Abstract
Core Binding Factor β (CBFβ) is complexed with the RUNX family of transcription factors in the nucleus to support activation or repression of genes related to bone (RUNX2), hematopoiesis (RUNX1) and gastrointestinal (RUNX3) development. Furthermore, RUNX proteins contribute to the onset and progression of different types of cancer. Although CBFβ localizes to cytoskeletal architecture, its biological role in the cytoplasmic compartment remains to be established. Additionally, the function and localization of CBFβ during the cell cycle are important questions relevant to its biological role. Here we show that CBFβ dynamically distributes in different stages of cell division and importantly is present during telophase at the midbody, a temporal structure important for successful cytokinesis. A functional role for CBFβ localization at the midbody is supported by striking defects in cytokinesis that include polyploidy and abscission failure following siRNA-mediated downregulation of endogenous CBFβ or overexpression of the inv(16) fusion protein CBFβ-SMMHC. Our results suggest that CBFβ retention in the midbody during cytokinesis reflects a novel function that contributes to epigenetic control.
Keywords: Midbody, Core Binding Factor β, Cytokines
Introduction
CBFβ binding to the Runt related (RUNX) family of transcription factors is required for fidelity of bone (RUNX2) (Komori et al., 1997), hematopoietic (RUNX1) (Wang et al., 1996) and gastrointestinal (RUNX3) (Li et al., 2002) development, proliferation and differentiation of respective lineages. The activity of RUNX transcription factors is primarily confined to the nucleus and bound to the nuclear scaffold (Stein et al., 2000; Zaidi et al., 2010). CBFβ is imported to the nucleus by associating with RUNX factors, as CBFβ lacks a nuclear localization signal (Adya et al., 1998). CBFβ dimerizes with RUNX proteins as a co-factor to enhance DNA binding affinity by stabilizing the interaction of the RUNX runt domain with DNA (Bravo et al., 2001; Nagata et al., 1999). Association of CBFβ with RUNX factors also confers protection against RUNX protein degradation (Huang et al., 2001). In humans, CBFβ resides in chromosome 16 q22 and produces two isoforms that differ in several amino acids at the carboxy terminus (Ogawa et al., 1993). The biological relevance of Cbfβ has been demonstrated in a knock-out mouse model that exhibits embryonic lethality due to defective fetal liver hematopoiesis and central nervous system bleeding recapitulating the Runx1 null phenotype (Sasaki et al., 1996; Wang et al., 1996). Conversely, heterozygous Cbfβ+/− knock-in mice survive gestation but die soon after birth with bone developmental defects that are comparable to but less severe than Runx2−/− mice (Kundu et al., 2002).
From a disease perspective, CBFβ is required to support transformation and tumor progression that includes tumorigenesis and invasion of breast, prostate and ovarian cancer (Davis et al., 2010; Mendoza-Villanueva et al., 2010). Acute Myeloid Leukemia of the M4E0 type is caused by a chromosomal translocation designated inversion (16) (Inv(16)) (p13q22), which gives rise to a fusion between CBFβ and the MYH11 gene encoding the smooth muscle myosin heavy chain (SMMHC) (Liu et al., 1993). The CBFβ-SMMHC chimeric protein interferes with the RUNX1 transcriptional program by sequestering RUNX1 from the nucleus (Adya et al., 1998; Kanno et al., 1998). The heavy chain tail of CBFβ-SMMHC also has transcriptional repressor activities and acts as a dominant inhibitor of the RUNX1/CBFβ complex (Lutterbach et al., 1999).
Interestingly, CBFβ is located primarily in the cytoplasmic compartment (Tanaka et al., 1997). Cytoplasmic CBFβ interacts with Filamin A, a cytoskeletal actin-binding protein (Yoshida et al., 2005). In mesenchymal stem cells, the small molecule Kartogenin has been shown to block sequestration of CBFβ by Filamin A and to direct chondrocyte differentiation by promoting the CBFβ/RUNX1 transcriptional program (Johnson et al., 2012).
The dynamics and biological as well as clinical relevance of CBFβ in the cytoplasm remain elusive. Here, we have investigated the subcellular localization of CBFβ during cell division in an effort to gain insight into regulatory functions for this molecule. Cell division is a highly regulated process that architecturally and dynamically includes DNA replication, packing DNA as chromatin, condensation of mitotic chromosomes, symmetric segregation of chromosomes to progeny cells and cell separation through cytokinesis. Cytokinesis involves the interplay of a spectrum of proteins and molecular pathways to ensure that dividing cells are separated into single and independent biological entities (reviewed in Lacroix and Maddox, 2012). Failure of cytokinesis elicits polyploidy that can lead to tumorigenesis (Fujiwara et al., 2005; Vinciguerra et al., 2010). The midbody, a dynamic structure formed within the cleavage furrow during cytokinesis is essential to promote the successful separation of daughter cells (Chen et al., 2013). In this study, we directly demonstrate that CBFβ associates with the midbody and has a role in cytokinesis. Our results suggest a novel function for CBFβ during cell division beyond transcriptional control.
Material and Methods
Cell culture and synchronization
SaOS-2 cells and HeLa cells were grown following recommended cell culture conditions from ATCC. Cells were blocked in mitosis by nocodazole treatment for 16 h followed by shake-off to detach mitotic cells. Cells were washed and replated in growth medium for mitotic release.
