Abstract
Representatives of all major metazoan lineages form biominerals. The molecular mechanisms that underlie this widespread and evolutionarily ancient ability are gradually being revealed for some lineages. However, until a wider range of metazoan biomineralization strategies are understood, the true diversity, and therefore the evolutionary origins of this process, will remain unknown. We have previously shown that the coralline demosponge, Astrosclera willeyana, in some way employs its endobiotic bacterial community to form its highly calcified skeleton. Here, using in situ hybridization and immunohistochemistry, we show that an ortholog of ATG8 (most likely a GABARAPL2/GATE-16 ortholog) is expressed in cells that construct the individual skeletal elements of the sponge. In TEM sections sponge cells can be observed to contain extensive populations of bacteria, and frequently possesses double-membrane structures which we interpret to be autophagosomes. In combination with our previous work, these findings support the hypothesis that the host sponge actively degrades a proportion of its bacterial community using an autophagy pathway, and uses the prokaryotic organic remains as a framework upon which calcification of the sponge skeleton is initiated.
Keywords: biomineralization, biocalcification, sponge, autophagy, symbiosis, symbiophagy, evolution, bacteria, GABARAPL2/GATE-16, ATG8
Introduction
The molecular mechanisms employed by animals to build their skeletons are of inherent interest to evolutionary biologists. This is because the fossil record of animal life tells us that most major metazoan lineages began to biomineralize relatively synchronously about 545 million years ago, and that this phenomenon closely coincides with an unprecedented diversification of body plans. While it is clear that the evolution of the ability to biomineralize did not cause the Cambrian Explosion per se (all of the major metazoan lineages must have been established prior to this great diversification), it is likely that the widespread adoption of various biomineralization strategies greatly supported the evolution of various body forms. Understanding the ways in which a diverse range of animals are able to biomineralize would help us to understand the origins of this ability, providing deep insight into one aspect of how the evolution of multicellular animal life was supported.
To this end we have been studying how the so-called “living fossil” Astrosclera willeyana makes its skeleton. A. willeyana is an informative animal to study with regard to the evolution of biomineralization for several reasons. First, it is a member of that group of animals (sponges) to diverge from the rest of the Metazoa. This means that any biomineralizing mechanisms present in both sponges and all other animals are likely to have also been present in the last common ancestor of the Metazoa. Extant sponges also display a wide repertoire of biomineralization modes (mineralogically and morphologically), and fossil evidence clearly indicates that their deep ancestors also did so.1-3 Furthermore, the body-plan of A. willeyana is unusual among extant sponges as it shares similarity with a now-extinct lineage of organisms known as stromatoporoids. Stromatoporoids were once abundant in the oceans of the Palaeozoic and Mesozoic, and formed extensive marine reefs that provided habitats for the ancestors of contemporary metazoans to occupy, similar to the ecological role that corals play today. While the morphological similarities between A. willeyana and the stromatoporoids are not considered to be adequate evidence for a direct phylogenetic relationship (i.e., it is unclear if A. willeyana is in fact a living stromatoporoid, or a close relative of this lineage),4 this possibility affords A. willeyana a special status with regards to its value as a model for studying the evolution of metazoan biomineralization mechanisms. Finally, A. willeyana has the extremely unusual ability of being able to deposit 2 distinct biominerals (siliceous spicules and a “massive” secondary calcium carbonate skeleton) in a spatially and temporally overlapping fashion.
Our previous work has allowed us to construct a working model of how calcification occurs in A. willeyana. For a detailed description of the morphological events that accompany this process see Wörheide.4 Briefly, the distal-most layer of cells in the animal (the ectosome) initiate the deposition of calcified “spherulites.” These spherulites increase in size and are moved proximally to the hyper-calcified “basal skeleton” where they fuse together and further increase in size by epitaxial growth along the c-axis of the aragonite crystal. “Large vacuole cells” are responsible for the initiation of spherulite formation in the ectosome, and the hypothesis proposed by Wörheide4 was that bacteriocytes (sponge cells containing large numbers of intracellular bacteria), may degrade their microbial communities and subsequently use these organic remains as a template upon which CaCO3 could be precipitated from solution, making the bacteriocyte a precursor state to the large vacuole cell state. We subsequently gathered biogeochemical evidence that supports this hypothesis by demonstrating that the organic material occluded within nascent and midterm spherulites is primarily of prokaryotic origin.5 In other words, the organic material that acts as a scaffold upon which biomineralization takes place in this animal is primarily derived from bacteria, and not from the sponge. Furthermore, our discovery of a gene product associated with spherulite formation that was horizontally transferred from a bacterium into the A. willeyana genome6 also serves to highlight the intimate and long standing association between A. willeyana, its microbial community, and the process of biomineralization. We have also demonstrated the involvement of an α carbonic anhydrase in the process of spherulite formation,7 indicating that deeply conserved metazoan skeleton-forming molecular mechanisms are also employed by A. willeyana.
