Abstract
Heterochromatin protein 1 (HP1) proteins were originally identified as critical components in heterochromatin mediated gene silencing and are now recognized to play essential roles in several other processes including gene activation. Several eukaryotes possess more than one HP1 paralog. Despite high sequence conservation, the HP1 paralogs achieve diverse functions. Further, in many cases, the same HP1 paralog is implicated in multiple functions. Recent biochemical studies have revealed interesting paralog-specific biophysical differences and unanticipated conformational versatility in HP1 proteins that may account for this functional promiscuity. Here, we review these findings and describe a molecular framework that aims to link the conformational flexibility of HP1 proteins observed in vitro with their functional promiscuity observed in vivo.
Keywords: HP1, Chromatin, Conformational flexibility, Functional versatility
The multiple functions of HP1 proteins
The assembly of DNA into higher-order chromatin is central to the spatial and temporal regulation of the eukaryotic genome1,2. Several studies spanning more than a century have revealed two broad classes of chromatin domains: euchromatin, which contains gene-rich regions of the genome and heterochromatin which contains gene-poor and transcriptionally repressed regions3–6. At the core of the most conserved form of heterochromatin lies the HP1 protein (heterochromatin protein 1)7. The first HP1, HP1a, was identified in Drosophila melanogaster and was shown to localize to heterochromatin and act as a dominant suppressor of position effect variegation (PEV)8,9, a phenomenon whereby non-uniform silencing of a euchromatic gene is translocated into a heterochromatic region10–13. PEV was shown to require the spread of silencing activity by HP1a together with di- and tri-methylation of lysine 9 on histone H3 (H3K9me2/3) 12–18.
Parallel to these genetic experiments, biochemical and structural characterization of HP1 proteins have helped provide molecular explanations for their roles in heterochromatin. These studies have identified multiple domains within HP1 proteins: the chromodomain (CD), which specifically recognizes the H3K9me2/3 mark, the chromoshadow domain (CSD), which forms a dimerization interface that recruits specific ligands, and a connecting hinge region, which interacts with nucleic acids16,17,19–22 (Figure 1). Mammalian HP1 proteins have further been shown to interact with H3K9 methyltransferase and to oligomerize beyond dimers23,24. Collectively, these studies have provided important starting points to explain how HP1 might participate in heterochromatin spread and chromatin condensation.
Figure 1. Domain map and sequence identity of human and fission yeast HP1 proteins.
a. Top: Domain map of human HP1α, HP1β, HP1γ and fission yeast Swi6. Bottom: Percentage sequence identity relative to human HP1α.
b. Top: Domain map of fission yeast Swi6 and Chp2. Bottom: Percentage sequence identity relative to Swi6.
c. Cartoon depicting the domain architecture of HP1 proteins.
For a, b and c, light green indicates the N-terminal extension (NTE), yellow the chromodomain (CD), brown the hinge (H) region, blue the chromoshadow domain (CSD) and light red the C-terminal extension (CTE), respectively. For c, only the CD, the H and the CSD are shown for clarity. Sequence identity in a and b was calculated using the Needleman-Wunsch alignment method.
While HP1 proteins were originally identified in the context of heterochromatin, as reflected by their name, it is now clear that this family of proteins has additional nuclear functions including transcriptional activation and elongation, sister chromatid cohesion, chromosome segregation, telomere maintenance, DNA repair, and RNA splicing25–39. Consistent with the role of HP1 proteins outside of heterochromatin, the H3K9me2/3 mark, which helps recruit HP1, proteins is also found in certain euchromatic regions25. The versatility of HP1 proteins can be explained in part by the fact that many eukaryotes have more than one HP1 paralog (Figure 1). Here, using language described in previous work, we use the term HP1 to define proteins containing a CD, a CSD, and a hinge region, and having homology to the originally identified Drosophila HP1a protein40,41. For example, humans possess three main HP1 paralogs: alpha (α), beta (β), and gamma (γ), encoded by the CBX5, CBX1 and CBX3 genes, respectively. Drosophila possesses at least five paralogs (a, b, c, d, and e), while the fission yeast S. pombe has two paralogs (Swi6 and Chp2). As with their functions, the cytological distribution of HP1 paralogs is often distinct. For example, human HP1α (hHP1α) and hHP1β primarily associate with heterochromatic regions of the genome, such as centromeres and telomeres, and help mediate transcriptional gene silencing. In contrast, hHP1γ largely localizes to euchromatic regions and plays roles in transcriptional elongation and RNA processing30,42,43. Similarly, Drosophila HP1a is mainly associated with heterochromatin, while Drosophila HP1c helps regulate the transcription of genes in euchromatin44.
Intriguingly, even though HP1 paralogs perform different functions, they share high sequence homology. For example, in humans, the CD and CSD of HP1α and hHP1γ show 71% and 87% sequence identity, respectively (Figure 1). Nonetheless, it appears that these small sequence differences are important, as domain-swapping experiments between different HP1 paralogs suggest that corresponding domains have specific and non-redundant functions45–47. Further, a single amino acid change in the CSD domain of Drosophila HP1a (dHP1a) has been shown to change its specificity for ligand recognition48,49. These observations have led to the hypothesis that small deviations in sequence can result in large biochemical differences between HP1 paralogs that, in turn, translate into significantly distinct biological functions 27,41,43,50,51. For these reasons, HP1 paralogs have previously been compared to histone variants where, despite 96% identity between H3.1 and H3.3 and 59% identity between H2A and H2AZ, the paralogs fulfill fundamentally different biological functions41,52. Just as with histone variants, the molecular basis for how small sequence changes in HP1 proteins cause large functional changes remains largely a mystery.
While the above examples provide a rationale for how related HP1 paralogs can perform different functions, accumulating data also indicates that the same HP1 paralog participates in mutliple functions. Consistent with this possibility, different populations of a given HP1 paralog, based on distinct on- and off-rates from chromatin, have been observed in mammalian and S. pombe cells53–56. A clear example is the multiplicity of roles attributed to the fission yeast HP1 protein Swi6, in gene silencing. Swi6 enables transcriptional silencing by recruiting silencing factors to reduce RNA polymerase occupancy 57–59 and post-transcriptional silencing by promoting the destruction of RNA transcripts 60,61. Surprisingly, Swi6 can also interact with anti-silencing proteins to limit heterochromatin spread62,63. Despite this accumulating evidence, how HP1 paralogs like Swi6 can fulfill multiple, and at times contrasting, roles is not well understood.
