Abstract
The proteins that comprise the Atg1 kinase complex constitute a key set of components that participate in macroautophagy (hereafter autophagy). Among these proteins, Atg13 plays a particularly important, although as yet undefined role, in that it is critical for the proper localization of Atg1 to the phagophore assembly site (PAS) and its efficient kinase activity. Atg13 is hyperphosphorylated in vegetative conditions when autophagy occurs at a basal level, and is largely dephosphorylated upon the induction of autophagy. Inhibitory phosphorylation of Atg13 reflects the activity of TOR complex 1 (TORC1) and protein kinase A. Accordingly, monitoring the phosphorylation state of Atg13 provides a convenient way to follow early steps of autophagy induction as well as the activity of some of the upstream nutrient-sensing kinases. However, the detection of Atg13 by western blot can be problematic. Here, we present a detailed protocol for sample preparation and detection of the Atg13 protein from yeast.
Keywords: Atg13, autophagy, TORC1, vacuole, yeast
1. Introduction
Autophagy is a catabolic process essential to recycle cellular components that allows survival under nutrient-limited conditions. It is also required to eliminate damaged organelles or other materials during homeostasis. During autophagy, the cargo to be eliminated is engulfed by a double-membrane compartment, the phagophore; upon completion the phagophore matures into an autophagosome, which transports the cargo to the vacuole, where it is degraded and recycled.
The formation of the autophagosome begins at the PAS, with the assembly of several autophagy-related (Atg) proteins and subsequent nucleation and expansion of the double membrane.1 The yeast Saccharomyces cerevisiae Atg1 kinase forms a complex with Atg13, Atg17, Atg29, and Atg31, which is critical for the induction of autophagy and PAS organization.2,3 Among these proteins, Atg13 plays an important role in the regulation of Atg1 catalytic activity and consequently in autophagy activation.4 In a nutrient-rich environment, Atg13 is directly phosphorylated on different residues by at least 2 nutrient-sensing kinases, TORC1 and protein kinase A (PKA).4-6 TORC1-mediated Atg13 hyperphosphorylation may inhibit the association between Atg13 and Atg1, which then leads to decreased Atg1 kinase activity and ultimately results in autophagy repression. According to this model, once Atg13 gets dephosphorylated, upon TORC1 inhibition, it can associate with Atg1, which in turn becomes fully active and triggers autophagy.4 The association between Atg1 and Atg13 has been considered important for autophagy induction; however, another study found that these proteins are constitutively associated, even in the presence of nutrients.7 This result suggests that the critical step for Atg1 kinase activation is the dephosphorylation of Atg13 instead of its association with Atg1.
Atg13 phosphorylation/dephosphorylation can therefore be used to follow early events in autophagy and also to monitor TORC1 and PKA signaling activities. Atg13 hyperphosphorylation mediated by TORC1 is easily detected on SDS-polyacrylamide gels, since it displays a slower migration when compared with the partially unphosphorylated form. Phosphorylation mediated by PKA can be detected by Atg13 immunoprecipitation followed by incubation with a specific antibody for PKA residues.6,8 However, the detection of Atg13 by western blot can be difficult if cells are lysed using conventional extraction protocols. Here, we describe a simple protocol for protein extraction and detection of Atg13 by western blot, which does not affect the phosphorylation status of Atg13. Therefore, this method can be used to easily monitor TORC1 activity.
2. Materials
2.1 Cells and culture
1. The yeast strain background depends on the experimental parameters; no particular strain is required for detecting the phosphorylated or dephosphorylated forms of Atg13. It is helpful to have an isogenic atg13∆ strain as a negative control.
2. Synthetic minimal medium (SD): 0.67% yeast nitrogen base without amino acids, 2% glucose and auxotrophic amino acids and vitamins as needed. This medium is stable for months at room temperature.
3. Starvation medium (SD-N): 0.17% yeast nitrogen base without ammonium sulfate or amino acids, containing 2% glucose. This medium is stable for months at room temperature.