To further investigate the association between A. willeyana, its microbial community, and the process of biomineralization, we decided to search for molecular markers that would indicate a mechanism of active degradation of intracellular microorganisms by the sponge host. Autophagy is an ancient eukaryotic process present in organisms ranging from yeasts to humans.8 Autophagy, or “self-eating,” allows cells to degrade and recycle organelles and proteins, and is active under a wide range of conditions including normal development, metamorphosis, and starvation.9,10 So-called “nonclassical” instances of autophagy are also activated as a line of defense against invading intracellular pathogens.11 These instances differ from “classical autophagy” in that the autophagy machinery is more specific in what it targets for degradation.12 With the discovery of highly conserved genes involved in the formation of the autophagosome,13 it is now possible to study the involvement of the autophagy pathway in non-model organisms for which few molecular resources exist. An illustrative example of this is provided by Dunn et al.,14 the first to show that autophagy is involved in the process of coral bleaching (the degradation and/or ejection of symbiotic zooxanthellae algae during periods of environmental stress to the coral host). Downs et al.15 subsequently investigated the relationship of autophagy and coral bleaching and termed the consumption of symbiotic algae by the coral host as “symbiophagy.”
The majority of the more than 30 AuTopahGy-related (ATG) genes originally identified in yeast16 have orthologs and paralogs in all metazoans studied to date.8 Some of these autophagy-associated genes are also involved in other cellular processes making their use as autophagy-specific markers problematic. However a subset of these 30 is considered critical for autophagosome formation in all cell types studied thus far;13 one of these core autophagy genes is ATG8. There is a single ATG8 gene in yeast, which has been duplicated more than once in the genomes of more complex metazoans to generate the GABARAP, GABARAPL2/GATE-16 and MAP1LC3 (LC3) genes. Atg8 is a ubiquitin-like protein that is involved in the formation and fusion of autophagosome membranes.17 In addition, autophagosomes possess double membranes, making it possible to identify them using TEM.18
Here we provide molecular, histochemical, and morphological evidence that suggests A. willeyana is actively degrading its intracellular microbial community using the autophagy pathway, and that bacteriocytes are most likely the precursors to the spherulite-forming large vacuolar cells. As far as we are aware, this is the first time an association between the process of autophagy and biomineralization has been made in a Metazoan.
Results and Discussion
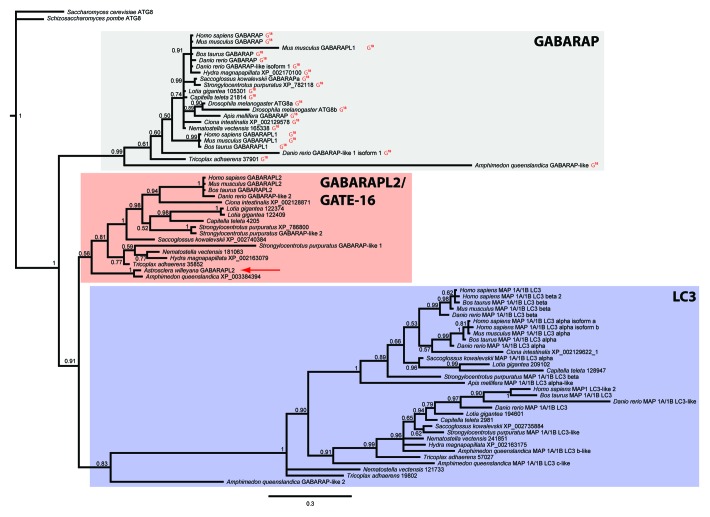
Degenerate and RACE-PCR-based methods allowed us to isolate a 707 bp putative full-length cDNA (GenBank with accession number KC567304) encoding a 117-residue protein with clear similarity to Atg8 proteins from a variety of other metazoans. The sequence-based maximum likelihood phylogenetic analysis Shpilka et al.19 performed for the Atg8 protein family is the first and only one reported in the scientific literature for an Atg8 ortholog. While our Bayesian analysis returns the same three major clades of metazoan Atg8 orthologs reported by Shpilka et al.19 (a GABARAPL2/GATE-16, GABARAP and LC3 clade; Fig. 1) the relationships of these three clades is fundamentally different. Shpilka et al.19 suggest that there are some defining sequence features of Atg8 orthologs such as an invariant glycine residue at position 18 in all GABARAP orthologs. All sequences with this invariant feature form a strongly supported clade in our tree with the same relative position as the GABARAPL2/GATE-16 clade presented by Shpilka et al.19 Conversely, their GABARAP clade occupies the same position as our GABARAPL2 clade (their Fig. 1 cf. our Fig. 1). The topology of our tree is therefore a GABARAP clade as an earlier branching sister clade to the GABARAPL2 + LC3 clades. Based on our phylogenetic analysis this Awi-Atg8 sequence falls within a clade of orthologs of GABARAPL2 (Fig. 1). From here on we will therefore refer to this sequence as Awi-GABARAPL2. Included in our phylogenetic analysis are 5 Atg8 orthologs identified from the genome of the demosponge Amphimedon queenslandica. The placement of these sponge sequences throughout our tree indicates that it was after the divergence of the last common ancestor of the Metazoa from its unicellular ancestors that the ATG8 gene family was duplicated and diversified (only one ATG8 gene is known from fungal genomes).