Our current understanding of the molecular basis for HP1 function is directly linked to the scope of the biophysical approaches that have been used to study HP1 behavior. In this review, we use recent results derived from biophysical studies of S. pombe, mammalian, and Drosophila HP1 proteins, in the context of a wealth of previous observations, to suggest molecular explanations for the differential functions of HP1 proteins. We describe how differences in the biophysical properties of HP1 paralogs can help explain their different biological roles. We further propose that conformational flexibility in HP1 proteins allows for their functional promiscuity in vivo.
Versatility of function from differences in sequence
The different HP1 domains (Figure 1) are thought to interact with specific regions of chromatin and with different ligands in a modular manner. Below we describe how sequence differences between these domains can alter the interactions made by the individual domains and thereby alter the functional properties of the corresponding HP1 proteins.
The chromodomain and the N-terminal extension (NTE)
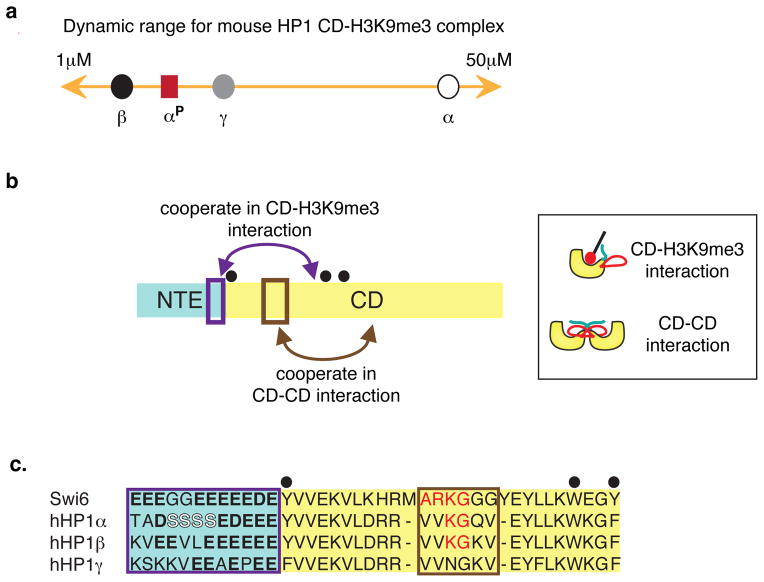
The chromodomain (CD) contains a specialized hydrophobic cage formed by aromatic residues that bind methyl marks on histones with high specificity but low affinity. The CD of many HP1 proteins has been shown to specifically bind to H3K9me2/316,17,64,65; however, depending on the HP1 paralog, the affinity for an H3K9me3 tail peptide can span approximately 5 to 40μM as demonstrated for mouse HP1 paralogs (Fig. 2A)66. This affinity difference provides a dynamic range that can be tuned in a paralog specific manner for different functions. For example, phosphorylation of serine residues in the NTE of mouse HP1α increases its affinity for H3K9me3 tail peptides by approximately 5-fold, bringing the overall affinity of the CD-H3K9me3 tail complex close to that of both mouse HP1β and mouse HP1γ66. Interestingly, in both mice and humans, these serine residues are only present in the HP1α paralog (Figs. 2B&C). In the HP1β and HP1γ paralogs, negatively charged glutamate residues replace some of the serines. Further, hHP1γ has fewer acidic residues in its NTE compared to hHP1α and hHP1β. Molecular dynamics studies suggest that these differences make the HP1γ CD-H3K9me3 complex more flexible than the HP1β CD-H3K9me3 complex67. The corresponding region of the NTE of the fission yeast homolog Swi6 does not contain the serines found in hHP1α, but instead contains several acidic residues that have been shown to stabilize binding to the H3K9me3 tail68.
Figure 2. Mechanisms controlling the functions of the chromodomain (CD).
a. Dynamic range of dissociation constants for binding of an H3K9me3 tail peptide to CDs from mouse HP1α (white circle, Kd=35 μM), HP1β (black circle, Kd=5.8 μM), HP1γ (grey circle, Kd=12 μM) and phosphorylated HP1α (red square, Kd=8.3 μM) as measured 67.
b. NTE is in light green and the CD in yellow. Left panel: Diagram indicates cooperation between NTE region (purple box) and the CD in mediating CD-H3K9me3 tail interactions and cooperation between the histone mimic loop (brown box) and the aromatic cage (black dots) in mediating CD-CD interactions. Right panel: Cartoons indicating the two types of CD mediated interactions observed in the context of Swi6. The ARK loop in the CD of Swi6 is in red. The H3 tail is in black with the H3K9me3 mark is shown as a red circle.
c. Sequence alignment between Swi6, hHP1α, hHP1β, hHP1γ indicates the conservation and divergence of the key residues that mediate the CD-H3K9me3 tail and CD-CD interactions. Interactions between the aromatic cage and H3K9me3 mark are conserved across all HP1 isoforms. The interaction between the NTE and the H3 tail has been shown for Swi6 and HP1α. Interaction between the ARK loop and the aromatic cage has been shown for Swi6. Color-coding is the same as the left panel of 2b. Negatively charged residues regulating the CD-H3K9me3 tail interactions in Swi6 and hHP1α are in bold. Phosphorylatable serines in hHP1α regulating its CD-H3K9me3 tail interaction are in white. The loop residues conserved with Swi6 are in red.
The above observations suggest that the presence of a negative charge in the NTE increases the affinity of HP1 proteins for methylated chromatin. Therefore, regulating the degree of negative charge via phosphorylation of the serines in hHP1α may allow the assembly of hHP1α on H3K9me2/3 chromatin to rapidly increase or decrease based on the activity of kinases or phosphatases, respectively. This property may be needed more for HP1α molecules, which are predominantly found in heterochromatic regions and may have to undergo coordinated assembly and disassembly during developmental transitions and the cell cycle. Interestingly, NTE phosphorylation of mammalian HP1α persists during the cell cycle66. We speculate, however, that small changes in the levels of phosphorylated HP1α may have large effects given the proposed cooperative nature of heterochromatin spread. The results with mammalian HP1α proteins are consistent with earlier in vivo studies in Drosophila showing that reversible phosphorylation of HP1a is essential for heterochromatin function69,70.
