 software, using a one-way ANOVA with Tukey’s multiple comparison posttests or a two-way ANOVA with Bonferroni posttests. Statistical significance was determined when p<0.05.
Results and Discussion
Design and characterization of a quenched fluorogenic MMP-sensitive peptide substrate
MMP degradable hydrogels are often used to culture a number of different cell types, including primary cells, progenitor cells, and cell lines. Cells interact with and degrade these matrices, allowing the cells to spread, proliferate, and migrate within the matrix. This remodeling of the matrix is typically inferred by observations of cell morphology and motility when encapsulated in these hydrogel systems; Figure 1 shows a phase contrast image of human mesenchymal stem cells (hMSCs) encapsulated within a PEG hydrogel crosslinked with MMP degradable peptide linkers. Prior to sufficient degradation and remodeling of the matrix (2 hrs post-encapsulation), hMSCs remain rounded (Fig. 1a). However, after 24 hrs of culture, the cells have remodeled the local surrounding matrix and are able to spread within the gel (Fig. 1b). While changes in cell morphology are readily visualized, characterizing the actual degradation of the gel by the resident cells can be difficult, especially as a function of time. Understanding the degradation properties of the gel is important for designing instructive degradable synthetic matrices for cell biology applications, where material properties dictate a number of cell functions. Additionally, for tissue engineering applications, careful control of material degradation is critical for determining the persistence time of the material in vivo, the ability of local or delivered cells to infiltrate, as well as the kinetics of growth factor or drug release.
Figure 1. hMSCs encapsulated in PEG-NB hydrogels.
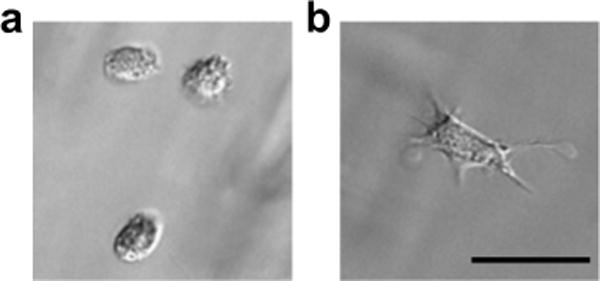
for (a) 2 hr and (b) 24 hr. Scale bar = 50 μm.
To better characterize the global MMP activity that drives the degradation and remodeling of synthetic hydrogels, we synthesized a cell culture platform with a peptide based MMP sensor covalently incorporated into the hydrogel to measure MMP activity in situ. Specifically, a quenched MMP sensitive peptide substrate was constructed, Dabcyl-GGPQG↓IWGQK-Fluorescein-AhxC (where ↓ denotes the cleavage site), using a peptide sequence based on the native collagen sequence, GPQG↓IAGQ. The tryptophan substitution for alanine increases the cleavage kinetics of the peptide [32]. This sequence is also often used as a peptide linker in PEG hydrogels to render the gels sensitive to cell mediated degradation, and is the same sequence used here to crosslink the hydrogels during polymerization [6,8,11]. Previously, this sequence was demonstrated to be sensitive to cleavage by a number of MMPs, including MMPs 1, 2, 3, 7, 8, and 9, making it an appropriate choice to investigate global MMP activity [33].
To this peptide sequence, an aminohexyl (Ahx) spacer and a C-terminal cysteine residue were added for covalent incorporation as a pendant group into synthetic PEG hydrogels (Fig. 2a). A fluorophore and a quencher, fluorescein and dabcyl, were coupled to the peptide on either side of the cleavage site. The proximity of the quencher and fluorophore when coupled to the intact peptide results in a significant decrease in fluorescence intensity of the fluorophore, i.e. quenching. Degradation of the peptide by MMPs separates the fluorophore from the quencher, resulting in a dramatic increase in fluorescence intensity, thus generating a signal corresponding to MMP activity. A control peptide, with the same sequence in a scrambled order (GGIQQWGPG), was constructed in a similar manner.
Figure 2. Design and characterization of fluorogenic peptides.
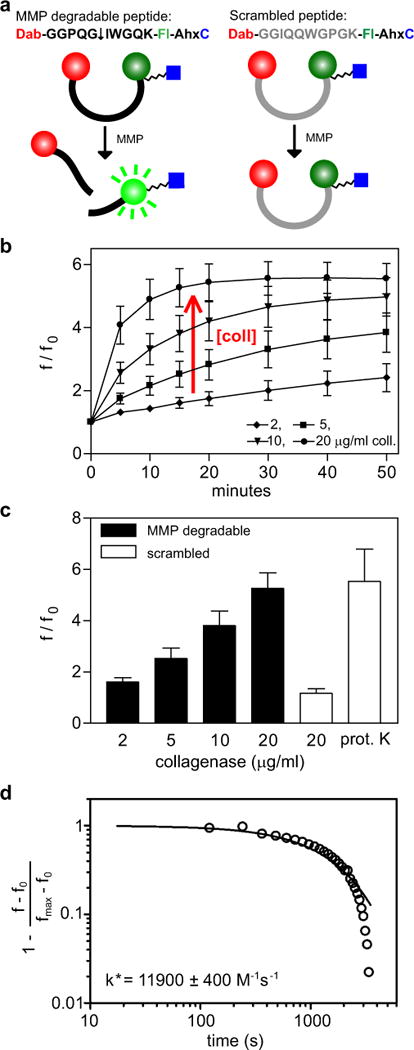
(a) Sequence and design of MMP degradable peptide and control scrambled peptide. MMP or collagenase cleavage of the degradable peptide (↓ indicates cleavage site), but not the scrambled control, separates the quencher, dabcyl (Dab), from the fluorophore, fluorescein (Fl), allowing excitation of the fluorophore. A C-terminal cysteine residue allows for coupling to PEG-norbornene. (b) Increase in fluorescence intensity (fluorescence/original fluorescence) of the MMP degradable peptide exposed to collagenase (coll) over 50 minutes. (c) MMP degradable and scrambled peptides exposed to collagenase or proteinase K (prot. K) for 15 min. (d) Representative experimental data and theoretical modeling of enzymatic degradation of the MMP degradable fluorogenic peptide by 5 μg/ml collagenase. n = 3 ± SEM.