Figure 1. Bayesian phylogenetic analysis of the relationships between a broad sample of metazoan Atg8 orthologs. All Atg8 sequences were identified from the complete or draft genomes of all species presented here (except for A. willeyana). The yeast Saccharomyces cerevisiae Atg8 sequence was set as an outgroup. The A. willeyana sequence (highlighted by a red arrow) falls within the “γ-aminobutyric acid receptor-associated protein” clade. Sequences possessing a glycine residue at position 18 (an invariant feature of all GABARAP proteins as identified by Shpilka et al.,19 are indicated with a red “G18.” This tree topology is a 50% majority rule consensus of 778 trees, which were sampled every 10,000 generations. Node values are Bayesian posterior probabilities.
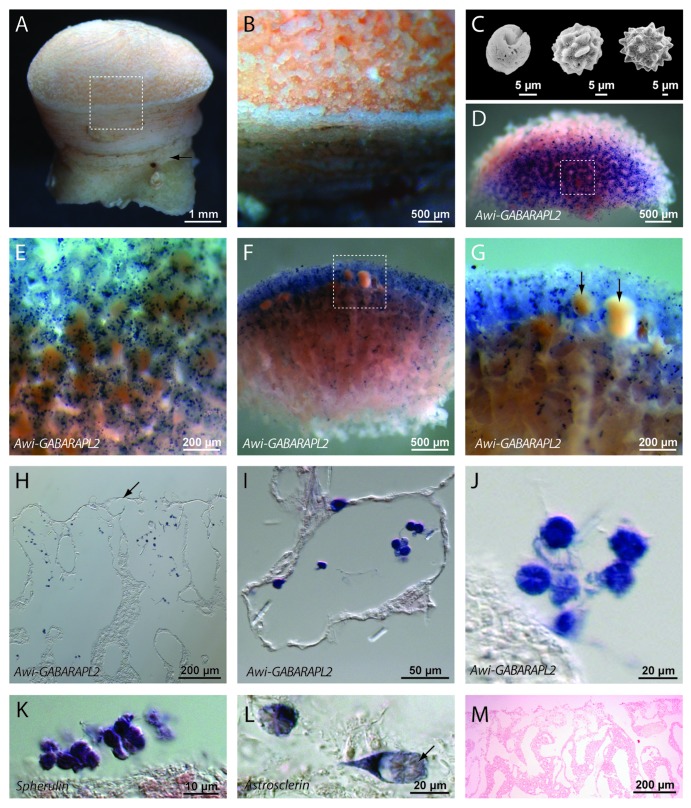
We also used the Awi-GABARAPL2 sequence to generate a complimentary DIG labeled ribo-probe in order to perform whole mount in situ hybridization (WMISH) in adult A. willeyana. Expression of Awi-GABARAPL2 is clearly restricted to cells in the adult sponge responsible for spherulite formation (Fig. 2). Awi-GABARAPL2 transcripts are not present in any other differentiated cell types, and are also absent from developing larvae (Fig. 2). Free-living, extracellular bacterial cells are occasionally visible in the WMISH processed and sectioned material, however these were never observed to be Awi-GABARAPL2+. Spherulite-forming cells are easily morphologically distinguishable and are the only cell type to express Awi-GABARAPL2.