4. YNB without amino acids (ForMedium, CYN0410).
5. YNB without ammonium sulfate or amino acids (ForMedium, CYN0501).
2.2 Plasmids
1. It is possible, but difficult to detect endogenous Atg13. As alternatives, we describe plasmids that can be used to simplify the detection of this protein. The plasmid pHC0783 encodes ATG13 with an N-terminal 3HA tag under the control of the endogenous ATG13 promoter in the pRS315 vector. YEp351[APG13] expresses ATG13 on a multicopy plasmid under the control of the ATG13 promoter.4 Plasmid pPHY2427 expresses a similar N-terminal 3HA tag under the control of the CUP1 promoter in the pRS426 vector (see Note 1).9
2.3 Buffers and solutions
1. MURB: 50 mM sodium phosphate, 25 mM MES (Research Organics, 0113M), pH 7.0, 1% SDS, 3 M urea, 0.5% 2-mercaptoethanol, 1 mM sodium azide.
2. NP-40 lysis buffer: 150 mM sodium chloride, 1.0% Tergitol solution Type NP-40 (nonyl phenoxypolyethoxylethanol/nonylphenol ethoxylate; Sigma, NP40S), 50 mM Tris, pH 8.0.
3. Protein sample buffer (PSB): 200 mM Tris-HCl, 8% SDS, 40% glycerol, 6% 2-mercaptoethanol, 0.4% bromophenol blue.
4. TTBS: 1% Tween 20 (Sigma, P1379), 50 mM Tris-buffered saline, pH 7.6.
2.4 Equipment and reagents
1. SDS-PAGE apparatus.
2. Western blot transfer apparatus (Bio-Rad Trans-Blot Semi-Dry Electrophoretic Transfer Cell, 170-3940 or the equivalent).
3. PVDF immobilon-P (Thermo-Fisher Scientific, IPVH00010).
4. Protease inhibitor: Complete protease inhibitor tablets, EDTA free (Roche Applied Sciences, 05056489001).
5. Phosphatase inhibitor: PhosSTOP (Roche Applied Sciences, 04906837001).
6. BCA protein assay kit (Thermo-Fisher Scientific, PI-23225).
7. Chemiluminescent detection kit (Fisher, Thermo Scientific SuperSignal West Pico Chemiluminescent Substrate, PI-34080; or EMD Millipore, Immobilon Western Chemiluminescent HRP Substrate, WBKLS0100).
2.5 Antibody
1. The generation of antiserum to Atg13 was described previously.10
2. Monoclonal anti-HA antibody (Sigma, H3663).
3. HRP-conjugated goat anti-rabbit IgG (Thermo-Fisher Scientific, ICN55676).
4. HRP-conjugated rabbit anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc., 315-035-003).
3. Methods
3.1 Cell growth
1. S. cerevisiae strains are grown at 30 °C to early mid-log phase at O.D600 = 0.7~1.0 in YPD or synthetic minimal medium containing the appropriate auxotrophic nucleosides and amino acids.
2. For starvation, cultured cells from step 1 are collected by centrifugation at 5,000 × g for 3 min and are washed once with water. Cells are typically cultured in SD-N for 1 h. The starvation time depends on the experiment (see Note 2).
3.2 Preparation of protein extract
1. For protein extraction, aliquots corresponding to 1.0 to 2.0 O.D.600 units (1 ml of cells at an O.D.600 = 1.0 is equivalent to 1 unit of cells) of the cultured cells are harvested by centrifugation at 5000 × g for 3 min in plastic 1.7 ml microcentrifuge tubes (see Note 3).
2. Trichloroacetic acid (TCA) is added to 10% final concentration, and the samples are incubated for 20 min (see Note 4) on ice or at −20 °C.
3. The precipitated proteins are pelleted by centrifugation at 15,000 × g for 3 min.
4. After washing the pellet twice with acetone, the pellet is air-dried and the dry cell pellet is then resuspended in MURB (see Note 5) and disrupted by vortex with approximately one-half volume of acid-washed glass beads (see Note 6),11 for 5 min. The samples are incubated at 70 °C for 10 min, and unlysed cells are removed by centrifugation at 10,000 × g for 1 min.
3.3 Western blot
1. Protein extracts equivalent to OD600 = 0.2 units of yeast cells are loaded onto gels and resolved by SDS-PAGE. For detecting phosphorylated Atg13, the protein extracts are run on 6~8% SDS-PAGE gels (see Note 7), approximately until the 50-kDa protein marker reaches the bottom of the gel. Usually the protein extract from an atg13∆ strain is loaded as a negative control for recognizing the specific Atg13 bands.
2. A standard western blotting procedure is performed using PVDF membrane.
3. Protein detection: After blotting, PVDF membranes are probed with antiserum against Atg13 (1:4,000 dilution) or with antibody against HA (1:5,000), by incubation at 4 °C overnight.