Primary and secondary antibodies
The following antibodies were used in this study: CBFβ rabbit polyclonal (ab33516, Abcam, Cambridge, MA, USA) and A303-547A, A303-548A, A303-549A, Bethyl Labs, Montgomery, TX, USA). Dilutions for ab33516 were IF (1:500) and WB (1:500). Dilutions and concentration used for CBFβ Bethyl antibodies were IF (1:1000), IP (5 µg) and WB (1:1,000). GFP rabbit polyclonal (ab290, Abcam, Cambridge, MA, USA), was used at dilutions for WB (1:1000) and IP (5 µl). RUNX2 (8G5) mouse monoclonal (MBL International, Woburn, MA, USA), dilution used for IF (1:600). Filamin A (EP2405Y) rabbit polyclonal (ab76289, Abcam, Cambridge, MA, USA) dilution was used for IF (1:1000). Beta-tubulin mouse monoclonal (T-4026, Sigma Aldrich, St. Louis, MO, USA), dilution used for IF (1:1000) and WB (1:1000). PRC1 goat polyclonal (K-18) (sc-9342, Santa Cruz Biotechnology, Santa Cruz, CA, USA), dilution used for IF (1:300). KIF4A goat polyclonal (ab3815, Abcam, Cambridge, MA, USA), dilution used for IF (1:300). MRLC3 goat polyclonal (sc9449, Santa Cruz Biotechnology, Santa Cruz, CA, USA), dilution used for WB (1:200). Secondary antibodies conjugated with HRP (Santa Cruz Biotechnology, Santa Cruz, CA, USA), dilution used for WB (1:5000). Secondary antibodies conjugated with Alexa fluor 488 or 594 (Life Technologies-Invitrogen, Carlsbad, CA, USA), were used at dilution IF (1:500).
Immunofluorescence microscopy
Cells grown on coverslips were fixed in PBS-PFA, 4% (15 min, RT), permeabilized in PBS-Triton X-100, 0.25% (15 min, RT) and blocked in PBS-BSA, 1% (1 h, RT). Primary antibodies were diluted in PBS-BSA, 1% and incubated for 1 h RT or overnight at 4°C. After successive washes in PBS, 1X, secondary antibodies coupled to Alexa Fluor 488 and/or 594 were added (1 h, RT in the dark). Samples were washed in PBS with a last wash in distilled water and mounted in Prolong-DAPI mounting reagent (Life Technologies-Invitrogen, Carlsbad, CA, USA). Cells were analyzed using an epifluorescence Zeiss microscope coupled with a Hammamatsu CCD camera. Images were acquired using the Zen 2011 imaging software (Carl Zeiss Microscopy, LLC, Thornwood, NY, USA) or ImageJ software (Macphotonics).
DNA plasmids, siRNA and transfection methods
CBFβ-GFP and CBFβ-MYH11-GFP were kindly supplied by Dr. Paul Liu (Adya et al., 1998). DNA Plasmids, human CBFβ siRNA and control siRNA (sc-37681 and c-37007, Santa Cruz Biotechnology, Santa Cruz, CA, USA) were using Lipofectamine 2000 or Oligofectamine, respectively, according to manufacturer’s conditions.
Co-immunoprecipitation and Western blot analysis
Cells were washed twice with ice-cold PBS and harvested in cold sonication buffer {150 mM NaCl, 50 mM Tris (pH 8), 1% NP-40, 0.5% Deoxycholate, 25 mM MG132, and 2× protease inhibitor mixture (Roche, COMPLETE-EDTA free)}. Cells were sonicated (QSonica Sonicator system fitted with a 1.6-mm tip) and centrifuged (9391 RCF for 5 min at 4°C). Lysates were incubated overnight at 4°C with specific antibodies or normal IgG (Millipore, Billerica, MA, USA). Lysates were then incubated with protein A/G beads (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 3 h, followed by 4 washes with ice cold wash buffer {50 mM NaCl, 20 mM Tris (pH 8.3), 0.5% Sodium deoxycholate, 0.5% Nonidet P-40, 2 mM EDTA, 25 µM MG132, and 2× protease inhibitor mixture}. Immunoprecipitated protein complexes were resolved by 12% SDS/PAGE and transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore, Billerica, MA, USA). Blots were incubated with different primary antibodies followed by incubation with HRP conjugated secondary antibodies. Chemiluminescence (Perkin-Elmer Life Sciences, Boston, MA, USA) was visualized by using the BioRad ChemiDoc SRS device.
Mass spectrometry and proteomic analysis
Silver-stained gel bands were excised and destained with a 50 µL 1:1 mixture of 30 mM potassium ferricyanide (Sigma, St. Louis, MO, USA) and 100 mM sodium thiosulfate (Sigma) in siliconized tubes until brownish stain disappeared (~5 min). The gel bands were then washed with water, followed by reduction and alkylation of disulfides with 10 mM dithiothreitol and 50 mM iodoacetamide, respectively. After washing with 40 mM NH4HCO3, the gel pieces were minced to 1 mm3, dehydrated with acetonitrile (CH3CN), dried in a Speedvac, and then digested with trypsin (Promega V511A, 5–20 µg/ml; to achieve trypsin:protein ratio of 1:20 to 1:100 w/w) for 16 h at 37°C. Peptides were extracted successively with 1% FA/50% CH3CN, 80%CH3CN/1% FA, and 100%CH3CN, and dried.
The dried peptides were reconstituted with 2.5% acetonitrile (CH3CN)/2.5% formic acid (FA) and analyzed by capillary LC-MS/MS on a linear ion trap (LTQ XL) mass spectrometer coupled to a Surveyor MS Pump Plus (Thermo Fisher Scientific, Waltham, MA, USA). Half of the digest was loaded directly onto a 100 µm × 120 mm capillary column packed with MAGIC C18 (5 µm particle size, 20 nm pore size, Michrom Bioresources, Auburn, CA, USA) at a flow rate of 500 nL/min, and peptides were separated by a gradient of 10–35% CH3CN /0.1% FA over 33 min, 35–100% CH3CN/0.1% FA in 0.1 min, and 100% CH3CN/0.1% FA in 7 min. Peptides were introduced into the linear ion trap via a nanospray ionization source. Mass spectrometry data were acquired in a data-dependent acquisition mode in which a survey scan from m/z 365–1600 was followed by 10 MS/MS scans of the most abundant ions. Dynamic exclusion was enabled (repeat count: 2; repeat duration: 30 sec; exclusion list size: 180; exclusion duration: 60 sec). The minimum signal threshold was 500. Production spectra were searched against the human subset of the International Protein Index (IPI) database (ver.3.87) containing sequences in forward and reverse orientations using the SEQUEST and MASCOT search engines embedded in the Proteome Discoverer 1.4 (Thermo Fisher Scientific, Waltham, MA, USA). The database was indexed with the following: full enzymatic activity and two missed cleavage sites allowed for trypsin; peptide MW of 350–5000 Da. Search parameters were as follows: mass tolerance of 2 Da and 0.8 Da for precursor and fragment ions, respectively; four differential PTMs allowed per peptide; dynamic modification on methionines (+15.9949 Da for oxidized methionine) and static modification on cysteines (+57.0215 Da for carbamidomethylated cysteine). Cross-correlation (XCorr) filters were applied to limit the false positive (FP) rates to less than 1% in the data sets.