Figure 2. An A. willeyana GABARAPL2 ortholog is expressed exclusively in spherulite-forming cells. (A) A lateral view of a mature A. willeyana illustrating the calcified stalk (arrow) upon which the living part of the animal grows. (B) A magnified view of the boxed region in (A) highlighting the individual spherulites that comprise the calcified skeleton. (C) A series of SEMs illustrating the growth and change in morphology of spherulites (reproduced from).7 (D) A representative “apical” view of a decalcified and sagittally sectioned A. willeyana individual following WMISH against Awi-GABARAPL2. (E) Magnification of the boxed region in (D) to illustrate the distribution and density of Awi-GABARAPL2-positive cells. (F) A representative lateral view of the sectioned face of a decalcified and sagittally sectioned A. willeyana individual following WMISH against Awi-GABARAPL2. Awi-GABARAPL2-positive cells are clearly most abundant in the distal-most spherulite-forming region of the sponge. (G) Magnification of the boxed region in (F) reveals an absence of these transcripts from developing larvae (arrows). The blue dot present on the left-most larvae in this panel is overlying the larva. (H) Paraffin sections of WMISH material show the location of Awi-GABARAPL2-positive cells in more detail. These cells are located just below the distal-most layer of ectosomal cells (indicated by an arrow). (I) A higher magnification of this sectioned material reveals the distinctive morphology of spherulite-forming cells, and the exclusive signal of Awi-GABARAPL2 in these cells. (J) A magnified view of several Awi-GABARAPL2-positive cells. The texture of the IOM (cf. M) that remains following decalcification is visible within these cells. (K) Cells with the same morphology as those expressing Awi-GABARAPL2 also express Spherulin, a gene that was horizontally transferred from a bacterium into the genome of A. willeyana and is most likely now involved in biomineralization in A. willeyana.6 The texture of the IOM that remains following decalcification is also visible within these cells (cf. M). (L) Cells with the same morphology as those expressing Awi-GABARAPL2 also express Astrosclerin, an α-CA homolog that is involved in biomineralization in A. willeyana.7 The texture of the IOM that remains following decalcification is clearly visible within these cells (arrow, cf. Fig. 4F). (M) A representative section of A. willeyana material that was subjected to WMISH using a sense GABARAPL2 control probe. This section was counterstained with Nuclear Fast Red.
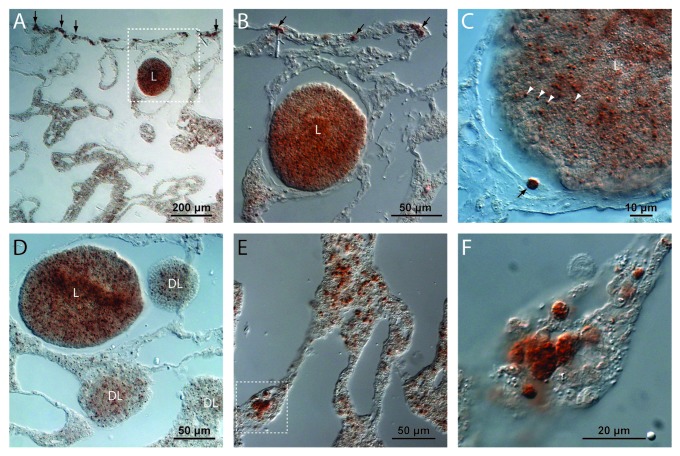
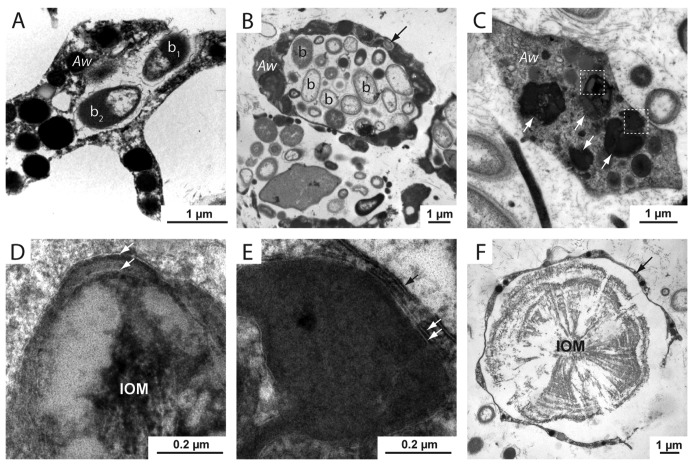
We also performed immunohistochemistry using an antibody directed against LC3A, another component of the autophagy pathway. Based on the recognizable morphology of spherulite-forming cells in A. willeyana, the spatial distribution of Awi-LC3A also indicates that the autophagy pathway is active in the same spherulite-forming cells that Awi-GABARAPL2 is expressed in Figure 3. Interestingly, this antibody also cross reacts with cells within larvae of various developmental stages (Fig. 3A–D). These LC3A+ larval cells are significantly smaller than the differentiated