For some HP1 proteins, the CD also helps mediate HP1 oligomerization, a key process in the spread of heterochromatin 24,68,71,72. In particular, recent work has shown that the fission yeast Swi6 CD contains a sequence (ARK94GGG) on a loop that resembles the amino acid sequence of the H3-tail surrounding the K9 position (ARK9STG) (Fig. 2B)68. Mutagenesis of the loop residues showed that the sequence promotes CD-CD interactions in solution suggesting that the loop-CD interaction mimics the H3K9me3-CD interaction68. Interestingly, in the context of a Swi6 dimer, the loop-CD interaction blocks H3K9 methyl mark recognition and higher-order oligomerization. Binding to methylated nucleosomes via the CD pays the energetic cost for switching Swi6 dimers to a spreading competent state68. In this state, the vacant CDs can then either directly bind adjacent nucleosomes or bind the CD of another dimer via the ARK94GGG loop (Figure 4).
Figure 4. Models for functional versatility of HP1 on chromatin.
a. Cartoon depicting how different HP1 conformations can associate with nucleosomes. The spreading competent and spreading limiting conformations are based on recent findings with Swi6.
b. Cartoon depicting how different conformations of HP1 on nucleosomes can recruit different protein partners and itself. The white, brown and green shapes refer to possible ligands of HP1 proteins bound to chromatin. The white shape is a CSD ligand, the brown shape is a putative interactor with the CD loop, and the green shape is a putative capping protein that stabilizes HP1 in a closed state.
c. Cartoon depicting different possible stoichiometries of HP1 molecules on a nucleosome.
The nucleosome depicted here contains a histone octamer (grey), DNA (black) and an H3K9me3 mark (red). Only the CD (yellow), the hinge region (black line), and the CSD (blue) of HP1 are shown for clarity. For a, and b, the depicted CD-CD interaction is mediated by the CD-ARK loop recently found to occur in Swi6 and described in Figure 2B. For clarity the ARK loop is not shown here.
While the lysine and proximal glycine in the histone mimic loop of Swi6 are conserved, the rest of the sequence degenerates in higher organisms (Fig. 2B). Thus, it remains unclear whether the specific mechanism of CD-CD interaction observed in the fission yeast Swi6 is conserved in higher eukaryotes. However, the conserved lysine in hHP1α and β is differentially modified: it is monomethylated in hHP1α and acetylated in hHP1β73. Human HP1γ, on the contrary, lacks the corresponding lysine. Post-translational modification of this conserved lysine has the potential to dramatically alter the basic biochemical properties of HP1α and β to promote a rapid switch between different biological functions. While HP1 proteins preferentially recognize trimethylated H3K9, the affinity of the CD for monomethylated H3K9 tails has been shown to be least 10-fold higher than unmethylated H3K9 tails in two different contexts 74,75. We therefore speculate that if the loop in hHP1α plays similar roles to that in Swi6, then monomethylation of the loop lysine would increase its affinity for another CD and energetically compensate for the loss of the remaining histone mimic residues found in Swi6. Monomethylation of the lysine in hHP1α, in principle, could promote CD-CD interactions between two hHP1α proteins enabling oligomerization or providing a non-histone binding target for other CD containing proteins (Figure 2B). As with phosphorylation, we speculate that such modulation may be needed more for hHP1α than other paralogs because the assembly and disassembly of hHP1α needs to be coordinately regulated over large stretches of the genome. The methylation status of the corresponding loop lysine in Swi6 is not known. Lysine acetylation, on the other hand, could allow hHP1β to interact with bromodomain containing proteins or could disrupt potential CD-CD interactions between HP1 molecules. Since hHP1β has both gene silencing and activating roles, acetylation may provide a means to switch between these two opposing roles by changing the oligomerization properties and binding partners of hHP1β. Analogously, we speculate that the absence of lysine in the corresponding loop region of hHP1γ may reduce its oligomerization across long stretches of chromatin, thereby enabling hHP1γ to function as a gene activator by exerting a more local effect.
The chromoshadow domain and the C-terminal extension (CTE)
The chromoshadow domain (CSD) is involved in homo-dimerization of HP1 proteins and is a hub for many partners of HP119,20. In Drosophila, swapping the CSD domains of dHP1a and dHP1c directs dHP1c to heterochromatin and dHP1a to euchromatin76. These results suggest that functional specificity of HP1 homologs in the cell relies, at least in part, on the ability of different CSD domains to mediate association with specific nuclear proteins.
In general, the CSD dimer of HP1 proteins is capable of reading PxVxL pentapeptide motifs, as well as sequences that partially deviate from it, such as the PGTVAL sequence on the αN1-helix of histone H3, the LSVKI sequence on the Drosophila HP2 protein, and the PRVKV sequence in the Drosophila PIWI protein20,48,49,77–79. Recognition of PXVXL-type sequences relies on a set of conserved residues in the CSD (Figure 3A). The valine (V) at the center of the PXVXL motif (position 0) binds a conserved hydrophobic pocket in the dimer interface formed by tyrosine 168 (Y168) and leucine (L172) (amino acid numbering for hHP1α). The plasticity of the binding site around the proline at position −2 allows other large hydrophobic groups, such as leucine or methionine, to substitute for it80 (Figure 3A). However, if recognition of a PxVxL type sequences relies on conserved residues in the CSD domains, how does a given CSD discriminate between different PxVxL sequences and how do different CSD domains achieve different sequence specificities? Recent biochemical and structural studies with peptide ligands in solution have provided some insights into this question.