The classification system called PANTHER (protein annotation through evolutionary relationship) (http://www.pantherdb.org/) was used to cluster the retrieved list of proteins from mass spectrometry analysis according to functionality (Mi et al., 2013).
Results
CBFβ is dynamically localized during cell division
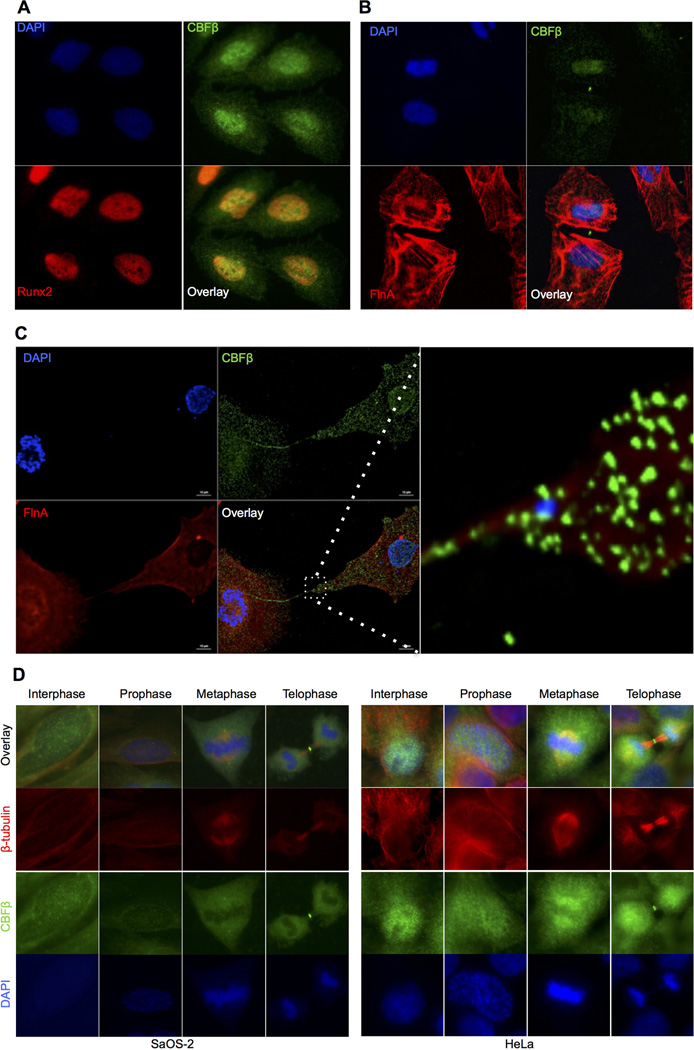
We have previously demonstrated that localization of RUNX2 to discrete subnuclear foci is required for bone-specific transcriptional control (Choi et al., 2001). Because CBFβ is an important heterodimeric partner of RUNX factors, we first determined its subcellular localization during interphase. Fig. 1A shows the characteristic subnuclear localization of RUNX2 in the osteosarcoma cell line SaOS-2. In agreement with previous reports, CBFβ is localized in both the cytoplasm and the nucleus of interphase cells (Tanaka et al., 1997; Yoshida et al., 2005) (Fig. 1A). The specificity of the antibody was confirmed by peptide competition (Supplementary Material Fig. S1A). We verified endogenous CBFβ localization with two different antibodies that recognize unique CBFβ epitopes (Fig. 1A and Supplementary Material Fig. S1B). Interestingly, CBFβ localizes to the midbody during telophase (Fig. 1B). Occasionally this structure appeared to be released to the extracellular space (less than 5%, n=100). We also infrequently observed a DNA-containing structure located in the abscission bridge during cytokinesis (less than 5%, n=100) where CBFβ is abundantly localized (Fig. 1C). In mitosis, DNA is normally condensed and equally segregated to progeny cells. In some events, DNA segregation fails and micronuclei are sometimes formed (Crasta et al., 2012). The formation of this DNA containing structure is often linked to defects in mitotic events and alterations in cytokinesis, which is associated with genomic instability and aneuploidy (Decordier et al., 2008).
Fig. 1.
Subcellular localization of CBFβ during the cell cycle. A: Immunofluorescence microscopy images of SaOS-2 cells in interphase for RUNX2 (red), CBFβ (green) and DAPI (blue). RUNX2 is mainly located in the nucleus whereas CBFβ shows both nuclear and cytoplasmic localization. B: Recently divided cells show localization of CBFβ in the nucleus and cytoplasm, but also unexpectedly concentrated in the midbody. Immunofluorescence staining with Filamin A to delimitate membrane boundaries shows that the midbody is released to the extracellular space. C: A small percentage of dividing cells exhibit a DNA-containing structure (stained with DAPI) located in the midbody together with CBFβ (dotted square in overlay, enlarged on the right). D: Subcellular localization of CBFβ in SaOS-2 (left panels) and HeLa cells (right panels), during the cell cycle. β-tubulin and DAPI immunofluorescence was used to determine cell cycle stages. CBFβ is localized in the midbody during telophase.