spherulite-forming LC3A+ adult cells (see Fig. 3C), and are unlikely to be involved in spherulite formation. Indeed, larvae of A. willeyana are unlikely to begin biocalcification prior to metamorphosis, which in turn occurs following release from the parent sponge and planktonic dispersal. Due to the presence of LC3A in these larval cells it appears as though the autophagy pathway may be playing an ontogenetic role in A. willeyana. In addition to our in situ and immuno results, TEM sections of A. willeyana adult material reveal an abundance of inter- and intracellular bacteria, and intracellular double-membrane structures (Fig. 4), which in other model systems can be used to identify autophagosomes.18
Figure 3. Immunocytochemistry reveals A. willeyana LC3A+ cells. (A) A low magnification image of a section illustrating the inner choanosome and the outer ectosome of A. willeyana. Near the outermost ectosome layer is a larva (L) containing LC3A-positive cells. Also clearly visible in this section are spherulite-forming ectosomal LC3A+ cells (vertical arrows). (B) A magnification of the boxed region in A detailing the LC3A+ cells in the larva (L), and the larger mature cells in the ectosome. (C) An adult LC3A+ cell (black arrow) in close proximity to a larva illustrates the clear difference in size between differentiated spherulite-forming cells and larval LC3A+ cells (white arrowheads). (D) Developing larvae (DL) and relatively mature larvae (L) contain LC3A+ cells suggesting that the process of autophagy is active throughout development in A. willeyana. However LC3A+ cells in larval stages do not have the same morphology as fully differentiated adult spherulite-forming cells, suggesting that these larval cells are not forming spherulites. (E) An image from the deeper choanosome layer where there can be many LC3A+ cells. (F) A magnified view of the boxed region in (E) illustrates the rounded morphology of these spherulite-forming cells.
Figure 4. TEM sections of A. willeyana tissue reveal a rich population of inter- and intracellular bacteria, and subcellular structures bound by multiple membranes. (A) An A. willeyana cell (Aw) in the process of phagocytosing a bacterial cell (b1). (B) A representative TEM section illustrating the abundant intracellular bacterial community of A. willeyana. Indicated are bacterial cells (b) located within a large membrane-bound space within the sponge cell (Aw). A bacterial cell located within the cytoplasm is indicated with an arrow. (C) An A. willeyana cell (Aw) containing likely autophagosomes (white arrows). (D) Magnification of the upper left-most boxed region in (C). A double membrane is indicated by 2 white arrows, and the heterogeneous IOM that will form the material upon which calcification will take place is clearly visible. (E) Magnification of the lower boxed section in (C) illustrates another double membrane enclosing an autophagosome (white arrows). The cell membrane is indicated by a black arrow. (F) A TEM section of decalcified A. willeyana tissue. Shown is a spherulite-forming cell (arrow) surrounding the insoluble organic material (IOM) that is normally occluded within the calcified spherulite. Reproduced from reference 7.