Cleavage of the fluorogenic peptide substrates was first characterized in solution with collagenase, a commercially available mixture of MMPs, by observing fluorescence generation measured with a microplate reader (excitation at 494 nm, emission at 521 nm). Cleavage of the MMP substrate by collagenase resulted in a linear increase in fluorescence at short times (<30 minutes) and plateaued at over a five-fold increase in fluorescence (f/f0 = fluorescence intensity/fluorescence intensity measured at time zero) (Fig. 2b). Increasing collagenase concentration increased the rate of the change in fluorescence intensity. To verify the specificity of our peptide probe for MMP activity, the scrambled fluorogenic substrate was also tested, and importantly, the scrambled substrate was less susceptible to cleavage by collagenase (Fig 2c). Incubation of the scrambled substrate with 20 μg/ml collagenase for 15 minutes generated a slight increase in fluorescence intensity (~15%) as compared to the MMP degradable substrate, which had over a five-fold (500%) increase in fluorescence. To investigate if the reduced fluorescence intensity observed with the scrambled substrate was due to reduced quenching of the fluorophore, the scrambled substrate was incubated with proteinase K, an enzyme with a broad specificity that cleaves peptide bonds after hydrophobic residues. Incubation with proteinase K resulted in a five-fold increase in fluorescence, confirming that the scrambled substrate was indeed quenched.
To investigate if the addition of fluorophores to the MMP cleavable peptide interfered with enzyme degradation, the degradation of the fluorogenic peptide substrate was modeled using Michaelis-Menten enzyme kinetics [34–36] and accounting for deactivation of the enzyme as a first-order decay (detailed in Supplementary information). Assuming an average molecular weight for collagenase of 100 kDa, values for k* ranged from 9 000 to 15 000 M−1 s−1 (Fig. 2d). These values for k* are the same order of magnitude as previously measured values reported for k* for purified MMPs [32] and indicate that the addition of fluorophores to the peptide did not appear to hinder the ability of MMPs to cleave the peptide in solution.
We next sought to integrate this fluorogenic substrate reporter system into a material to create functional gels that would enable quantitative measurements of MMP activity of encapsulated cells. Inclusion of a thiol-containing cysteine residue in the design of the MMP degradable fluorogenic substrate allows facile incorporation into a number of synthetic hydrogel systems, including photoinitiated thiol-ene reactions [27,28] as well as Michael-type additions with vinyl sulfones [6,37] or acrylate moieties [38,39]. Here, a thiol-ene poly(ethylene glycol) (PEG) hydrogel system was used, where a functionalized PEG (PEG-norbornene – PEG-NB) can be crosslinked with a bis-cysteine degradable peptide in the presence of a photoinitiator and upon exposure to an appropriate wavelength of light (Fig. 3) [28]. The MMP degradable fluorogenic substrate and a cell adhesion peptide, CRGDS, were incorporated using a similar thiol-ene reaction. Since the light and active radicals generated during photoinitiated polymerizations have the potential to adversely affect the fluorophores coupled to the fluorogenic peptide substrate, we performed solution studies mimicking the photopolymerization reaction and observed no adverse effects on the fluorogenic peptide substrate (Supplementary Fig. 1).
Figure 3. Fluorogenic substrate functionalization of PEG-NB hydrogel.
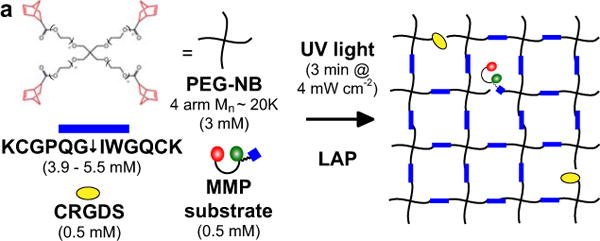
(a) Hydrogel formation by polymerization of PEG-NB (4 arm MW~20 000), MMP degradable crosslinker (KCGPQG↓IWGQCK), cell adhesion peptide (CRGDS), MMP fluorogenic peptide, and photoinitiator (LAP) exposed to 4 mW cm−2 of 365 nm light for 3 minutes.
The fluorogenic peptide substrate was next incorporated as a pendant functionality of the gel at a concentration of 0.5 mM, which is approximately 4% of the total norbornene functionalities that exist at the thiol:ene ratio used for gel formation. Selecting the peptide concentration is a balance between achieving the desired sensitivity for monitoring degradation, but not significantly altering the hydrogel structure when studying cells in their microenvironment. To measure changes in fluorescence intensity throughout the hydrogel during MMP degradation, area scans of the wells were performed using a microplate reader. As a general observation, the fluorescence intensity was localized to the hydrogel at early time points, as the pendant peptide sequence became cleaved (Fig. 4a). However, over time during collagenase degradation, the fluorescence increases and becomes more diffuse throughout the well as the structure of the gel itself is eroded, releasing the fluorogenic substrate into the buffer solution (Fig. 4a). Thus, to capture changes in global fluorescence, the average fluorescence intensity of the area scan of the entire well was used. PEG-NB hydrogels containing the fluorogenic peptide substrate were exposed to a range of collagenase concentration from 1.25 to 500 μg/ml, and increased fluorescence was observed over three orders of magnitude of collagenase concentration (Fig. 4b,c).
Figure 4. Sensitivity of fluorogenic substrate functionalized hydrogels.