Figure 3. Mechanisms controlling the functions of the chromoshadow (CSD).
a. Structure of the CSD domain of mouse HP1β (blue) in complex with the CAF (PXVXL) peptide (red), constructed using PDB file 1S4Z [Thiru et al. EMBO J.23, 489–99 (2004)] 86. The numbering of the PXVXL-containing ligand is relative to the central valine (V) and the +5, +6, −6 and −7 residues are indicated with red circles. The residues in the CSD dimer that specifically interact with the different positions in the PXVXL peptide are shown as green and grey circles. For clarity, the CSD residues that interact with position 0 of a PXVXL peptide are shown only on one of the two dimers as yellow circles.
b. Cartoon depicting how the CSD domain of human HP1α, β and γ differentially recognizes the PGTVAL motif on histone H3 and the phosphorylation of H3Y41. For clarity, only one copy of the PGTVAL sequence on the nucleosome is shown.
c. Dynamic range of CSD-CSD dimerization constants measured for human HP1β (black circle), Swi6 (black circle) and dHP1a (white circle).
d. Diagram indicating functional and structural cooperation between the CTE region (brown box) and the CSD of HP1 proteins in mediating CSD-CSD and CSD-ligand interactions. The region of CSD containing phosphorylatable serines in dHP1a (purple box) regulates the CSD-CSD dimerization and CSD-ligand interaction. The sequence of this region is compared with that in human HP1 proteins below.
e. Cartoon depicting how, in Swi6, the CD can contribute to strengthening the dimerization of the full-length protein. The CD-CD interaction shown here is the one shown in figure 2b (inset box) and is mediated by the ARK loop in Swi6.
The residues upstream and downstream of the PXVXL pentapeptide motif (Figure 3A, −5 to −7 and +5 and +6, respectively) have been shown to act as key specificity determinants80,81. This is best illustrated by recent studies with dHP1a, which prefers to bind the HP2 protein over the PIWI protein. This preference relies, in part, on a conserved leucine in the CSD, which recognizes amino acids at positions −7/−6 and +5/+6 with respect to the LSVKI sequence in the HP2 protein48,49. These residues are missing in the amino acids flanking the PRVKV sequence in the PIWI protein. As a result, the CSD of dHP1a binds the LSVKI containing sequence in the HP2 protein 50-fold tighter than the PRVKV containing sequence in the PIWI protein. Substitution of this leucine with a lysine reduces the affinity of dHP1a CSD approximately 20-fold for the LSVKI containing sequence in HP2, while reducing the affinity of the PRVKV containing sequence in PIWI by approximately 1.6-fold49. These differential effects reduce the preference of the CSD for the LSVKI vs. PRVKV containing sequence and demonstrate how a single amino acid can be responsible for dHP1a’s partner specificity. Similarly, hHP1α and γ, but not β, bind to the PGTVAL motif on the αN1–helix of H3 (Figure 3B). HP1α additionally recognizes tyrosine 41 (Y41), which is two residues upstream from the PGTVAL motif. Phosphorylation of Y41 inhibits binding by HP1α to the PGTVAL motif in H3 77,78. In contrast, hHP1γ does not seem to require Y41 for binding the PGTVAL region and therefore it has been suggested that binding of HP1γ to H3 may be insensitive to Y41 phosphorylation77,78 (Figure 3B). Although the molecular nature of such specificity remains elusive, the results make sense from a biological perspective as phosphorylation of this tyrosine in H3 is correlated with increased transcription. Furthermore, these results imply that in the context of chromatin, non-histone ligands of the CSD domains of hHP1α and γ would have to compete with the PGTVAL sequence in histone H3.
Since ligand recognition by the CSD requires CSD dimerization, the strength of dimerization can further tune the affinity for ligand. If the cellular concentration of a particular HP1 paralog is lower that its dissociation constant (Kd) for dimerization, then a regulatory process that strengthens dimerization or increases local concentration can increase the affinity of the CSD interface for its ligands. The dimerization values of CSD-CSD domains from different HP1 paralogs range from low nanomolar for the CSD in Swi6 and hHP1β to micromolar for the CSD in dHP1a (Fig. 3C) 48,49,68,72,80. Analogous to how the NTE in the CD regulates CD-CD and CD-H3K9me3 tail interactions, the CTE in the CSD domains of Drosophila HP1 paralogs has been shown to regulate interactions between two CSDs as well as interactions between the CSD-CSD dimer and its ligands (Fig. 3D). The CTE length can be quite variable and is only 4 residues long in dHP1a, while 87 residues long in dHP1b. In dHP1b, the CTE significantly impairs CSD-CSD dimerization, lowering the dimerization affinity by approximately 100-fold, while the CTE in dHP1a contributes by increasing its affinity for specific CSD ligands48,49. The dimerization of Swi6 can also be controlled by the CD-CD interface as described above (Figure 3E), resulting in a 10-fold increase in dimerization of the full-length Swi6 relative to the CSD domain alone68. The CD-CD interface can thus provide an additional location from which to regulate the dimerization of the CSD-CSD interface. This mechanism is in contrast to dHP1a for which the dimerization of the CSD-CSD interface and the full-length protein are similar, suggesting that other domains do not participate in the dimerization of the full-length dHP1a protein48.
Specific post-translational modifications in the CSD can also modulate ligand affinity. For example, phosphorylation of serines in the dHP1a CSD decreases dimerization by ~6-fold but increases overall binding to PXVXL containing ligands by ~10-fold48. This argues that the affinity of the phosphorylated dimer is ~ 60-fold tighter for the PXVXL ligands.
The Hinge region
Compared to the CD and CSD, the hinge region is more diverse and less conserved among HP1 proteins. The length of the hinge region also varies substantially between paralogs (Figure 1). The hinge region contains a nuclear localization sequence and is the site of several different post-translational modifications identified in HP1 from phosphorylation to SUMOylation73,82–84. Analogous to the CD and CSD, the hinge is implicated in interactions with different types of ligands including RNA and DNA, histone deacetylases, the inner centromere protein (INCENP), and components of the Origin Recognition Complex (ORC)21,22,60,68,85–87, and these interactions are thought to help confer specific functions on the HP1 protein.