It must be pointed out that our interpretation of an association between biomineralization and autophagy in A. willeyana is based on correlative associations rather than functional experiments. Knocking down Awi-GABARAPL2 protein production by delivery of double-stranded RNAs, morpholinos or a pharmaceutical inhibitor of autophagy would be predicted to inhibit biomineralization in A. willeyana, and would provide a functional link between the 2 processes. However the biology of this animal makes such functional assays extremely difficult; A. willeyana is exclusively found in Indo-Pacific marine caves and cannot be maintained under in vitro conditions, therefore there currently exists no system for gene expression manipulation in this animal. Furthermore the growth rate of this animal is extremely slow (230 μm per year),4 making any kind of manipulative experiment focused on rates of biomineralization in A. willeyana intractable. Nonetheless, our previous work provides support for the association between autophagy and biomineralization that we propose here. First, Wörheide’s detailed description of A. willeyana’s morphology using a variety of techniques (staining with fluorescent and nonfluorescent dyes, histological sections, electron microscopy, and X-ray) has identified the few cell types that exist in this animal.4 Spherulite-forming cells can be recognized in sections of GABARAPL2 in situ and LC3A immunohistochemistry material because the insoluble organic material of the spherulite remains visible (Fig. 2J–L; Fig. 3F). These cells are also located in the correct position within the sponge as would be expected for a role in biomineralization as described by Wörheide.4 Spherulite-forming Awi-GABARAPL2+ and LC3+ cells also appear to be the same as those that express the biomineralization genes Astrosclerin (a homolog of an α-carbonic anhydrase which is involved in spherulite formation),7 and Spherulin (a gene that was horizontally transferred from a proteobacterium into A. willeyana’s genome and is now involved in spherulite formation).6 Importantly, the protein products of these 2 previously reported biomineralization genes were isolated from purified calcified spherulites. Consistent with all of these observations is the additional fact that the organic material that can be isolated from within purified A. willeyana spherulites is primarily of prokaryotic (most likely bacterial) origin.5 Taking all of these observations together leads us to propose that Awi-GABARAPL2 and Awi-LC3A most likely have the same spatial expression profiles as Astrosclerin and Spherulin (Fig. 2), and that autophagy is linked with the process of biomineralization in A. willeyana. Further work to elucidate the role of autophagy in A. willeyana should include isolation of additional autophagy pathway components and their spatial localization, and inhibition or induction of autophagy using pharmaceutical reagents. However, this last exercise would require both the establishment of an in vitro culturing system for A. willeyana (an exercise that has many technical challenges) and confirmation that drugs developed for the modulation of the autophagy pathway in other organisms also function as expected in the deeply evolutionarily divergent A. willeyana.
We previously suggested that the use of degraded intracellular microbes as an organic matrix upon which to nucleate the growth of CaCO3 crystals might represent an “evolutionary fast track” to a mode of biomineralization.5 This is because the complex biomineralizing organic matrix synthesized de novo by “more complex” metazoans (such as molluscs, urchins, and vertebrates) did not need to be evolved by A. willeyana. Apparently A. willeyana has exapted an ancestral autophagy pathway in order to actively degrade its symbiotic bacterial community and to generate a biomineralizing organic matrix. This scenario serves to further highlight both the importance and deep history of the association between sponges and microbes, and also extends the list of physiological roles that autophagy is associated with. Whether this link between symbiophagy and biomineralization exists in extant eumetazoans, or once existed in their ancestors, remains an intriguing open question for evolutionary-biomineralization research.
Materials and Methods
RNA extraction, PCR, cloning, and probe generation
Total RNA extraction was performed using Trizol according to the manufacturer’s instructions. cDNA synthesis was performed as previously described in Jackson et al.6 Degenerate primers designed to bind conserved coding regions of ATG8 were used to amplify a 132 bp fragment of Awi-GABARAPL2. This fragment was cloned into the pGEM-T Easy TA vector (Promega, A1360) and sequenced using standard molecular methods and reagents. From this sequence forward and reverse gene specific primers were then designed for RACE-PCR in order to isolate the full-length Awi-GABARAPL2 transcript. All primer sequences are shown in Table 1. A 582 bp PCR product (generated with the specific primers) with flanking SP6 and T7 promoters was used as a template to generate a DIG labeled ribo-probe complimentary to Awi-GABARAPL2. An alignment of all 5 A. queenslandica ATG8 paralogs revealed an average percent identity of 49% at the nucleotide level. Given this low similarity and the in situ hybridization conditions we used (50% formamide, 825 mM NaCl and 50 °C) it is unlikely that the complementary Awi-GABARAPL2 probe we employed would be recognizing other A. willeyana ATG8 paralogs; however, we cannot rule out this possibility.
Table 1. Primers used to isolate a degenerate fragment of Awi-GABARAPL2, and to perform 5′ and 3′ RACE reactions.