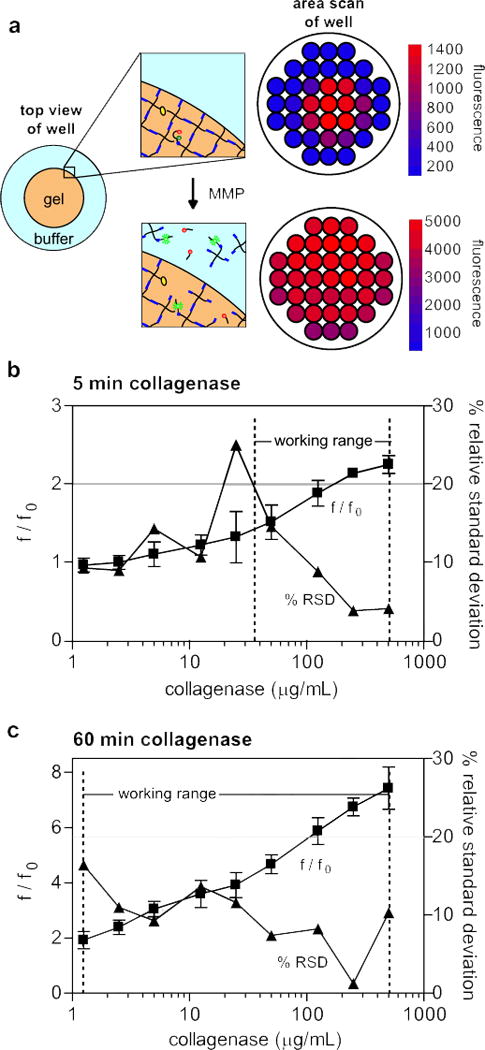
(a) Before exposure to MMPs or collagenase, the fluorogenic substrate is localized within the hydrogel, as reflected by a microplate reader area scan of a PEG-NB hydrogel swelled for 2 hrs in PBS post-gelation. After 60 minutes of collagenase degradation, the MMP fluorogenic peptide and the hydrogel structure are degraded, and increased diffuse fluorescence is observed. Calibration curve (f/f0
 ) and precision profile (%RSD
) and precision profile (%RSD
 ) for PEG-NB hydrogels with 0.5 mM MMP fluorogenic peptide at (b) 5 minutes and (c) 60 minutes of collagenase degradation. Working range indicated for the range of collagenase concentrations where the relative standard deviation is less than 20%. n = 3 ± SD.
) for PEG-NB hydrogels with 0.5 mM MMP fluorogenic peptide at (b) 5 minutes and (c) 60 minutes of collagenase degradation. Working range indicated for the range of collagenase concentrations where the relative standard deviation is less than 20%. n = 3 ± SD.
To assess the sensitivity of this functional material system as an assay for MMP activity, the calibration curve and precision profiles were plotted for fluorescence measurements of fluorogenic substrate functionalized hydrogels incubated with collagenase. The precision profile was determined by calculating the relative standard deviation (RSD), i.e. the standard deviation divided by the mean and multiplied by 100% [40]. The working range was defined as the range where the relative standard deviation is less than 20%. At 5 minutes, the working range was observed to be between 35 and 500 μg/ml of collagenase (Fig. 4b). Increasing incubation time decreased the variance observed and increased the sensitivity. At 60 minutes, the RSD was below 20% for all concentrations tested, with a working range of 1.25 to 500 μg/ml collagenase (Fig. 4c). Previous studies have demonstrated that 300 μg/mL collagenase will completely degrade similar hydrogel networks in 24 hrs [35]; thus, we believe an upper detection limit of 500 μg/ml of collagenase will be sufficient for most cell culture conditions where the gels persist for greater than a week in culture. The lower limit of detection necessary for cell studies is more difficult to estimate, as the total MMP enzyme activity is not known and may change dramatically with culture conditions (i.e., cell density, growth factors, material properties). However, a detectable range of three orders of magnitude of enzyme concentration at sixty minutes suggests that at even longer time scales (>24 hrs) the fluorogenic substrate functionalized gels will provide detectable fluorescent signals for a variety of culture conditions, even at very minimal enzyme concentration. The ability of our system to capture MMP activity in situ from the initial encapsulation through the extended culture times provides a distinct advantage over other methods to measure MMP activity and should prove useful for studying cell-mediated degradation and remodeling of degradable hydrogels.
Quantification of cellular MMP activity within fluorogenic substrate-functionalized hydrogels
We next sought to test the ability to in situ observe and characterize MMP activity of cells encapsulated in this “reporter” hydrogel culture system. Human mesenchymal stem cells (hMSC) were encapsulated in the PEG-NB hydrogels with the fluorogenic peptide substrate. The encapsulated cells were incubated for 2 hrs in full serum media, and then the media was exchanged for low serum (1%) media for the duration of the experiment to minimize cleavage of the fluorogenic substrate by serum components, which increases the background signal. After 24 hrs, over an 8 fold increase in fluorescence intensity was observed indicating cell secretion of active MMPs and degradation of the matrix (Fig. 5a). To verify that this increase in fluorescence was due specifically to MMP activity of the hMSCs, several control conditions were investigated. hMSCs were encapsulated in PEG-NB hydrogels functionalized with the scrambled substrate, shown above to be less sensitive to cleavage by MMPs, to quantify nonspecific peptide cleavage. Fluorescence measurements of the scrambled substrate functionalized gels did not show a statistically significant increase in fluorescence over the control gels (gels functionalized with the MMP substrate without cells), indicating the peptide sequence was critical for the increased fluorescence observed with the MMP fluorogenic substrate (Fig. 5a). In addition, incubation with a general MMP inhibitor, GM 6001, also reduced the increased fluorescence intensity observed with hMSCs encapsulated in the functionalized hydrogels to levels similar to the control, further demonstrating the cleavage specificity of the fluorogenic substrate by MMPs. Fluorogenic substrate functionalized gels without cells cultured under identical conditions were observed to have an approximately two-fold increase in fluorescence, likely due to proteases present in the serum in the culture media. However, this increase was much less than that observed in gels with encapsulated cells.