Because of its weak sequence conservation and its variable length, the hinge region represents an important source of functional differentiation between HP1 paralogs (Figure 1). Human HP1α and γ have different affinities for RNAs in vitro and these could arise due to differences in the hinge regions of these proteins21. However, the molecular basis of this difference is not fully understood because the critical charged residues responsible for nucleic acid binding (KRK and KKK) in the hinge region are conserved between all three mammalian HP1s. Additionally, in vivo, site-specific post-translational modifications have been suggested to account for paralog-specific nucleic acid binding. For example, SUMOylation of residue 84 in the hinge region of mouse HP1α appears to promote the association of HP1α with RNA transcripts and specifically target HP1α, but not HP1β or γ, to pericentromeric regions of the genome82. In the case of human HP1γ, phosphorylation of serine 83 in its hinge region increases its interaction with Ku70, a central DNA repair protein, and increases its specific localization to euchromatin88.
Finally, results with Swi6 raise the possibility that binding via the hinge region to DNA and RNA may have dramatically different biochemical effects. DNA enhances binding of Swi6 to the H3K9me3 mark while RNA antagonizes binding to the H3K9me3 mark 60,68 suggesting that differential recognition of DNA and RNA by the hinge region may allow Swi6 to switch between its roles in transcriptional and post-transcriptional silencing. This switch may occur because transcriptional silencing requires Swi6 to be associated with H3K9 methylated chromatin while post-transcriptional silencing entails dissociation of Swi6-RNA complexes from chromatin60.
Together, the data presented in this section show how small changes in the sequences between HP1 paralogs may enable different biological functions by affecting ligand recognition of the individual domains.
Versatility of function from conformational flexibility
Numerous enzymes and scaffold proteins involved in orchestrating more than one signaling event display some degree of functional promiscuity89–93. It has been hypothesized that single proteins that pilot signaling networks achieve their different functions by exhibiting some degree of conformational flexibility89–93, which allows one molecule to generate diverse functional states that can be regulated by specific protein partners. The binding of these protein partners ultimately promotes a conformational change in the signaling molecule resulting in the switching on or off of a particular cascade of events in a network. The magnitude of conformational diversity of signaling molecules can range from small fluctuations of side chains to more global tertiary structure rearrangements. In either case, the conformational flexibility provides a mechanism by which a single protein can achieve functional versatility. For example, the adenovirus early region 1A (E1A) oncoprotein is involved in cell cycle regulation and epigenetic cell reprogramming and interacts with diverse protein partners by means of its intrinsic structural adaptability. However, binding of any one specific molecular partner of EA1 results in its allosteric activation toward only one specific signaling pathway94. We propose that conceptually similar mechanisms might generate functional promiscuity within a single HP1 protein.
Conformational flexibility within an HP1 protein
NMR-based structural studies of human HP1β not only reveal that the hinge region is highly flexible and unfolded, but that the CD and CSD show significant conformational dynamics in solution95. In particular, the CD appears to sample more than one structural state in addition to the canonical H3K9me3 binding conformation95. However, the roles of the other conformations remain unknown. Nonetheless, these observations raise the possibility that ligands other than chromatin may stabilize alternative conformations and promote specific functional states.
In the case of Swi6, at least two distinct structural states have been identified: a closed and an open state68. Mediated by the CD-CD interaction described above (Fig. 2B), the closed state inhibits binding of the H3K9 methyl mark while the open state is capable of H3K9 methyl mark binding and spreading across chromatin via oligomerization (Figures 4A &B). We speculate that different conformations of Swi6 allow for different types of interactions with chromatin and other ligands. For example, while the spreading competent state may promote Swi6 assembly on chromatin, the closed, auto-inhibited conformation may serve to disengage Swi6 from chromatin. Binding by RNA appears to disengage Swi6 from methylated chromatin in vivo60, and we speculate that RNA binding drives Swi6 into the closed state. The closed state may also be stabilized by other chromatin binding proteins that might serve to cap heterochromatin spreading at boundary regions, therefore inhibiting heterochromatin assembly by promoting Swi6 auto-inhibition. For example, it has been shown that the anti-silencing protein Epe1 needs to be recruited by Swi6 to limit heterochromatin spread62,63. Given that the closed state does not recognize the H3K9me mark, it is also possible that this conformation binds euchromatin through interactions with the nucleosomal DNA and possibly the PGTVAL sequence in histone H3 (Figure 4).
Conformational flexibility of HP1 domains on a nucleosome
The modular nature in which individual HP1 domains bind a nucleosome raises the possibility that different types of HP1-nucleosome complexes are formed depending upon which HP1 domains are engaged with the nucleosome and which HP1 domains are engaged with other ligands (Figure 4B). In the case of Swi6, four molecules bind one H3K9 methylated nucleosome72. This architecture allows the formation of two unoccupied CDs that can bind nearby nucleosomes (Figures 4A & B). The interaction of Swi6 with a nucleosome is stabilized by multiple contacts made through the CD, the hinge region, and the CSD. Together, these interactions increase the stability of a Swi6-H3K9me3 nucleosomes complex approximately 100-fold over a Swi6-H3K9me3 tail peptide complex (Kd=10μM vs. 100 nM, respectively for peptide versus nucleosome)68. Studies of hHP1α on tetranucleosomes analogously suggest that the hinge and the CSD both help recognize methylated nucleosomes96. We hypothesize that the presence of multiple contacts between HP1 molecules and a nucleosome provides sufficient binding energy to allow individual domains to interact with other ligands while remaining associated with chromatin. Thus, a CSD dimer could interact with a PxVxL-containing ligand, while the CD and hinge region provide the contacts to remain bound to chromatin. Similarly, the histone mimic sequence in the CD of Swi6 could interact with other CD containing proteins while Swi6 remains bound to chromatin. Such mechanisms would allow different types of functions to be recruited to the HP1-nucleosome complex and help explain how the same HP1 paralog can perform different biological roles.
In contrast to Swi6, NMR-based studies imply that only one dimer of human HP1β binds per nucleosome97. Further, hHP1β seems to bind methylated nucleosomes with an affinity similar to methylated H3 tail peptides97, arguing that unlike Swi6 and hHP1α, hHP1β only contacts the H3K9me3 tail and no other nucleosomal surface. In this context, one can imagine that the CSD-CSD dimer interface of hHP1β is perhaps more readily available for binding PxVxL–containing ligands than the CSD-CSD interface of hHP1α.