We further explored the endogenous localization of CBFβ by immune-fluorescence microscopy during cell division in SaOS-2 cells that express RUNX2 and in HeLa cells that do not express RUNX activity (Fig. 1D). Interphase cells exhibit both nuclear and cytoplasmic CBFβ staining. In prophase, CBFβ is uniformly diffuse throughout the cell. In metaphase, CBFβ primarily co-localizes with beta-tubulin in the cytoplasmic compartment. A small fraction of CBFβ consistently remains associated with mitotic chromosomes as well as with the centrosomes during spindle formation. During telophase, CBFβ localizes throughout the cell. Importantly, a strong CBFβ signal is evident at the midbody. The localization of CBFβ in the midbody was verified in somatic and cancer cell lines (Supplementary Material Fig. S2).
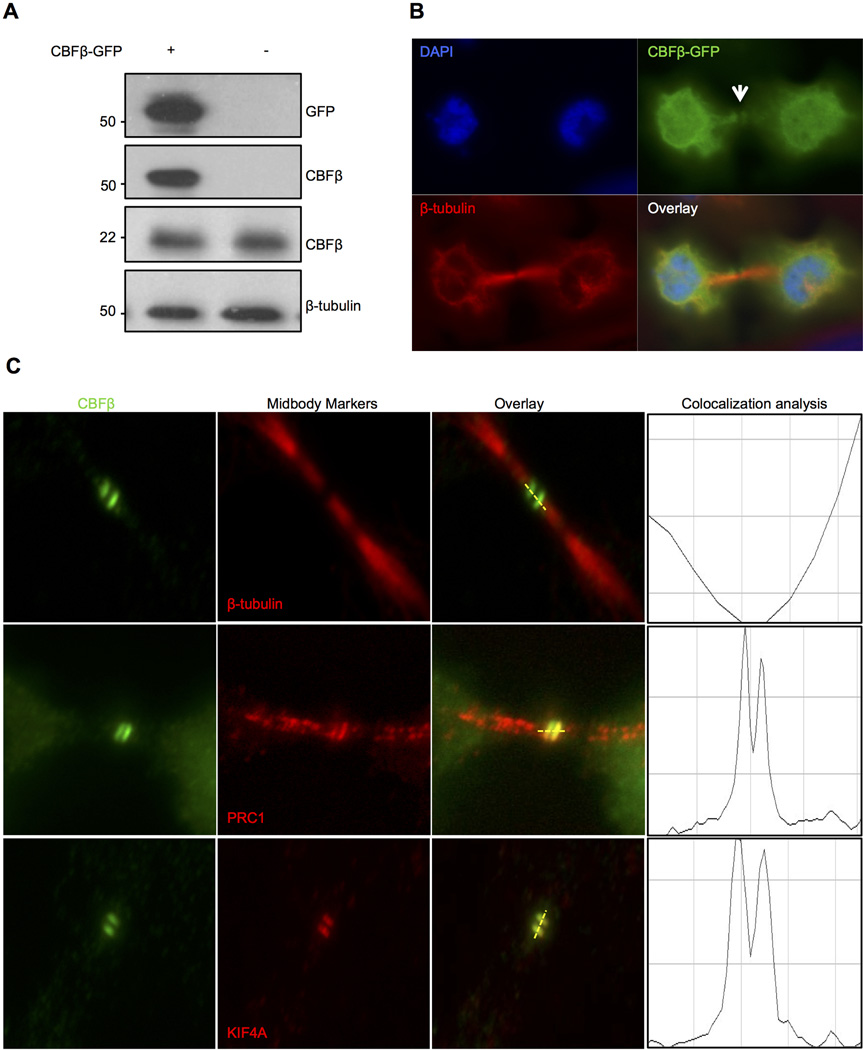
CBFβ is associated in the central region of the midbody
To verify the association of CBFβ with the midbody, we expressed CBFβ fused with GFP (CBFβ-GFP). Antibodies against GFP and CBFβ were used to verify that the CBFβ-GFP protein was of the expected size (Fig. 2A). Cells transfected with CBFβ-GFP exhibited the expected subcellular distribution of CBFβ, and in dividing cells GFP fluorescence was detected in the midbody (Fig. 2B). The midbody is a transient, complex and well-organized structure formed immediately after midzone formation and furrow ingression and is important to localize abscission (Hu et al., 2012). Because the midbody is divided into the bulge, dark zone and flanking zone (see diagram in Supplementary Material Fig. S3A), we evaluated the compartmentalization of CBFβ within this structure relative to the localization of midbody-specific markers. Cells were stained for Protein Regulator of Cytokinesis 1 (PRC1) and the chromosome-associated kinesin KIF4A, which mark the dark zone and delimitate the flanking zone, and β-tubulin that identifies the boundaries of the flanking zone but is excluded from the dark zone and bulge (Fig. 2C; entire images shown in Supplementary Material Fig. S3B). CBFβ staining did not co-localize with β-tubulin in the midbody but co-localized with the flanking region and the dark zone (see KIF4A and PRC1 staining). From this analysis we conclude that CBFβ is located in the central region of the midbody.
Fig. 2.
CBFβ is localized with midbody-related proteins. A: HeLa cells were transfected with CBFβ-GFP. Western blot analysis using anti-GFP and anti-CBFβ antibodies shows the expected size of the exogenous protein (~54Kda) (upper 2 panels). β-tubulin was used as a loading control and total cellular levels of CBFβ were not altered by expression of CBFβ-GFP (lower 2 panels). B: HeLa cells transfected with CBFβ-GFP were fixed and CBFβ was detected by GFP fluorescence. White arrow shows that exogenous Cbfβ is located to the midbody region. C: Fixed cells were labeled for midbody markers by immunofluorescence. Right panels show line scans depicting co-localization (yellow merged signal). Cbfβ co-localizes with PRC1 and KIF4A but not with β-tubulin in the midbody, suggesting that CBFβ is specifically located in the central region of the midbody.