Figure 5. Measurement of MMP activity of hMSCs.
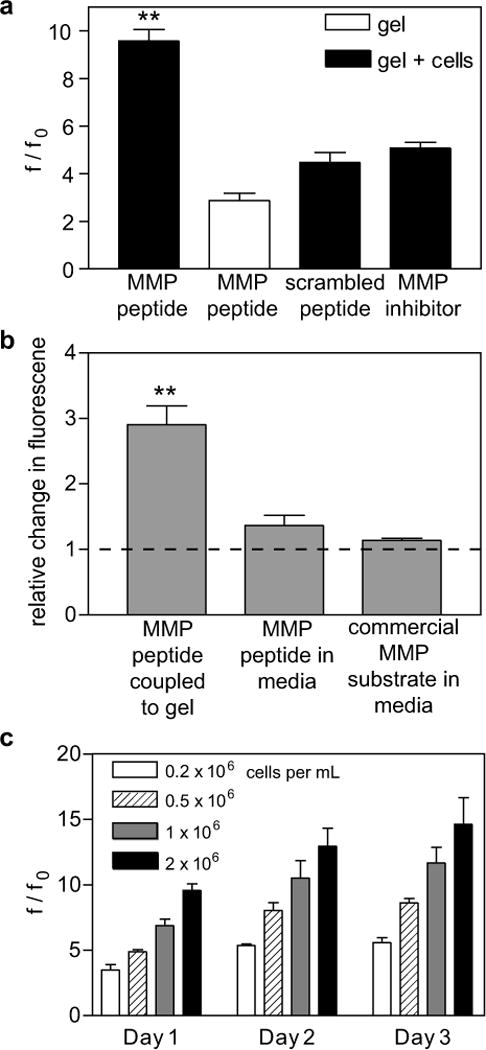
(a) Change in fluorescence intensity of MMP fluorogenic peptide or scrambled peptide by hMSCs encapsulated in PEG-NB hydrogels, treated with DMSO control or an MMP inhibitor (GM6001), or a hydrogel in media without cells, cultured for 24 hrs. (b) Fold change in fluorescence relative to control gels without cells of MMP fluorogenic substrates in cell culture conditions for 24 hrs. hMSCs were encapsulated in a PEG-NB with MMP fluorogenic peptide coupled to PEG-NB or an equivalent amount of the MMP peptide or a commercial substrate added to the media. Dashed line indicates baseline level of control gels without cells. (c) Degradation of MMP degradable peptide by hMSCs encapsulated at different seeding densities and fluorescence intensity measured every 24 hrs for three days. n = 3 ± SEM, ** p < 0.01.
Covalent coupling of the fluorogenic peptide to the hydrogel network enables monitoring of MMP activity in situ (i.e., that the peptide does not diffuse away during extended culture times) giving better insight into the degradation environment experienced by the same peptide sequence used to crosslink the degradable hydrogel. In contrast, commercially available MMP cleavable fluorogenic substrates do not contain the chemical functionalities necessary for covalent incorporation into hydrogel systems and are typically used to assay MMP activity in conditioned media that is removed from the cell culture. To compare our fluorogenic substrate to commercially available materials and to determine if the covalent coupling is necessary to observe cell-mediated MMP activity, the fluorogenic MMP degradable peptide substrate was coupled to the PEG-NB hydrogel during polymerization, or an equivalent amount (25 nmoles) per culture condition of fluorogenic substrate (the MMP fluorogenic peptide or a commercial substrate) was added to the cell culture media after polymerization. After 24 hrs, the fluorogenic substrates added solubly to the cell culture media showed a large increase in fluorescence that was not significantly different between the control condition (gel with no cells) and the encapsulated cell condition. However, a three-fold increase in fluorescence intensity was observed in the PEG-NB hydrogels with the coupled fluorogenic MMP degradable peptide substrate as compared to the control condition (Fig. 5b). Increased fluorescence of the soluble substrates as compared to the tethered substrate in the control conditions suggests non-specific cleavage of the soluble peptides. The decreased degradation of the coupled fluorogenic substrate could possibly be attributed to increased stabilization due to covalently attaching the substrate to the PEG hydrogel, similar to the stabilization of proteins covalently linked to PEG molecules observed by others [41,42]. Although MMP activity could potentially be detected with higher concentrations of soluble peptide, these results suggest that covalent coupling of the peptide probe to the hydrogel network enhances sensitivity and decreases non-specific degradation of the fluorogenic peptide substrate.
Fluorogenic MMP substrate functionalized hydrogels were then used to measure MMP activity of hMSCs at different seeding densities over time. hMSCs were encapsulated in PEG-NB hydrogels functionalized with the MMP degradable substrate over a range of seeding densities from 0.2 to 2 × 106 cells/mL and fluorescence intensity observed. As expected, increasing seeding density increased MMP activity as indicated by higher fluorescence measurements (Fig. 5c). However, a two fold increase in the seeding density did not result in two times the increase in fluorescence activity, possibility indicating a role for cell-cell interactions or paracrine signaling in regulating MMP activity. The functionalized hydrogel reporter system also enables measurements to be taken over time for the same samples. Fluorescence increased with culture time, and did not appear to plateau even after three days in culture. Collectively, these results demonstrate the feasibility of using the fluorogenic peptide substrate to measure MMP activity of encapsulated cells in 3D culture over time.
Sensing the regulation of MMP activity by soluble factors
A number of growth factors and cytokines can induce MMP expression (e.g., TNF-α, EGF, bFGF, IL-1, and TGF-β). For example, TGF-β has been demonstrated to increase MMP activity in mammary epithelial cells in traditional two-dimensional culture [43,44]. However, how TGF-β induced MMP expression translates to matrix degradation and MMP activity levels in a 3D microenvironment is less well understood.