The above observations illustrate how the same HP1 molecule has the potential to adopt different functional conformations in vivo. In addition, the different stoichiometries adopted by different HP1 paralogs on chromatin impose different constraints on the ability of these molecules to oligomerize and recruit other factors. Finally, given that the CSD can dimerize and, in certain cases like Swi6, the CD can dimerize, it is possible that different HP1 paralogs form heterodimers. Indeed, co-immunoprecipitation-based studies have suggested that mammalian HP1 paralogs can form heterodimers in solution as well as on chromatin98. The formation of such heterodimers can further diversify the structures that can be adopted by HP1-chromatin complexes and the types of ligands that are recruited to these complexes. However, it remains to be determined what regulates the formation of various HP1-nucleosome complexes and what their functions are in the cell.
Concluding remarks
While HP1 proteins were originally thought to be structural proteins important for the integrity of heterochromatin, a wealth of data has shown that these proteins are also active participants in diverse nuclear activities. The studies reviewed here clearly show that the diversity in the HP1 sequences and conformations can help explain their different biological functions. It is possible to imagine HP1 proteins as chromatin adapter proteins that enhance the signaling of histone modifications by providing additional functionalities to the chromatin platform (Figure 4B). Similar to histone variants and histone modifications, these functionalities can be regulated by HP1 paralogs and post-translational modifications. However, compared to histone variants and histone modifications, we know substantially less about the molecular impact of HP1 paralogs and HP1 modifications. The biophysical studies carried out on HP1 proteins to date have been critical for bridging the gap between biological phenomena and molecular mechanism. Since the biological roles played by HP1 proteins are being shown to be increasingly complex, it is even more critical to understand how HP1 paralogs engage with chromatin at a structural and thermodynamic level. This is because understanding the molecular basis for HP1 function requires, as a first step, an understanding of the types of interactions made between HP1 proteins and methylated chromatin. This process can be accelerated by increasing the types of quantitative biochemical methods available to study HP1 interactions and by high-resolution structures of HP1-chromatin complexes. As we learn more about the biochemical properties of different HP1 paralogs, the types of thermodynamic concepts described here can clarify the functions of HP1 proteins in vivo.
Highlights.
HP1 proteins play diverse roles invivo
Functional versatility can arise from small sequence differences between HP1 paralogs
Functional versatility may arise from conformational flexibility of an HP1 protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cavalli G. Chromatin as a eukaryotic template of genetic information. Curr Opin Cell Biol. 2002;14:269–278. doi: 10.1016/s0955-0674(02)00324-1. [DOI] [PubMed] [Google Scholar]
- 2.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 3.Flemming W. Beitraege zur Kenntniss der Zelle und ihrer Leb- enserscheinungen. Arch Mikroskop. 1878;16:302–436. [Google Scholar]
- 4.Flemming W. Zellsubstanz, Kern und Zelltheilung. 1882. [Google Scholar]
- 5.Heitz E. Das Heterochromatin der Moose, 1. Jahrb Wiss Bot. 1928;69:762–818. [Google Scholar]
- 6.Heitz E. Heterochromatin, Chromocentren, Chromomeren. Ber Dtsch Bot Ges. 1929;47:274–284. [Google Scholar]
- 7.Grewal SIS, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 8.James T, Elgin S. Identification of a Nonhistone Chromosomal Protein Associated with Heterochromatin in Drosophila melanogaster and Its Gene. Mol Cell Biol. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eissenberg JC, et al. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1990;87:9923–7. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller HJ, Altenburg E. The Frequency of Translocations Produced by X-Rays in Drosophila. Genetics. 1930;15:283–311. doi: 10.1093/genetics/15.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spofford JB. Single-locus modification of position-effect variegation in Drosophila melanogaster. I White variegation. Genetics. 1967;57:751–66. doi: 10.1093/genetics/57.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark RF, Elgin SC. Heterochromatin protein 1, a known suppressor of position-effect variegation, is highly conserved in Drosophila. Nucleic Acids Res. 1992;20:6067–74. doi: 10.1093/nar/20.22.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Platero JS, Hartnett T, Eissenberg JC. Functional analysis of the chromo domain of HP1. EMBO J. 1995;14:3977–3986. doi: 10.1002/j.1460-2075.1995.tb00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallrath LL. Unfolding the mysteries of heterochromatin. Curr Opin Genet Dev. 1998;8:147–153. doi: 10.1016/s0959-437x(98)80135-4. [DOI] [PubMed] [Google Scholar]
- 15.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 16.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 17.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–20. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 18.Peters AHFM, et al. Loss of the Suv39h Histone Methyltransferases Impairs Mammalian Heterochromatin and Genome Stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 19.Cowieson NP, Partridge JF, Allshire RC, McLaughlin PJ. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr Biol. 2000;10:517–525. doi: 10.1016/s0960-9822(00)00467-x. [DOI] [PubMed] [Google Scholar]
- 20.Smothers JF, Henikoff S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr Biol. 2000;10:27–30. doi: 10.1016/s0960-9822(99)00260-2. [DOI] [PubMed] [Google Scholar]
- 21.Muchardt C, et al. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 2002;3:975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meehan RR, Kao C-F, Pennings S. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. EMBO J. 2003;22:3164–74. doi: 10.1093/emboj/cdg306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto K, Sonoda M. Self-interaction of heterochromatin protein 1 is required for direct binding to histone methyltransferase, SUV39H1. Biochem Biophys Res Commun. 2003;301:287–292. doi: 10.1016/s0006-291x(02)03021-8. [DOI] [PubMed] [Google Scholar]
- 24.Yamada T, Fukuda R, Himeno M, Sugimoto K. Functional domain structure of human heterochromatin protein HP1(Hsalpha): involvement of internal DNA-binding and C-terminal self-association domains in the formation of discrete dots in interphase nuclei. J Biochem. 1999;125:832–7. doi: 10.1093/oxfordjournals.jbchem.a022356. [DOI] [PubMed] [Google Scholar]