| Primer name | Primer sequence (5′ to 3′) | Translation |
|---|---|---|
| ATG8 degenerate forward | GARAARGCMATGTTYYTGTTTGT | EKAMFLFV |
| ATG8 degenerate reverse | CCRAAWGTRTTYTCTCCRCTGTAAG | AYSGENTFG |
| Awi-GABARAPL2-specific forward | ACGTGATAATTGCCGAGCGG | located in 5′ UTR |
| Awi-GABARAPL2-specific reverse | GTTCTGGGGAAAAGGTCCAGTGGT | located in 3′ UTR |
Whole mount in situ hybridization
WMISH was performed as previously described.7 Briefly, adult A. willeyana specimens were first fixed for 30 min in a 4% paraformaldehyde-buffered solution (0.5 M MOPS, 10 mM MgSO4, 5 mM EDTA, 2.5 M NaCl). Samples were then dehydrated and stored at −20 °C. When ready for processing, samples were equilibrated to room temperature and rehydrated into 1× phosphate-buffered saline (PBS) and then decalcified in a solution of 350 mM EDTA, 4% paraformaldehyde and 1× PBS. Once thoroughly decalcified, the material was washed in 1× PBS-Tween and stepped into hybridization buffer (0.75 M sodium chloride, 75 mM sodium citrate, 5 mM EDTA, 50% formamide, 50 μg/ml heparin, 0.1% Tween-20, 0.1 mg/ml yeast total RNA). Samples were slowly brought to the hybridization temperature (50 °C) and prehybridized for 3 to 4 h. Sense and anti-sense probes were denatured at 75 °C for 10 min and added to the prepared tissue. Hybridization was performed at 50 °C for 12 to 16 h. Samples were washed at 50 °C with an increasingly stringent series of washes consisting of 3 washes with 4× wash (50% formamide, 4× SSC, 0.1% Tween-20), 3 washes with 2× wash (50% formamide, 2× SSC, 0.1% Tween-20) and 3 washes with 1× wash (50% formamide, 1× SSC, 0.1% Tween-20). All wash solutions were brought to 60 °C prior to use. Samples were then brought to room temperature and washed several times with 1× SSC, 0.1% Tween-20. To prevent nonspecific binding of the anti-DIG antibody, tissue was blocked for 3 to 4 h at room temperature with a solution of 1× Roche blocking reagent. The DIG hapten was detected by incubation in a solution of anti-DIG fab fragments (Roche, 11214667001) diluted 1:5000 in 1× Roche blocking reagent for 12 to 16 h. Unbound antibody was removed by extensive washing in PBS-Tween. Colorimetric detection of DIG bound alkaline phosphatase conjugated fragments was performed in a solution of alkaline phosphatase detection buffer and NBT/BCIP (Roth, 4421 and A155 respectively) as per manufacturer’s instructions. The alkaline phosphatase reaction was allowed to proceed until signal intensity reached an appropriate strength, and then stopped with several washes in stop buffer (0.1 M glycine pH 2). The tissue was then washed in PBTw, dehydrated through an ethanol series, and photographed whole mount, and then sectioned at 5 μm following embedding in paraffin. Control sections (hybridized with a sense probe) were processed as above and counter stained with Nuclear Fast red (Roth, N069).
Immunohistochemistry
A. willeyana material was fixed, stored, decalcified, dehydrated, embedded in paraffin, and sectioned as described above. Sections were deparaffinized in 3 washes of xylene and then rehydrated into water. Antigen unmasking was performed in TE (10 mM Tris pH 9, 1 mM EDTA) at 95 °C for 18 min. Sections were then allowed to cool to room temperature for 30 min and then incubated in 3% H2O2 for 10 min. Slides were then briefly washed in H2O and then once in wash buffer (TBST: 10 mM Tris pH 7.6, 15 mM NaCl, 0.1% Tween-20) for 5 min. The tissue was then blocked in 5% normal goat serum in TBST at room temperature for 1 h, and then an anti LC3A antibody (Cell Signaling Technology, 4599) at a range of dilutions (1:800 to 1:6,400) was applied and incubated at 4 °C overnight. Sections were then washed three times in TBST for 5 min each, and the signal developed using Cell Signaling Technology’s “Signal Boost Detection Reagent” (8114P) and their DAB chromogen reagent (11725P) as per the provided instructions. All dilutions of the primary antibody gave signals, but the 1:800 dilution yielded the highest signal to noise ratio. Sections were washed in water, and a coverslip mounted with Aquafix.
Phylogenetic analysis
Protein orthologs of ATG8 were isolated from the complete genome sequences of the following organisms: yeast (Saccharomyces cerevisiae and Schizosaccharomyces pombe), choanoflagellate (Monosiga brevicollis), human, mouse (Mus musculus), cow (Bos taurus), acorn worm (Saccoglossus kowalevskii), snail (Lottia gigantea), worm (Capitella teleta), fruit fly (Drosophila melanogaster), honey bee (Apis mellifera), sponge (Amphimedon queenslandica) and placozoan (Trichoplax adhaerens). Sequences were aligned using Muscle18 as implemented by Seaview.20 Conserved sites were selected using G-Blocks21 as implemented in Seaview. Full-length sequences, their accession numbers and the full-length and G-Blocked alignments are available in the supplementary material. A Bayesian analysis of the G-Blocked alignment was performed using a parallelized version of MrBayes (v 3.1.2) installed on a Linux cluster with the following parameters: lset rates = gamma; prset aamodelpr = mixed; mcmcp nruns = 2 ngen = 20000000 printfreq = 10000 samplefreq = 10000 nchains = 4 savebrlens = yes temp = 0.2 stoprule = yes. 1,038 trees were returned and the first 25% discarded as burnin. These were summarized to generate a 50% majority-rule consensus tree. Samples for TEM and SEM were fixed and processed as described in Wörheide.4
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are indebted to Valérie Pierrefite who shared unpublished data, and encouraged us to search for evidence of autophagy in A. willeyana. This work was supported by Deutsche Forschungsgemeinschaft (DFG) funding to DJJ through the CRC for Geobiology and the German Excellence Initiative, and to GW (Wo896/7 “Deep DownUnder”).