Here, we investigated the effect of TGF-β on MMP activity of MCF10As, a human mammary epithelial cell line, encapsulated in 3D PEG-NB hydrogels using the newly developed fluorogenic peptide substrate. Because TGF-β can also have cytostatic effects, MMP activity was normalized to metabolic activity using a commercially available reagent, alamarBlue, which utilizes a non-fluorescent molecule that is reduced in the mitochondria of living cells to become highly fluorescent. The metabolic activity reagent and the fluorogenic MMP substrate have distinct fluorescence spectrums (excitation/emission: 560nm/590nm and 494nm/521nm, respectively); thus, we were able to measure MMP activity and metabolic activity with the same well concurrently, and demonstrating the facile nature of the peptide reporter approach in combination with other standard cellular assays.
Using our newly developed functionalized hydrogel system, a significant increase in MMP activity was observed after 24 hrs of TGF-β1 treatment (Fig. 6a). A similar increase in MMP activity was also observed using a standard MMP activity assay, zymography. After 24 hrs of TGF-β1 treatment, conditioned media was collected, and MMP activity of the gelatinases (MMP-2 and 9) was analyzed by gelatin zymography. An approximate three-fold increase in pro-MMP-2 (latent MMP-2, 72 kDa) and a two-fold increase of the active form of MMP-2 (63 kDa) were observed by zymography (Fig. 6b). Additionally, a band corresponding to MMP-9 (84 kDa) was observed with TGF-β treatment, but not in the control condition in two out of three experiments. While TGF-β1 treatment was observed to increase MMP activity using both our newly developed functionalized hydrogels and zymography, a smaller increase in MMP activity was observed with the functionalized gels (1.5 fold) than with zymography (2–3 fold). This difference may be due to the fact that the fluorogenic substrate is cleaved by a number of MMPs, but zymography can only assess two members of the MMP family. It is not known what fraction of the cleavage of the MMP degradable peptide is due to MMP-2 and MMP-9, therefore direct conclusions cannot be drawn from zymography data to the extent of degradation of the matrix. However, utilizing the fluorogenic substrate functionalized gels may provide more accurate insight into the global degradation of the gel, as it measures the cleavage of the same peptide sequence as that used to crosslink the gel.
Figure 6. TGF-β1 stimulation of MMP activity of MCF10As.
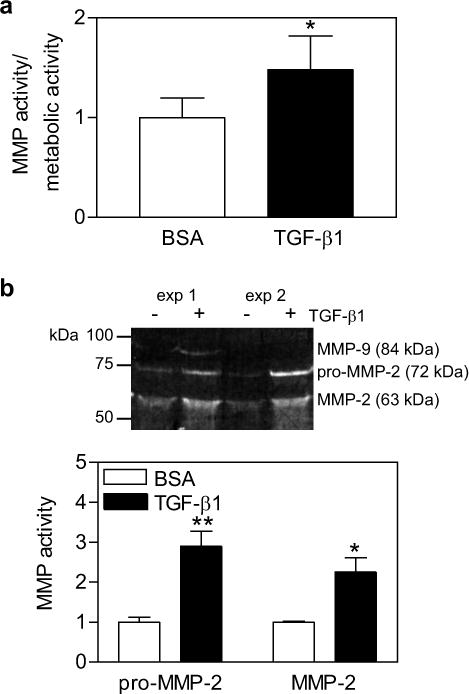
(a) MMP activity normalized to metabolic activity of MCF10As encapsulated in PEG-NB hydrogels, treated with 1 ng/ml TGF-β1 or BSA (control) for 24 hrs. (b) Representative gelatin zymogram of two experiments and quantification of conditioned media from MCF10As encapsulated in PEG-NB hydrogels and treated with TGF-β1 for 24 hrs. n = 3 ± SEM,* p < 0.05, ** p < 0.01 as compared to control condition.
Monitoring MMP activity as a function of microenvironmental properties
In addition to soluble factors, material properties can regulate a number of cell functions, including proliferation, apoptosis, and differentiation [45–47]. Extending these observations to 3D studies has been more difficult, where degradability of the matrix is necessary for cell functions, such as proliferation and migration, but this degradation also simultaneously changes the material properties. Material properties, such as elasticity, can be robustly tuned in synthetic hydrogels a priori; however, characterizing how these properties change with time due to MMP activity of encapsulated cells has remained challenging. Towards a better understanding of how MMP activity is regulated in 3D, we investigated whether elasticity would affect MMP activity of hMSCs, as elasticity is known to influence cell spreading and other functional properties of hMSCs (e.g. proliferation, differentiation) [48,45,49,50]. To tune hydrogel elasticity, the crosslinking density of the PEG-NB hydrogels was varied. Specifically, by increasing the concentration of peptide crosslinker while holding the concentration of PEG-NB constant, the shear modulus of the material, as measured by rheology, also increased, resulting in a range of shear moduli from 200 to 1200 Pa (Fig. 7a). Previous studies have shown this range of moduli to be relevant for a number of 3D cell behaviors, including cell spreading, differentiation, migration, and morphogenesis [51–55].
Figure 7. Effect of hydrogel crosslinking on MMP activity.