- 25.Vakoc CR, Mandat Sa, Olenchock Ba, Blobel Ga. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–91. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 26.Mateescu B, Bourachot B, Rachez C, Ogryzko V, Muchardt C. Regulation of an inducible promoter by an HP1beta-HP1gamma switch. EMBO Rep. 2008;9:267–72. doi: 10.1038/embor.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon SH, Workman JL. HP1c casts light on dark matter. Cell Cycle. 2011;10:625–630. doi: 10.4161/cc.10.4.14796. [DOI] [PubMed] [Google Scholar]
- 28.Nonaka N, et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 29.Yamagishi Y, Sakuno T, Shimura M, Watanabe Y. Heterochromatin links to centromeric protection by recruiting shugoshin. Nature. 2008;455:251–255. doi: 10.1038/nature07217. [DOI] [PubMed] [Google Scholar]
- 30.Hayakawa T, Haraguchi T, Masumoto H, Hiraoka Y. Cell cycle behavior of human HP1 subtypes: distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase. J Cell Sci. 2003;116:3327–38. doi: 10.1242/jcs.00635. [DOI] [PubMed] [Google Scholar]
- 31.Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 2004;23:2651–63. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perrini B, et al. HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol Cell. 2004;15:467–76. doi: 10.1016/j.molcel.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 33.Canudas S, et al. A role for heterochromatin protein 1γ at human telomeres. Genes Dev. 2011;25:1807–19. doi: 10.1101/gad.17325211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dinant C, Luijsterburg MS. The emerging role of HP1 in the DNA damage response. Mol Cell Biol. 2009;29:6335–40. doi: 10.1128/MCB.01048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luijsterburg MS, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol. 2009;185:577–86. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soria G, Almouzni G. Differential contribution of HP1 proteins to DNA end resection and homology-directed repair. Cell cycle. 2013;12:422–9. doi: 10.4161/cc.23215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loomis RJ, et al. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell. 2009;33:450–61. doi: 10.1016/j.molcel.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saint-André V, Batsché E, Rachez C, Muchardt C. Histone H3 lysine 9 trimethylation and HP1γ favor inclusion of alternative exons. Nat Struct Mol Biol. 2011;18:337–44. doi: 10.1038/nsmb.1995. [DOI] [PubMed] [Google Scholar]
- 39.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levine MT, et al. Phylogenomic Analysis Reveals Dynamic Evolutionary History of the Drosophila Heterochromatin Protein 1 (HP1) Gene Family. PLoS Genet. 2012;8:e1002729. doi: 10.1371/journal.pgen.1002729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vermaak D, Malik HS. Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu Rev Genet. 2009;43:467–92. doi: 10.1146/annurev-genet-102108-134802. [DOI] [PubMed] [Google Scholar]
- 42.Smallwood A, et al. CBX3 regulates efficient RNA processing genome-wide. Genome Res. 2012;22:1426–36. doi: 10.1101/gr.124818.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Billur M, Bartunik HD, Singh PB. The essential function of HP1 beta: a case of the tail wagging the dog? Trends Biochem Sci. 2010;35:115–23. doi: 10.1016/j.tibs.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 44.Kwon SH, et al. Heterochromatin protein 1 (HP1) connects the FACT histone chaperone complex to the phosphorylated CTD of RNA polymerase II. Genes Dev. 2010;24:2133–45. doi: 10.1101/gad.1959110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smothers JF, Henikoff S. The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol Cell Biol. 2001;21:2555–2569. doi: 10.1128/MCB.21.7.2555-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kato M, et al. Functional domain analysis of human HP1 isoforms in Drosophila. Cell Struct Funct. 2007;32:57–67. doi: 10.1247/csf.06032. [DOI] [PubMed] [Google Scholar]
- 47.Sadaie M, et al. Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol Cell Biol. 2008;28:6973–6988. doi: 10.1128/MCB.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mendez DL, et al. The HP1a disordered C terminus and chromo shadow domain cooperate to select target peptide partners. Chembiochem. 2011;12:1084–96. doi: 10.1002/cbic.201000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mendez DL, Mandt RE, Elgin SCR. Heterochromatin Protein 1a (HP1a) Partner Specificity is Determined by Critical Amino Acids in the Chromo Shadow Domain and C-terminal Extension. J Biol Chem. 2013;228:22315–23. doi: 10.1074/jbc.M113.468413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lomberk G, Wallrath L, Urrutia R. The Heterochromatin Protein 1 family. Genome Biol. 2006;7:228. doi: 10.1186/gb-2006-7-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fanti L, Pimpinelli S. HP1: a functionally multifaceted protein. Curr Opin Genet Dev. 2008;18:169–74. doi: 10.1016/j.gde.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 52.Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–75. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 53.Cheutin T, et al. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Sci (New York, NY) 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 54.Schmiedeberg L, Weisshart K, Diekmann S, Meyer Zu, Hoerste G, Hemmerich P. High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol Biol Cell. 2004;15:2819–2833. doi: 10.1091/mbc.E03-11-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Müller KP, et al. Multiscale analysis of dynamics and interactions of heterochromatin protein 1 by fluorescence fluctuation microscopy. Biophys J. 2009;97:2876–2885. doi: 10.1016/j.bpj.2009.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheutin T, Gorski S, May K. In vivo dynamics of Swi6 in yeast: evidence for a stochastic model of heterochromatin. Mol Cell Biol. 2004;24:3157–3167. doi: 10.1128/MCB.24.8.3157-3167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SIS. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell. 2005;20:173–185. doi: 10.1016/j.molcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 58.Sugiyama T, et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 59.Haldar S, Saini A, Nanda JS, Saini S, Singh J. Role of Swi6/HP1 self-association-mediated recruitment of Clr4/Suv39 in establishment and maintenance of heterochromatin in fission yeast. J Biol Chem. 2011;286:9308–20. doi: 10.1074/jbc.M110.143198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keller C, et al. HP1(Swi6) Mediates the Recognition and Destruction of Heterochromatic RNA Transcripts. Mol Cell. 2012;47:215–227. doi: 10.1016/j.molcel.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 61.Iida T, Nakayama J, Moazed D. siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol Cell. 2008;31:178–89. doi: 10.1016/j.molcel.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zofall M, Grewal SIS. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell. 2006;22:681–692. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 63.Braun S, et al. The Cul4-Ddb1(Cdt)2 ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell. 2011;144:41–54. doi: 10.1016/j.cell.2010.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nielsen PR, et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 65.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Sci (New York, NY) 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 66.Hiragami-Hamada K, et al. N-terminal phosphorylation of HP1{alpha} promotes its chromatin binding. Mol Cell Biol. 2011;31:1186–200. doi: 10.1128/MCB.01012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Machado MR, Dans PD, Pantano S. Isoform-specific determinants in the HP1 binding to histone 3: insights from molecular simulations. Amino Acids. 2010;38:1571–81. doi: 10.1007/s00726-009-0371-3. [DOI] [PubMed] [Google Scholar]