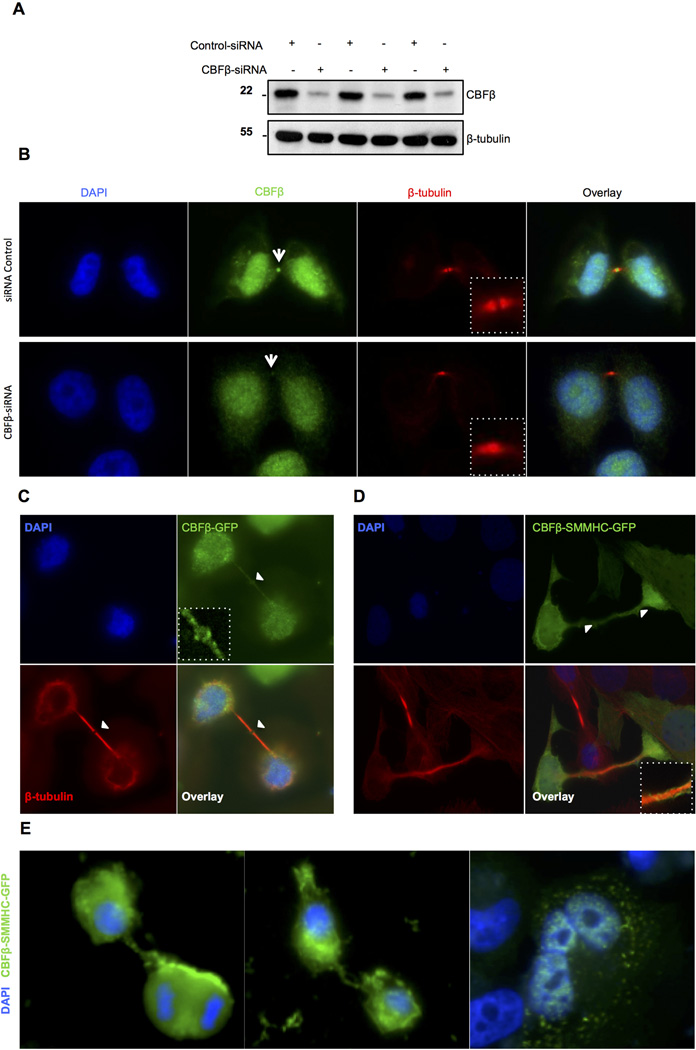
CBFβ depletion affects midbody architecture
Based on the localization of CBFβ in the midbody, the question arises whether CBFβ plays a direct role in maintaining midbody structure. Therefore, we analyzed the localization of crucial components of the midbody after CBFβ depletion (Fig. 3). Transfection of CBFβ-siRNA resulted in significantly decreased levels of CBFβ in comparison to control siRNA-transfected cells (Fig. 3A). In control siRNA-treated cells β-tubulin exhibited characteristic labeling in the midbody without labeling the dark zone, as antibodies do not penetrate the dense component of the central region (Hu et al., 2012) (Fig. 3B). In contrast, CBFβ-siRNA knockdown cells showed continuous tubulin staining along the midbody with no dark zone. Mis-localization of PRC1 was also evident in CBFβ-siRNA knockdown cells versus control-siRNA cells (Supplementary Material Fig. S4A, compare with Fig. 2C). Normally, PRC1 is distributed in the midbody and also present in the dark zone, whereas CBFβ depleted cells showed PRC1 uniformly distributed along the midbody with scarce evidence of the dark zone. Rescue experiments showed that over-expression of CBFβ-GFP in CBFβ-siRNA knockdown cells decreased the number of mis-localization events (approximately 80% and 60% for tubulin and PRC1, respectively) (Supplementary Material Fig. S4B). Taken together, these results indicate that midbody structure is impaired in the absence of CBFβ.
Fig. 3.
CBFβ is involved in midbody structure and cytokinesis. A: siRNA mediated downregulation of endogenous CBFβ in HeLa cells. Western blot shows consistent downregulation by CBFβ siRNA in comparison to scrambled control siRNA (n=3). Tubulin was used as a loading control. B: Depletion of CBFβ causes defects in the midbody structure. Cells transfected with CBFβ siRNA lose CBFβ staining and exhibit altered distribution of β-tubulin. The parallel plate observed in control cells is replaced by continuous tubulin staining across the midbody (see insets). C: CBFβ-GFP shows normal localization in the midbody (white arrowhead and inset). β-tubulin is shown for comparison. D: Presence of CBFβ/SMMHC-GFP elicits failure of midbody abscission. White arrowheads show that Inv(16) results in altered midbody structure with no evidence of the dark zone (see inset). E: SaOS-2 cells expressing Cbfβ-Inv(16)-GFP failed to undergo proper cytokinesis; di-, tri- or tetra-nucleated cells are present under these conditions. Nuclei were stained with DAPI.
The leukemogenic fusion protein CBFβ-SMMHC causes cytokinesis failure
Expression of the CBFβ-SMMHC fusion protein in leukemias of the M4E0 subtype provides mechanistic insight into the contribution of CBFβ to biological control and aberrations associated with tumorigenesis. We examined the subcellular distribution of CBFβ-SMMHC tagged with GFP (Fig. 3D, E). In control cells, CBFβ-GFP localized in the central region of the midbody, whereas the tubulin signal was detected along the midbody but was absent from the dark zone (Fig. 3C, white arrowhead; see inset). In contrast, when CBFβ-SMMHC-GFP was present, we detected cytokinesis failure (Fig. 3D). Cells in interphase appeared to be connected by a thicker and distorted bridge comprised of tubulin and surrounded with Cbfβ-SMMHC (white arrowheads, see inset). CBFβ-SMMHC-GFP expressing cells that are joined together apparently retain competency to divide mitotically (Fig. 3E), as less than 20% of the progeny cells are bi-, tri- or tetra-nucleated (n=100). Our results suggest that CBFβ-SMMHC impairs a final step in cytokinesis.