Glossary
Abbreviations:
- ATG
autophagy
- EDTA
ethylenediaminetetraacetic acid
- GABARAPL2
γ-aminobutyric acid receptor-associated protein-like 2
- GATE-16
Golgi-associated ATPase enhancer of 16 kDa
- PBS
phosphate-buffered saline
- WMISH
whole mount in situ hybridization
References
- 1.Millet J, Kiessling W. First record of coralline demosponges in the Pleistocene: implications for reef ecology. Coral Reefs. 2009;28:867–70. doi: 10.1007/s00338-009-0549-x. [DOI] [Google Scholar]
- 2.Reitner J, Worheide G. Non-lithistid fossil Demospongiae - origins of their palaeobiodiversity and highlights in history of preservation. In: Hooper, JNA, van Soest, RWM, (s). Systema Porifera: A guide to the classification of sponges. New York:Kluwer Academic/Plenum Publishers, 2002:52-68 [Google Scholar]
- 3.Uriz M-J. Mineral skeletogenesis in sponges. Can J Zool. 2006;84:322–56. doi: 10.1139/z06-032. [DOI] [Google Scholar]
- 4.Wörheide G. The reef cave dwelling ultraconservative coralline demosponge Astrosclera willeyana Lister 1900 from the Indo-Pacific - Micromorphology, Ultrastructure, Biocalcification, Isotope Record, Taxonomy, Biogeography, Phylogeny. Facies. 1998;38:1–88. doi: 10.1007/BF02537358. [DOI] [Google Scholar]
- 5.Jackson DJ, Thiel V, Wörheide G. An evolutionary fast-track to biocalcification. Geobiology. 2010;8:191–6. doi: 10.1111/j.1472-4669.2010.00236.x. [DOI] [PubMed] [Google Scholar]
- 6.Jackson DJ, Macis L, Reitner J, Wörheide G. A horizontal gene transfer supported the evolution of an early metazoan biomineralization strategy. BMC Evol Biol. 2011;11:238. doi: 10.1186/1471-2148-11-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson DJ, Macis L, Reitner J, Degnan BM, Wörheide G. Sponge paleogenomics reveals an ancient role for carbonic anhydrase in skeletogenesis. Science. 2007;316:1893–5. doi: 10.1126/science.1141560. [DOI] [PubMed] [Google Scholar]
- 8.Hughes T, Rusten TE. Origin and evolution of self-consumption: autophagy. Adv Exp Med Biol. 2007;607:111–8. doi: 10.1007/978-0-387-74021-8_9. [DOI] [PubMed] [Google Scholar]
- 9.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 10.Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 2005;12(Suppl 2):1535–41. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- 11.Dorn BR, Dunn WA, Jr., Progulske-Fox A. Bacterial interactions with the autophagic pathway. Cell Microbiol. 2002;4:1–10. doi: 10.1046/j.1462-5822.2002.00164.x. [DOI] [PubMed] [Google Scholar]
- 12.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–62. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 14.Dunn SR, Schnitzler CE, Weis VM. Apoptosis and autophagy as mechanisms of dinoflagellate symbiont release during cnidarian bleaching: every which way you lose. Proc Biol Sci. 2007;274:3079–85. doi: 10.1098/rspb.2007.0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Downs CA, Kramarsky-Winter E, Martinez J, Kushmaro A, Woodley CM, Loya Y, Ostrander GK. Symbiophagy as a cellular mechanism for coral bleaching. Autophagy. 2009;5:211–6. doi: 10.4161/auto.5.2.7405. [DOI] [PubMed] [Google Scholar]
- 16.Klionsky DJ, Cregg JM, Dunn WA, Jr., Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–45. doi: 10.1016/S1534-5807(03)00296-X. [DOI] [PubMed] [Google Scholar]
- 17.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–78. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 18.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 2011;12:226. doi: 10.1186/gb-2011-12-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–4. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 21.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–52. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.