- 68.Canzio D, et al. A conformational switch in HP1 releases auto-inhibition to drive heterochromatin assembly. Nature. 2013;496:377–81. doi: 10.1038/nature12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao T, Eissenberg JC. Phosphorylation of heterochromatin protein 1 by casein kinase II is required for efficient heterochromatin binding in Drosophila. J Biol Chem. 1999;274:15095–100. doi: 10.1074/jbc.274.21.15095. [DOI] [PubMed] [Google Scholar]
- 70.Zhao T, Heyduk T, Eissenberg JC. Phosphorylation site mutations in heterochromatin protein 1 (HP1) reduce or eliminate silencing activity. J Biol Chem. 2001;276:9512–8. doi: 10.1074/jbc.M010098200. [DOI] [PubMed] [Google Scholar]
- 71.Wang G, et al. Conservation of heterochromatin protein 1 function. Mol Cell Biol. 2000;20:6970–6983. doi: 10.1128/mcb.20.18.6970-6983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Canzio D, et al. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol Cell. 2011;41:67–81. doi: 10.1016/j.molcel.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.LeRoy G, et al. Heterochromatin protein 1 is extensively decorated with histone code-like post-translational modifications. Mol Cell Proteomics. 2009;8:2432–42. doi: 10.1074/mcp.M900160-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SIS. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell. 2005;20:173–85. doi: 10.1016/j.molcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 75.Fischle W, et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–81. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smothers J, Henikoff S. The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol Cell Biol. 2001;21:2555–2569. doi: 10.1128/MCB.21.7.2555-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dawson Ma, et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–22. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lavigne M, et al. Interaction of HP1 and Brg1/Brm with the globular domain of histone H3 is required for HP1-mediated repression. PLoS Genet. 2009;5:e1000769. doi: 10.1371/journal.pgen.1000769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Richart AN, Brunner CIW, Stott K, Murzina NV, Thomas JO. Characterization of the chromoshadow domain-mediated binding of heterochromatin protein 1 α (HP1α) to histone H3. J Biol Chem. 2012;287:18730–18737. doi: 10.1074/jbc.M111.337204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brasher SV, et al. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–97. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thiru A, et al. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 2004;23:489–99. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maison C, et al. SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat Genet. 2011;43:220–227. doi: 10.1038/ng.765. [DOI] [PubMed] [Google Scholar]
- 83.Shimada A, et al. Phosphorylation of Swi6/HP1 regulates transcriptional gene silencing at heterochromatin. Genes Dev. 2009;23:18–23. doi: 10.1101/gad.1708009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Badugu R, Yoo Y, Singh PB, Kellum R. Mutations in the heterochromatin protein 1 (HP1) hinge domain affect HP1 protein interactions and chromosomal distribution. Chromosoma. 2005;113:370–84. doi: 10.1007/s00412-004-0324-2. [DOI] [PubMed] [Google Scholar]
- 85.Zhang CL, McKinsey TA, Olson EN. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol Cell Biol. 2002;22:7302–7312. doi: 10.1128/MCB.22.20.7302-7312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ainsztein aM, Kandels-Lewis SE, Mackay aM, Earnshaw WC. INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J Cell Biol. 1998;143:1763–74. doi: 10.1083/jcb.143.7.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Badugu R, Shareef MM, Kellum R. Novel Drosophila heterochromatin protein 1 (HP1)/origin recognition complex-associated protein (HOAP) repeat motif in HP1/HOAP interactions and chromocenter associations. J Biol Chem. 2003;278:34491–8. doi: 10.1074/jbc.M305262200. [DOI] [PubMed] [Google Scholar]
- 88.Lomberk G, Bensi D, Fernandez-Zapico ME, Urrutia R. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat Cell Biol. 2006;8:407–15. doi: 10.1038/ncb1383. [DOI] [PubMed] [Google Scholar]
- 89.James LC, Tawfik DS. Conformational diversity and protein evolution--a 60-year-old hypothesis revisited. Trends Biochem Sci. 2003;28:361–8. doi: 10.1016/S0968-0004(03)00135-X. [DOI] [PubMed] [Google Scholar]
- 90.Khersonsky O, Roodveldt C, Tawfik DS. Enzyme promiscuity: evolutionary and mechanistic aspects. Curr Opin Chem Biol. 2006;10:498–508. doi: 10.1016/j.cbpa.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 91.Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–7. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 92.Wrabl JO, et al. The role of protein conformational fluctuations in allostery, function, and evolution. Biophys Chem. 2011;159:129–41. doi: 10.1016/j.bpc.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Good MC, Zalatan JG, Lim Wa. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–6. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ferreon ACM, Ferreon JC, Wright PE, Deniz Aa. Modulation of allostery by protein intrinsic disorder. Nature. 2013;498:390–394. doi: 10.1038/nature12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Munari F, Rezaei-Ghaleh N, Xiang S, Fischle W, Zweckstetter M. Structural plasticity in human heterochromatin protein 1β. PLoS One. 2013;8:e60887. doi: 10.1371/journal.pone.0060887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mishima Y, et al. Hinge and chromoshadow of HP1α participate in recognition of K9 methylated histone H3 in nucleosomes. J Mol Biol. 2012;425:54–70. doi: 10.1016/j.jmb.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 97.Munari F, et al. Methylation of Lysine 9 in Histone H3 Directs Alternative Modes of Highly Dynamic Interaction of Heterochromatin Protein hHP1β with the Nucleosome. J Biol Chem. 2012;287:33756–65. doi: 10.1074/jbc.M112.390849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nielsen aL, et al. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol Cell. 2001;7:729–39. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]