4. Wash the membranes 3 times with TTBS, then incubate for 1 h with HRP-conjugated goat anti-rabbit IgG (1:10,000 dilution) or with HRP-conjugated rabbit anti-mouse IgG (1:10,000 dilution), depending on the primary antibody used.
5. The signal for Atg13 is detected using a chemiluminescent kit.
Results and Discussion
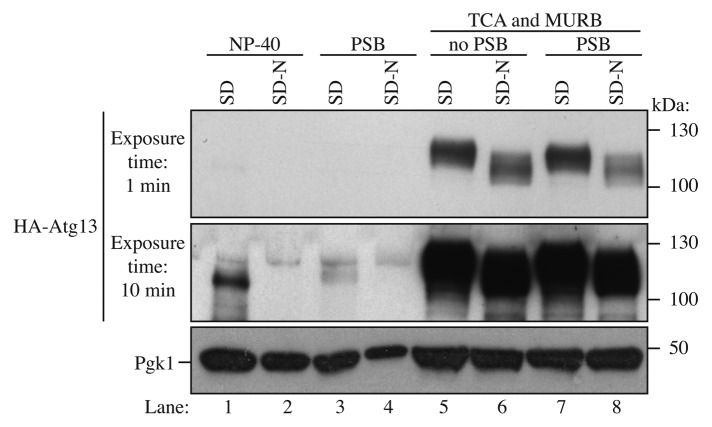
Atg13 can be detected as hyperphosphorylated and dephosphorylated species, and the dephosphorylation occurs within 1 min of shifting cells from vegetative to starvation conditions (Fig. 1). The specific migration pattern of Atg13 depends on many factors including the method of sample preparation, the acrylamide percentage of the resolving gel, the expression level of the protein, and the affinity of the antibody/antiserum. Overexpressed Atg13 can typically be detected as multiple bands (Fig. 2). HA-Atg13 can also be detected when expressed from the endogenous ATG13 promoter (Fig. 3). We found that endogenous or overexpressed Atg13 could be readily detected when protein extracts were prepared using TCA precipitation followed by resuspension in MURB as described in this paper (Figs. 1 and 3). In contrast, when examining HA-Atg13 expressed from the ATG13 promoter, extraction using NP-40 lysis buffer (see Note 8) or PSB was much less efficient (Fig. 3, lane 1–4; see Note 9). The addition of PSB was not necessary to load the samples or to improve detection of Atg13 when using the TCA-MURB method (Fig. 3, lane 5–8); however, the addition of bromophenol blue to MURB can be helpful for loading the samples.
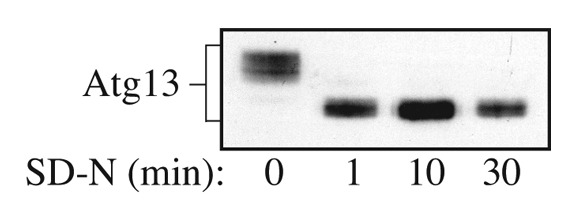
Figure 1. Detection of overexpressed Atg13 using rabbit polyclonal antiserum. Yeast cells overexpressing Atg13 were grown in SD and then shifted to SD-N at time 0. Samples were collected at the indicated time points and analyzed by SDS-PAGE and western blot using antiserum to Atg13. This figure is a modification of data previously published in reference 8, and is reproduced by permission of the American Society for Biochemistry and Molecular Biology and Elsevier, copyright 2000.
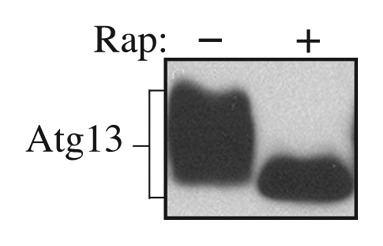
Figure 2. Wild-type yeast cells expressing Atg13 under the endogenous promoter from a multicopy plasmid (YEp351[APG13]) were grown in SD for 8 h and treated with rapamycin for 15 min as indicated. Protein extracts were analyzed by western blot using anti-Atg13 serum.