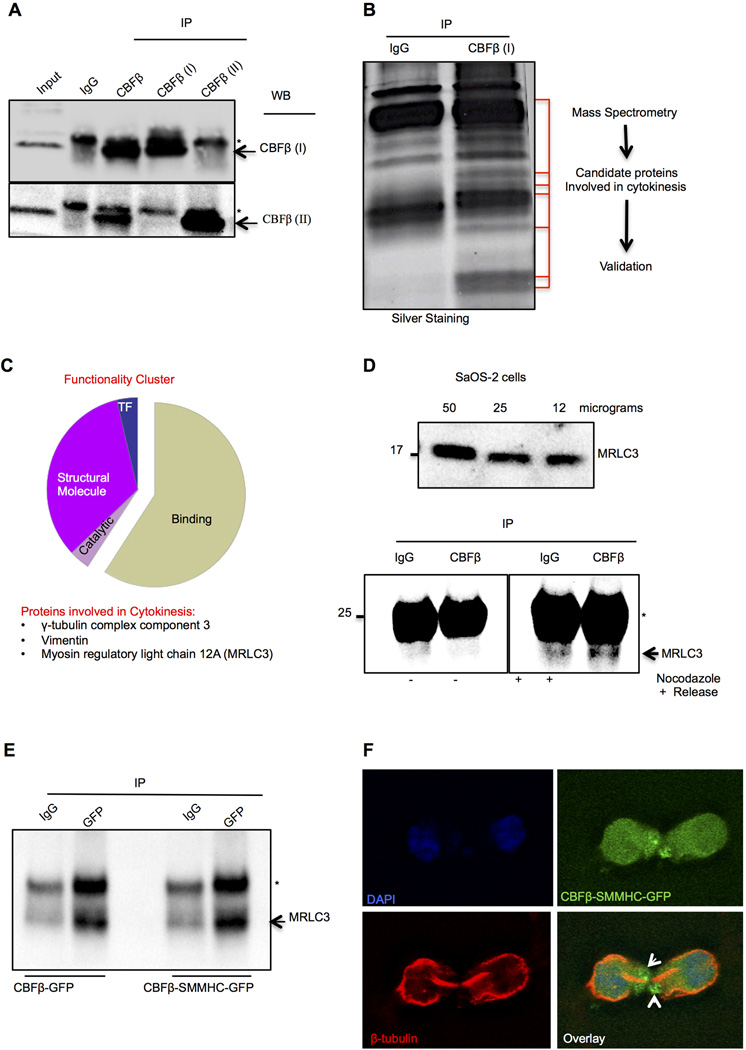
CBFβ complexes with MRLC3, a component required for cytokinesis
CBFβ interaction with RUNX proteins and with cytoskeletal proteins has been widely documented (Bravo et al., 2001; Kundu et al., 2002; Tanaka et al., 1998; Tanaka et al., 1997; Yoshida et al., 2005). To date, 38 proteins have been shown to interact with CBFβ using different approaches and different experimental models (Supplementary Material Fig. S5 and Table S1). However, none of these proteins has a known role in cytokinesis. Therefore, to identify CBFβ interacting proteins, we performed co-immunoprecipitation experiments using whole cell lysates and three different antibodies directed against CBFβ (Pan, isoform I and isoform II). Specificity of the antibodies was demonstrated by immunoprecipitating with each and probing with the CBFβ isoform I and isoform II antibodies (Fig. 4A). Proteins immunoprecipitated with the CBFβ isoform I antibody were then subjected to PAGE-SDS, in-gel trypsin digestion and analysis by mass spectrometry with normal IgG used as control (Fig. 4B). A total of 49 proteins were identified, and those clustered in the “structural molecules” category were searched for proteins involved in cytokinesis (Fig. 4C). Our analysis retrieved 3 candidate proteins: gamma tubulin complex component 3 (Shu et al., 1995), vimentin (Goto et al., 2003) and myosin regulatory light chain 3 (MRLC3) (Komatsu et al., 2000). Because myosin plays essential roles during mitosis and cell division, we focused on MRLC3. Fig. 4D (top panel) demonstrates detectable levels of MRLC3 in SaOS-2 cells. To verify the results obtained by mass spectrometry analysis, we performed immunoprecipitation for CBFβ in SaOS-2 cells. Nocodazole was used to enrich the mitotic population; untreated cells were used as control. Western blotting of lysates from nocodazole-released cells showed that MRLC3 co-immunoprecipitates with CBFβ. We were unable to detect CBFβ /MRLC3 complex in cells that were not mitotically enriched (Fig. 4D, bottom panel). This could be due to sensitivity of the assay and the amount of starting material.
Fig. 4.
The cytokinesis related Myosin Regulatory Light 3 Chain co-immunoprecipitates with CBFβ. A: Whole cell lysates from SaOS-2 cells were immunoprecipitated using 3 different CBFβ antibodies recognizing unique epitopes/isoforms. Western blot with CBFβ 1 and CBFβ 2 isoforms shows specificity; arrowheads and asterisk indicate CBFβ and IgG light chain, respectively. B: Proteomic analysis of CBFβ interacting proteins. CBFβ and IgG immunoprecipitates were resolved by PAGE-SDS; the bands indicated in red were subjected to mass spectrometry analysis. C: Identified proteins complexed with CBFβ were clustered into functional categories. Three structural proteins were found to be involved in the midbody and cytokinesis. D: Myosin Regulatory Light Chain 3 (MRLC3) is endogenously expressed in SaOS-2 cells (top panel) and co-immunoprecipitates with CBFβ from lysates of nocodazole-blocked and released cells. E: Expression of CBFβ-GFP or CBFβ-SMMHC-GFP and subsequent immunoprecipitation with GFP antibody. Western blot shows that MRLC3 is complex with CBFβ and Cbfβ-SMMHC; IgG light chain is denoted by (*). F: Immunofluorescence microscopy of Cbfβ-SMMHC-GFP and tubulin. Absence in the central region of the midbody and accumulation of CBFβ-SMMHC in regions adjacent to the midbody is shown (white arrows).