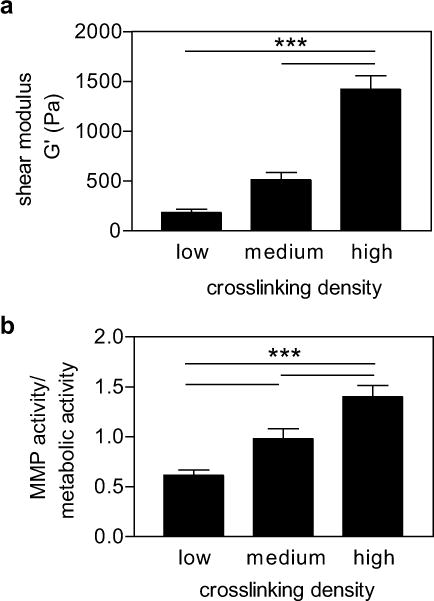
(a) Shear modulus of PEG-NB hydrogels with increasing crosslinking density. (b) MMP activity normalized to metabolic activity of hMSCs encapsulated in varying stiffness hydrogels. n = 3 ± SEM, *** p < 0.001.
hMSCs were next encapsulated in functionalized hydrogels of varying moduli and the MMP activity quantified at 24 hours. Higher MMP activity was observed in stiffer hydrogels, nearly three times higher than hMSCs cultured in the softest gels (Fig. 7b). As a control, cell number was also quantified by DNA content in addition to metabolic activity. After fluorescence intensity was measured, the PEG-NB hydrogels were degraded by collagenase and centrifuged to recover the encapsulated cells. The cells were lysed, and DNA content measured. A similar trend of increasing MMP activity in stiffer hydrogels was also observed when MMP activity was normalized to DNA content (Supplementary Fig. 2), confirming the effect of matrix stiffness on MMP activity. These observations highlight the importance of observing matrix degradation and MMP activity in situ: degradation of these materials was not uniform in all culture conditions, and the rate of degradation changed depending on biochemical and biophysical cues from the 3D microenvironment. By measuring MMP activity in situ of the same degradable peptide sequence, one can better characterize this cell-mediated matrix remodeling. For future studies, it will be interesting to correlate the observed changes in MMP activity with changes in material properties as a function of time and spatial location.
Conclusions
Coupling of a quenched MMP sensitive fluorogenic peptide to a hydrogel provides a simple method for quantitation of cellular MMP activity within synthetic hydrogels. Using a standard plate reader in a microwell format, changes in fluorescence intensity were observed over several orders of magnitude of enzyme concentration while still being sensitive enough to detect as little as 2 μM of enzyme. Furthermore, covalent tethering of the fluorogenic substrate to a PEG hydrogel enabled kinetic measurements of MMP activity in cell-laden hydrogels over several days in culture. Importantly, addition of a soluble cue, TGF-β1, increased MMP activity in mammary epithelial cells, which was confirmed by zymography. Use of this system also provided new insight into how material properties affect cell-mediated degradation of synthetic hydrogels, as we observed increasing MMP activity with increasing matrix stiffness for hMSCs cultured in these gel environments. In summary, functionalization of synthetic hydrogels with our newly developed fluorogenic MMP substrate provides a valuable tool to characterize the cell-mediated degradation properties of synthetic degradable hydrogels that is not easy to observe in solution or with select recombinant MMPs. Given the broad importance of MMPs, we anticipate that this method could easily be extended to other MMP sensitive peptide sequences, and should allow unique experiments studying the dynamics of cellular activity in 3D hydrogel culture.
Supplementary Material
Collagenase degradation of MMP degradable peptide pre-exposed for 3 minutes to 4 mW cm−2 UV light; LAP; UV light and LAP; or UV light, LAP, PEG-NB, and cysteine. n = 3 ± SEM, ** p < 0.01 as compared to other pretreatment conditions.
Normalization of MMP activity to cell number as measured by metabolic activity or DNA content. n = 3 ± SEM.
Acknowledgments
The authors thank Kelly Schultz for helpful discussions and manuscript editing. Funding for these studies was provided in part from the Howard Hughes Medical Institute and grants from the National Institutes of Health (R01DE016523).
Abbreviations
- 2D
two dimensional
- 3D
three dimensional
- ECM
extracellular matrix
- hMSC
human mesenchymal stem cells
- LAP
lithium phenyl-2,4,6-trimethylbenzoylphosphinate
- MMP
matrix metalloproteinase
- PEG-NB
poly(ethylene glycol)-norbornene
- RSD
relative standard deviation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tibbitt MW, Anseth KS. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol Bioeng. 2009;103:655–63. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lutolf MP, Gilbert PM, Blau HM. Designing materials to direct stem-cell fate. Nature. 2009;462:433–41. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hersel U, Dahmen C, Kessler H. RGD modified polymers: biomaterials for stimulated cell adhesion and beyond. Biomaterials. 2003;24:4385–415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 4.West JL, Hubbell JA. Polymeric biomaterials with degradation sites for proteases involved in cell migration. Macromolecules. 1999;32:241–4. [Google Scholar]
- 5.Mann BK, Gobin AS, Tsai AT, Schmedlen RH, West JL. Smooth muscle cell growth in photopolymerized hydrogels with cell adhesive and proteolytically degradable domains: synthetic ECM analogs for tissue engineering. Biomaterials. 2001;22:3045–51. doi: 10.1016/s0142-9612(01)00051-5. [DOI] [PubMed] [Google Scholar]
- 6.Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, et al. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: engineering cell-invasion characteristics. Proc Natl Acad Sci USA. 2003;100:5413–8. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loessner D, Stok KS, Lutolf MP, Hutmacher DW, Clements JA, Rizzi SC. Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials. 2010;31:8494–506. doi: 10.1016/j.biomaterials.2010.07.064. [DOI] [PubMed] [Google Scholar]