Figure 3. Comparison between different methods to detect Atg13. Wild-type cells (BY4741) were transformed with a vector expressing HA-Atg13 under its own promoter (pHC078). Cultures were grown until mid-log phase in SD and starved for nitrogen 2 h (SD-N). Lanes 1 and 2 correspond to cells that were lysed with NP-40 buffer and disrupted with glass beads; cells were harvested by centrifugation, resuspended in NP-40 lysis buffer (200 µl) containing protease and phosphatase inhibitors, followed by glass bead lysis and centrifugation at 700 × g for 3 min. Proteins were quantified by the BCA assay. The same amount of protein was loaded onto the gel. Lanes 3 and 4 correspond to cells that were disrupted by vortex with glass beads and protein sample buffer (PSB). Lanes 5 to 8 samples were prepared using the TCA-MURB method described in this paper. The protein extracts of lane 7 and 8 were mixed with PSB.
We conclude that this is a very simple and fast protocol to detect Atg13 phosphorylation, providing information about autophagy and nutrient-sensing pathways.
Notes
1. In our experience, efficient expression from the CUP1 promoter will occur in most standard media without the addition of extra copper, which can be toxic to cells at high concentrations. The addition of 10–100 µM copper will further derepress the promoter.
2. Dephosphorylation of Atg13 upon autophagy induction, and rephosphorylation following a return to vegetative conditions is extremely rapid, occurring within 1 min.
3. When the volume of the culture corresponding to 1 or 2 ODs is greater than 1 ml, cells are pelleted and resuspended in 1 ml of rich medium or distilled water.
4. We recommend a minimum of 20 min incubation on ice; longer times may improve the efficiency of precipitation.
5. To help visualize the loading of samples, 0.4% bromophenol blue can be added to MURB.
6. The optimal volume of glass beads may need to be determined empirically. Approximately one-half to one volume of glass beads usually works well, but it is critical that the volume not exceed the liquid volume (i.e., the beads should not rise above the meniscus of the sample). An excess of beads can reduce lysis efficiency and result in sample loss.
7. 10% SDS-PAGE gels can also be used depending on the difference in phosphorylation among the samples.
8. Tergitol solution Type NP-40 is not the same as Nonidet P-40 (octyl phenoxypolyethoxylethanol), although both are often written as NP-40. We have not evaluated the efficacy of Nonidet P-40 for the detection of Atg13 by western blot.
9. Lysis buffer containing Tween 20 has been used to immunoprecipitate Atg13;12 however, we have not made a direct comparison to determine the efficiency of solubilization and recovery of Atg13 when lysing in the presence of only Tween 20 vs. MURB. For immunoprecipitation, we use MURB to lyse the cells (~7 OD600 units) and extract proteins as described in this protocol. The resuspension is then diluted in ~5 volumes of IP buffer (50 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 150 mM NaCl, 0.05% Tergitol solution Type NP-40 or 50 mM Tris-HCl, pH 7.5, 0.1 mM EDTA, 150 mM NaCl, 0.5% Tween 20), followed by a standard immunoprecipitation protocol. It should be noted that although both Tween 20 and Tergitol are nonionic detergents, they do not have identical properties.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Drs Jiefei Geng, Kai Mao, and Zhifen Yang for helpful comments. This work was supported by NIH grant GM053396 to DJK. We thank Fundação para a Ciência e Tecnologia and Tiago Outeiro for supporting PA (SFRH/BI/5177/2011) and LMF (SFRH/BD/36065/2007).
Glossary
Abbreviations:
- Atg
autophagy-related
- PAS
phagophore assembly site
- PKA
protein kinase A
- PSB
protein sample buffer
- TCA
trichloroacetic acid
- TORC1
TOR complex 1
References
- 1.Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194:341–61. doi: 10.1534/genetics.112.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161–70. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:668–81. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–13. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–58. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci U S A. 2009;106:17049–54. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kraft C, Kijanska M, Kalie E, Siergiejuk E, Lee SS, Semplicio G, Stoffel I, Brezovich A, Verma M, Hansmann I, et al. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 2012;31:3691–703. doi: 10.1038/emboj.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deminoff SJ, Howard SC, Hester A, Warner S, Herman PK. Using substrate-binding variants of the cAMP-dependent protein kinase to identify novel targets and a kinase domain important for substrate interactions in Saccharomyces cerevisiae. Genetics. 2006;173:1909–17. doi: 10.1534/genetics.106.059238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeh YY, Wrasman K, Herman PK. Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics. 2010;185:871–82. doi: 10.1534/genetics.110.116566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott SV, Nice DC, III, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275:25840–9. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 11.Cheong H, Klionsky DJ. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 2008;451:1–26. doi: 10.1016/S0076-6879(08)03201-1. [DOI] [PubMed] [Google Scholar]
- 12.Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16:2544–53. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]