However, MRLC3 co-immunoprecipated from lysates of HeLa cells transfected with CBFβ-GFP or CBFβ-SMMHC-GFP (Fig. 4E), suggesting that CBFβ-SMMHC also complexes with MRLC3. Because presence of CBFβ-SMMHC increased the rate of cytokinesis failure (see Fig. 3D,E), we explored the localization of CBFβ-SMMHC in cells undergoing cytokinesis (Fig. 4F). In contrast to CBFβ (see Fig. 3C for comparison), CBFβ-SMMHC was absent from the central region of the midbody (delimitated by tubulin staining) (Fig. 4F), but accumulated in regions adjacent to this structure (white arrows). Our results suggest that CBFβ interacts with proteins involved in cytokinesis; furthermore, CBFβ-SMMHC is absent from the midbody and may cause sequestration of midbody-related proteins important for completion of cytokinesis.
Discussion
Here we have presented evidence that CBFβ is localized in the midbody, a structure important for cytokinesis in several culture models. Interestingly, Ito and colleagues have reported the localization of RUNX3, a well-established interacting partner of CBFβ, at the midbody (Chuang et al., 2012). We were unable to detect RUNX2 at this structure in SaOS-2 cells. Significantly, we have detected CBFβ association with the midbody in HeLa cells (Fig. 1D) and embryonic stem cells (not shown) that do not express RUNX factors. Our finding that downregulation of CBFβ results in defects in midbody architecture reinforces the importance of regulating distribution of CBFβ during cytokinesis. Furthermore, the mutant version of CBFβ, CBFβ-SMMHC causes cytokinesis failure and polyploidy. However, further studies will be required to address the biological impact of CBFβ-SMMHC in the cytokinesis machinery of patients with Acute Myeloid Leukemia of the M4E0 type.
The proteomic analysis of CBFβ interacting proteins performed in this study revealed a set of proteins involved in cytokinesis. The presence of MRLC3 in a complex with CBFβ and with CBFβ-SMMHC further extends the regulatory implications for CBFβ in the context of cytokinesis. Given that CBFβ-SMMHC is able to sequester RUNX proteins in the cytoplasmic compartment (Adya et al., 1998), the possibility arises that CBFβ-SMMHC may also complex and sequester non-RUNX related proteins such as MRLC3 and thus decrease rates of successful cytokinesis. The extent of the compartmentalization and dynamics of cellular RUNX/CBFβ proteins with the midbody is yet to be established. Our results provide further understanding of the retention of regulatory proteins during mitosis to support biological activities in progeny cells. Because the midbody is retained by only one of the progeny cells, our results also imply a novel dimension of selectivity for proteins involved in gene expression during cytokinesis. Our findings suggest that CBFβ may play a role in cytokinesis through association with the midbody and might reflect a novel mechanism that contributes to epigenetic control.
Supplementary Material
Acknowledgements
We thank UVM Mass Spectrometry Core, especially Julia Ganister Fields and Ying Wai Lam, for their assistance with mass spec analysis and suggestions, Paul Liu (for the GFP plasmids used in this study), Philip Tai and Kaleem Zaidi for helpful comments and suggestions.
Contract grant sponsor: National Institutes of Health; Contract grant number: P01 CA082834; Contract grant sponsor: National Institutes of Health; Contract grant number: P01 AR048814; Contract grant sponsor: National Institute of General Medical Sciences; Contract grant number: P20 GM103449
Footnotes
The authors declare no conflict of interest.
Literature Cited
- Adya N, Stacy T, Speck NA, Liu PP. The leukemic protein core binding factor beta (CBFbeta)-smooth-muscle myosin heavy chain sequesters CBFalpha2 into cytoskeletal filaments and aggregates. Mol Cell Biol. 1998;18:7432–7443. doi: 10.1128/mcb.18.12.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J, Li Z, Speck NA, Warren AJ. The leukemia-associated AML1 (Runx1)--CBF beta complex functions as a DNA-induced molecular clamp. Nat Struct Biol. 2001;8:371–378. doi: 10.1038/86264. [DOI] [PubMed] [Google Scholar]
- Chen CT, Ettinger AW, Huttner WB, Doxsey SJ. Resurrecting remnants: the lives of post-mitotic midbodies. Trends Cell Biol. 2013;23:118–128. doi: 10.1016/j.tcb.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Pratap J, Javed A, Zaidi SK, Xing L, Balint E, Dalamangas S, Boyce B, van Wijnen AJ, Lian JB, Stein JL, Jones SN, Stein GS. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc Natl Acad Sci U S A. 2001;98:8650–8655. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LS, Lai SK, Murata-Hori M, Yamada A, Li HY, Gunaratne J, Ito Y. RUNX3 interactome reveals novel centrosomal targeting of RUNX family of transcription factors. Cell Cycle. 2012;11:1938–1947. doi: 10.4161/cc.20278. [DOI] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JN, Rogers D, Adams L, Yong T, Jung JS, Cheng B, Fennell K, Borazanci E, Moustafa YW, Sun A, Shi R, Glass J, Mathis JM, Williams BJ, Meyers S. Association of core-binding factor beta with the malignant phenotype of prostate and ovarian cancer cells. J Cell Physiol. 2010;225:875–887. doi: 10.1002/jcp.22298. [DOI] [PubMed] [Google Scholar]