- 8.Miller JS, Shen CJ, Legant WR, Baranski JD, Blakely BL, Chen CS. Bioactive hydrogels made from step-growth derived PEG-peptide macromers. Biomaterials. 2010;31:3736–43. doi: 10.1016/j.biomaterials.2010.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moon JJ, Saik JE, Poché RA, Leslie-Barbick JE, Lee S-H, Smith AA, et al. Biomimetic hydrogels with pro-angiogenic properties. Biomaterials. 2010;31:3840–7. doi: 10.1016/j.biomaterials.2010.01.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adelöw C, Segura T, Hubbell JA, Frey P. The effect of enzymatically degradable poly(ethylene glycol) hydrogels on smooth muscle cell phenotype. Biomaterials. 2008;29:314–26. doi: 10.1016/j.biomaterials.2007.09.036. [DOI] [PubMed] [Google Scholar]
- 11.Benton JA, Fairbanks BD, Anseth KS. Characterization of valvular interstitial cell function in three dimensional matrix metalloproteinase degradable PEG hydrogels. Biomaterials. 2009;30:6593–603. doi: 10.1016/j.biomaterials.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson SB, Lin C-C, Kuntzler DV, Anseth KS. The performance of human mesenchymal stem cells encapsulated in cell-degradable polymer-peptide hydrogels. Biomaterials. 2011;32:3564–74. doi: 10.1016/j.biomaterials.2011.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung EH, Gilbert M, Virdi AS, Sena K, Sumner DR, Healy KE. Biomimetic artificial ECMs stimulate bone regeneration. J Biomed Mater Res A. 2006;79:815–26. doi: 10.1002/jbm.a.30809. [DOI] [PubMed] [Google Scholar]
- 14.He X, Jabbari E. Material properties and cytocompatibility of injectable MMP degradable poly(lactide ethylene oxide fumarate) hydrogel as a carrier for marrow stromal cells. Biomacromolecules. 2007;8:780–92. doi: 10.1021/bm060671a. [DOI] [PubMed] [Google Scholar]
- 15.Phelps EA, Landázuri N, Thulé PM, Taylor WR, García AJ. Bioartificial matrices for therapeutic vascularization. Proc Natl Acad Sci USA. 2010;107:3323–8. doi: 10.1073/pnas.0905447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salimath AS, Phelps EA, Boopathy AV, Che P, Brown M, García AJ, et al. Dual delivery of hepatocyte and vascular endothelial growth factors via a protease-degradable hydrogel improves cardiac function in rats. PLoS ONE. 2012;7:e50980. doi: 10.1371/journal.pone.0050980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas TL, Stitelman D, Davis SJ, Apte SS, Madri JA. Egr-1 mediates extracellular matrix-driven transcription of membrane type 1 matrix metalloproteinase in endothelium. J Biol Chem. 1999;274:22679–85. doi: 10.1074/jbc.274.32.22679. [DOI] [PubMed] [Google Scholar]
- 18.Takahra T, Smart DE, Oakley F, Mann DA. Induction of myofibroblast MMP-9 transcription in three-dimensional collagen I gel cultures: regulation by NF-κB, AP-1 and Sp1. The International Journal of Biochemistry & Cell Biology. 2004;36:353–63. doi: 10.1016/s1357-2725(03)00260-7. [DOI] [PubMed] [Google Scholar]
- 19.Hotary KB, Allen ED, Brooks PC, Datta NS, Long MW, Weiss SJ. Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell. 2003;114:33–45. doi: 10.1016/s0092-8674(03)00513-0. [DOI] [PubMed] [Google Scholar]
- 20.Lee S-H, Miller JS, Moon JJ, West JL. Proteolytically degradable hydrogels with a fluorogenic substrate for studies of cellular proteolytic activity and migration. Biotechnology Progress. 2005;21:1736–41. doi: 10.1021/bp0502429. [DOI] [PubMed] [Google Scholar]
- 21.Sameni M, Cavallo-Medved D, Dosescu J, Jedeszko C, Moin K, Mullins SR, et al. Imaging and quantifying the dynamics of tumor-associated proteolysis. Clin Exp Metastasis. 2009;26:299–309. doi: 10.1007/s10585-008-9218-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garamszegi N, Garamszegi SP, Scully SP. Matrix metalloproteinase-1 contribution to sarcoma cell invasion. J Cell Mol Med. 2011 doi: 10.1111/j.1582-4934.2011.01402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S-H, Moon JJ, Miller JS, West JL. Poly(ethylene glycol) hydrogels conjugated with a collagenase-sensitive fluorogenic substrate to visualize collagenase activity during three-dimensional cell migration. Biomaterials. 2007;28:3163–70. doi: 10.1016/j.biomaterials.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Packard BZ, Artym VV, Komoriya A, Yamada KM. Direct visualization of protease activity on cells migrating in three-dimensions. Matrix Biol. 2009;28:3–10. doi: 10.1016/j.matbio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeForest CA, Polizzotti BD, Anseth KS. Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments. Nat Mater. 2009;8:659–64. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bremer C, Bredow S, Mahmood U, Weissleder R, Tung CH. Optical imaging of matrix metalloproteinase-2 activity in tumors: feasibility study in a mouse model. Radiology. 2001;221:523–9. doi: 10.1148/radiol.2212010368. [DOI] [PubMed] [Google Scholar]
- 27.Aimetti AA, Machen AJ, Anseth KS. Poly(ethylene glycol) hydrogels formed by thiol-ene photopolymerization for enzyme-responsive protein delivery. Biomaterials. 2009;30:6048–54. doi: 10.1016/j.biomaterials.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS. A versatile synthetic extracellular matrix mimic via thiol-norbornene photopolymerization. Advanced Materials. 2009;21:5005–10. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 30.Carman ML, Estes TG, Feinberg AW, Schumacher JF, Wilkerson W, Wilson LH, et al. Engineered antifouling microtopographies – correlating wettability with cell attachment. Biofouling. 2006;22:11–21. doi: 10.1080/08927010500484854. [DOI] [PubMed] [Google Scholar]
- 31.Hawkes SP, Li H, Taniguchi GT. Zymography and reverse zymography for detecting MMPs and TIMPs. Methods Mol Biol. 2010;622:257–69. doi: 10.1007/978-1-60327-299-5_16. [DOI] [PubMed] [Google Scholar]
- 32.Patterson J, Hubbell JA. Enhanced proteolytic degradation of molecularly engineered PEG hydrogels in response to MMP-1 and MMP-2. Biomaterials. 2010;31:7836–45. doi: 10.1016/j.biomaterials.2010.06.061. [DOI] [PubMed] [Google Scholar]
- 33.Nagase H, Fields GB. Human matrix metalloproteinase specificity studies using collagen sequence-based synthetic peptides. Biopolymers. 1996;40:399–416. doi: 10.1002/(SICI)1097-0282(1996)40:4%3C399::AID-BIP5%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 34.Rice MA, Sanchez-Adams J, Anseth KS. Exogenously triggered, enzymatic degradation of photopolymerized hydrogels with polycaprolactone subunits: experimental observation and modeling of mass loss behavior. Biomacromolecules. 2006;7:1968–75. doi: 10.1021/bm060086+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schultz KM, Anseth KS. Monitoring degradation of matrix metalloproteinases-cleavable PEG hydrogels via multiple particle tracking microrheology. Soft Matter. 2013;9:1570. [Google Scholar]
- 36.Cheng Y, Prud’homme RK. Enzymatic degradation of guar and substituted guar galactomannans. Biomacromolecules. 2000;1:782–8. doi: 10.1021/bm005616v. [DOI] [PubMed] [Google Scholar]
- 37.Metters A, Hubbell J. Network formation and degradation behavior of hydrogels formed by Michael-type addition reactions. Biomacromolecules. 2005;6:290–301. doi: 10.1021/bm049607o. [DOI] [PubMed] [Google Scholar]
- 38.Rydholm AE, Bowman CN, Anseth KS. Degradable thiol-acrylate photopolymers: polymerization and degradation behavior of an in situ forming biomaterial. Biomaterials. 2005;26:4495–506. doi: 10.1016/j.biomaterials.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 39.Salinas CN, Anseth KS. The enhancement of chondrogenic differentiation of human mesenchymal stem cells by enzymatically regulated RGD functionalities. Biomaterials. 2008;29:2370–7. doi: 10.1016/j.biomaterials.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ekins R, Edwards P. Point on the meaning of “sensitivity. Clinical Chemistry. 1997;43:1824–31. [PubMed] [Google Scholar]
- 41.Gaertner HF, Puigserver AJ. Increased activity and stability of poly(ethylene glycol)-modified trypsin. Enzyme and Microbial Technology. 1992;14:150–5. doi: 10.1016/0141-0229(92)90174-m. [DOI] [PubMed] [Google Scholar]
- 42.Rawat S, Raman Suri C, Sahoo DK. Molecular mechanism of polyethylene glycol mediated stabilization of protein. Biochem Biophys Res Commun. 2010;392:561–6. doi: 10.1016/j.bbrc.2010.01.067. [DOI] [PubMed] [Google Scholar]
- 43.Kim E-S, Kim M-S, Moon A. TGF-beta-induced upregulation of MMP-2 and MMP-9 depends on p38 MAPK, but not ERK signaling in MCF10A human breast epithelial cells. Int J Oncol. 2004;25:1375–82. [PubMed] [Google Scholar]
- 44.Kim E-S, Sohn Y-W, Moon A. TGF-beta-induced transcriptional activation of MMP-2 is mediated by activating transcription factor (ATF)2 in human breast epithelial cells. Cancer Lett. 2007;252:147–56. doi: 10.1016/j.canlet.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 45.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 46.Klein EA, Yin L, Kothapalli D, Castagnino P, Byfield FJ, Xu T, et al. Cell-cycle control by physiological matrix elasticity and in vivo tissue stiffening. Curr Biol. 2009;19:1511–8. doi: 10.1016/j.cub.2009.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leight JL, Wozniak MA, Chen S, Lynch ML, Chen CS. Matrix rigidity regulates a switch between TGF-β1-induced apoptosis and epithelial-mesenchymal transition. Mol Biol Cell. 2012;23:781–91. doi: 10.1091/mbc.E11-06-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kong HJ, Polte TR, Alsberg E, Mooney DJ. FRET measurements of cell-traction forces and nano-scale clustering of adhesion ligands varied by substrate stiffness. PNAS. 2005;102:4300–5. doi: 10.1073/pnas.0405873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 2010;9:518–26. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bian L, Hou C, Tous E, Rai R, Mauck RL, Burdick JA. The influence of hyaluronic acid hydrogel crosslinking density and macromolecular diffusivity on human MSC chondrogenesis and hypertrophy. Biomaterials. 2013;34:413–21. doi: 10.1016/j.biomaterials.2012.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mason BN, Starchenko A, Williams RM, Bonassar LJ, Reinhart-King CA. Tuning three-dimensional collagen matrix stiffness independently of collagen concentration modulates endothelial cell behavior. Acta Biomaterialia. 2013;9:4635–44. doi: 10.1016/j.actbio.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Her GJ, Wu H-C, Chen M-H, Chen M-Y, Chang S-C, Wang T-W. Control of three-dimensional substrate stiffness to manipulate mesenchymal stem cell fate toward neuronal or glial lineages. Acta Biomaterialia. 2013;9:5170–80. doi: 10.1016/j.actbio.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 53.Zaman MH, Trapani LM, Sieminski AL, Siemeski A, Mackellar D, Gong H, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci USA. 2006;103:10889–94. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ehrbar M, Sala A, Lienemann P, Ranga A, Mosiewicz K, Bittermann A, et al. Elucidating the role of matrix stiffness in 3D cell migration and remodeling. Biophysical Journal. 2011;100:284–93. doi: 10.1016/j.bpj.2010.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–54. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 56.Mandl I, MacLennan JD, Howes EL, DeBellis RH, Sohler A. Isolation and characterization of proteinase and collagenase from cl. histolyticum J Clin Invest. 1953;32:1323–9. doi: 10.1172/JCI102861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Collagenase degradation of MMP degradable peptide pre-exposed for 3 minutes to 4 mW cm−2 UV light; LAP; UV light and LAP; or UV light, LAP, PEG-NB, and cysteine. n = 3 ± SEM, ** p < 0.01 as compared to other pretreatment conditions.
Normalization of MMP activity to cell number as measured by metabolic activity or DNA content. n = 3 ± SEM.