- Decordier I, Cundari E, Kirsch-Volders M. Survival of aneuploid, micronucleated and/or polyploid cells: crosstalk between ploidy control and apoptosis. Mutat Res. 2008;651:30–39. doi: 10.1016/j.mrgentox.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- Goto H, Yasui Y, Kawajiri A, Nigg EA, Terada Y, Tatsuka M, Nagata K, Inagaki M. Aurora-B regulates the cleavage furrow-specific vimentin phosphorylation in the cytokinetic process. J Biol Chem. 2003;278:8526–8530. doi: 10.1074/jbc.M210892200. [DOI] [PubMed] [Google Scholar]
- Hu CK, Coughlin M, Mitchison TJ. Midbody assembly and its regulation during cytokinesis. Mol Biol Cell. 2012;23:1024–1034. doi: 10.1091/mbc.E11-08-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Shigesada K, Ito K, Wee HJ, Yokomizo T, Ito Y. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 2001;20:723–733. doi: 10.1093/emboj/20.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Zhu S, Tremblay MS, Payette JN, Wang J, Bouchez LC, Meeusen S, Althage A, Cho CY, Wu X, Schultz PG. A stem cell-based approach to cartilage repair. Science. 2012;336:717–721. doi: 10.1126/science.1215157. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Kanno T, Sakakura C, Bae SC, Ito Y. Cytoplasmic sequestration of the polyomavirus enhancer binding protein 2 (PEBP2)/core binding factor alpha (CBFalpha) subunit by the leukemia-related PEBP2/CBFbeta-SMMHC fusion protein inhibits PEBP2/CBF-mediated transactivation. Mol Cell Biol. 1998;18:4252–4261. doi: 10.1128/mcb.18.7.4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu S, Yano T, Shibata M, Tuft RA, Ikebe M. Effects of the regulatory light chain phosphorylation of myosin II on mitosis and cytokinesis of mammalian cells. J Biol Chem. 2000;275:34512–34520. doi: 10.1074/jbc.M003019200. [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Kundu M, Javed A, Jeon JP, Horner A, Shum L, Eckhaus M, Muenke M, Lian JB, Yang Y, Nuckolls GH, Stein GS, Liu PP. Cbfbeta interacts with Runx2 and has a critical role in bone development. Nat Genet. 2002;32:639–644. doi: 10.1038/ng1050. [DOI] [PubMed] [Google Scholar]
- Lacroix B, Maddox AS. Cytokinesis, ploidy and aneuploidy. J Pathol. 2012;226:338–351. doi: 10.1002/path.3013. [DOI] [PubMed] [Google Scholar]
- Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, Kim HM, Kim WJ, Yamamoto H, Yamashita N, Yano T, Ikeda T, Itohara S, Inazawa J, Abe T, Hagiwara A, Yamagishi H, Ooe A, Kaneda A, Sugimura T, Ushijima T, Bae SC, Ito Y. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Liu P, Tarle SA, Hajra A, Claxton DF, Marlton P, Freedman M, Siciliano MJ, Collins FS. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- Lutterbach B, Hou Y, Durst KL, Hiebert SW. The inv(16) encodes an acute myeloid leukemia 1 transcriptional corepressor. Proc Natl Acad Sci U S A. 1999;96:12822–12827. doi: 10.1073/pnas.96.22.12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Villanueva D, Deng W, Lopez-Camacho C, Shore P. The Runx transcriptional co-activator, CBFbeta, is essential for invasion of breast cancer cells. Mol Cancer. 2010;9:171. doi: 10.1186/1476-4598-9-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Gupta V, Sorce D, Kim WY, Sali A, Chait BT, Shigesada K, Ito Y, Werner MH. Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain. Nat Struct Biol. 1999;6:615–619. doi: 10.1038/10658. [DOI] [PubMed] [Google Scholar]
- Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, Shigesada K. Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology. 1993;194:314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Yagi H, Bronson RT, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci U S A. 1996;93:12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu HB, Li Z, Palacios MJ, Li Q, Joshi HC. A transient association of gamma-tubulin at the midbody is required for the completion of cytokinesis during the mammalian cell division. J Cell Sci. 1995;108(Pt 9):2955–2962. doi: 10.1242/jcs.108.9.2955. [DOI] [PubMed] [Google Scholar]
- Stein GS, van Wijnen AJ, Stein JL, Lian JB, Montecino M, Choi J, Zaidi K, Javed A. Intranuclear trafficking of transcription factors: implications for biological control. J Cell Sci. 2000;113(Pt 14):2527–2533. doi: 10.1242/jcs.113.14.2527. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Fujii M, Hayashi K, Chiba N, Akaishi T, Shineha R, Nishihira T, Satomi S, Ito Y, Watanabe T, Satake M. The chimeric protein, PEBP2 beta/CBF beta-SMMHC, disorganizes cytoplasmic stress fibers and inhibits transcriptional activation. Oncogene. 1998;17:699–708. doi: 10.1038/sj.onc.1201985. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Watanabe T, Chiba N, Niki M, Kuroiwa Y, Nishihira T, Satomi S, Ito Y, Satake M. The protooncogene product, PEBP2beta/CBFbeta, is mainly located in the cytoplasm and has an affinity with cytoskeletal structures. Oncogene. 1997;15:677–683. doi: 10.1038/sj.onc.1201235. [DOI] [PubMed] [Google Scholar]
- Vinciguerra P, Godinho SA, Parmar K, Pellman D, D'Andrea AD. Cytokinesis failure occurs in Fanconi anemia pathway-deficient murine and human bone marrow hematopoietic cells. J Clin Invest. 2010;120:3834–3842. doi: 10.1172/JCI43391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N, Ogata T, Tanabe K, Li S, Nakazato M, Kohu K, Takafuta T, Shapiro S, Ohta Y, Satake M, Watanabe T. Filamin A-bound PEBP2beta/CBFbeta is retained in the cytoplasm and prevented from functioning as a partner of the Runx1 transcription factor. Mol Cell Biol. 2005;25:1003–1012. doi: 10.1128/MCB.25.3.1003-1012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino MA, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat Rev Genet. 2010;11:583–589. doi: 10.1038/nrg2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.